Abstract
Purpose
We examined hospital use of the epidermal growth factor receptor (EGFR) assay for lung cancer patients. Our goal was to inform the development of a model to predict T3 translation of guideline-directed, molecular diagnostic tests.
Methods
This was a retrospective observational study. Using logistic regression, we analyzed the association between likelihood to order the EGFR assay and hospital’s institutional and regional characteristics.
Results
Significant institutional predictors included: Affiliation with an academic medical center (odds ratio [OR], 1.48; 95% CI, 1.20–1.83), Participation in an NCI clinical research cooperative group (OR, 2.06, 1.66–2.55), PET scan (OR, 1.44, 1.07–1.94) and cardio thoracic surgery (OR, 1.90, 1.52–2.37) services. Significant regional predictors included: Metropolitan county (OR, 2.08, 1.48 to 2.91), Above average education (OR, 1.46, 1.09 to 1.96), Above average income (OR, 1.46, 1.04–2.05). Distance from an NCI cancer center was a negative predictor (OR, 0.996, 0.995–0.998), a 34% decrease in likelihood for every 100 miles.
Conclusion
In 2010, 12% of US acute care hospitals ordered the EGFR assay, suggesting most lung cancer patients did not have access to this test. This case study illustrated the need for: 1) Increased dissemination and implementation research. 2) Interventions to improve adoption of guideline-directed, molecular diagnostic tests by community hospitals.
Keywords: equity, access, lung, cancer, genomics
INTRODUCTION
Molecular diagnostic (MDx) tests are a key component of personalized cancer genomics. These tests identify mutations that drive cancer progression. Specific mutations predict response to therapies that may improve progression-free survival for patients.1–3 Recent reviews of cancer genomics cite widespread use of molecular diagnostics.4–6 However, little evidence supports these claims, particularly in community hospitals where 85% of cancer patients obtain care.7 While underuse may be due to an uncertain evidence base,8–10 delays translating healthcare innovations to community hospitals is a longstanding problem. In cancer genomics, these delays may widen the gap in cancer disparities.9,11,12
Historically, translational research has been focused on discovery and development of early stage innovations.13 Recently, public health researchers have proposed dividing translational research into four stages: T0–T4.14 Bench to commercial product activities take place in stages T0–T2. Dissemination and implementation research is conducted in T3. Health outcomes research is in T4. The majority of funding for cancer genomics has been directed toward T0–T2 research, the discovery and development of applications, with less than 2% of funding directed toward T3 research, evaluating bedside implementation and dissemination.15 This dynamic has contributed to limited empirical data evaluating ‘real world’ use of genomic applications in patient care. With only half of patients in the United States (US) benefitting from advances incorporated into guidelines,16 there is a growing need to fund more studies to analyze dissemination, implementation, and health outcomes of cancer genomics.17 This T3 research analyzes dissemination of a T0–T2 translational advance in lung cancer.
The history of lung cancer molecular diagnostics provides a context for understanding barriers to T3 translation. In 2004, researchers at a National Cancer Institute cancer center (NCI CC) discovered a link between epidermal growth factor receptor (EGFR) mutations in lung tumors and the likelihood to respond to tyrosine kinase inhibitors (TKIs), a class of oral anti-cancer drugs.18,19 Erlotinib, an EGFR TKI, was approved for treatment of patients with non-small cell lung cancer (NSCLC). The drug’s labeling cited prolonged survival for EGFR positive patients. This significant T0–T2 innovation set the stage for greater understanding of lung cancer molecular biology.
The EGFR assay was commercialized in 2005.20 By 2007, National Comprehensive Cancer Network (NCCN) encouraged oncologists to consider molecular tests to identify patients who may benefit from EGFR TKIs. However, specific recommendations for EGFR mutation analysis were not included in American Society of Clinical Oncology (ASCO) and NCCN guidelines until April 2010.21,22 This was due to the immature state of lung cancer genomics and a lack of consensus about which diagnostic technology to use.
The EGFR assay is a high complexity test that requires a laboratory to be accredited by the Clinical Laboratory Improvement Amendments (CLIA) program for cytogenetic testing. A limited number of large academic medical centers and NCI CCs are CLIA accredited to conduct high complexity testing. These centers perform EGFR mutation analysis within their own institutions using laboratory developed tests (LDTs).23 However, less than 1% of laboratories are accredited to conduct genetic testing.24 The vast majority of hospitals order molecular diagnostic testing by sending patients’ paraffin embedded tissue slides to commercial reference laboratories.
