Abstract
The Southern house mosquito Culex quinquefasciatus has the largest repertoire of odorant receptors (ORs) of all mosquitoes and dipteran species whose genomes have been sequenced to date. Previously, we have identified and de-orphanized two ORs expressed in female antennae, CquiOR2 and CquiOR10, which are sensitive to oviposition attractants. In view of a new nomenclature for the Culex genome (VectorBase) we renamed these ORs as CquiOR21 (formerly CquiOR10) and CquiOR121 (CquiOR2). In addition, we selected ORs from six different phylogenetic groups for deorphanization. We cloned four of them by using cDNA from female antennae as a template. Attempts to clone CquiOR87 and CquiOR110 were unsuccessful either because they are pseudogenes or are not expressed in adult female antennae, the main olfactory tissue. By contrast, CquiOR1, CquiOR44, CquiOR73, and CquiOR161 were highly expressed in female antennae. To de-orphanize these ORs, we employed the Xenopus oocyte recording system. CquiORx-CquiOrco-expressed oocytes were challenged with a panel of 90 compounds, including known oviposition attractants, human and vertebrate host odorants, plant kairomones, and naturally occuring repellents. While CquiOR161 did not respond to any test compound in two different laboratories, CquiOR1 showed the features of a generic OR, with strong responses to 1-octen-3-ol and other ligands. CquiOR44 and CquiOR73 showed preference to plant-derived terpenoids and phenolic compounds, respectively. While fenchone was the best ligand for the former, 3,5-dimethylphenol elicited the strongest responses in the latter. The newly de-orphanized ORs may be involved in reception of plant kairomones and/or natural repellents.
Keywords: Culex quinquefasciatus, CquiOR21, CquiOR121, CquiOR1, CquiOR44, CquiOR73, CquiOR161
1. Introduction
The Southern house mosquito, Culex quinquefasciatus Say, has the largest repertoire of odorant receptors (ORs) of all dipteran species whose genomes have been hitherto sequenced (Arensburger et al., 2010) and may possess one of the most, if not the most, acute olfactory system in mosquitoes for the reception of host-derived compounds, such as nonanal (Syed and Leal, 2009). Several species of Culex, including Cx. quinquefasciatus, blood feed on birds and humans and serve as bridge vectors of West Nile virus in the United States (Andreadis, 2012). Throughout the world, Culex mosquitoes are pathogen vectors for human diseases, including filariasis and various types of encephalitis. Understanding how they perceive the world through small, signal-carrying molecules (semiochemicals) may lead us to discover novel repellents for reducing bites and disease transmission as well as “green chemicals” for monitoring and controlling mosquito populations. Only two Culex ORs have been de-orphanized (Hughes et al., 2010; Pelletier et al., 2010) to date. Our initial approach was based on the identification of ORs in the Culex genome that share high amino acid identity with orthologs from the malaria mosquito, Anopheles gambiae. We have demonstrated that these ORs were sensitive to compounds known to be oviposition attractants for Culex mosquitoes (Blackwell et al., 1993; Leal et al., 2008; Mboera et al., 2000; Millar et al., 1992). This approach has limitations as orthologs may be involved only in the detection of common ligands, and the chemical ecology of the malaria and the Southern house mosquitoes differ. For the current study we selected putative Culex quinquefasciatus ORs from six phylogenetic groups, five of which with no Anopheles gambiae orthologs. Following cloning, quantitative PCR analysis was performed to confirm expression in female antennae, and then the ORs were co-expressed with the obligatory co-receptor Orco in Xenopus oocytes for de-orphanization. As reported here, we have identified one OR that responds to multiple compounds and another that did not respond to any compound tested, in addition to an OR displaying stronger responses to plant-derived, natural mosquito repellents, and another sensitive to phenolic compounds, particularly eugenol.
2. Materials and methods
2.1 Phylogenetic analysis of mosquito ORs
Amino acid sequences of mosquito ORs were combined to create an entry file for phylogenetic analysis in Mega 5.05 (Tamura et al., 2011). An unrooted consensus neighbor joining tree was calculated at default settings with pairwise gap deletions. Branch support was assessed by bootstrap analysis based on 1000 replicates. Seventy-six Anopheles gambiae, ninety-nine Aedes aegypti and one-hundred-thirty Culex quinquefasciatus ORs were included in this analysis. Sequence alignments were performed with ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Sequences available in databases were screened for full-length functional ORs based on multiple alignments and prediction of transmembranes. Partial sequences, truncated sequences, and pseudogenes, based on current OR genes annotations, were omitted (AgamOR81; AaegOR6, 12, 18, 22, 29, 32, 35, 38, 39, 51, 54, 57, 64, 68, 73, 77, 82, 83, 86, 91, 97, 108, 112, 116, 118, 120, 126, 127, 128, 129, 130, 131; CquiOR3, 8, 9, 15, 17, 19, 26, 31, 33, 34, 35, 41, 49, 59, 66, 74, 76, 94, 100, 101, 102, 103, 104, 105, 111, 119, 124, 125, 129, 133, 134, 135, 138, 139, 140, 144, 147, 152, 158, 159, 160, 167, 168, 170, 172, 174, 176, 177, 178, 179, 180).
