Abstract
Medulloblastoma (MB) is the most common malignant brain tumor in children. While aggressive surgery, radiation, and chemotherapy have improved outcomes, survivors suffer severe long-term side effects, and many patients still succumb to their disease. For patients whose tumors are driven by mutations in the Sonic hedgehog (SHH) pathway, SHH antagonists offer some hope. However, many SHH-associated MBs do not respond to these drugs, and those that do may develop resistance. Therefore, more effective treatment strategies are needed for both SHH and non-SHH-associated MB. One such strategy involves targeting the cells that are critical for maintaining tumor growth, known as tumor-propagating cells (TPCs). We previously identified a population of TPCs in tumors from patched mutant mice, a model for SHH-dependent MB. These cells express the surface antigen CD15/SSEA-1 and have elevated levels of genes associated with the G2/M phases of the cell cycle. Here, we show that CD15+ cells progress more rapidly through the cell cycle than CD15- cells and contain an increased proportion of cells in G2/M, suggesting that they might be vulnerable to inhibitors of this phase. Indeed, exposure of tumor cells to inhibitors of Aurora and Polo-like kinases, key regulators of G2/M, induces cell cycle arrest, apoptosis and enhanced sensitivity to conventional chemotherapy. Moreover, treatment of tumor-bearing mice with these agents significantly inhibits tumor progression. Importantly, cells from human patient-derived MB xenografts are also sensitive to Aurora and Polo-like kinase inhibitors. Our findings suggest that targeting G2/M regulators may represent a novel approach for treatment of human MB.
Keywords: Medulloblastoma, Cancer stem cell, Aurora kinase, Polo-like kinase, Hedgehog
Introduction
Medulloblastoma (MB) is the most common malignant pediatric brain tumor, with the majority of cases occurring in children under the age of 15 (1). MB patients are commonly treated with surgery, radiation, and chemotherapy, but survivors suffer severe side effects, including cognitive and developmental deficits and an increased risk of secondary tumors later in life (2, 3). Therefore, alternative approaches to treatment of MB are essential.
Recent genomic analyses have identified four major subtypes of MB that differ from one another in terms of gene expression, DNA copy number and mutation, epidemiology and prognosis. Although the genetic drivers of these subtypes are not fully understood, one group of tumors – representing ~25% of MB cases – is characterized by activation of the SHH signaling pathway. In some cases, this activation can be attributed to mutation or amplification of known pathway components, including the membrane proteins Patched (PTCH1) and Smoothened (SMO), the cytoplasmic regulator Suppressor of Fused (SUFU), and the transcription factors GLI1 and GLI2; however, in many cases, the basis for SHH pathway activation remains unclear (4).
Patients with SHH-associated tumors have a variable prognosis, with a subset faring poorly despite aggressive therapy (4). The development of small-molecule inhibitors of the SHH pathway has offered some hope for these patients (5, 6). These agents represent one of the first classes of targeted therapies for MB, but they are effective only in patients with SHH-associated tumors. Importantly, most of these compounds act on SMO and are thus unlikely to be active against tumors driven by mutations in downstream components such as SUFU and GLI (7). Moreover, recent studies in both patients and animals have shown that PTCH- and SMO-driven tumors that initially respond to SHH antagonists quickly develop resistance (6, 8). Thus, more innovative approaches to therapy are required for SHH-associated MB.
One approach to improving treatment of MB may involve targeting tumor-propagating cells (TPCs). TPCs, often called cancer stem cells, are operationally defined as the cells within a tumor that are capable of regenerating the tumor upon transplantation into a naïve host. TPCs have been identified in multiple tumor types, including those of the brain, breast, prostate, colon, pancreas, liver, lung, and skin, among others (9–16). The ability of TPCs to regenerate tumors has led to the notion that these cells are responsible for tumor resistance and recurrence after therapy. Indeed, TPCs have been shown to display resistance to both chemotherapy and radiation (17–19). Given this capacity for evading standard therapies and regenerating tumors, identification of therapeutic approaches to target and eliminate these cells could substantially improve patient outcomes.
We recently identified a population of TPCs in patched heterozygous mice, a widely studied mouse model of SHH-associated MB (20). These cells, which can be identified based on their expression of the cell surface carbohydrate antigen CD15/SSEA-1, are not multipotent and cannot form neurospheres, but are uniquely capable of propagating tumors following transplantation. When CD15+ cells are transplanted into the cerebella of naïve mice, 100% of recipients develop tumors, whereas CD15- cells never generate tumors. Expression profiling revealed that CD15+ cells display decreased expression of genes associated with differentiation and elevated expression of genes associated with proliferation. CD15 is also found in a subset of human MBs, and patients whose tumors express high levels of a CD15-associated gene signature have a poorer prognosis.
Because CD15+ cells are critical for tumor propagation, we hypothesized that further understanding the properties of these cells might enable us to identify vulnerabilities that could be targeted by therapeutic intervention. Here, we report that CD15+ cells from patched mutant tumors display elevated expression of genes encoding regulators of G2 and M phases of the cell cycle and a corresponding over-representation of cells in G2/M phase. Furthermore, inhibition of Aurora kinases (Aurk) or Polo-like kinases (Plk), important G2/M regulators, inhibits proliferation in vitro and blocks tumor growth in vivo. Therefore, targeting TPCs through inhibition of G2/M regulators may represent a novel approach for improving treatment of patients with this disease.
Materials and Methods
Mice
All animal experiments were performed according to protocols approved by the Institutional Animal Care and Use Committees of Duke University and the Sanford-Burnham Medical Research Institute. Germline patched heterozygous mutant mice (21) were maintained by breeding with 129X1/SvJ or C57BL/6 mice from The Jackson Laboratory (Bar Harbor, ME). Conditional Math1-CreER; Ptcflox/flox mice (22) were treated with 0.8 mg of tamoxifen (T5648, Sigma, St. Louis, MI) in 40 µl of corn oil at post-natal day 4 to generate tumors 10–16 weeks later. CD-1 Nu/Nu mice were obtained from Charles River Laboratories (Wilmington, MA), and NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NOD scid gamma; NSG) mice were obtained from The Jackson Laboratory.
Human tumor isolation and propagation
Human MB tissue for patient-derived xenografts was obtained from surgical resection of tumors at Duke University Medical Center (Durham, NC) or Rady Children’s Hospital (San Diego, CA). All procedures using human tissue were approved by the Institutional Review Boards of the respective institutions. Upon retrieval, the tissue was mechanically dissociated into a single-cell suspension, then immediately injected into the cerebella of NSG mice. When the mice became symptomatic, the tumors were again dissociated into single-cell suspensions and then re-transplanted back into the cerebella of naïve hosts to establish a propagated line for each patient-derived xenograft.
Molecular classification of human tumors
The tumors were assigned molecular subgroups using a class prediction algorithm, Prediction Analysis for Microarrays (PAM) (23), as implemented in the pamr R package (v 1.51). The RNA expressions of subgroup-specific markers were measured by a NanoString assay (24) and subsequently used as features for class prediction. Predicted subgroups with confidence probabilities higher than established thresholds (24) were considered bona fide subgroup assignments. Principal component analyses (PCA) plots were generated by PCA on the training data. The resulting Eigen vectors were used to project the expression profiles of the classified samples onto the vector space spanned by the first two Eigen vectors of the training data. The background confidence score gradient was generated using 200 replicates of the training data with Gaussian noise and subsequently smoothed by Nadaraya-Watson normalization (fields v6.7.6 R package).
Chemicals
The Aurk inhibitors VX-680, PHA-739358, SNS-314, CYC116, AT9283, MLN8237, PHA-680602, CCT129202, ENMD-2076, and AZD1152-HQPA, the Plk inhibitors BI-2536, BI-6727, GSK461364, and ON-01910, and the chemotherapeutic agents Vincristine, Cisplatin, and Cyclophosphamide were obtained from Selleck Chemicals (Houston, TX). The SHH antagonist NVP-LDE225 was kindly provided by Novartis (Boston, MA).
Tumor cell isolation and culture
Tumors were obtained from germline patched heterozygous or conditional Math1-CreER; Ptcflox/flox mice, and each experiment was performed multiple times using cells isolated from each strain. The complete tumor dissociation procedure has previously been described (20, 22). Briefly, tumors were digested in a papain solution to obtain a single-cell suspension, then centrifuged through a 35%-65% Percoll gradient. Cells from the 35%–65% interface were suspended in Dulbecco’s Phosphate-Buffered Saline (DPBS) plus 5% Fetal Bovine Serum (FBS) for cell sorting or in NB-NS21 (Neurobasal with 1 mM sodium pyruvate, 2 mM L-glutamine, penicillin/streptomycin, and NS-21 supplement (25)) plus 1% FBS (Invitrogen) for culture. The cells were plated on growth factor-reduced matrigel-(BD Biosciences, La Jolla, CA) coated plates.
Cell sorting
To obtain CD15+ and CD15− cell populations, cells were stained with control mouse IgM or anti-CD15 (clone MMA, BD Biosciences) antibodies, followed by anti-mouse IgM-phycoerythrin (PE) (Jackson ImmunoResearch, West Grove, PA). The cells were then sorted on a FACSVantage or FACSVantage SE DiVa flow cytometer (BD Biosciences). After sorting, the cells were pelleted and resuspended in NB-NS21 culture media or frozen until use for expression analysis.
