Abstract
Introduction
Adequate tumor acquisition is essential to identify somatic molecular alterations in non-small-cell lung cancer (NSCLC), such as EGFR mutations and ALK translocations. The success and failure rates for tumor genotyping of tissue obtained from fine needle aspirates of nodal tissue using a convex probe endobronchial ultrasound (CP-EBUS) and other diagnostic modalities in routine NSCLC care have not been described.
Methods
Clinicopathologic data, tumor genotype success and failure rates were retrospectively compiled and analyzed from 207 patient-tumor samples sent for routine tumor genotype in clinical practice, including 42 patient-tumor samples obtained from hilar or mediastinal lymph nodes using CP-EBUS.
Results
The median age was 65 years, 62.3% were women, 77.8% were white, 26.6% were never smokers, 73.9% had advanced NSCLC and 84.1% had adenocarcinoma histology. Tumor tissue was obtained from CP-EBUS-derived hilar or mediastinal nodes in 42 cases (20.2% of total). In this latter cohort, the overall success rate for EGFR mutation analysis was 95.2%, for KRAS mutation 90.5%, and for ALK FISH 90.5%. In the complete 207 tumors, the success rate for EGFR was 92.3%, for KRAS 91.8%, and for ALK 89.9%. The failure rates were not significantly different when comparing CP-EBUS-derived nodal tissue versus all other samples or versus surgical biopsies of mediastinal nodes, but were significantly lower than image-guided percutaneous transthoracic core-needle biopsies.
Conclusions
The success rate of multiple tumor genomic analyses techniques for EGFR, KRAS and ALK gene abnormalities using routine lung cancer tissue samples obtained from hilar or mediastinal lymph nodes by means of CP-EBUS exceeds 90%, and this method of tissue acquisition is not inferior to other specimen types. Tumor genotype techniques are feasible in most CP-EBUS-derived samples and therefore further expansion of routine tumor genotype for the care of patients with NSCLC may be possible using targeted sample acquisition through CP-EBUS.
Keywords: lung cancer, non-small-cell lung cancer, endobronchial ultrasound, EBUS, mediastinoscopy, never smokers, epidermal growth factor receptor, EGFR, anaplastic lymphoma kinase, ALK, KRAS, tumor genotype, failure, bone specimen, core biopsy, transbronchial needle aspiration, computed tomography, molecular testing
INTRODUCTION
Non-small-cell lung cancers (NSCLCs) comprise a heterogeneous group of cancers characterized by different genomic abnormalities (1). In some NSCLCs, driver oncogenes have been identified; with the best studied including v-ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS), epidermal growth factor receptor (EGFR) and anaplastic lymphoma kinase (ALK). Specifically, mutations in EGFR and rearrangements involving ALK are part of the pathogenesis of NSCLCs (predominantly in never and/or light smokers) and predict improved outcomes with therapy based on tyrosine kinase inhibitors (TKIs). Evidence-based practice guidelines now mandate adequate tissue acquisition for molecular studies aimed at identifying the aforementioned aberrations (2).
Over the last decade, the development of convex probe endobronchial ultrasound (CP-EBUS) with capability to obtain real-time visualization of mediastinal and hilar lymph nodes as well as guide transbronchial needle aspiration (TBNA) has revolutionized the diagnosis and staging of lung cancer. CP-EBUS offers a less invasive alternative for histologic sampling of these nodal stations when compared to mediastinoscopy, and evidence-based guidelines of diagnosis and staging of lung cancer now suggest the use of CP-EBUS-TBNA for the confirmation of suspected N2 and N3 disease. Several large prospective studies have demonstrated the sensitivity of CP-EBUS-TBNA to be equivalent to mediastinoscopy (3), the current gold standard, in indicating the true pathologic N stage. However, in the case of restaging, CP-EBUS-TBNA allows for multiple, repeat biopsy events in a minimally invasive way, and thus confers an advantage over mediastinoscopy (4). When compared to other clinical staging strategies, CP-EBUS has diagnostic accuracy of 98% versus 72.5% for PET and 60.8% for computed tomography (5). CP-EBUS-TBNA is an emerging method of tissue acquisition in NSCLC.