2.2 Insects
Culex quinquefasciatus mosquitoes used in this study were from a laboratory colony maintained at UC Davis. This colony was initiated with adult mosquitoes from a colony maintained by A.J.C. at the Kearney Agricultural Center, University of California, and started from mosquitoes collected in Merced, CA in the 1950s. In Davis, mosquitoes were kept in an insectary at 27±1°C, under a photoperiod of 16:8 h (L:D) for the last 3 years.
2.3. Cloning of OR genes from Cx. quinquefasciatus
Total RNA was extracted from one thousand 1–5-day-old female Cx. quinquefasciatus antennae with TRIzol reagent (Invitrogen, Carlsbad, CA). Antennal cDNA was synthesized from 1 μg of antennal total RNA using SMARTer™ RACE cDNA amplification kit according to manufacturer’s instructions (Clontech, Mountain View, CA). To clone their ORFs into pGEMHE vector, PCR was performed with the following gene specific primers with restriction endonuclease sites (nucleotides upstream of the restriction sites were omitted for brevity): CquiOR1 Fwd-XmaI (underlined) primer 5′ –CCCGGGATGAAATTCGCTCCGCTCCAG-3′ and Rev-XbaI (underlined) primer, 5′-TCTAGATCAGATTCTTTCCTTCAGCAC -3′; CquiOR44 Fwd-XmaI (underlined) primer, 5′-CCCGGGGGGAATGGACACCTGTGCGCATCAG-3′ and Rev-HindIII (underlined) primer, 5′-AAGCTTGGGTTATTTCGTCACCTCGAGCAG -3′; CquiOR73 Fwd-XmaI (underlined) primer, 5′-CCCGGGACCATGTCGTCCATCAACCTTCCAT-3′ and Rev- HindIII (underlined) primer, 5′-AAGCTTGCTCTAGA TCATTCCTCTGCGTAGAGCTGTTG-3′; CquiOR87 Fwd-XmaI (underlined) primer, 5′-CCCGGGGGGAATGAATGACAGTTACAATGTTG-3′ and Rev-XbaI (underlined) primer, 5′-TCTAGAGCCTACATTTTGCTCCCCATC-3′; CquiOR110 Fwd (1)-XmaI (underlined) primer, 5′-CCCGGGGGGAATGGGAATTACCTGTAGTTG-3′, Rev (1)-XbaI (underlined) primer, 5′-TCTAGAGCTTACTCAAACACGCTGAG-3′; CquiOR110 Fwd (2)-XmaI (underlined) primer, 5′-CCCGGGGGGAATGGACTTGAGCTTCATGTTG -3′, Rev (2)-XbaI (underlined) primer, 5′-TCTAGAGCTTAATGTCCCCACGGTAGAAC -3′; and CquiOR161 Fwd-XmaI (underlined) primer, 5′-CCCGGGGATGGCCAACCGAAGAAAGCTC -3′ and Rev-HindIII (underlined) primer, 5′-AAGCTTTTACATATTTTGCAACATCAT -3′.
PCR amplifications were performed using Pfu Ultra II polymerase (Stratagene, La Jolla, CA) under the following condition: 5 cycles of 94°C for 30 s, 57°C for 30 s, 72°C for 3 min, and 30 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 3 min, and then 72°C for 10 min. PCR products were purified using QIAquick Gel Extraction kit (Qiagen, Valencia, CA), ligated into EcoRV site of pBluescript SK (+) (Stratagene) using T4 DNA ligase (Promega, Madison, WI) and transformed using One Shot TOP 10 competent cells (Invitrogen, Carlsbad, CA). After screening colonies, plasmids were extracted using the QIAprep Spin Miniprep kit (Qiagen) and sequenced by ABI 3730 automated DNA sequencer at Davis Sequencing (Davis, CA). Plasmids were digested with appropriate restriction enzymes (20 U/μl) for 2 h at 37°C. Digested products were purified using QIAquick Gel Extraction kit (Qiagen), ligated into pGEMHE, and transformed using One Shot TOP 10 competent cells (Invitrogen). Plasmids were extracted using the QIAprep Spin Miniprep kit (Qiagen) and sequenced by ABI 3730 automated DNA sequencer at Davis Sequencing (Davis, CA) for confirmation.