Real-time PCR
Real-time PCR was performed to examine the mRNA expression levels of AurkA, AurkB, and Plk1 in the CD15+ and CD15− populations. mRNA was prepared using an RNeasy kit (QIAGEN Inc., Valencia, CA), and real-time PCR was performed using the QuantiTect SYBR Green RT-PCR kit (QIAGEN). Each reaction consisted of 10 ng of the appropriate RNA, 12.5 µl of 2X QuantiTect SYBR Green RT-PCR Master Mix, 1.25 µl of a 10 µM stock of the appropriate forward and reverse primers, 0.25 µl of QuantiTect RT mix, and RNase-free water in a total volume of 25 µl. The following primer sequences were used: Aurora kinase A, F: GTTCCCTTCGGTCCGAAA, R: AATCATTTCCGGAGGCTG; Aurora kinase B, F: TCAGAAGGAGAACGCCTACCC, R: GACTCTCTGGGACAACGTGTT; Polo-like kinase 1, F: ACGTCGTAGGCTTCCATGAC, R: CTCGTTCAGGAAGAGGTTGC; and Actin, F: TATTGGCAACGAGCGGTTCC, R: GGCATAGAGGTCTTTACGGATGTC. Duplicate reactions were prepared without the QuantiTect RT mix to confirm the absence of genomic DNA contamination. The following reaction conditions were run on a Bio-Rad C1000 Thermal Cycler and CFX96 Real-time System (Bio-Rad Laboratories, Hercules, CA): reverse transcription at 50°C for 30 minutes, HotStarTaq DNA Polymerase activation at 95°C for 15 minutes, followed by 40 cycles of 94°C for 15 seconds, 60°C for 30 seconds, and 72°C for 30 seconds. Each reaction was analyzed in triplicate using the delta Ct method to determine the level of expression of each gene in the CD15+ population relative to the CD15− population in each tumor.
BrdU and cell cycle analysis
To monitor cell cycle kinetics, tumor cells were first sorted into CD15+ and CD15− populations as described above. After sorting, 2 million cells per well were plated into 24-well plates in NB-NS21 culture media. The cells were pulsed with BrdU for 30 minutes, then washed with media to remove any remaining BrdU. Cells were collected immediately after the pulse (“30 minutes”), or 6, 12, or 24 hours later, then fixed and stained using the FITC BrdU Flow Kit (BD Biosciences) and 7-Aminoactinomycin (7-AAD) according to the manufacturer’s instructions. For cell cycle analysis of cells that were not labeled with BrdU, the same kit was used for fixation, permeabilization, and 7-AAD staining, but the anti-BrdU staining step was eliminated. The analysis was performed using a FACScan or FACSCanto flow cytometer (BD Biosciences) and FlowJo v.7.6.4 software (Tree Star, Inc., Ashland, OR).
CFSE analysis
As an alternative approach to measure the timing of cell divisions in CD15+ and CD15− populations, patched mutant tumor cells were sorted, then labeled with 1 µM carboxyfluorescein diacetate (CFSE) at a density of 1 × 106 cells/ml using the CellTrace CFSE Cell Proliferation kit (Invitrogen). The cells were then cultured for 48, 72, or 96 hours, fixed with 2% PFA, and CFSE fluorescence was analyzed using a FACSCanto flow cytometer (BD Biosciences) and FlowJo v.7.6.4 software (Tree Star, Inc).
Western blotting
To assess the levels of Histone H3 phosphorylation following treatment with inhibitors, cells were cultured in 24-well plates at a density of 2.5 million cells per well in the presence of the indicated concentrations of DMSO, VX-680, or BI-2536. Cells were then lysed in RIPA buffer (Millipore, Billerica, MA) containing 1 mM sodium orthovanadate, 2 mM sodium fluoride (both from Sigma, St. Louis, MO), and Complete, Mini, EDTA-free protease inhibitor tablets (Roche Applied Science, Indianapolis, IN). Proteins (30 µg) were resolved by SDS-PAGE and transferred to nitrocellulose membranes (Invitrogen), which were then probed with antibodies against phospho-Histone H3 (Ser 10) (Millipore), total Histone H3 (Cell Signaling Technology, Danvers, MA), Actin (Santa Cruz Biotechnology, Santa Cruz, CA), or GAPDH (Cell Signaling Technology), followed by goat anti-rabbit antibodies conjugated to IRdye 680 (Rockland, Gilbertsville, PA). Proteins were detected using the Odyssey imaging system (LI-COR, Lincoln, NE).
Proliferation and apoptosis assays
To examine the effects of inhibitors on proliferation, tumor cells from patched mutant mice or patient-derived xenografts were isolated as described above and plated in 96-well plates at a density of 0.2 million cells per well. Cells were cultured in the presence of the indicated concentrations of inhibitors for 48 hours in triplicate wells, then pulsed with [methyl-3H]thymidine (Amersham/GE Healthcare, Piscataway, NJ) and cultured for an additional 16–18 hours. Cells were harvested onto filters using a Mach IIIM Manual Harvester 96 (Tomtec, Hamden, CT), and incorporated radioactivity was quantified by liquid scintillation spectrophotometry on a Wallac MicroBeta scintillation counter (PerkinElmer, Waltham, MA).
To measure the effects of inhibitors in combination with radiation, patched mutant tumor cells were plated in 96-well plates at a density of 0.2 million cells per well and cultured in the presence of DMSO, 10 nM BI-2536, or 30 nM VX-680. After 24 hours, cells were subjected to 0, 0.25, 0.5, or 1 Gy radiation using a Gammacell 40 Exactor (Low-dose cesium 137 irradiator) (Best Theratronics Ltd., Ottawa, Ontario, Canada). The cells were then cultured for an additional 24 hours, and [methyl-3H]thymidine assays were performed as described above.
To determine whether Aurk or Plk inhibitors induce apoptosis in vitro, tumor cells from patched mutant mice were isolated as described above and plated in 48-well plates at a density of 0.5 million cells per well. Cells were cultured in the presence of the indicated concentrations of inhibitors for 48 hours, then collected and suspended in 100 µl Annexin-binding buffer containing 5 µL of Annexin V-FITC (BD Biosciences). The cells were incubated at room temperature for 15 minutes, 400 µL of Annexin-binding buffer was added, and the percentage of Annexin V-FITC-bound cells was analyzed using a FACSCanto flow cytometer.
In vivo drug administration
To assess the effects of Aurk or Plk inhibition on tumor growth, 8 million cells from patched mutant mice were suspended in 50% NB-NS21/50% growth-factor reduced matrigel and subcutaneously injected in a total volume of 100 µl into the flanks of CD-1 Nu/Nu mice. Tumors were measured using calipers, and tumor volumes were calculated using the formula volume = 0.52 x length x width2 (26). Drug treatment was initiated when tumors reached a volume of approximately 150 mm3. Animals were treated with 50 mg/kg BI-2536 (suspended in 0.1 N HCl, then diluted in saline) twice weekly via tail vein. Animals treated with PHA-739358 (30 mg/kg in 5% dextrose) were injected intraperitoneally twice daily. The animals were sacrificed when the largest tumor volume in the cohort exceeded 2 cm3. After sacrifice, tumors were collected, weighed and photographed.
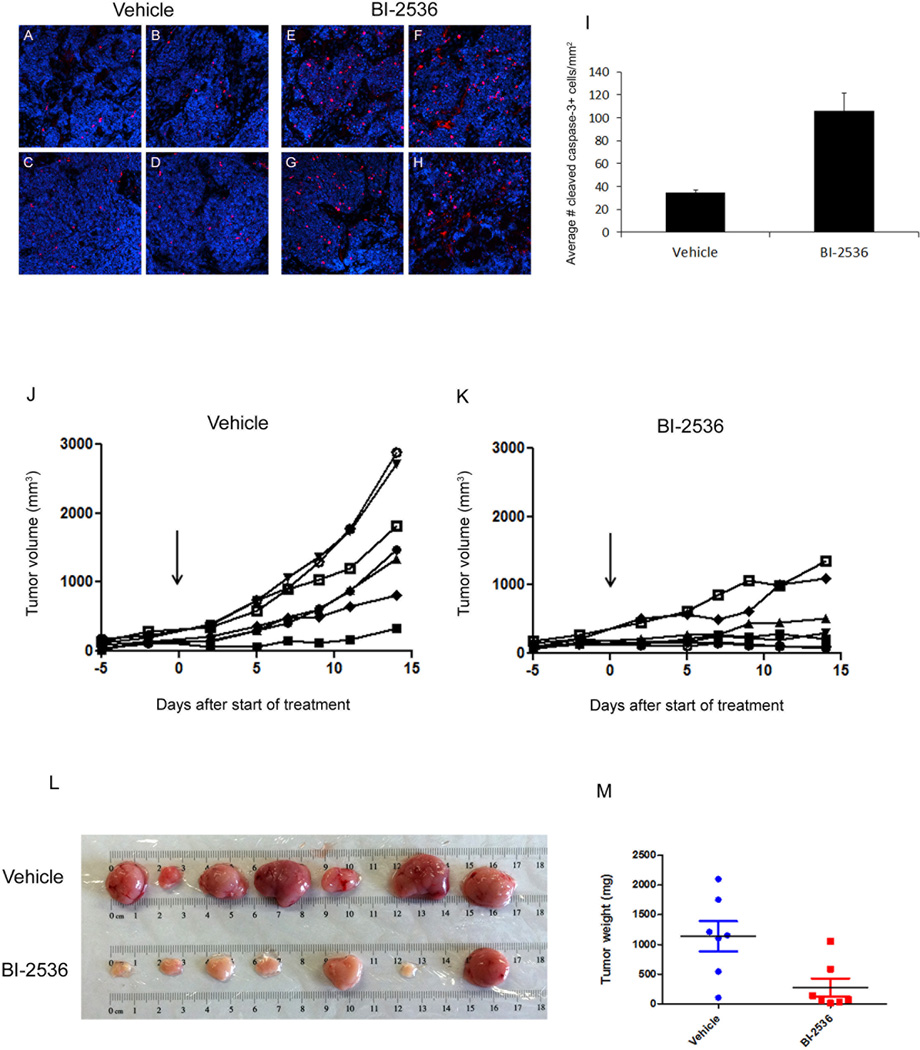
To determine whether Plk inhibition induces apoptosis in vivo, tumor-bearing mice were treated with a single dose of vehicle or BI-2536, and tumors were harvested 24 hours later and fixed overnight in 4% PFA. After fixation, samples were transferred to a solution of 30% sucrose (Sigma) for 2 days, then embedded in Tissue-Tek Optimal Cutting Temperature (OCT) compound (Sakura Finetek USA, Inc., Torrance, CA). Samples were then stored at −80°C until sectioning (12 µm) on a Leica CM3050S Cryostat (Leica Microsystems, Inc., Buffalo Grove, IL). For immunostaining, tumor sections were blocked and permeabilized for 1 hour with PBS containing 0.1% Triton X-100 and 1% normal goat serum, stained with anti-cleaved caspase-3 antibodies (Cell Signaling Technology) overnight at 4°C, and incubated with Alexa Fluor-594 anti-rabbit IgG (1:200) secondary antibodies for 45 minutes at room temperature. Sections were counterstained with 4',6-diamidino-2-phenylindole (DAPI; Invitrogen) and mounted with Fluoromount-G (SouthernBiotech, Birmingham, AL). Images were acquired using a Zeiss LSM 700 confocal microscope (Carl Zeiss Microscopy, LLC, Thornwood, NY). Quantitation of apoptosis was performed by acquiring images of 6 representative regions from 4 tumors from vehicle-treated mice and 4 tumors from BI-2536-treated mice and analyzing the number of cleaved caspase-3-positive cells relative to the DAPI-occupied area within each image using Image Pro-Plus 7.0 software (Media Cybernetics, Inc., Rockville, MD).