In the face of these two groundbreaking scientific advances – the characterization of the predictive value of driver oncogenes and the development of CP-EBUS as a method of tissue acquisition - it is evident that the combined use of CP-EBUS-TBNA and genotyping will likely impact the diagnosis, staging and treatment of non-small cell lung cancer. In this study, we aim to describe our experience in 207 patients with NSCLC who underwent diverse diagnostic procedures for genotypic analyses with a focus on the group that had tumor acquisition through CP-EBUS-TBNA.
MATERIALS AND METHODS
Patient selection
Patients with a diagnosis of lung cancer, who were seen by our providers and whose tumors were genotyped for at least EGFR mutations were retrospectively identified through an ongoing Institutional Review Board (IRB) approved protocol at Beth Israel Deaconess Medical Center (BIDMC2009-P-000182). Patients and tumor pairs were excluded if genotyping of at least EGFR mutation, KRAS mutation and ALK translocations were not performed. There were 207 patient-tumor specimens that were submitted for these multiple tumor genotype techniques between 2007 and 2012. The data cut off for analyses was December 19th, 2012. Study data were collected and managed using REDCap electronic data capture tools hosted at BIDMC.
Tumor processing and genotype
Surgical (i.e., either incisional or excisional biopsies that required a surgical procedure) and core needle biopsies were processed using standard histopathologic techniques: 10% neutral buffered formalin fixation and paraffin-embedding. Cell aspirates or cell concentrates from fluid samples were collected into a methanol-water fixative (CytoLyt, Hologic Corp., Marlborough, MA), with residual material used to create a cell block using a plasma-thrombin method prior to formalin-fixation and paraffin embedding. Once a diagnosis was established on histologic and/or immunohistiologic slides, the residual material in the formalin-fixed paraffin-embedded (FFPE) tissue blocks were submitted for molecular analysis. When multiple tissue blocks were available, the one with the highest tumor cellularity was chosen, without additional tumor microdissection or enrichment. Molecular analysis of tumor specimens was performed at a commercial vendor, Integrated Oncology (LabCorp, Esoterix Genetic Laboratories, LLC). DNA was isolated and EGFR exons 18 to 21 sequenced. For KRAS mutation analysis, exon 2 was amplified and subjected to single nucleotide primer xtension to detect mutations at codons 12 and 13. ALK translocation status was analyzed using the Vysis ALK Break-Apart fluorescence in situ hybridization (FISH) probe (Abbott Molecular, Inc., Des Plaines, IL). Failure of the assays was defined as insufficient material to isolate DNA or inability to complete sequencing for EGFR and KRAS mutations, and lack of hybridization signals after two attempts for ALK FISH.
Data collection
The site of biopsy (lung, lymph node, pleura, bone, brain, liver, pericardium, or adrenal) and the type of biopsy (surgical specimen [both excisional or incisional], core needle biopsy or cell block from aspirate/fluid) were extracted from the medical record. Slides from all specimens that failed molecular testing, as well as a subset of the successfully genotyped cases were re-reviewed by a pathologist (PAV), with data compiled on tissue preparation, tumor content, and other histological features annotated.
EBUS technique and tumor collection with TBNA
The CP-EBUS bronchoscope used for tissue acquisition was a 7.5 MHz Olympus (BF-UC160F-OL8; Olympus America Inc., Center Valley, PA), fitted with color Doppler ultrasound capability. A 21 g needle was used to obtain TBNA samples. Two to eight passes (usually 3 passes) per lymph node were obtained. Lymph node stations reachable by CP-EBUS included paratracheal lymph nodes (2R, 2L, 4L, 4R), subcarinal (7), and the hilar, interlobar and lobar lymph nodes (10R, 10L, 11R, 11L, 12R, 12L). The lymph node aspirates were placed in methanol-water fixative and prepared as described above. At our institution, all CP-EBUS-TBNA cases were done by experienced interventional pulmonologists. As previously described, the learning curve on CP-EBUS-TBNA is likely to require 50 or more supervised cases in order to accomplish a systematic mediastinal staging (8).