There was debate about which patients to test and whether payers should reimburse for testing. Early research indicated incidence of EGFR mutations varied with patient ethnicity, gender, and smoking status.25 However, a recent study reported that 57% of mutations would be missed if only clinical characteristics were used.26 By 2008, several third party payers, including Medicare, reimbursed for the assay. Yet, there was confusion about which billing codes to use and concerns that, if the test was ordered within 14 days of an inpatient stay, it would be considered part of the Diagnosis Related Group (DRG) bundled payment. These factors may have contributed to delays implementing the EGFR assay. A 2010 NCCN survey reported that only 21–41% of oncologists conducted EGFR mutation testing.27
Current ASCO and NCCN guidelines call for NSCLC patients with adenocarcinoma, large cell carcinoma, or “not otherwise specified” to undergo EGFR mutation testing.22,28 This is approximately 68% or 138,462 of newly diagnosed lung cancer patients per year.
Publications describing the incidence of EGFR mutations in Blacks illustrate how differences in access to cancer genomics may exacerbate disparities. Studies published in 2005 and 2009 analyzed 93 tumors from Blacks and found a 2% incidence rate of EGFR mutations.29,30 This contributed to a misperception that Blacks have lower incidence of EGFR mutations. Although Blacks have the highest incidence, morbidity, and mortality of lung cancer, they have been under-represented in lung cancer research. This is due, in part, to racial, regional, and socioeconomic differences in use of biopsies.16,31 Recent studies report the incidence of EGFR mutations in Blacks is similar to Caucasians.32,33
This example demonstrates the need for empirical, population level data to inform the translational research process. By examining factors associated with hospital use of the EGFR assay, we hope to inform the development of a model to predict T3 translation of cancer genomic applications.
MATERIALS AND METHODS
This was a retrospective cross sectional study. We merged one proprietary dataset with seven public datasets. Genzyme Genetics, which owned the rights to distribute the EGFR assay, provided a dataset containing name, city, state, zip code, and number of EGFR assays sold to each hospital, laboratory, or outpatient clinic in the US during calendar year 2010. Public datasets included:
Centers for Medicare and Medicaid Services (CMS) Provider of Service (POS) file, which contained hospital characteristics, including academic medical affiliation and services offered.
CMS CLIA specialty file, which identified accreditation to condct cytogenetic testing.
NCI Provider of Services (POS) file indentified hospital participation in NCI clinical research cooperative groups.
Census population file, which provided county-level sociodemographic indicators.
National Program of Cancer Registries (NPCR) and Centers for Disease Control and Prevention (CDC) State Cancer Profiles, which provided county-level lung cancer incidence.
National Institute of Standards and Technology (NIST) file, which provided zip-code longitude and latitude.
Federal Information Processing Standards (FIPS) file which provided the county code.
We aggregated and summarized orders at the hospital level, recording each hospital’s Medicare provider number from the CMS POS file. The aggregated dataset was merged with public datasets using hospital number, zip code, and FIPS code as unique identifiers for each US hospital and county. The unit of observation for multivariate analysis was the non-federal acute care hospital. The outcome variable was whether a hospital ordered the EGFR assay, treated dichotomously and coded as 1 if it ordered one or more assay.
Most independent variables were dichotomous, coded as 1 if the hospital or region had the characteristic. Number of lung cancer cases per county, percentage of the county population that was Black, and hospital distance to an NCI CC were treated as continuous variables. Hospital distance to an NCI CC was measured in miles, calculated using the longitude and latitude of each institution. To accurately interpret the odds ratio (OR) of distance, we obtained the logit coefficient and multiplied it by increments of 100 miles.
The dataset contained orders from 27 of the 60 NCI CCs. However, several NCI CCs confirmed use of LDTs to identify EGFR mutations. For consistency, NCI CCs orders were excluded from the dataset. We also examined the CLIA specialty file to evaluate potential use of LDT by other hospitals.
Statistics
We conducted univariate and bivariate analyses, including t-tests for continuous variables and chi-square for categorical variables. Statistically significant explanatory variables (p-values < 0.05) were included in the logistic regression model. Number of lung cancer cases and percent Black, which were not statistically significant in bivariate analysis, were retained as control variables. We performed multivariate logistic regression to identify characteristics associated with a hospital’s ordering an EGFR assay. We performed sensitivity analysis to ensure that observed effects were not an artifact of modeling. This included using different cut points for independent variables and estimates of LDT use. STATA statistical program version 12.0 (STATA Corp, College Station, TX) was used for analyses.
RESULTS
In 2010, 743 institutions ordered 7,958 EGFR assays (Table 1), which represented 5.7% of newly diagnosed, guideline-directed patients. Non-federal acute care hospitals accounted for 76% of tests ordered. Federal hospitals, NCI CCs, independent pathology labs, outpatient ambulatory cancer centers and physician offices accounted for the remainder. Of 4,781 acute care hospitals in the database, 12% ordered the assay and 148 hospitals ordered only 1 assay.
Table 1.