Results
CD15+ cells display elevated expression of G2/M regulators
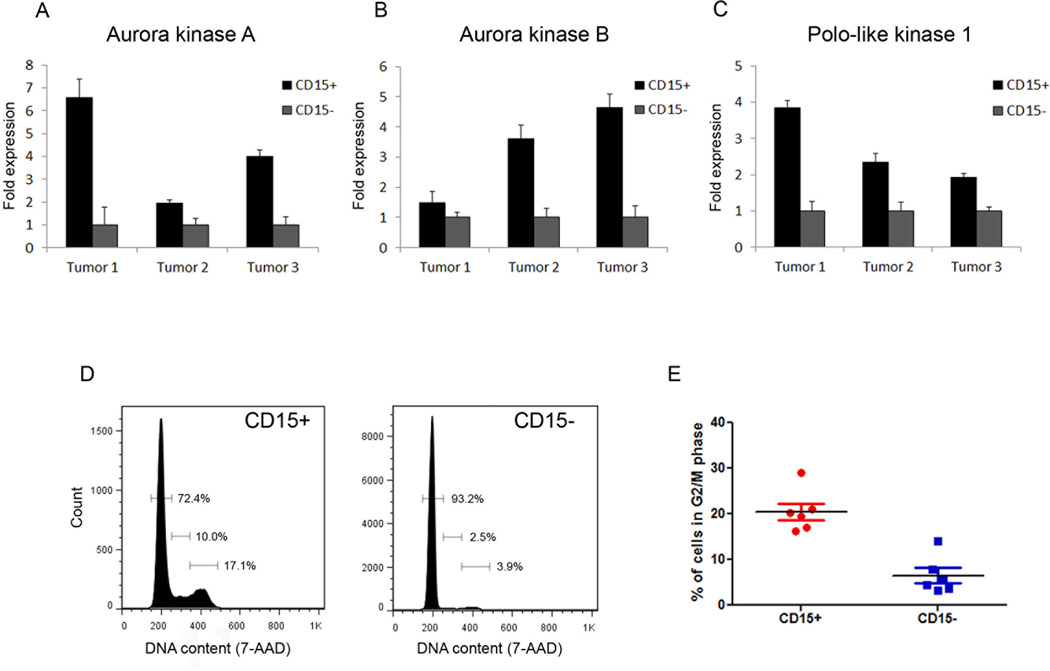
To gain insight into the mechanisms underlying tumor propagation by CD15+ cells, we previously compared their gene expression profiles to those of CD15− cells from the same tumors (20). Our analysis revealed that CD15+ cells express elevated levels of cell cycle regulators, and in particular, regulators of G2/M. To validate these data, we analyzed expression of several of these regulators by real-time RT-PCR. As shown in Figure 1A–C, expression of Aurora kinase A (AurkA), Aurora kinase B (AurkB), and Polo-like kinase 1 (Plk1) was significantly higher in the CD15+ population compared to the CD15− population in each tumor examined (n=3). These results suggest that CD15+ and CD15− cells can be distinguished based on their expression of G2/M regulators.
Figure 1. CD15+ cells display elevated expression of Aurora and Polo-like kinases and an increased proportion of cells in G2/M.
(A–C) Real-time RT-PCR analysis of AurkA (A), AurkB (B), and Plk1 (C) expression in CD15+ cells and CD15− cells from patched mutant tumors (n=3). (D) Flow cytometric analysis of DNA content in CD15+ and CD15− cells from a representative tumor. (E) Quantitation of the percentage of cells in G2/M phase in the CD15+ and CD15− populations from 6 separate tumors.
CD15+ cells are enriched in G2/M
The differential expression of G2/M regulators in CD15+ and CD15− cells suggested that these populations might differ in terms of cell cycle distribution. To examine this, we performed cell cycle analysis on freshly isolated CD15+ and CD15− cells. Analysis of multiple tumors (n=6) indicated that compared to the CD15− population, the CD15+ population contains a significantly higher proportion of cells in G2/M phase; ~20% of CD15+ cells reside in G2/M, compared to ~5% of CD15− cells (Figure 1D, E). A similar, but less pronounced, enrichment was seen in the proportion of CD15+ cells in S phase (data not shown). These data suggest that the elevated expression of G2/M regulators correlates with an increased proportion of CD15+ cells in G2/M phase.
CD15+ cells progress more rapidly through the cell cycle than CD15− cells
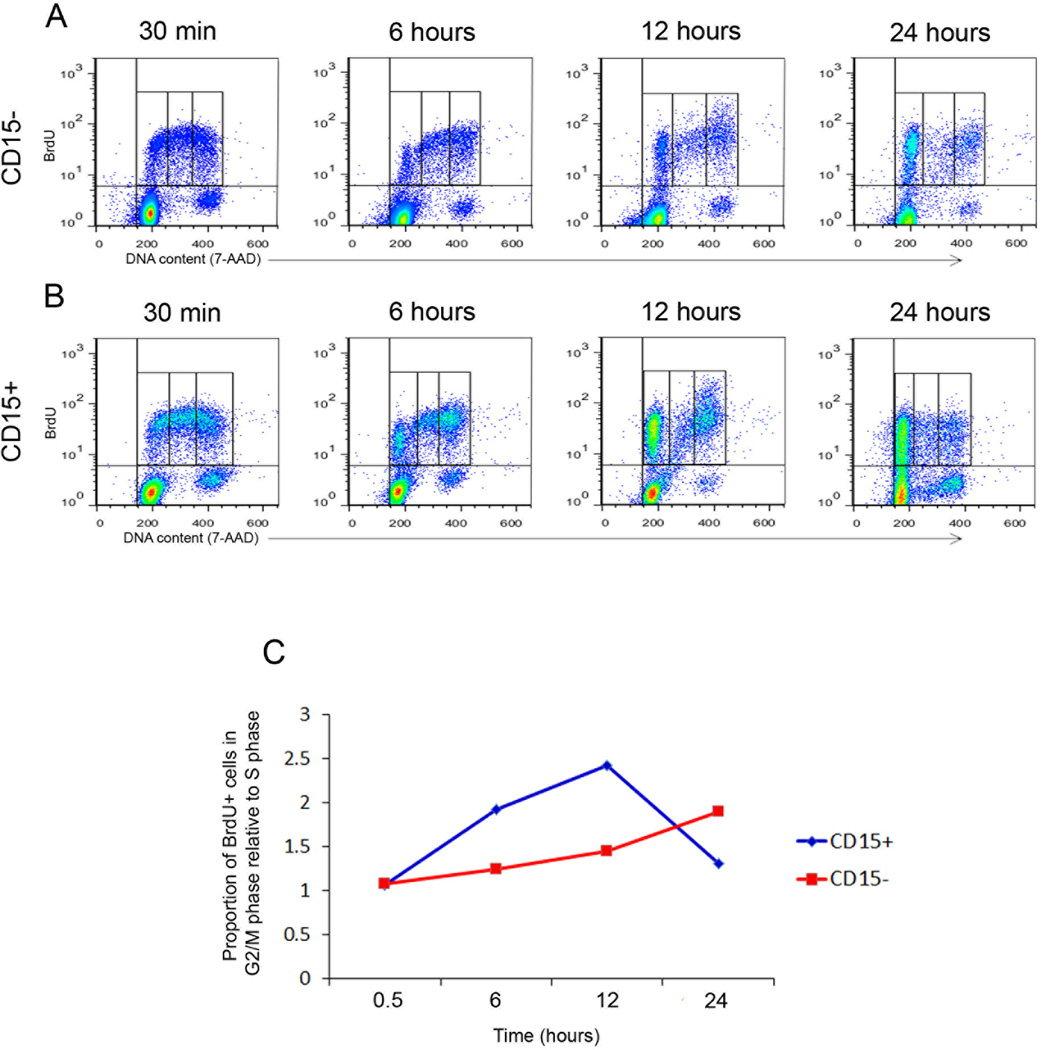
The increased proportion of CD15+ cells in G2/M phase could be explained by differences in cell cycle kinetics between the CD15+ and CD15− populations. To address this possibility, we pulse-labeled cells with BrdU and followed their progression through the cell cycle. CD15+ and CD15− cells from patched mutant tumors were cultured in the presence of BrdU for 30 minutes, and then washed and collected immediately for cell cycle analysis or cultured for an additional 6, 12, or 24 hours. As shown in Figure 2A and B, BrdU was incorporated into both CD15+ and CD15− cells; approximately 27% of the CD15+ cells incorporated the BrdU label, while only 7% of the CD15− cells were labeled. These data suggest that the over-representation of CD15+ cells in G2/M phase may result, in part, from an increased percentage of cells transiting through the cycle.