Statistical methods
Fisher’s exact test was performed to compare categorical variables. A p-value < 0.05 was considered statistically significant. All p-values we reported were two-sided. We performed our statistical analyses with STATA version 12 (STATA Corp, College Station, TX).
RESULTS
Patient and tumor characteristics
Table 1 summarizes the clinical and pathological characteristics of the 207 patient-tumor pairs that were included in our cohort and highlights the characteristics of the 42 samples (42/207 [20.2%]) that were obtained from EBUS-derived mediastinal or hilar nodes (Table 1).
Table 1.
Baseline patient and tumor characteristics of the overall cohort and of the CP-EBUS-TBNA cohort
| All patients (n=207) |
CP-EBUS-TBNA lymph node cohort (n=42) |
|
|---|---|---|
|
| ||
| Age at the time of biopsy median(range) |
65(29-89) | 61.5(39-84) |
|
| ||
| Women n(%) | 129(62.3) | 28(66.7) |
|
| ||
| Race n(%) | ||
| White | 161(77.8) | 34(81.0) |
| Asian | 19(9.18) | 2(4.76) |
| Black | 17(8.21) | 2(4.76) |
| Others | 10(4.83) | 4(9.52) |
|
| ||
| Smoking status n(%) | ||
| Current smoker | 42(20.3) | 10(23.8) |
| Former smoker | 110(53.1) | 23(54.8) |
| Never smoker | 55(26.6) | 9(21.4) |
|
| ||
| Stage n(%) | ||
| I | 14(6.76) | 0(0) |
| II | 13(6.28) | 1(2.38) |
| III | 27(13.0) | 7(16.7) |
| IV | 153(73.9) | 34(81.0) |
|
| ||
| Histology n(%) | ||
| Adenocarcinoma | 174(84.1) | 36(85.7) |
| Squamous cell carcinoma | 9(4.35) | 0(0) |
| NSCLC(NOS) | 22(10.6) | 6(14.3) |
| Others | 2(0.97) | 0(0) |
|
| ||
| Anatomic site of biopsy n(%) | ||
| Bone | 13(6.28) | 0(0) |
| Brain | 17(8.21) | 0(0) |
| Liver | 3(1.45) | 0(0) |
| Lung | 98(47.3) | 0(0) |
| Lymph node | 51(24.6) | 42(100) |
| Pleura | 23(11.1) | 0(0) |
| Others | 2(0.97) | 0(0) |
|
| ||
| Type of biopsy n(%) | ||
| Core needle biopsy | 39(18.8) | 0(0) |
| Surgical biopsy | 95(45.9) | 0(0) |
| Cell block from FNA | 61(29.5) | 42(100) |
| Cell block from fluid | 12(5.80) | 0(0) |
|
| ||
|
EGFR mutation analysis Success n (%, [95% CI]) |
191(92.3 [87.5-95.4]) | 40(95.2 [82.6-99.2]) |
| Positive/Mutated | 32(15.5) | 5(11.9) |
| Negative/Wild-type | 159(76.8) | 35(83.3) |
| Failure | 16(7.73) | 2(4.8) |
|
| ||
|
KRAS mutation analysis Success n (%, [95% CI]) |
190(91.8 [87.0-95.0]) | 38(90.5 [76.5-96.9]) |
| Positive/Mutated | 65(31.4) | 18(42.9) |
| Negative/Wild-type | 125(60.4) | 20(47.6) |
| Failure | 17(8.21) | 4(9.5) |
|
| ||
|
ALK FISH analysis Success n (%, [95% CI]) |
186(89.9 [84.7-93.5]) | 38(90.5 [76.5-96.9]) |
| Positive | 11(5.31) | 1(2.4) |
| Negative | 175(84.5) | 37(88.1) |
| Failure | 21(10.1) | 4(9.5) |
CI, confidence interval
Success and failure rates of tumor genotype techniques
In these 207 tumors, all three tests were ordered simultaneously. The success rate for EGFR mutation analysis was 92.3%, for KRAS mutation 91.8%, and for ALK FISH 89.9%.