Number and type of institutions ordering EGFR assay
| Type of Institution | Sites (n=743) |
Assays (n=7958) |
||
|---|---|---|---|---|
| Acute care hospitals, No. (%) | 593 | (80) | 6074 | (76) |
| Federal hospitals (Veterans Administration), No. (%) | 15 | (2) | 93 | (1) |
| NCI Cancer Centers, No. (%) | 27 | (4) | 1019 | (13) |
| Pathology labs, No. (%) | 60 | (8) | 522 | (7) |
| Independent outpatient oncology clinics or MDs, No. (%) | 48 | (6) | 250 | (3) |
Four NCI CCs had contracted with Genzyme to be the exclusive provider of EGFR testing. These orders (588) accounted for more than half the total NCI CC orders. We used these data to develop an estimate of EGFR mutation analysis conducted by NCI CCs (Supplement 1). Although it was not possible to estimate use of LDTs by non-NCI CCs, analysis of the CLIA specialty file revealed that only 4% (203) of US hospitals are accredited for cytogenetic testing. Of these 203 hospitals, 52 were NCI CCs. Of the 151 non-NCI hospitals accredited to conduct cytogenetic testing, 37 (18%) had ordered the EGFR assay.
State Analysis
Table 2 summarizes by state the number of hospitals, NCI CCs, cytogenetic hospital labs, and EGFR assays ordered. We estimated the percentage of lung cancer cases tested. Approximately 4.4% of guideline-directed patients accessed the Genzyme EGFR assay through acute care hospitals. Estimates suggested that approximately 5.3% of patients accessed EGFR mutation analysis at NCI CCs, many of which were likely through LDTs.
No hospitals in Alaska ordered the assay. One hospital per state in Arkansas, Montana, New Mexico, South Dakota, Vermont, and Wyoming ordered the assay. California, Florida, Illinois, New York, and Pennsylvania had the greatest number of hospitals ordering the assay. These are also the most populous states with the highest number of lung cancer cases. Between 4.4% to 10.52% of guideline-directed lung cancer cases in these states were tested for EGFR mutations with the Genzyme assay. However, these states also have the most hospitals accredited for cytogenetic testing. Patients may have had EGFR mutation analysis through LDTs. North Dakota had the highest percentage (17.6%) of lung cancer cases tested. It has neither an NCI CC nor a hospital accredited to conduct cytogenetic testing. Three North Dakota hospitals that ordered the assay had academic medical school affiliations, participated in NCI cooperative groups, offered cardiothoracic surgery, and were located in metropolitan counties with above average education and income. These are all characteristics we hypothesized to predict likelihood of ordering the assay.
County Analysis
There are 2,496 counties with acute care hospitals. Of these, 349 (14%) have hospitals which ordered the EGFR assay; 96 (4%) have hospitals accredited to conduct cytogenetic testing; 50 (2%) have NCI CCs. Most hospitals that ordered the EGFR assay, 522 (88%), are located within metropolitan counties; 497 (84%) are located in counties with above average education level; and 511 (86%) are located in counties with above average income.
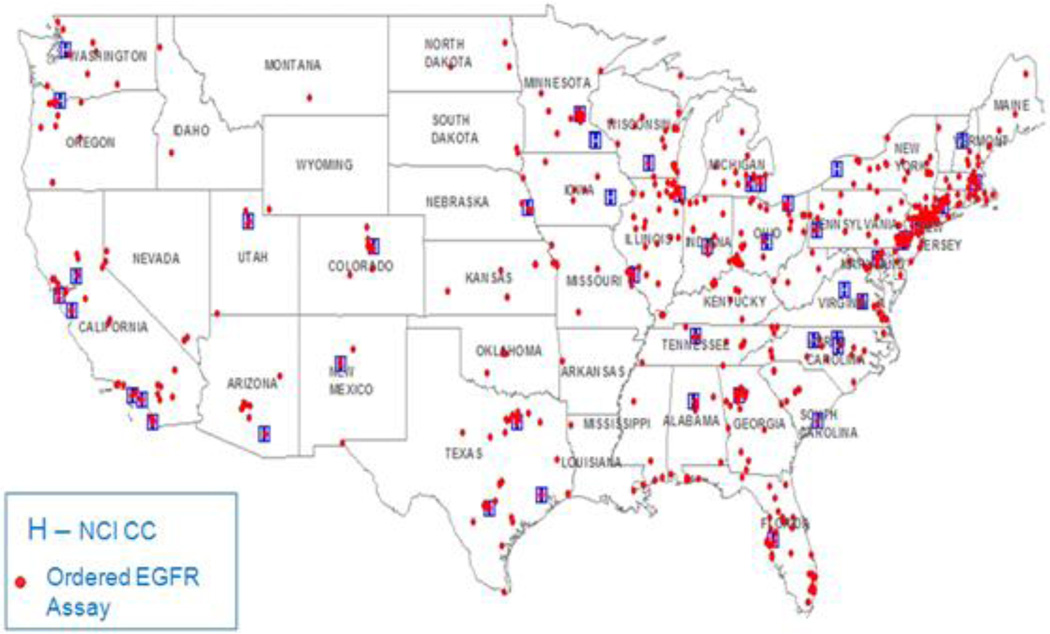
Diffusion of the EGFR assay appeared to emanate from NCI CCs (Figure 1). This made NCI counties particularly relevant. In NCI counties, 19% of hospitals ordered the assay versus 11% of hospitals in non-NCI counties. NCI CCs are located in metropolitan counties with above average education and income. Education and income differences that exist between NCI and non-NCI counties are mirrored when ‘EGFR counties’ were compared to ‘non-EGFR counties.’ In EGFR counties, 24.4% of the population had bachelor degree education and 21.2% earned at least $75,000 per household annually. In non-EGFR counties 16% of the population had bachelor degree education and 13.3% of the population earned at least $75,000. There were also racial differences between NCI and non NCI counties. NCI CCs are located in urban cities where 20% of the population is Black. In non NCI counties, 9% of the population is Black. Less racial difference existed between EGFR counties and non-EGFR counties. In EGFR counties, 12% of the population was Black. In non EGFR counties, only 9% of the population was Black.