Figure 2. CD15+ cells progress more rapidly through the cell cycle than CD15− cells.
CD15+ and CD15− cells from germline patched mutant tumors were pulsed with BrdU for 30 minutes, washed, and then analyzed immediately (30 min) or cultured for an additional 6, 12 or 24 hours. (A, B) Flow cytometric analysis of BrdU (Y-axis) and DNA content (X-axis) of CD15− (A) and CD15+ (B) cells at the indicated time points. (C) Ratio of [percentage of cells in G2/M]:[percentage of cells in S phase] among the BrdU+ population at each time point. The percentage of CD15+ cells in G2/M phase increases much more rapidly than the percentage of CD15− cells in G2/M between 30 minutes and 12 hours, and CD15+ cells exit G2/M phase (indicated by drop in the G2/M to S ratio) sooner than CD15− cells.
To assess the kinetics with which each population proceeds through S phase and into G2/M, we examined the ratio of BrdU+ cells with 4N DNA (G2/M phase) to those with DNA content > 2N and < 4N (S phase) at each time point (Figure 2C and Supplemental Figure 1A–E). Thirty minutes after the BrdU pulse, the CD15+ and CD15− populations included similar proportions of cells in S and G2/M (G2/M:S ratios = 1.05 and 1.07, respectively). However, as early as 6 hours after the BrdU pulse, CD15+ and CD15− cells began to exhibit differences in cell cycle distribution. In the CD15− population, the G2/M:S ratio increased slowly, to 1.24 at 6 hours, 1.45 at 12 hours, and 1.89 at 24 hours. In contrast, this ratio increased much more rapidly in the CD15+ population, reaching 1.92 at 6 hours and 2.42 at 12 hours. At 24 hours, the G2/M:S ratio in the CD15+ population dropped sharply, to 1.3, as many CD15+ cells exited G2/M and re-entered G1. Thus, CD15− cells require 24 hours to accumulate in G2/M phase at levels similar to those reached by the CD15+ cells within 6 hours. Based on these results, we conclude that CD15+ cells move through the cell cycle more rapidly than CD15− cells and that this rapid progression may also contribute to the over-representation of CD15+ cells in G2/M phase.
As an alternative approach to comparing the cell cycle kinetics of CD15+ and CD15− populations, we followed cells after labeling with carboxyfluorescein diacetate succinimidyl ester (CFSE). CFSE is equally distributed between daughter cells during division, such that a 50% reduction in fluorescence corresponds to one cell division. Thus, the number of divisions a population of cells has undergone can be determined by counting the number of CFSE fluorescence peaks. As shown in Supplemental Figure 1F, CD15+ and CD15− cells incorporated similar levels of the CFSE label at t=0. By comparing the median fluorescence of the last peak (lowest fluorescence) in the CD15+ and CD15− populations to the peak with the highest fluorescence in the CD15− population (representing undivided cells) at 48, 72, and 96 hours, we found that the majority of the CD15+ population had undergone more divisions than the CD15− population at each time point (Supplemental Figure 1G–I). Furthermore, closer inspection of the CFSE fluorescence at 96 hours after labeling revealed that approximately 86% of the CD15+ population had undergone 4 cell divisions, suggesting an average cell cycle time of 24 hours (Supplemental Figure 1J–L). In contrast, only 5% of the CD15− population had undergone 4 divisions, while the remaining cycling cells were relatively equally distributed between 1, 2, and 3 divisions, suggesting an average cell cycle time of 60 hours. Therefore, these data further support the conclusion that the CD15+ population from patched mutant tumors contains a greater proportion of cycling cells and that these cells progress through the cell cycle more rapidly than CD15− cells.
Targeting G2/M regulators blocks progression through the cell cycle, inhibits proliferation and induces apoptosis
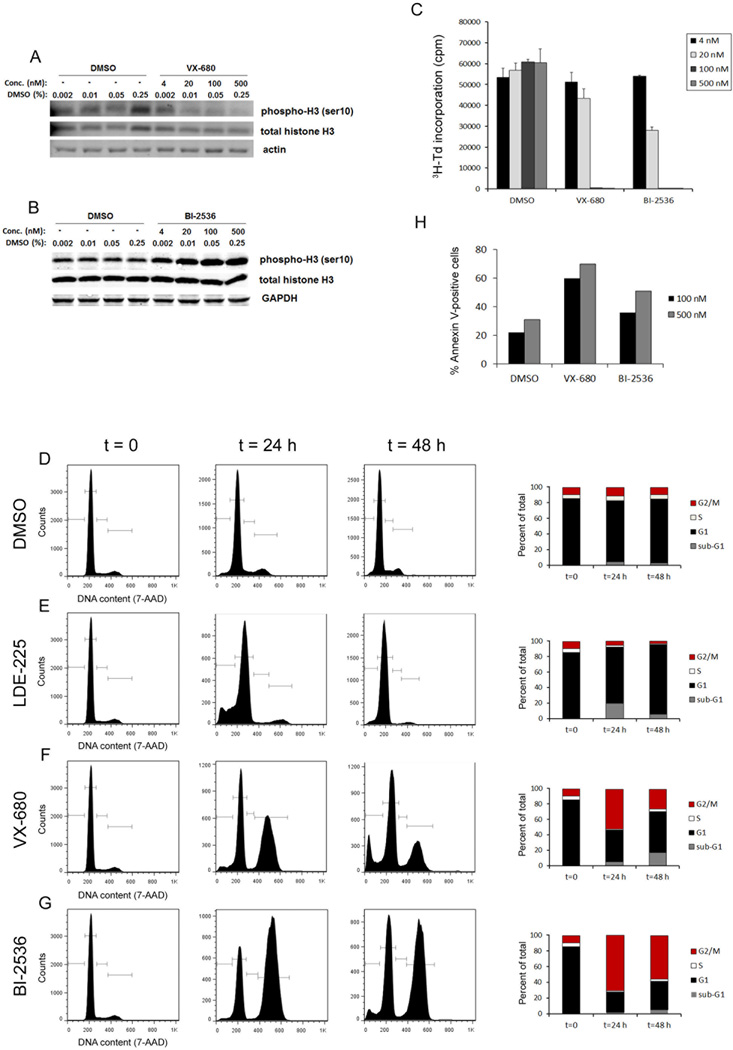
The increased proportion of CD15+ cells in G2/M phase led us to hypothesize that patched mutant tumors might be sensitive to inhibitors of regulators of G2/M progression. To address this possibility, we treated tumor cells with the Aurk inhibitor VX-680 (Tozasertib) and the Plk inhibitor BI-2536, and examined phosphorylation of Histone H3 on Serine 10. This residue is a direct target of AurkB (27, 28), and as shown in Figure 3A, the Aurk inhibitor VX-680 potently blocked its phosphorylation. Paradoxically, inhibitors of Plk1 have been reported to promote increased phosphorylation of Histone 3 on Serine 10 (29, 30); consistent with this, we observed increased levels of phospho-H3Ser10 and decreased levels of Serine 46 phosphorylation of TCTP (translationally controlled tumor protein, a direct Plk1 substrate) upon treatment with BI-2536 (Figure 3B and data not shown). These data suggest that inhibitors of Aurora and Polo-like kinases are active in patched mutant tumor cells.
Figure 3. Aurk and Plk inhibitors block proliferation and cell cycle progression and induce apoptosis.
Cells from conditional patched mutant tumors were cultured in the presence of the indicated concentrations of VX-680, BI-2536, or the corresponding percentages of DMSO. (A, B) Western blot analysis of the levels of phospho-Histone H3 (Ser10) after 6 hours of treatment with VX-680 (A) or BI-2536 (B). Note that Aurk inhibition decreases phosphorylation whereas Plk inhibition increases it. (C) Effects of Aurk and Plk inhibitors on proliferation. Cells were cultured with DMSO, VX-680 or BI-2536, pulsed with tritiated thymidine (3H-Td) at 48 hours, and harvested at 66 hours for analysis of 3H-Td incorporation. Data represent means of triplicate wells ± SEM. Treatment with 20 nM, 100 nM and 500 nM BI-2536 or VX-680 significantly inhibited 3H incorporation compared to corresponding DMSO controls (p < 0.01, based on paired 2-tailed t-test). (D–G) Flow cytometric analysis of DNA content, and graphical representation of percentages of cells in each cell cycle phase, after treatment with DMSO (D), 100 nM LDE-225 (E), 100 nM VX-680 (F), or 100 nM BI-2536 (G). (H) Effects of Aurk and Plk inhibitors on apoptosis. Cells were cultured with DMSO, VX-680 or BI-2536 for 48 hours, then analyzed for Annexin V-FITC binding. Graph represents percentage of Annexin V-positive cells under each condition in a representative experiment.