2.4. Quantitative analysis of OR gene expression (qPCR)
Antennae from 3–5 day old 100 female and 100 male Cx. quinquefasciatus were dissected and collected in DEPC-water on ice using a stereo microscope (Zeiss, Stemi DR 1663, Germany). Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA). cDNA was synthesized from 0.5 μg of total RNA using RT-for-PCR kit according to the manufacturer’s instructions (Clontech, Mountain View, CA). Real-time quantitative PCR (qPCR) was carried out by using a CFX96 TouchTM Real-Time PCR Detection System (Bio-Rad, Hercules, CA) and SsoAdvanced SYBR Green Supermix (Bio-Rad, Hercules, CA): final volume 20 μl, including 200 nM gene specific primers and approximately 50 ng of cDNA. CquiRpS7 gene was used as reference. The primers were designed by Primer 3 program (http://frodo.wi.mit.edu/) and IDT online server (http://www.idtdna.com/scitools/Applications/RealTimePCR/). CquiOR1 forward and reverse; 5′-TCCGGAAAGGAAGATCATTG -3′ and 5′-CGTTACAAACTCGGGACGAT -3′; CquiOR44 forward and reverse; 5′-AGTGGCACAGTGAGATGCAG -3′ and 5′-CACCTCGAGCAGAAACATCA -3′; CquiOR73 forward and reverse; 5′-CTGGGTATGCTGAGGAACTTC-3′ and 5′-GCAGCCAGATCCAAAAGTTG -3′; CquiOR161 forward and reverse; 5′-GTCCAGAGCTGGATCCTCAG -3′ and 5′-AGCGAAAAGGCAAAGTTGAA -3′; CquiRpS7 forward and reverse; 5′-ATCCTGGAGCTGGAGATGA -3′and 5′-GATGACGATGGCCTTCTTGT -3′. Reactions were run with the following standard program: 95°C for 30 s, 39 cycles of 95°C for 5 s, 55°C for 10 s, 72°C for 30 s, melt curve of 65°C to 95°C, increment 0.5°C, 5 s. Data were analyzed using the 2−ΔΔCT method using Bio-Rad CFX Manager 2.1 software.
2.5. In vitro transcription, oocyte microinjection and electrophysiology
In vitro transcription of cRNAs was performed by using a mMESSAGE mMACHINE T7 Kit (Ambion) according to the manufacturer’s protocol. Briefly, plasmids were linearized with NheI or SphI, and capped cRNAs were transcribed using T7 RNA polymerase. The cRNAs were purified with LiCl precipitation solution and re-suspended in nuclease-free water at a concentration of 200 μg/ml and stored at 80°C in aliquots. RNA concentrations were determined by UV spectrophotometry. cRNA were microinjected (2 ng of CquiORX cRNA and 2 ng of CquiOrco cRNA) into stage V or VI Xenopus laevis oocytes (EcoCyte Bioscience, Austin TX). The oocytes were then incubated at 18°C for 3–7 days in modified Barth’s solution [in mM: 88 NaCl, 1 KCl, 2.4 NaHCO3, 0.82 MgSO4, 0.33 Ca(NO3)2, 0.41 CaCl2, 10 HEPES, pH 7.4] supplemented with 10 μg/ml of gentamycin, 10 μg/ml of streptomycin and 1.8 mM sodium pyruvate. The two-electrode voltage clamp (TEVC) was employed to detect inward currents. Oocytes were placed in perfusion chamber and challenged with a panel of 90 compounds in a random order (flow rate was 10 ml/min). Chemical-induced currents were amplified with an OC-725C amplifier (Warner Instruments, Hamden, CT), voltage held at −70 mV, low-pass filtered at 50 Hz and digitized at 1 kHz. Data acquisition and analysis were carried out with Digidata 1440A and software pCLAMP 10 (Molecular Devices, LLC, Sunnyvale, CA).