Figure 1.
Acute care hospitals that ordered the EGFR assay in 2010
Bivariate Analysis of Hospital Characteristics
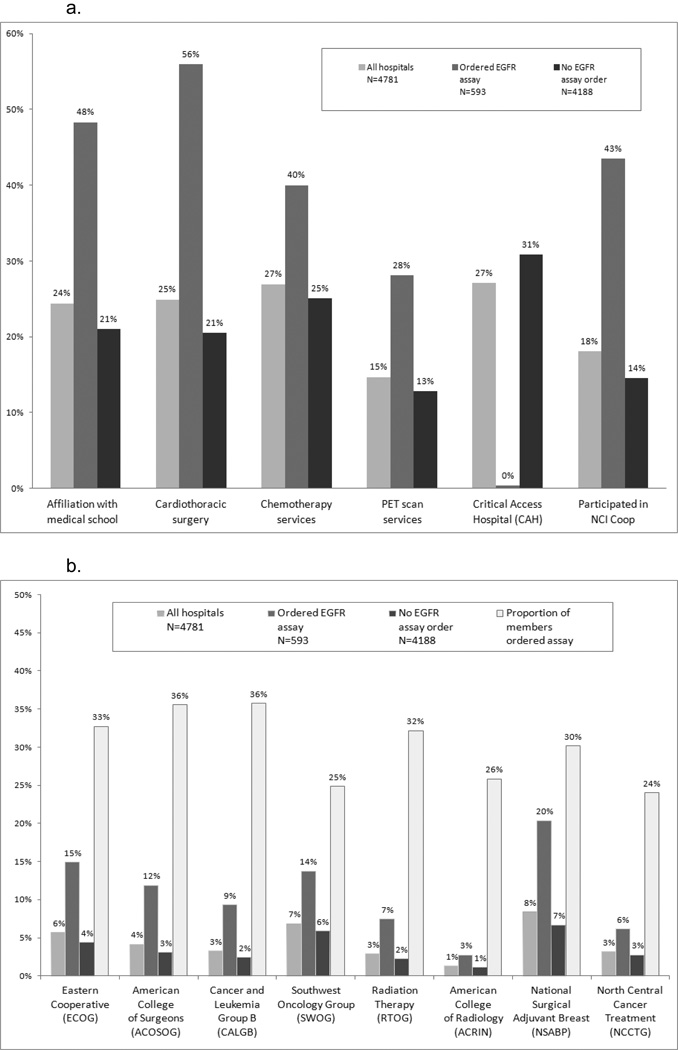
Cardiothoracic surgery appeared to be the strongest predictor of the hospital ordering the EGFR assay; 56% of hospitals that ordered the assay reported this service (Figure 2a) versus 21% of hospitals not ordering it. Only 2 hospitals designated by Medicare as Critical Access ordered the EGFR assay. These types of hospitals are usually located in rural areas.
Figure 2.
a. Hospital characteristics by status of ordering EGFR assay
b. Membership in cooperative group by status of ordering EGFR assay
It was hypothesized that cooperative groups which participated in EGFR TKI trials would have a higher percentage of hospitals that ordered the assay. Figure 2b illustrates that American College of Surgeons (ACOSOG) and Cancer and Leukemia Group B (CALGB) had the highest percentage of members (36%) that ordered the assay. Yet, the difference between cooperative groups was relatively small. National Surgical Adjuvant Breast (NSABP), the largest cooperative group with 398 hospital members, had the most members that ordered the EGFR assay, which may reflect the established role of molecular markers in breast cancer.