To determine the effects of Aurk and Plk inhibitors on proliferation, we performed 3H-thymidine incorporation assays. As shown in Figure 3C, treatment with 100 nM or 500 nM VX-680 or BI-2536 caused nearly complete inhibition of proliferation. To define the IC50 values for VX-680 and BI-2536, we treated cells with increasing concentrations of these compounds (0.15 nM - 1.5 µM) and measured 3H-thymidine incorporation. As shown in Supplemental Figure 2A and B, the IC50 values for VX-680 and BI-2536 were 23 nM and 4.5 nM, respectively. These values are consistent with previously reported IC50 values for these drugs in other types of tumor cells (28, 29). To further validate the anti-proliferative effects of Aurk and Plk inhibition, we assessed the sensitivity of patched mutant tumor cells to additional Aurk or Plk inhibitors. As shown in Supplemental Figure 2C and D, multiple Aurk and Plk inhibitors displayed potent anti-proliferative effects. These data confirm that patched mutant tumor cells are vulnerable to small molecule-mediated inhibition of Aurk or Plk activity.
To assess the effects of these inhibitors on cell cycle progression, we treated patched mutant tumor cells with VX-680 or BI-2536 for 24 or 48 hours. For comparison, we also treated cells with the SHH antagonist NVP-LDE225 (LDE-225), which is currently in clinical trials for the treatment of SHH-associated MB (31). As shown in Figure 3D and E, exposure to LDE-225 caused a progressive decrease in the number of cells in G2/M and a concomitant accumulation of cells in G1. In contrast, both VX-680 and BI-2536 markedly increased the number of cells in G2/M, while decreasing the G1 population (Figure 3D, F, and G). Treatment with each of these inhibitors also caused an increase in the proportion of cells with < 2N DNA, most likely representing apoptotic cells. Consistent with this, VX-680 and BI-2536 each increased the percentage of Annexin V-labeled tumor cells (Figure 3H). These data suggest that patched mutant tumor cells are sensitive to Aurk or Plk inhibition and that the effects of these inhibitors on the cell cycle are distinct from those induced by inhibitors of the SHH pathway.
Plk inhibition cooperates with SHH antagonist, conventional chemotherapy and radiation
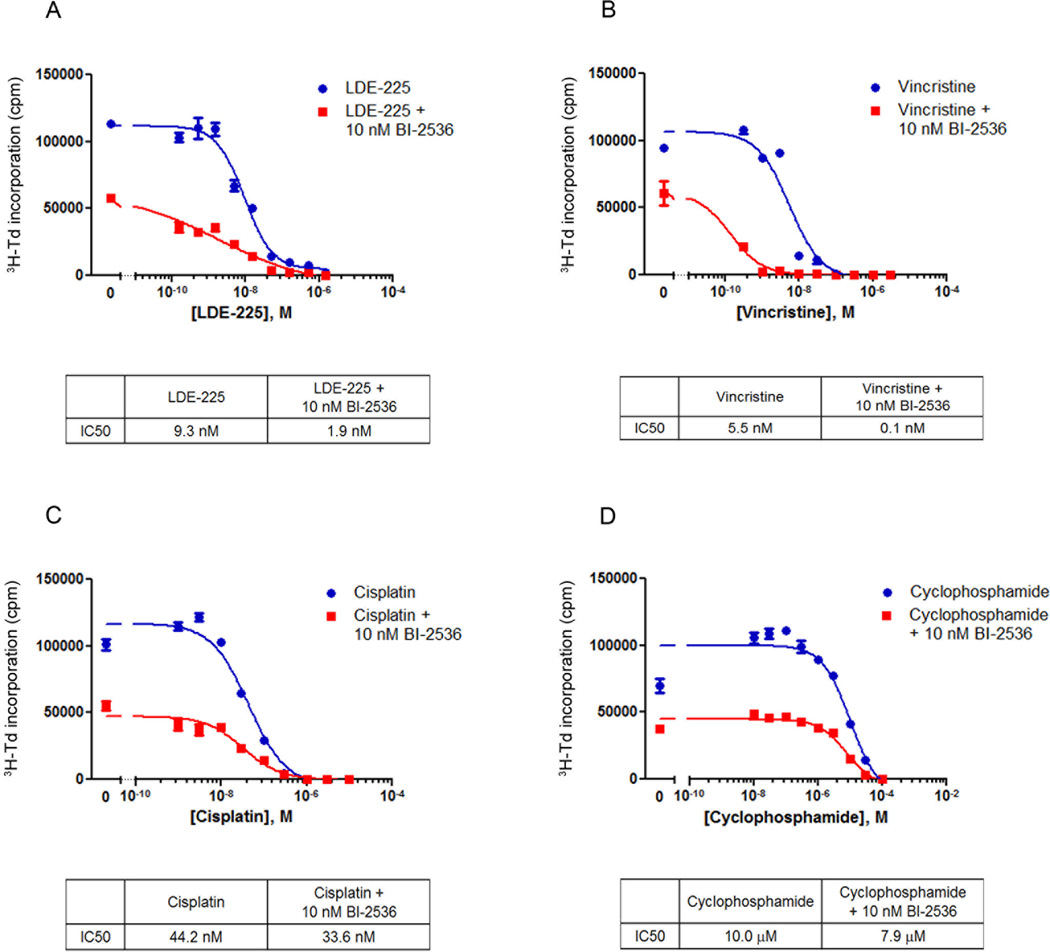
Given the distinct effects of LDE-225 and VX-680 or BI-2536 on cell cycle progression, we hypothesized that these drugs might exert complementary or cooperative effects. To address this possibility, we treated tumor cells with increasing concentrations (0.15 – 1500 nM) of LDE-225 either with or without 10 nM BI-2536, a concentration of BI-2536 that caused minimal inhibition of proliferation on its own. As shown in Figure 4A, treatment with LDE-225 alone inhibited proliferation at concentrations above 15 nM, with an IC50 of ~9 nM. However, concomitant treatment with 10 nM BI-2536 enhanced the inhibition of proliferation at all concentrations of LDE-225 and caused the IC50 for LDE-225 to shift to ~2 nM, suggesting that BI-2536 cooperates with LDE-225 to inhibit proliferation. Similar cooperation was observed between VX-680 and LDE-225 (Supplemental Figure 3A).
Figure 4. Plk inhibitor cooperates with SHH antagonist and chemotherapeutic agents.
Conditional patched mutant tumor cells were treated with increasing concentrations of the SHH antagonist LDE-225 (A) or the chemotherapeutic agents Vincristine (B), Cisplatin (C), or Cyclophosphamide (D), alone or in combination with 10 nM BI-2536. Cells were cultured for 48 hours, pulsed with 3H-Td, and harvested for analysis of 3H-Td incorporation at 66 hours. Data represent means of triplicate samples ± SEM. IC50 values were calculated using the log(inhibitor) vs. response equation (Y=Bottom + (Top-Bottom)/(1+10^((X-LogIC50))) in GraphPad Prism software.
To determine whether G2/M inhibitors can also cooperate with conventional chemotherapeutic agents, we treated patched mutant tumor cells with 10 nM BI-2536 in combination with Vincristine, Cisplatin, and Cyclophosphamide, chemotherapeutic agents that are currently being used to treat human MB (7). Addition of BI-2536 dramatically increased the sensitivity of the tumor cells to Vincristine; while the IC50 value for Vincristine alone was approximately 5 nM, the IC50 value for Vincristine combined with BI-2536 was approximately 0.1 nM (Figure 4B). Similar, but less dramatic, cooperation was observed with Cisplatin (IC50 = 44 nM for Cisplatin alone and 34 nM for Cisplatin + BI-2536) and with Cyclophosphamide (IC50 = 10 µM for Cyclophosphamide alone and 8 µM for Cyclophosphamide + BI-2536) (Figure 4C and D). VX-680 also cooperated with Vincristine and Cyclophosphamide (Supplemental Figure 3B–D).
To determine whether the G2/M inhibitors can cooperate with radiotherapy (another component of standard MB therapy that is associated with substantial toxicity), we treated patched mutant tumor cells with 10 nM BI-2536 or 30 nM VX-680 in combination with increasing doses (0, 0.25, 0.5, 1 Gy) of radiation. Both BI-2536 and VX-680 significantly enhanced the sensitivity of the cells to radiation (Supplemental Figure 4A and B). These data suggest that the addition of BI-2536 or VX-680 can lower the concentrations of chemotherapy or radiation required for effective inhibition of tumor cell proliferation.
Inhibition of G2/M regulators blocks tumor growth in vivo
Given the strong anti-proliferative effects of the Aurk and Plk inhibitors in vitro, we next questioned whether inhibition of G2/M regulators could affect tumor growth in vivo. Because the Plk inhibitor BI-2536 has demonstrated promising results in clinical trials completed thus far (32–34), we prioritized this compound for our in vivo studies. patched mutant tumor cells were implanted subcutaneously into the flanks of Nu/Nu mice, and two weeks later, mice were treated with either vehicle or BI-2536 (50 mg/kg via tail vein). Tumors were harvested 24 hours later, and sections from 4 independent tumors per condition were stained with antibodies specific for cleaved caspase-3 (CC3). As shown in Figure 5A–I, tumors from BI-2536-treated animals contained significantly more CC3+ cells compared to tumors from vehicle-treated animals (106 CC3+ cells/mm2 with BI-2536 vs. 34 CC3+ cells/mm2 with vehicle). These data suggest that treatment with the Plk inhibitor causes apoptosis in vivo.