2.6 Panel of odorants
Oocytes expressing test ORs were challenged with a panel of 90 compounds, including known mosquito oviposition attractants, plant and vertebrate host kairomones, and natural repellents: 1-hexanol, 1-octanol, (E)-2-hexen-1-ol, (Z)-2-hexen-1-ol, 1-hexen-3-ol, 1-heptene-3-ol, 3-octanol, 1-octen-3-ol (Kline et al., 1990), 3-octyn-1-ol, 1-octyn-3-ol, 1-nonanol, 1-hexadecanol, 2-phenoxyethanol, 2,3-butanediol, ethyl acetate, propyl acetate, butyl acetate, pentyl acetate, hexyl acetate, octyl acetate, decyl acetate, (E)-2-hexenyl acetate, (Z)-3-hexenyl acetate, ethyl lactate, methyl propionate, ethyl propionate, methyl butyrate, ethyl 3-hydroxyhexanoate, methyl salicylate, 2-heptanone, 2-nonanone, 2-undecanone, cyclohexanone, acetophenone, 6-methyl-5-hepten-2-one (Birkett et al., 2004; Logan et al., 2009; Logan et al., 2010), 2-butoxylacetone, 2-tridecanone, 2,3-butanedione, ethyl stearate, methyl myristate, γ-valerolactone, γ-hexalactone, γ-octalactone, γ-decalactone, (5R,6S)-6-acetoxy-5-hexadecanolide (MOP) (Laurence and Pickett, 1982), 2-undecanone, propanal, pentanal, hexanal, heptanal, octanal, nonanal (Leal et al., 2008; Syed and Leal, 2009), decanal, undecanal, phenylacetaldehyde, furfural, trans-2-methyl-2-butenal, benzaldehyde, phenol, 2-methylphenol, 3-methylphenol, 4-methylphenol, 4-ethylphenol, 3,5-dimethylphenol, 2,3-dimethylphenol, 2-methoxy-4-propylphenol, guaiacol, indole, 3-methylindole (=skatole) (Blackwell et al., 1993; Leal et al., 2008; Millar et al., 1992; Olagbemiro et al., 2004), butylamine, heptylamine, octylamine, trimethylamine (Leal et al., 2008), nonanoic acid, (±)-lactic acid, geraniol, nerol, geranylacetone (Logan et al., 2009; Logan et al., 2010), trans-p-menthane-3,8-diol, cis-p-menthane-3,8-diol (Paluch et al., 2010), geranyl acetate, (±)-linalool (Choi et al., 2002), (−)-fenchone, (+)-fenchone, (±)-thujone, linalool oxide, (±)-eucalyptol, eugenol (Kafle and Shih, 2013), and (±)-citronellal (Paluch et al., 2010).
3. Results and discussion
3.1 Reconciling Culex OR nomenclature
Prior to publication of the Cx. quinquefasciatus genome (Arensburger et al., 2010), we identified and de-orphanized two ORs from the Southern house mosquito. We named them CquiOR2 (Pelletier et al., 2010) and CquiOR10 (Hughes et al., 2010) based on their high amino acid identity with AgamOR2/AaegOR2 and AgamOR10/AaegOR10 from the mosquitoes Anopheles gambiae and Aedes (Stegomyia) aegypti, respectively. RT-PCR analysis showed that CquiOR2 and CquiOR10 genes are expressed exclusively in olfactory tissues. While neither was detected in non-olfactory tissues from adult females, CquiOR2 was expressed only in antennae, whereas CquiOR10 was expressed mainly in antennae and secondarily in maxillary palps (Pelletier et al., 2010). We then demonstrated with the Xenopus oocyte recording system that CquiOR2 responded to various compounds with indole being the best ligand (Pelletier et al., 2010), whereas CquiOR10 was narrowly tuned to the oviposition attractant skatole (Hughes et al., 2010). CquiOR2 and CquiOR10 shared high amino acid identity with two annotated ORs in the genome of Cx. quinquefasciatus: CquiOR121 (VectorBase, CPIJ802644; formerly CPIJ014392) and CquiOR21 (VectorBase, CPIJ801844; formerly CPIJ002479; previously named CqOR2 in VectorBase), respectively. CquiOR2 and CquiOR121 differ in 4 residues, Glu- vs Gln-89, Phe- vs Val-171, Lys- vs Glu-235, and Asp- vs Glu-301. They may be isoforms caused by single nucleotide polymorphism (SNPs) differences. Cx. quinquefasciatus and related Culex pipiens complex mosquitoes have a very high densities of SNPs, in fact more than any other mosquito thus far studied (Lee et al., 2012). It is worth mentioning that the genome was sequenced from the Johannesburg strain (Arensburger et al., 2010), whereas we cloned the genes (Hughes et al., 2010; Pelletier et al., 2010) using cDNA template from a California strain. CquiOR21 is one residue shorter than CquiOR10 and these proteins differ in two residues: Ala-345 followed by Ile-346 in CquiOR21 and Ile-345-Thr-Val-347 in CquiOR10 (Hughes et al., 2010). The “skipped” threonine (Thr-346) residue could be an error of annotation given that Ile-346 in CquiOR21 (VectorBase) overlaps with an intron splice site, whereas the other differences could be due to polymorphism, including one possible SNP (Val-347 vs. Ile-346). In summary, we assume that CquiOR121 and CquiOR21 in VectorBase are isoforms of CquiOR2 (GenBank, ADF42901) and CquiOR10 (ADF42902), respectively. They might be alleles from the same genes from different populations. Thus, we wish to reconcile these discrepancies in the Culex OR nomenclature by renaming our previously identified CquiORs as CquiOR121 (=CquiOR2) and CquiOR21 (=CquiOR10).