Prediction Model
The relationship between the independent variables and likelihood to order the EGFR assay persisted when we conducted multivariate logistic regression analysis. Table 3 summarizes the odds ratios and 95% confidence intervals of each variable. Significant institutional predictors of ordering the assay included: affiliation with an academic medical center (OR =1.48), participation in an NCI cooperative group (OR=2.06), PET scan services (OR=1.44) and cardiothoracic surgery services (OR =1.90); PET scan services, which are not routinely used in lung cancer, may be a proxy for early adopters of technology.
Table 3.
OR and CIs for Hospitals that Ordered the EGFR Assay
| Institution ordered EGFR assay | OR | P>|z| | [95% CI] | |
|---|---|---|---|---|
| Distance to NCI CC* | 0.996 | 0.00 | 0.995 | 0.998 |
| Affiliated w/ academic medical center* | 1.48 | .00 | 1.20 | 1.83 |
| NCI clinical research cooperative* | 2.06 | .00 | 1.66 | 2.55 |
| Positron emission tomography (PET scans)* | 1.44 | .02 | 1.07 | 1.94 |
| Cardiothoracic surgery* | 1.90 | .00 | 1.52 | 2.37 |
| Inpatient chemotherapy | 1.15 | .30 | .88 | 1.50 |
| Within metropolitan county* | 2.08 | .00 | 1.48 | 2.91 |
| Education at BS level above mean* | 1.46 | .01 | 1.09 | 1.96 |
| Income over $75,000 above mean* | 1.46 | .03 | 1.04 | 2.05 |
| % Black | 1.30 | .48 | .63 | 2.67 |
| Annual lung cancer cases | 1.00 | .86 | 1.00 | 1.00 |
Abbreviations: OR, odds ratio. CI, confidence interval. EGFR, epidermal growth factor receptor. NCI, National Cancer Institute. CC, cancer center.
Distance to NCI CC was calculated using the longitude and latitude of each institution. This was a continuous variable measured in miles.
Significant at P<.05
Significant regional predictors included metropolitan county (OR=2.08) and above average education and income (OR=1.46). Distance from an NCI CC was a negative predictor for ordering the assay. Analyzed independently, the logit coefficient of distance was −.008 (OR=0.992). To interpret the coefficient, we multiplied by 100 miles and exponentiated it to get a value of 0.449. Hospitals located 100 miles away from an NCI CC had a 55% decreased likelihood of ordering the EGFR assay. Hospitals located 500 miles or more away from an NCI CC had less than a 2% likelihood of ordering the assay.
In the multivariate analysis, the effect of distance was reduced but still significant (OR=0.996, −0.004 logit coefficient). Holding hospital and regional characteristics constant, hospitals located 100 miles away from an NCI CC had a 33% decreased likelihood of ordering the EGFR assay. When the distance was increased to 500 miles, other things equal, the likelihood decreased to 14%. Annual number of lung cancer cases, inpatient chemotherapy, and race were not statistically significant predictors of ordering the assay.
DISCUSSION
This study is the first to use empirical data to analyze dissemination of the EGFR assay. In 2010, the EGFR assay was underutilized. Only 12% of non-federal hospitals ordered the assay. This study supports recent findings that institutional and regional characteristics predict hospital use of innovations in cancer care.11,34 The number of lung cancer cases in the county did not predict whether hospitals ordered the assay or the rate of testing.
Although the EGFR assay was commercialized in 2005,18,19 routine testing was not recommended by ASCO and NCCN guidelines until 2010. This was an important factor impeding dissemination. As a result, underutilization was an expected finding. Advances in biomarker clinical trial design, regulatory, and reimbursement processes have improved the pace at which lung cancer molecular diagnostics are included in guidelines. This has increased use 35 but continued analysis is needed to ensure that patients at community and critical access hospitals access molecular diagnostics.
There are inherent limitations to hospital rather than patient level data. Yet, hospital data, which are not protected by the Health Insurance Portability and Accountability Act, allow for timely identification of areas where evidence-based recommendations for genetic testing are not implemented.
The dataset did not capture laboratory developed EGFR mutation analysis conducted as part of clinical trials or by hospitals accredited for cytogenetic testing. With NCI use representing 13% of Genzyme’s EGFR assays, it is important to understand LDT use. However, 85% of cancer patients are treated in community hospitals and less than 2% of lung cancer patients enroll in clinical trials.36 Therefore, we believe a very small percentage of patients accessed LDTs.
The dataset did not consider patient level variables, test characteristics, or processes of care that that may explain use. A robust T3 research agenda requires analysis of population level claims data, examination of medical records, and interviews with various stakeholders to elucidate factors influencing dissemination and implementation. Yet, even with population level claims data, if MDx tests are bundled in the DRG payment, it will be difficult to identify patients who underwent EGFR mutation analysis.