Figure 5. Plk inhibitor induces apoptosis and blocks tumor growth in vivo.
Mice bearing subcutaneous allografts of conditional patched mutant tumor cells were treated twice weekly with vehicle (saline) or 50 mg/kg BI-2536. (A–I) 24 hours after a single dose of vehicle or BI-2536, tumors were harvested and stained with cleaved caspase-3 antibodies to detect apoptotic cells and DAPI to detect nuclei. Representative images from 4 independent vehicle-treated (A–D) and BI-2536-treated (E–H) mice are shown. (I) Cleaved caspase-3+ cells were quantitated and normalized to the DAPI-positive area in 6 regions from each vehicle-treated and BI-2536-treated tumor. Graph displays the average number of cleaved caspase-3+ cells per mm2 of DAPI. BI-2536-treated tumors contained significantly more cleaved caspase-3-positive cells relative to vehicle-treated tumors (p=0.0038, paired two-tailed t-test). (J, K) Tumor volume (mm3) was measured using calipers. Arrow indicates start of treatment, and each line represents an individual mouse. (L) Images of tumors. (M) Tumor weights. Each point represents a single tumor, and grey lines represent mean tumor weights, which were significantly different between vehicle and BI-2536 treated mice (p < 0.05, based on paired two-tailed t-test).
To assess the effects of Plk inhibition on tumor growth, tumor-bearing mice were treated twice weekly with vehicle or BI-2536 for 2 weeks. As shown in Figure 5J and K, BI-2536 dramatically inhibited tumor growth, as measured by tumor volume over time. Upon harvesting the tumors (~2.5 weeks after starting treatment), marked differences in tumor size and weight were observed (Figure 5L and M). Overall, tumors from the BI-2536-treated mice were significantly smaller and weighed less than tumors from the vehicle-treated mice. The Aurk inhibitor PHA-739358 also blocked tumor growth and led to a reduction in tumor size and weight (Supplemental Figure 5A, B, and C). Collectively, these data suggest that inhibition of G2/M regulators can effectively block tumor progression in vivo.
Aurk and Plk inhibitors suppress growth of human SHH-associated MB
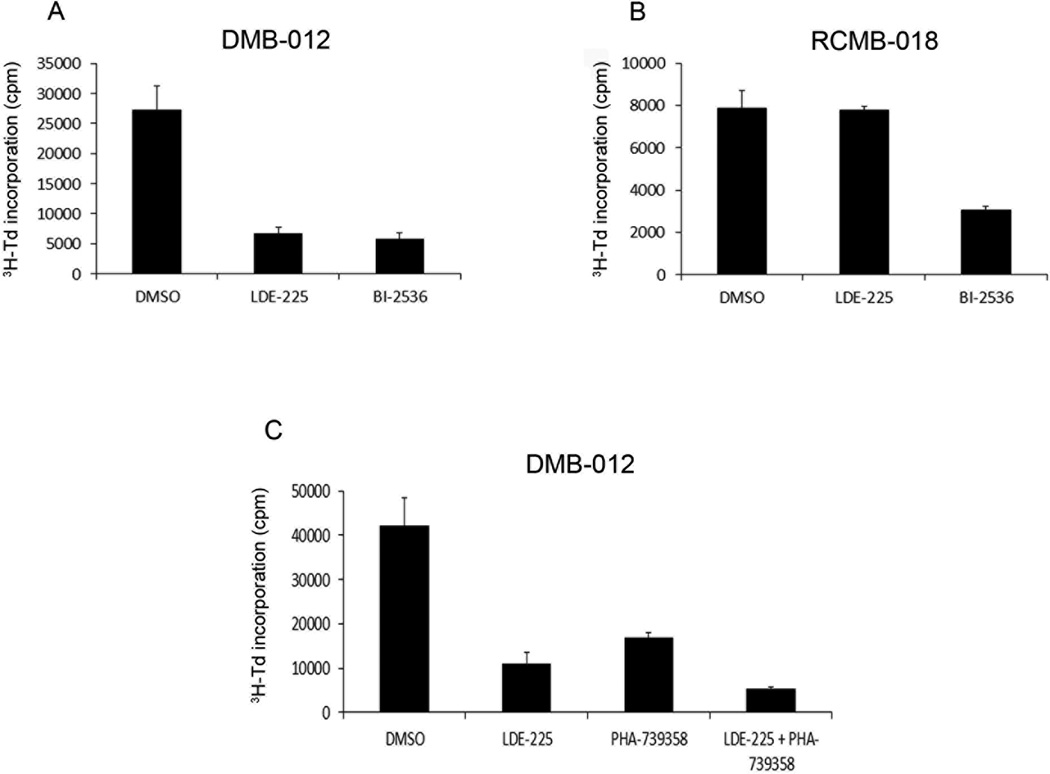
The studies above focused on tumors from patched mutant mice. To determine whether G2/M inhibitors might also be effective against human MB, we used cells from patient-derived xenografts that were molecularly classified as SHH-associated MB (Supplemental Figure 6A and B). As shown in Figure 6A, treatment of cells from the human SHH-associated MB xenograft DMB-012 with BI-2536 caused a marked inhibition of proliferation, comparable to that seen with the SHH antagonist LDE-225. In addition, RCMB-018 cells, derived from a SHH-associated MB that is insensitive to LDE-225 (due to amplification of SHH pathway components downstream of SMO), were also inhibited by BI-2536 (Figure 6B). These data suggest that G2/M inhibitors might be useful for treating human SHH-associated tumors, including those that display resistance to SHH antagonists.
Figure 6. Aurk and Plk inhibitors suppress proliferation of human SHH-associated MB.
(A, B) Cells from patient-derived xenografts of SHH-associated MB that are sensitive (DMB-012, panel A) or resistant (RCMB-018, panel B) to SHH antagonists were treated with DMSO (0.25%), LDE-225 (500 nM), or BI-2536 (500 nM). Cells were pulsed with 3H-Td after 48 hours and harvested for analysis of 3H-Td incorporation at 66 hours. In DMB-012, LDE-225 and BI-2536 significantly inhibited 3H incorporation compared to DMSO control (p < 0.01 based on paired two-tailed t-test). In RCMB-018, BI-2536 caused significant inhibition (p = 0.01), whereas LDE-225 did not (p = 0.89). (C) DMB-012 cells were cultured in the presence of LDE-225 (100 nM), PHA-739358 (100 nM) or the combination of LDE-225 + PHA-739358, and assayed for 3H–Td incorporation as described above. Data represent means of triplicate samples ± SEM. 3H-Td incorporation in the presence of LDE-225 + PHA-739358 was significantly lower than in the presence of LDE-225 alone (p = 0.05) or PHA-739358 alone (p = 0.004).
To address whether Aurk inhibition and SHH pathway antagonism cooperate in human MB, we treated DMB-012 cells with intermediate concentrations of the Aurk inhibitor PHA-739358 and the SHH antagonist LDE-225. As shown in Figure 6C, treatment with 100 nM LDE-225 alone or 100 nM PHA-739358 alone caused inhibition of proliferation, as expected. However, treatment with 100 nM LDE-225 together with 100 nM PHA-739358 further inhibited proliferation beyond that of either compound alone. These data suggest that, similar to the results observed in cells from patched mutant tumors, inhibition of G2/M regulators can cooperate with SHH pathway antagonism to block the growth of human SHH-associated MB.
Discussion
Although treatment for MB has significantly improved survival in recent years, patients often suffer severe side effects, and better treatment strategies are still required. Targeting MB TPCs represents one approach to improving treatment. Using the patched mutant mouse (a robust model of SHH-associated MB), we have demonstrated that CD15+ TPCs disproportionately reside in G2/M phase of the cell cycle and that inhibition of Aurora kinases or Polo-like kinases using clinically relevant agents can inhibit tumor growth in vitro and in vivo. In addition, we have shown that these inhibitors can block the growth of cells from patient-derived xenografts of human SHH-associated MB, including those that are resistant to SHH antagonists. Our data suggest that incorporating Aurk or Plk inhibitors into MB therapy could lead to improvements in treatment outcome for patients with SHH-associated tumors.
Our previous studies indicated that CD15+ TPCs from patched mutant tumors display elevated expression of G2 and M phase cell cycle regulators (20). Here, we confirmed increased expression of AurkA, AurkB, and Plk1. Each of these serine/threonine kinases plays a distinct role in G2/M phase progression (35). AurkA is involved in centrosome duplication, bipolar spindle assembly, and entry into mitosis, while AurkB functions in chromatin modification, microtubule-kinetochore attachment, spindle assembly checkpoint activation, and cytokinesis. Plk1 is involved in centrosome separation, spindle assembly and maturation, cytokinesis, and exit from mitosis. Overexpression of each of these kinases has been associated with poor prognosis in multiple tumor types, leading to the notion that elevated expression of these proteins might promote tumor growth (36–42). However, given the elevated expression of multiple G2/M regulators in CD15+ cells, we hypothesized that the expression profile reflected a general property of the CD15+ population, rather than a reliance on elevated expression of a single kinase to drive tumorigenicity. Cell cycle analysis of CD15+ and CD15− populations from multiple tumors demonstrated an increased percentage of the CD15+ population residing in G2/M phase compared to the CD15− population, suggesting that the elevated expression of G2/M regulators in the CD15+ population is likely a result, rather than a cause, of the increased percentage of cells residing in G2/M.