3.2 Current phylogenetic relationship of mosquito ORs
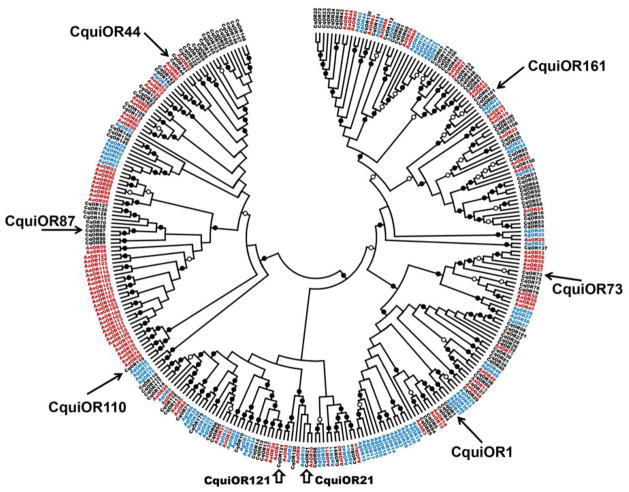
We have revised our previous phylogenetic analysis of mosquito ORs (Pelletier et al., 2010) in view of the annotation of the Culex genome (Arensburger et al., 2010), the update to Cx. quinquefasciatus gene sets (VectorBase), corrections of annotation mistakes (Pitts et al., 2011) and identification of pseudogenes. With these corrections, our estimate of 158 (Pelletier et al., 2010) and a later report of 180 putative OR genes (Arensburger et al., 2010) are now updated to 130 putative OR genes in the Cx. quinquefasciatus genome, whereas Ae. aegypti has 99 putative OR genes and An. gambiae 76 ORs. Despite significant reduction, Culex has still the largest repertoire of ORs of all dipteran species examined to date, as was previously suggested (Arensburger et al., 2010). The observed Culex/Aedes and Aedes/Culex specific expansions (Pelletier et al., 2010) remain valid, as does the Anopheles specific expansion (Fig. 2). In an attempt to identify Culex ORs, we selected 6 putative ORs, five of which with no An. gambiae orthologs and two from these Culex-Aedes expansions, to clone and de-orphanize.
Fig. 2.
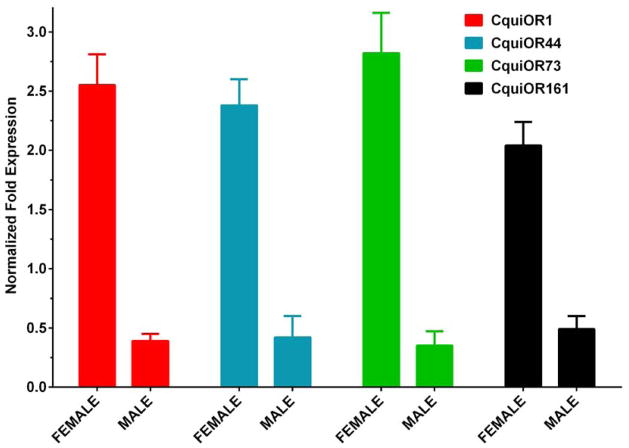
Quantitative PCR data. Comparison of expression of CquiOR1 (red bars), CquiOR44 (blue), CquiOR73 (green), and CquiOR161 (black) transcripts in female and male antennae. Data normalized to the expression of CquiRpS7. N=3.
3.3. Cloning of CquiOR genes and quantitative analysis
Previously we identified two CquiOR genes, CquiOR21 and CquiOR121 (Fig. 1, bottom of the figure). We used the odorant response profiles of An. gambiae ORs (Carey et al., 2010; Wang et al., 2010) to lead us to orthologous ORs in the genome of Cx. quinquefasciatus. Here, we attempted a different approach, i.e., by selecting 6 ORs in the phylogenetic tree, 5 of themwith no An. gambiae orthologs. Starting from the left of the tree (Fig. 1), they are: CquiOR44 (=CPIJ802556), CquiOR87 (=CPIJ802589), CquiOR110 (=CPIJ802608), CquiOR1 (=CPIJ802517), CquiOR73 (=CPIJ802564), and CquiOR161 (=CPIJ802651). Attempts to clone CquiOR87 and CquiOR110 were unrewarding thus suggesting that these genes are not expressed in adult female antennae. We successfully cloned the other genes and their sequences have been deposited in GenBank (CquiOR1, KF032022; CquiOR44, KF032024; CquiOR73, KF032023; CquiOR161, KF032025).
Fig. 1.
Phylogenetic relationships of mosquito ORs. Cx. quinquefasciatus ORs are in black, An. gambiae ORs are in blue and Ae. aegypti ORs are in red. Black and white circles indicate bootstrap values at specific nodes (black: 94–100%; white: 79–93%).
Quantitative PCR (qPCR) analysis showed that, not surprisingly, CquiOR1, CquiOR44, CquiOR73, and CquiOR161 were more highly expressed in female antennae (Fig. 2), but our analyses were not designed to quantify their expression levels. Thus, we proceeded to de-orphanize the newly cloned ORs with a panel of 90 compounds, including oviposition attractants, plant-derived kairomones, repellents from natural sources, and mosquito attractants.