The significant underutilization of the EGFR assay emphasizes the need for increased investment in dissemination and implementation (T3) and health outcomes (T4) research. Given the potential improvements in health outcomes that genetic tests offer, methods for timely T3 and T4 research are needed to increase the evidence base. This study provides an example of potential public-private collaborations that can inform implementation of guideline-directed genetic testing. This study informed dissemination of the anaplastic lymphoma kinase fusion gene (ALK) translocation assay, which lead to increased use of both ALK and EGFR MDx tests.35
Although T3 research is rare in cancer genomics, it is more robust in other areas of medicine and public health.17 Very little published research occurs in the T2 phase and beyond. Investment has been focused on commercialization of innovations without assuring that those innovations are translated to patient care. Even for well-established genetic tests such as BRCA1/2, discovered in 1994, it took until 2005 for it to become recommended by the US Preventive Services Task Force. Disparities in access to BRCA1/2 counseling/testing persist.12 Real world outcome data have been slow to develop.37 Recent ENCODE publications 38 illustrate that rapid development of genomic applications in T0/T1 continues. Since September 2009, CancerGEM Kb, a CDC/NCI collaborative database, has tracked more than 270 newly developed tests.39 Additional T2 research is sorely needed to evaluate these applications. For those with demonstrable clinical utility, T3–T4 research is needed to achieve maximum population health impact. Studying factors that accelerate or impede translation is essential for the success of cancer genomic medicine.40
Cancer genomics is maturing.4 A growing number of clinically useful molecular tests are being developed. Technologies are allowing for higher throughput genomic screening at competitive pricing. Academic medical centers are implementing next generation sequencing into routine cancer care. As T0–T2 innovations are commercialized, guidelines need to be developed to encourage hospitals and manufacturers to make utilization data available to facilitate dissemination, implementation, and health outcomes research. A robust translational research agenda will increase the pace of dissemination, decrease disparities in access that currently exist for patients seeking care in community and critical access hospitals, while improving quality and outcome analyses.
Supplementary Material
Acknowledgement
Julie Lynch has received funding from Center for Health Quality, Outcomes and Economic Research, a Veterans Administration Health Services Research & Development Center of Excellence; Department of Education GAANN #P200A060243;National Institute of Health DFCI/UMB U54 CA156734-01, Susan G. Komen Foundation, and the Lynch Foundation.
Footnotes
Previously presented: Methodology and preliminary results were previously presented as a poster at 10th International Congress on Targeted Anticancer Therapies, Amsterdam, March 8–10, 2012
Publisher's Disclaimer: Disclaimers: The opinions in this article reflect those of the authors and do not necessarily reflect the official position of the Department of Health and Human Services or the Veterans Health Administration. Genzyme Genetics did not sponsor this study. It had no role in data analysis, interpretation, or manuscript writing.
Author contributions:
Conception and design: Julie Lynch
Collection and assembly of data: Julie Lynch
Data analysis and interpretation: All authors
Manuscript writing: All authors
Final approval of manuscript: All authors
Conflicts of interest statement: Glenn Miller reports the following conflicts: He is currently Vice President and Head of Strategy, Portfolio and Alliance for Personalized Healthcare & Biomarkers at AstraZeneca Pharmaceuticals, LP. He was formerly Vice President and General Manager of a division of Genzyme Genetics, Genzyme Analytical Services. All other authors report no conflicts of interest.
Contributor Information
Julie A. Lynch, RN Veterans Health Administration, University of Massachusetts Boston & Dana Farber Cancer Institute.
Muin J. Khoury, Office of Public Health Genomics, CDC; Epidemiology and Genomics Research Program, NCI.
Ann Borzecki, Veterans Health Administration.
Jerry Cromwell, University of Massachusetts Boston & Research Triangle Institute.
Laura L. Hayman, RN University of Massachusetts Boston.
Pat Reid Ponte, RN Dana Farber Cancer Institute
Glenn A. Miller, Genzyme Genetics* (currently AstraZeneca Pharmaceutical LP).
Christopher S. Lathan, Dana Farber Cancer Institute.