In the 42 lymph node metastases obtained through EBUS-TBNA, the success rate for EGFR mutation analysis was 95.2%, for KRAS mutation 90.5%, and for ALK FISH 90.5% (Table 1).
CP-EBUS-derived nodal tissue compared to other tissue acquisition techniques
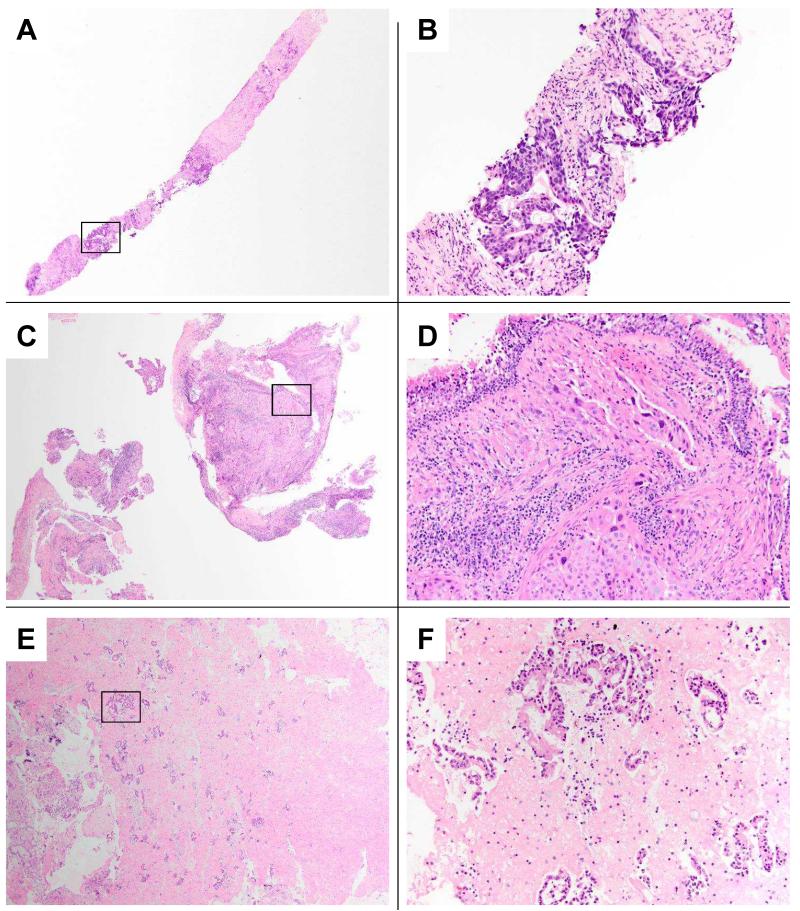
To determine if CP-EBUS-derived nodal tissue had similar or different success rates when compared to other possible methods of tumor acquisition, we first compared CP-EBUS-derived nodal tissue to all remaining samples. Typical histologic examples of image-guided percutaneous transthoracic core-needle biopsies, bronchoscopic-guided lung biopsies, and cell blocks prepared from CP-EBUS-TBNAs of mediastinal lymph nodes are compared in Figure 1.
FIGURE 1. Histopathology of different minimally invasive methods for thoracic tissue sampling.
A-B: Image-guided percutaneous transthoracic core-needle biopsies from a left upper lobe lung mass. The low power field highlights one of two needle core biopsies obtained, both demonstrating significant fibrosis with intervening areas of invasive adenocarcinoma. On higher power, the tumor cells demonstrate a glandular morphology, with associated chronic inflammation. Cytologic rapid on-site evaluation of touch prep slides from the core biopsies confirmed adenocarcinoma, and subsequent tumor genotyping successfully identified a deletion mutation in exon 19 of the EGFR gene.