3.4. De-orphanization of CquiORs
We subcloned CquiOR1, CquiOR44, CquiOR73, and CquiOR161 into pGEMHE, expressed them along with the obligatory co-receptor CquiOrco in Xenopus oocytes, and then performed electrophysiological recordings by subjecting oocytes to our panel of test compounds. CquiOR1CquiOrco-expressing oocytes behaved like a generic OR (Fig. 3), i.e., an OR that does not have a specific ligand, but responds to multiple compounds. Albeit responses were small in general, the strongest current amplitudes were recorded when CquiOR1 was challenged with 1-hexanol, 1-octen-3-ol, 2-phenoxyethanol, or benzaldehyde (Fig. 3, Fig. 4). Likewise, CquiOR44 was activated by multiple odorants at low level, but interestingly the strongest responses were recorded when CquiOR44•CquiOrco-expressing oocytes were challenged with plant kairomones (Fig. 3), including known natural repellents like p-menthane-3,8-diol (Paluch et al., 2010) and eucalyptol (Omolo et al., 2004). The most active ligand was fenchone (Fig. 4), but there was apparently no chiral discrimination as responses to (+)- and (−)-fenchone did not differ.
Fig. 3.
Electrophysiological recording from oocytes expressing candidate CquiORs along with co-receptor CquiOrco. Traces obtained with oocytes expressing CquiOR1 (red), CquiOR44 (blue), CquiOR73 (green), and CquiOR161 (black). CquiOR 1 and CquiOR44 behave like generic ORs, although CquiOR44 is more tuned to terpenoid compounds, particularly fenchone.
Fig. 4.
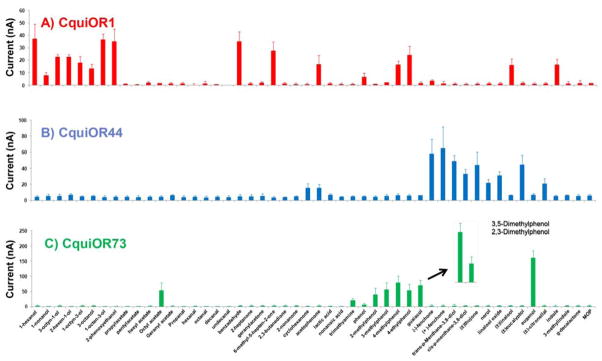
Quantification of current responses of oocytes expressing Culex ORs. CquiOR1 (red bars), CquiOR44 (blue), and CquiOR73 (green). Mean ± SEM, N=3–5
When challenged with the same panel of compounds CquiOR73•CquiOrco-expressing oocytes responded differently. Robust responses were seen with eugenol, smaller responses to phenolic compounds, particularly 4-methylphenol (Fig. 4), and no significant response to the majority of compounds in the panel, except for octyl acetate. Then, we repeated these experiments by focusing on phenolic compounds, including dimethylphenols (Fig. 4). These experiments showed strong responses elicited by 3,5-dimethylphenol (Fig. 3), stronger than those generated by other phenolic compounds, including methylphenols, but eugenol was the best ligand identified for this OR (Fig. 4). Based on these experiments we concluded that CquiOR73 is an eugenol-detecting OR, but the significance of a receptor tuned to phenolic compounds remains an interesting topic for future research. It did not escape our attention, however, that eugenol has been identified as a plant-derived insect repellent (Kafle and Shih, 2013).
Lastly, we attempted to de-orphanize CquiOR161, but in marked contrast to the above-mentioned ORs, it did not respond to any of the test compounds. Despite several attempts at the UC Davis laboratory, CquiOR161 remained silent. We then re-tested this OR in the UM laboratory with a panel of compounds, which, in addition to the compounds already tested at UC Davis, had the following compounds: 1-methylindole, 2-methylindole, 4-methylindole, 5-methylindole, 6-methylindole, 7-methylindole, 3-octanone, 2-tridecanone, 1-dodecanol, 4-propylbenzaldehyde, methyl benzoate, 2-ethoxythiazole, 2-isobutylthiazole, (+)-carvone, isoamylacetate, heptanoic acid, octanoic acid, decanoic acid, undecanoic acid, 2-acetylthiophene, and 2-butoxyethanol. None of these ligands activated CquiOR161•CquiOrco-expressing oocytes. As a positive control, CquiOR1•CquiOrco-expressing oocytes in the UM laboratory gave medium to large responses when challenged with indole, 4-ethylphenol, 4-methylphenol, phenol, acetophenone, benzaldehyde, and 6-methyl-5-hepten-2-one. Although we cannot rule out the possibility that we did not challenge CquiOR161 with the right ligand, this seems unlikely as in both labs we subjected oocytes expressing the receptor to all currently known odorants with physiological and/or ecological significance in Culex mosquitoes.