REFERENCES
- 1.Woodcock J, Woosley R. The FDA critical path initiative and its influence on new drug development. Annu Rev Med. 2008;59:1–12. doi: 10.1146/annurev.med.59.090506.155819. [DOI] [PubMed] [Google Scholar]
- 2.Tevak A, Kondratovich MV, Mansfield E. US FDA and personalized medicine: In vitro diagnostic regulatory perspective. Personalized Medicine. 2010;7(5):517–530. doi: 10.2217/pme.10.53. http://dx.doi.org/10.2217/pme.10.53. [DOI] [PubMed] [Google Scholar]
- 3.Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12(2):175–180. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]
- 4.Haber DA, Gray NS, Baselga J. The evolving war on cancer. Cell. 2011;145(1):19–24. doi: 10.1016/j.cell.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 5.Offit K. Personalized medicine: New genomics, old lessons. Hum Genet. 2011;130(1):3–14. doi: 10.1007/s00439-011-1028-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Auffray C, Caulfield T, Khoury MJ, Lupski JR, Schwab M, Veenstra T. Looking back at genomic medicine in 2011. Genome Med. 2012;4(1):9. doi: 10.1186/gm308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.National Cancer Institute. [Accessed 08/03, 2012];NCI community cancer centers program (NCCCP) fact sheet. http://ncccp.cancer.gov/Media/FactSheet.htm. Updated 2011.
- 8.Olson Steve, Adam C. Genome-based diagnostics: Clarifying pathways to clinical use: Workshop report. The National Academies Press; 2012. Berger R, Roundtable on Translating Genomic-Based Research for Health, Board on Health Sciences Policy, Institute of Medicine. http://www.nap.edu/openbook.php?record_id=13359. [Google Scholar]
- 9.Hernandez LM Institute of Medicine (U.S.). Roundtable on Translating Genomic-Based Research for Health., Institute of Medicine (U.S.) Board on Health Sciences Policy. Diffusion and use of genomic innovations in health and medicine workshop summary. Updated 2008. [Google Scholar]
- 10.Fiore LD, D'Avolio LW. Detours on the road to personalized medicine: Barriers to biomarker validation and implementation. JAMA. 2011;306(17):1914–1915. doi: 10.1001/jama.2011.1605. [DOI] [PubMed] [Google Scholar]
- 11.Carpenter WR, Meyer AM, Wu Y, et al. Translating research into practice: The role of provider-based research networks in the diffusion of an evidence-based colon cancer treatment innovation. Med Care. 2012;50(8):737–748. doi: 10.1097/MLR.0b013e31824ebe13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levy DE, Byfield SD, Comstock CB, et al. Underutilization of BRCA1/2 testing to guide breast cancer treatment: Black and hispanic women particularly at risk. Genet Med. 2011;13(4):349–355. doi: 10.1097/GIM.0b013e3182091ba4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woolf SH. The meaning of translational research and why it matters. JAMA. 2008;299(2):211–213. doi: 10.1001/jama.2007.26. [DOI] [PubMed] [Google Scholar]
- 14.Khoury MJ, Gwinn M, Yoon PW, Dowling N, Moore CA, Bradley L. The continuum of translation research in genomic medicine: How can we accelerate the appropriate integration of human genome discoveries into health care and disease prevention? Genet Med. 2007;9(10):665–674. doi: 10.1097/GIM.0b013e31815699d0. [DOI] [PubMed] [Google Scholar]
- 15.Schully SD, Benedicto CB, Khoury MJ. How can we stimulate translational research in cancer genomics beyond bench to bedside? Genet Med. 2012;14(1):169–170. doi: 10.1038/gim.2011.12. [DOI] [PubMed] [Google Scholar]
- 16.Welch HG, Sharp SM, Gottlieb DJ, Skinner JS, Wennberg JE. Geographic variation in diagnosis frequency and risk of death among medicare beneficiaries. JAMA. 2011;305(11):1113–1118. doi: 10.1001/jama.2011.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glasgow RE, Vinson C, Chambers D, Khoury MJ, Kaplan RM, Hunter C. National institutes of health approaches to dissemination and implementation science: Current and future directions. Am J Public Health. 2012;102(7):1274–1281. doi: 10.2105/AJPH.2012.300755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 19.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 20.Genzyme Genetics. [Accessed 12/4, 2011];Genzyme launches exclusive lung cancer test. http://www.genzyme.com/corp/investors/GENZ%20PR-092705.asp.
- 21.Ettinger DS, Akerley W, Bepler G, et al. Non-small cell lung cancer. J Natl Compr Canc Netw. 2010;8(7):740–801. doi: 10.6004/jnccn.2010.0056. [DOI] [PubMed] [Google Scholar]
- 22.ASCO. [Accessed September 30, 2011];Provisional clinical opinion recommending EGFR mutation testing prior to use of first line targeted drugs for advanced lung cancer. http://www.asco.org/ASCOv2/Press+Center/Latest+News+Releases/ASCO+News/ASCO+Issues+Provisional+Clinical+Opinion+Recommending+EGFR+Mutation+Testing+Prior+to+Use+of+First-Line+Targeted+Drugs+for+Advanced+Lung+Cance. Updated 2010.