Multiple factors could contribute to the accumulation of CD15+ cells in G2/M. One explanation is that a greater overall percentage of CD15+ cells transit through the cell cycle, while CD15− cells remain largely stationary in G0/G1 phase. This notion is supported by the observation that the CD15+ population also contains a greater fraction of cells in S phase than the CD15− population. Another possible explanation for the accumulation in G2/M could be that the CD15+ cells arrest in these phases of the cell cycle. To address this possibility, we performed BrdU labeling and cell cycle analysis to monitor the position of the BrdU-labeled cells in the cell cycle over time. We observed greater incorporation of the BrdU label in CD15+ cells, which again suggested that greater numbers of CD15+ cells transit through the cell cycle. However, examination of the BrdU-labeled cells over time indicated that CD15+ cells do not arrest in G2/M, but actually progress more rapidly through the cell cycle than CD15− cells. Similar results were observed using CFSE cell division analysis. While these data do not exclude the possibility of transient checkpoint activation and/or cell cycle arrest, they indicate that cell cycle arrest is not a primary contributor to the over-representation of CD15+ cells in G2/M phase. Furthermore, our data suggest that both increased numbers of cycling cells and an increased pace of progression through the cell cycle contribute to the over-representation of CD15+ cells in G2/M phase.
We speculated that the increased residency of CD15+ cells in G2/M phase could represent a vulnerability of these cells that could be targeted through inhibition of G2/M regulators. Because small molecule inhibitors of the Aurora kinases and Polo-like kinases have shown promising efficacy in Phase I and Phase II clinical trials for other tumor types, we selected the Aurk inhibitor VX-680 and the Plk inhibitor BI-2536 for evaluation in our studies (32, 33, 43–45). Our data demonstrate that patched mutant tumors are indeed sensitive to Aurk or Plk inhibition; both VX-680 and BI-2536 effectively blocked proliferation in vitro.
One principal aim of this study was to identify approaches that might enhance current MB therapy. SHH antagonists have recently been developed for treatment of human SHH-associated MB (5). Both patients and mice who receive these antagonists initially respond to treatment, but they quickly develop resistance (6, 8, 46). Previous studies have shown that SHH signaling regulates the transition between the G1 and S phases of the cell cycle (47–50); consistent with these observations, our data indicate that the SHH antagonist LDE-225 causes accumulation of cells in G1. In contrast, the Aurk and Plk inhibitors cause accumulation in G2/M phase. Given these distinct mechanisms of cell cycle inhibition, we speculated that blocking G2/M progression might represent an additional point of intervention to target the cells that escape sensitivity to SHH antagonists. Our data demonstrate that the combination of LDE-225 plus BI-2536 or VX-680 has a greater inhibitory effect than treatment with any compound alone. These data suggest that clinical combination of SHH antagonists plus Aurk or Plk inhibitors might enhance the efficacy of therapy and prevent the acquired resistance to SHH antagonists.
Although the development of SHH antagonists has provided additional options for therapy, most MB patients are still treated with conventional chemotherapy and radiotherapy. However, these treatments are extremely toxic and cause significant side effects (3). We questioned whether addition of Aurk or Plk inhibitors might allow for a reduction in the dose of chemotherapy or radiotherapy while maintaining the efficacy of treatment. Our data demonstrate that the combination of BI-2536 or VX-680 with chemotherapeutic agents (Vincristine, Cisplatin, or Cyclophosphamide) or radiation is more effective than chemotherapy or radiation alone. Previous studies have shown similar effects of combined treatment of established MB cell lines with chemotherapy or radiation plus Aurk or Plk inhibitors (51–53). Together, these studies suggest that incorporating Aurk or Plk inhibitors into MB therapy might enable a reduction in the doses of chemotherapy/radiotherapy and thereby reduce the long-term side-effects associated with these treatments.
The anti-proliferative effects of the Aurk and Plk inhibitors in vitro prompted us to examine the effects of the inhibitors in vivo. Our data indicate that treatment of mice harboring subcutaneous allografts of patched mutant tumors with BI-2536 promotes apoptosis and blocks tumor growth in vivo. These data validate the notion that targeting a vulnerability of the TPC population using inhibitors of G2/M regulators can block in vivo growth of SHH-associated tumors and suggest that inhibition of Plk may represent a viable approach for MB treatment.
By examining the TPC population in the patched mutant mouse model of MB, we have identified an opportunity for therapeutic intervention. Our data indicate that cells from patient-derived xenografts of human SHH-associated MB are also sensitive to Aurk or Plk inhibition. Importantly, tumor cells that are insensitive to SHH antagonists maintain sensitivity to BI-2536, validating the notion that treatment with the Plk inhibitor may represent an approach to overcome therapeutic resistance to SHH antagonist therapy. In addition, similar to the patched mutant tumor cells, treatment of human tumor cells with an Aurk inhibitor plus LDE-225 blocks proliferation more effectively than either compound alone. These data suggest that mouse and human SHH-associated MB are sensitive to Aurk or Plk inhibition and that targeting these G2/M regulators may represent an approach to prevent or overcome resistance to SHH antagonists. Therefore, our data strongly support the notion that incorporating Aurk or Plk inhibitors into therapeutic strategies may improve the outcome of treatment for patients with SHH-associated MB.
Supplementary Material
Acknowledgements
The authors thank Buddy Charbono for technical support with animal experiments, Lili Lacarra and Sonja Brun for help with animal colony maintenance, Beth Harvat, Mike Cook, Jonna Hurtado and Yoav Altman for assistance with flow cytometry, the DNA Microarray Facility at Duke University for microarray processing, Sri Gururangan and Roger McLendon for facilitating access to patient samples, and Sandi Dunn for sharing unpublished data and helpful discussions. We are grateful to Novartis for providing NVP-LDE225.
Financial Support: These studies were supported by R01-CA122759 from NCI and R01 NS052323 from NINDS, as well as pilot funds from Golfers Against Cancer and the Pediatric Brain Tumor Foundation Institute at Duke University. RWR is the recipient of a Leadership Award (CIRM LA1-01747) from the California Institute for Regenerative Medicine.
Footnotes
Conflict of Interest: The authors disclose no potential conflicts of interest.
Author contributions:
Conception and design of experiments: S.L. Markant, R.J. Wechsler-Reya
Development of methodology and acquisition of data: S.L. Markant, L.A. Esparza, J. Sun, K.L. Barton, L.M. McCoig
Provision and analysis of patient-derived xenografts: G.A. Grant, J.R. Crawford, M.L. Levy, M. Remke, D. Shih, P.A. Northcott, M.D. Taylor
Analysis and interpretation of data: S.L. Markant, L.A. Esparza, R.J. Wechsler-Reya
Writing, review, and revision of manuscript: S.L. Markant, R.J. Wechsler-Reya
References
- 1.Merchant TE, Pollack IF, Loeffler JS. Brain tumors across the age spectrum: biology, therapy, and late effects. Semin Radiat Oncol. 2010;20:58–66. doi: 10.1016/j.semradonc.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crawford JR, MacDonald TJ, Packer RJ. Medulloblastoma in childhood: new biological advances. Lancet Neurol. 2007;6:1073–1085. doi: 10.1016/S1474-4422(07)70289-2. [DOI] [PubMed] [Google Scholar]
- 3.Pollack IF. Multidisciplinary management of childhood brain tumors: a review of outcomes, recent advances, and challenges. J Neurosurg Pediatr. 2011;8:135–148. doi: 10.3171/2011.5.PEDS1178. [DOI] [PubMed] [Google Scholar]
- 4.Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, et al. Molecular Subgroups of Medulloblastoma: The Current Consensus. Acta Neuropathologica. 2012 doi: 10.1007/s00401-011-0922-z. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Metcalfe C, de Sauvage FJ. Hedgehog fights back: mechanisms of acquired resistance against Smoothened antagonists. Cancer Res. 2011;71:5057–5061. doi: 10.1158/0008-5472.CAN-11-0923. [DOI] [PubMed] [Google Scholar]