In conclusion, we have cloned four ORs, which are enriched in female mosquito antennae. Despite several attempts, one of them, CquiOR161, was silent as it did not respond to any of ligands tested. By contrast, CquiOR1 showed behavior of a generalist OR as it responded to various compounds, including alcohols and ketones of biological significance. Another OR, CquiOR73, was more tuned to phenolic compounds, with eugenol, which is the major constituent of clover oil and has mosquito repellent activity, being the best ligand. Lastly, CquiOR44 showed robust responses only to plant-derived terpenoid compound, particularly fenchone. The newly de-orphanized ORs might be involved in the detection of plant-derived kairomones and/or repellents.
Highlights.
Phylogenetic relationships of mosquito ORs are revised
SNPs and polymorphisms are identified in Cx. quinquefasciatus ORs
Four Culex ORs enriched in female antennae are cloned and three ORs are de-orphanized
Two Culex ORs are sensitive to plant-derived repellents
Acknowledgments
Research reported in this publication was supported by the National Institutes of Health under awards R01AI095514 from the National Institute of Allergy and Infectious Diseases (to W.S.L.) and RO1DC011091 from the National Institute on Deafness and Other Communicative Disorders (to C.W.L.). The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH. F.R.S. (Universidade de São Paulo, Campus of Piracicaba) received an undergraduate scholarship from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) under a FIPSE-CAPSE sponsored US-Brazil Higher education Consortium Program. FZ sabbatical leave at UC Davis was supported in part by the China Scholarship Council.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andreadis TG. The contribution of Culex pipiens complex mosquitoes to transmission and persistence of West Nile virus in North America. Journal of the American Mosquito Control Association. 2012;28:137–151. doi: 10.2987/8756-971X-28.4s.137. [DOI] [PubMed] [Google Scholar]
- Arensburger P, Megy K, Waterhouse RM, Abrudan J, Amedeo P, Antelo B, Bartholomay L, Bidwell S, Caler E, Camara F, Campbell CL, Campbell KS, Casola C, Castro MT, Chandramouliswaran I, Chapman SB, Christley S, Costas J, Eisenstadt E, Feschotte C, Fraser-Liggett C, Guigo R, Haas B, Hammond M, Hansson BS, Hemingway J, Hill SR, Howarth C, Ignell R, Kennedy RC, Kodira CD, Lobo NF, Mao C, Mayhew G, Michel K, Mori A, Liu N, Naveira H, Nene V, Nguyen N, Pearson MD, Pritham EJ, Puiu D, Qi Y, Ranson H, Ribeiro JM, Roberston HM, Severson DW, Shumway M, Stanke M, Strausberg RL, Sun C, Sutton G, Tu ZJ, Tubio JM, Unger MF, Vanlandingham DL, Vilella AJ, White O, White JR, Wondji CS, Wortman J, Zdobnov EM, Birren B, Christensen BM, Collins FH, Cornel A, Dimopoulos G, Hannick LI, Higgs S, Lanzaro GC, Lawson D, Lee NH, Muskavitch MA, Raikhel AS, Atkinson PW. Sequencing of Culex quinquefasciatus establishes a platform for mosquito comparative genomics. Science. 2010;330:86–88. doi: 10.1126/science.1191864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkett MA, Agelopoulos N, Jensen KM, Jespersen JB, Pickett JA, Prijs HJ, Thomas G, Trapman JJ, Wadhams LJ, Woodcock CM. The role of volatile semiochemicals in mediating host location and selection by nuisance and disease-transmitting cattle flies. Medical and veterinary entomology. 2004;18:313–322. doi: 10.1111/j.0269-283X.2004.00528.x. [DOI] [PubMed] [Google Scholar]
- Blackwell A, Mordue AJ, Hansson BS, Wadhams LJ, Pickett JA. A behavioral and electrophysiological study of oviposition cues for Culex quinquefasciatus. Physiol Entomol. 1993;18:343–348. [Google Scholar]
- Carey AF, Wang G, Su CY, Zwiebel LJ, Carlson JR. Odorant reception in the malaria mosquito Anopheles gambiae. Nature. 2010;464:66–71. doi: 10.1038/nature08834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WS, Park BS, Ku SK, Lee SE. Repellent activities of essential oils and monoterpenes against Culex pipiens pallens. Journal of the American Mosquito Control Association. 2002;18:348–351. [PubMed] [Google Scholar]
- Hughes DT, Pelletier J, Luetje CW, Leal WS. Odorant receptor from the southern house mosquito narrowly tuned to the oviposition attractant skatole. Journal of chemical ecology. 2010;36:797–800. doi: 10.1007/s10886-010-9828-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafle L, Shih CJ. Toxicity and repellency of compounds from clove (Syzygium aromaticum) to red imported fire ants Solenopsis invicta (Hymenoptera: Formicidae) Journal of economic entomology. 2013;106:131–135. doi: 10.1603/ec12230. [DOI] [PubMed] [Google Scholar]