- 23.Dacic S. EGFR assays in lung cancer. Adv Anat Pathol. 2008;15(4):241–247. doi: 10.1097/PAP.0b013e31817bf5a9. [DOI] [PubMed] [Google Scholar]
- 24.Yost J. [Accessed 08/12, 2012];CLIA and genetic testing oversight. http://www.genome.gov/Pages/About/OD/ReportsPublications/June2008_YostHoL.pdf. Published August 2011. Updated 2008.
- 25.Fukuoka M, Yano S, Giaccone G, et al. Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced Non–Small-cell lung cancer. Journal of Clinical Oncology. 2003;21(12):2237–2246. doi: 10.1200/JCO.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 26.D'Angelo SP, Pietanza MC, Johnson ML, et al. Incidence of EGFR exon 19 deletions and L858R in tumor specimens from men and cigarette smokers with lung adenocarcinomas. Journal of Clinical Oncology. 2011;29(15):2066–2070. doi: 10.1200/JCO.2010.32.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li E. [Accessed February, 2011];NCCN trends™ survey and data: EGFR mutation testing practices. http://www.nccn.org/about/news/ebulletin/2010-11-01/patient_advocacy.asp. Updated 2010.
- 28.Febbo PG, Ladanyi M, Aldape KD, et al. NCCN task force report: Evaluating the clinical utility of tumor markers in oncology. J Natl Compr Canc Netw. 2011;9(Suppl 5):S1–S32. doi: 10.6004/jnccn.2011.0137. quiz S33. [DOI] [PubMed] [Google Scholar]
- 29.Yang SH, Mechanic LE, Yang P, et al. Mutations in the tyrosine kinase domain of the epidermal growth factor receptor in non-small cell lung cancer. Clin Cancer Res. 2005;11(6):2106–2110. doi: 10.1158/1078-0432.CCR-04-1853. [DOI] [PubMed] [Google Scholar]
- 30.Leidner RS, Fu P, Clifford B, et al. Genetic abnormalities of the EGFR pathway in african american patients with non-small-cell lung cancer. J Clin Oncol. 2009;27(33):5620–5626. doi: 10.1200/JCO.2009.23.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lathan CS, Neville BA, Earle CC. The effect of race on invasive staging and surgery in non-small-cell lung cancer. J Clin Oncol. 2006;24(3):413–418. doi: 10.1200/JCO.2005.02.1758. [DOI] [PubMed] [Google Scholar]
- 32.Cote ML, Haddad R, Edwards DJ, et al. Frequency and type of epidermal growth factor receptor mutations in african americans with non-small cell lung cancer. J Thorac Oncol. 2011;6(3):627–630. doi: 10.1097/JTO.0b013e31820a0ec0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reinersman JM, Johnson ML, Riely GJ, et al. Frequency of EGFR and KRAS mutations in lung adenocarcinomas in african americans. J Thorac Oncol. 2011;6(1):28–31. doi: 10.1097/JTO.0b013e3181fb4fe2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Onega T, Duell EJ, Shi X, Demidenko E, Goodman D. Influence of place of residence in access to specialized cancer care for african americans. J Rural Health. 2010;26(1):12–19. doi: 10.1111/j.1748-0361.2009.00260.x. [DOI] [PubMed] [Google Scholar]
- 35.Lynch J, Lathan C. Differences in utilization of ALK rearrangement FISH analysis and EGFR assay. J Clin Oncol. 2012;30(suppl) abstr e18067. [Google Scholar]
- 36.Murthy VH, Krumholz HM, Gross CP. Participation in cancer clinical trials: Race-, sex-, and age-based disparities. JAMA. 2004;291(22):2720–2726. doi: 10.1001/jama.291.22.2720. [DOI] [PubMed] [Google Scholar]
- 37.Domchek SM, Friebel TM, Singer CF, et al. Association of risk-reducing surgery in BRCA1 or BRCA2 mutation carriers with cancer risk and mortality. JAMA. 2010;304(9):967–975. doi: 10.1001/jama.2010.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ecker JR, Bickmore WA, Barroso I, Pritchard JK, Gilad Y, Segal E. Genomics: ENCODE explained. Nature. 2012;489(7414):52–55. doi: 10.1038/489052a. http://dxdoiorg/101038/489052a. [DOI] [PubMed] [Google Scholar]
- 39.Center for Disease Control and Prevention and National Cancer Institute. CancerGEM KB. [Accessed 08/12, 2012];Cancer Genomic Evidence-based Medicine Knowledge Base. Web site. http://www.hugenavigator.net/CancerGEMKB/home.do. Updated 2012.
- 40.Khoury MJ, Clauser SB, Freedman AN, et al. Population sciences, translational research, and the opportunities and challenges for genomics to reduce the burden of cancer in the 21st century. Cancer Epidemiol Biomarkers Prev. 2011;20(10):2105–2114. doi: 10.1158/1055-9965.EPI-11-0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.