- 6.Rudin CM, Hann CL, Laterra J, Yauch RL, Callahan CA, Fu L, et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. The New England journal of medicine. 2009;361:1173–1178. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramaswamy V, Northcott PA, Taylor MD. FISH and chips: the recipe for improved prognostication and outcomes for children with medulloblastoma. Cancer Genet. 2011;204:577–588. doi: 10.1016/j.cancergen.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Buonamici S, Williams J, Morrissey M, Wang A, Guo R, Vattay A, et al. Interfering with resistance to smoothened antagonists by inhibition of the PI3K pathway in medulloblastoma. Sci Transl Med. 2010;2:51ra70. doi: 10.1126/scitranslmed.3001599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 11.O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 12.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 13.Xin L, Lawson DA, Witte ON. The Sca-1 cell surface marker enriches for a prostate-regenerating cell subpopulation that can initiate prostate tumorigenesis. Proc Natl Acad Sci U S A. 2005;102:6942–6947. doi: 10.1073/pnas.0502320102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bertolini G, Roz L, Perego P, Tortoreto M, Fontanella E, Gatti L, et al. Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proc Natl Acad Sci U S A. 2009;106:16281–16286. doi: 10.1073/pnas.0905653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai P, et al. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell. 2008;13:153–166. doi: 10.1016/j.ccr.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 16.Schober M, Fuchs E. Tumor-initiating stem cells of squamous cell carcinomas and their control by TGF-beta and integrin/focal adhesion kinase (FAK) signaling. Proc Natl Acad Sci U S A. 2011;108:10544–10549. doi: 10.1073/pnas.1107807108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alison MR, Lin WR, Lim SM, Nicholson LJ. Cancer stem cells: in the line of fire. Cancer Treat Rev. 2012;38:589–598. doi: 10.1016/j.ctrv.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 18.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 19.Liu G, Yuan X, Zeng Z, Tunici P, Ng H, Abdulkadir IR, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Read TA, Fogarty MP, Markant SL, McLendon RE, Wei Z, Ellison DW, et al. Identification of CD15 as a Marker for Tumor-Propagating Cells in a Mouse Model of Medulloblastoma. Cancer Cell. 2009;15:1–13. doi: 10.1016/j.ccr.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 22.Yang ZJ, Ellis T, Markant SL, Read TA, Kessler JD, Bourboulas M, et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell. 2008;14:135–145. doi: 10.1016/j.ccr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Northcott PA, Shih DJ, Remke M, Cho YJ, Kool M, Hawkins C, et al. Rapid, reliable, and reproducible molecular sub-grouping of clinical medulloblastoma samples. Acta Neuropathol. 2012;123:615–626. doi: 10.1007/s00401-011-0899-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y, Stevens B, Chang J, Milbrandt J, Barres BA, Hell JW. NS21: re-defined and modified supplement B27 for neuronal cultures. J Neurosci Methods. 2008;171:239–247. doi: 10.1016/j.jneumeth.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jensen MM, Jorgensen JT, Binderup T, Kjaer A. Tumor volume in subcutaneous mouse xenografts measured by microCT is more accurate and reproducible than determined by 18F-FDG-microPET or external caliper. BMC Med Imaging. 2008;8:16. doi: 10.1186/1471-2342-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu JY, Sun ZW, Li X, Reuben M, Tatchell K, Bishop DK, et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102:279–291. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- 28.Harrington EA, Bebbington D, Moore J, Rasmussen RK, Ajose-Adeogun AO, Nakayama T, et al. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat Med. 2004;10:262–267. doi: 10.1038/nm1003. [DOI] [PubMed] [Google Scholar]
- 29.Steegmaier M, Hoffmann M, Baum A, Lenart P, Petronczki M, Krssak M, et al. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr Biol. 2007;17:316–322. doi: 10.1016/j.cub.2006.12.037. [DOI] [PubMed] [Google Scholar]
- 30.Fink J, Sanders K, Rippl A, Finkernagel S, Beckers TL, Schmidt M. Cell type-- dependent effects of Polo-like kinase 1 inhibition compared with targeted polo box interference in cancer cell lines. Molecular cancer therapeutics. 2007;6:3189–3197. doi: 10.1158/1535-7163.MCT-07-0048. [DOI] [PubMed] [Google Scholar]
- 31.Miller-Moslin K, Peukert S, Jain RK, McEwan MA, Karki R, Llamas L, et al. 1-amino-4-benzylphthalazines as orally bioavailable smoothened antagonists with antitumor activity. J Med Chem. 2009;52:3954–3968. doi: 10.1021/jm900309j. [DOI] [PubMed] [Google Scholar]
- 32.Hofheinz RD, Al-Batran SE, Hochhaus A, Jager E, Reichardt VL, Fritsch H, et al. An open-label, phase I study of the polo-like kinase-1 inhibitor, BI 2536, in patients with advanced solid tumors. Clin Cancer Res. 2010;16:4666–4674. doi: 10.1158/1078-0432.CCR-10-0318. [DOI] [PubMed] [Google Scholar]
- 33.Sebastian M, Reck M, Waller CF, Kortsik C, Frickhofen N, Schuler M, et al. The efficacy and safety of BI 2536, a novel Plk-1 inhibitor, in patients with stage IIIB/IV non-small cell lung cancer who had relapsed after, or failed, chemotherapy: results from an open-label, randomized phase II clinical trial. J Thorac Oncol. 2010;5:1060–1067. doi: 10.1097/JTO.0b013e3181d95dd4. [DOI] [PubMed] [Google Scholar]
- 34.Mross K, Frost A, Steinbild S, Hedbom S, Rentschler J, Kaiser R, et al. Phase I dose escalation and pharmacokinetic study of BI 2536, a novel Polo-like kinase 1 inhibitor, in patients with advanced solid tumors. J Clin Oncol. 2008;26:5511–5517. doi: 10.1200/JCO.2008.16.1547. [DOI] [PubMed] [Google Scholar]
- 35.Lens SM, Voest EE, Medema RH. Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat Rev Cancer. 2010;10:825–841. doi: 10.1038/nrc2964. [DOI] [PubMed] [Google Scholar]
- 36.Neben K, Korshunov A, Benner A, Wrobel G, Hahn M, Kokocinski F, et al. Microarray-based screening for molecular markers in medulloblastoma revealed STK15 as independent predictor for survival. Cancer Res. 2004;64:3103–3111. doi: 10.1158/0008-5472.can-03-3968. [DOI] [PubMed] [Google Scholar]
- 37.Ali HR, Dawson SJ, Blows FM, Provenzano E, Pharoah PD, Caldas C. Aurora kinase A outperforms Ki67 as a prognostic marker in ER-positive breast cancer. Br J Cancer. 2012;106:1798–1806. doi: 10.1038/bjc.2012.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lehman NL, O'Donnell JP, Whiteley LJ, Stapp RT, Lehman TD, Roszka KM, et al. Aurora A is differentially expressed in gliomas, is associated with patient survival in glioblastoma and is a potential chemotherapeutic target in gliomas. Cell Cycle. 2012;11:489–502. doi: 10.4161/cc.11.3.18996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang X, Wang D, Wang Y, Zhou Z, Zhang J, Li J. Expression of Aurora Kinase A and B in chondrosarcoma and its relationship with the prognosis. Diagn Pathol. 2012;7:84. doi: 10.1186/1746-1596-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin ZZ, Jeng YM, Hu FC, Pan HW, Tsao HW, Lai PL, et al. Significance of Aurora B overexpression in hepatocellular carcinoma. Aurora B Overexpression in HCC. BMC Cancer. 2010;10:461. doi: 10.1186/1471-2407-10-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.King SI, Purdie CA, Bray SE, Quinlan PR, Jordan LB, Thompson AM, et al. Immunohistochemical detection of Polo-like kinase-1 (PLK1) in primary breast cancer is associated with TP53 mutation and poor clinical outcom. Breast Cancer Res. 2012;14:R40. doi: 10.1186/bcr3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng MW, Wang BC, Weng ZQ, Zhu XW. Clinicopathological significance of Polo-like kinase 1 (PLK1) expression in human malignant glioma. Acta Histochem. 2012;114:503–509. doi: 10.1016/j.acthis.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 43.Cheung CH, Coumar MS, Chang JY, Hsieh HP. Aurora kinase inhibitor patents and agents in clinical testing: an update (2009–10) Expert Opin Ther Pat. 2011;21:857–884. doi: 10.1517/13543776.2011.574614. [DOI] [PubMed] [Google Scholar]
- 44.Chopra P, Sethi G, Dastidar SG, Ray A. Polo-like kinase inhibitors: an emerging opportunity for cancer therapeutics. Expert Opin Investig Drugs. 2010;19:27–43. doi: 10.1517/13543780903483191. [DOI] [PubMed] [Google Scholar]
- 45.Traynor AM, Hewitt M, Liu G, Flaherty KT, Clark J, Freedman SJ, et al. Phase I dose escalation study of MK-0457, a novel Aurora kinase inhibitor, in adult patients with advanced solid tumors. Cancer Chemother Pharmacol. 2011;67:305–314. doi: 10.1007/s00280-010-1318-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yauch RL, Dijkgraaf GJ, Alicke B, Januario T, Ahn CP, Holcomb T, et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science. 2009;326:572–574. doi: 10.1126/science.1179386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adolphe C, Hetherington R, Ellis T, Wainwright B. Patched1 functions as a gatekeeper by promoting cell cycle progression. Cancer Res. 2006;66:2081–2088. doi: 10.1158/0008-5472.CAN-05-2146. [DOI] [PubMed] [Google Scholar]
- 48.Cayuso J, Ulloa F, Cox B, Briscoe J, Marti E. The Sonic hedgehog pathway independently controls the patterning, proliferation and survival of neuroepithelial cells by regulating Gli activity. Development. 2006;133:517–528. doi: 10.1242/dev.02228. [DOI] [PubMed] [Google Scholar]
- 49.Kenney AM, Rowitch DH. Sonic hedgehog promotes G(1) cyclin expression and sustained cell cycle progression in mammalian neuronal precursors. Mol Cell Biol. 2000;20:9055–9067. doi: 10.1128/mcb.20.23.9055-9067.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oliver TG, Grasfeder LL, Carroll AL, Kaiser C, Gillingham CL, Lin SM, et al. Transcriptional profiling of the Sonic hedgehog response: a critical role for N-myc in proliferation of neuronal precursors. Proc Natl Acad Sci U S A. 2003;100:7331–7336. doi: 10.1073/pnas.0832317100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muscal JA, Scorsone KA, Zhang L, Ecsedy JA, Berg SL. Additive effects of vorinostat and MLN8237 in pediatric leukemia, medulloblastoma, and neuroblastoma cell lines. Investigational new drugs. 2012 doi: 10.1007/s10637-012-9831-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harris PS, Venkataraman S, Alimova I, Birks DK, Donson AM, Knipstein J, et al. Polo-like kinase 1 (PLK1) inhibition suppresses cell growth and enhances radiation sensitivity in medulloblastoma cells. BMC Cancer. 2012;12:80. doi: 10.1186/1471-2407-12-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.El-Sheikh A, Fan R, Birks D, Donson A, Foreman NK, Vibhakar R. Inhibition of Aurora Kinase A enhances chemosensitivity of medulloblastoma cell lines. Pediatr Blood Cancer. 2010;55:35–41. doi: 10.1002/pbc.22465. [DOI] [PubMed] [Google Scholar]
- 54.Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29:1408–1414. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.