- Kline DL, Wood JR, Morris CD. Evaluation of 1-octen-3-ol as an attractant for Coquillettidia perturbans, Mansonia spp. and Culex spp. associated with phosphate mining operations. Journal of the American Mosquito Control Association. 1990;6:605–611. [PubMed] [Google Scholar]
- Laurence BR, Pickett JA. erythro-6-Acetoxy-5-hexadecanolide, the major compound of a mosquito attractant pheromone. Journal Chemical Society, Chemical communications. 1982:59–60. doi: 10.1007/BF00988573. [DOI] [PubMed] [Google Scholar]
- Leal WS, Barbosa RM, Xu W, Ishida Y, Syed Z, Latte N, Chen AM, Morgan TI, Cornel AJ, Furtado A. Reverse and conventional chemical ecology approaches for the development of oviposition attractants for Culex mosquitoes. PloS one. 2008;3:e3045. doi: 10.1371/journal.pone.0003045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Seifert SN, Nieman CC, McAbee RD, Goodell P, Fryxell RT, Lanzaro GC, Cornel AJ. High degree of single nucleotide polymorphisms in California Culex pipiens (Diptera: Culicidae) sensu lato. Journal of medical entomology. 2012;49:299–306. doi: 10.1603/me11108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan JG, Seal NJ, Cook JI, Stanczyk NM, Birkett MA, Clark SJ, Gezan SA, Wadhams LJ, Pickett JA, Mordue AJ. Identification of human-derived volatile chemicals that interfere with attraction of the Scottish biting midge and their potential use as repellents. Journal of medical entomology. 2009;46:208–219. doi: 10.1603/033.046.0205. [DOI] [PubMed] [Google Scholar]
- Logan JG, Stanczyk NM, Hassanali A, Kemei J, Santana AE, Ribeiro KA, Pickett JA, Mordue Luntz AJ. Arm-in-cage testing of natural human-derived mosquito repellents. Malaria journal. 2010;9:239. doi: 10.1186/1475-2875-9-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mboera LE, Takken W, Mdira KY, Pickett JA. Sampling gravid Culex quinquefasciatus (Diptera: Culicidae) in Tanzania with traps baited with synthetic oviposition pheromone and grass infusions. Journal of medical entomology. 2000;37:172–176. doi: 10.1603/0022-2585-37.1.172. [DOI] [PubMed] [Google Scholar]
- Millar JG, Chaney JD, Mulla MS. Identification of oviposition attractants for Culex quinquefasciatus from fermented Bermuda grass infusions. Journal of the American Mosquito Control Association. 1992;8:11–17. [PubMed] [Google Scholar]
- Olagbemiro TO, Birkett MA, Mordue Luntz AJ, Pickett JA. Laboratory and field responses of the mosquito, Culex quinquefasciatus, to plant-derived Culex spp. oviposition pheromone and the oviposition cue skatole. Journal of chemical ecology. 2004;30:965–976. doi: 10.1023/b:joec.0000028461.86243.19. [DOI] [PubMed] [Google Scholar]
- Omolo MO, Okinyo D, Ndiege IO, Lwande W, Hassanali A. Repellency of essential oils of some Kenyan plants against Anopheles gambiae. Phytochemistry. 2004;65:2797–2802. doi: 10.1016/j.phytochem.2004.08.035. [DOI] [PubMed] [Google Scholar]
- Paluch G, Bartholomay L, Coats J. Mosquito repellents: a review of chemical structure diversity and olfaction. Pest management science. 2010;66:925–935. doi: 10.1002/ps.1974. [DOI] [PubMed] [Google Scholar]
- Pelletier J, Hughes DT, Luetje CW, Leal WS. An odorant receptor from the southern house mosquito Culex pipiens quinquefasciatus sensitive to oviposition attractants. PloS one. 2010;5:e10090. doi: 10.1371/journal.pone.0010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts RJ, Rinker DC, Jones PL, Rokas A, Zwiebel LJ. Transcriptome profiling of chemosensory appendages in the malaria vector Anopheles gambiae reveals tissue- and sex-specific signatures of odor coding. BMC genomics. 2011;12:271. doi: 10.1186/1471-2164-12-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed Z, Leal WS. Acute olfactory response of Culex mosquitoes to a human- and bird-derived attractant. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18803–18808. doi: 10.1073/pnas.0906932106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular biology and evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Carey AF, Carlson JR, Zwiebel LJ. Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:4418–4423. doi: 10.1073/pnas.0913392107. [DOI] [PMC free article] [PubMed] [Google Scholar]