C-D: Bronchoscopy-guided lung biopsies of a right middle lobe lung mass. The low power field in panel C shows two of the six airway tissue biopsies obtained. The higher power view demonstrates a poorly differentiated adenocarcinoma with cytoplasmic mucin vacuoles infiltrating submucosal lymphatic spaces, with surrounding bronchial associated lymphoid tissue and overlying respiratory epithelium. Tumor genotyping successfully identified a mutation in exon 20 of the EGFR gene.
E-F: Cell block preparation from an CP-EBUS-TBNA of mediastinal lymph nodes. The low power field view in panel E, covering approximately 25% of the cell block area, highlights the cellular specimen with numerous clusters of tumor cells in a background of eosinophilic fibrin clot. On higher power the background lymphocytes confirm lymph node sampling, and the tumor morphology is that of a moderately differentiated adenocarcinoma. Tumor genotyping was successful.
Panels A, C, and E 40× original magnification; panels B, D, and F 200× original magnification representing boxed area in prior panel; Hematoxylin and Eosin stains.
CP-EBUS-derived nodal tissues were obtained from N1, N2, and/or N3 nodes, the median number of FNA passes per node was 3.28 (range 2-8) and there were no reported complications. As indicated in Table 2, the failure rates were not significantly different between CP-EBUS-derived nodal tissue versus all other samples for EGFR mutation, for KRAS mutation, and for ALK FISH (Table 2).
Table 2.
Success and failure rates of genotype tests comparing CP-EBUS-derived lymph node cell blocks and other sources of tissue acquisition
| EGFR mutation analysis | |||||
|---|---|---|---|---|---|
| CP-EBUS- derived node cell blocks |
All other methods of tissue acquisition |
Mediastinal/hi lar nodes from surgical biopsies |
Lung core biopsies (bronchos copy) |
Lung core needle biopsies (image- guided) |
|
| Success n (%) | 40 (95.2%) | 151 (91.5%) | 8 (100%) | 12 (86%) | 6(55%) |
| Failure n (%) | 2 (4.8%) | 14 (8.5%) | 0 (0%) | 2 (14%) | 5(45%) |
| Total | 42 | 165 | 8 | 14 | 11 |
| p-value | ref | 0.54 | 1 | 0.26 | 0.003 |
| ALK FISH analysis | |||||
| CP-EBUS- derived node cell blocks |
All other methods of tissue acquisition |
Mediastinal/hi lar nodes from surgical biopsies |
Lung core biopsies (bronchos copy) |
Lung core needle biopsies (imageg- guided) |
|
| Success n (%) | 38 (90.5%) | 148 (89.7%) | 8 (100%) | 12(86%) | 6(55%) |
| Failure n (%) | 4 (9.5%) | 17 (10.3%) | 0 (0%) | 2 (14%) | 5(45%) |
| Total | 42 | 165 | 8 | 14 | 11 |
| p-value | ref | 1 | 1 | 0.63 | 0.01 |
| KRAS mutation analysis | |||||
| CP-EBUS- derived node cell blocks |
All other methods of tissue acquisition |
Mediastinal/hi lar nodes from surgical biopsies |
Lung core biopsies (bronchos copy) |
Lung core needle biopsies (image- guided) |
|
| Success n (%) | 38 (90.5%) | 152 (92.1%) | 8 (100%) | 13 (93%) | 8(73%) |
| Failure n (%) | 4 (9.5%) | 13 (7.9%) | 0 (0%) | 1 (7%) | 3(27%) |
| Total | 42 | 165 | 8 | 14 | 11 |
| p-value | ref | 0.75 | 1 | 1 | 0.15 |
To better match the tissue of origin (thoracic lymph nodes), we compared the success and failure rates of CP-EBUS-TBNA to those of mediastinal or hilar nodes acquired through surgical biopsies (either through mediastinoscopy or thoracic resections). There were no failures in the eight nodal tissues acquired through surgical technique. However, the success rates were not significantly different between CP-EBUS-derived nodal tissue versus these eight surgical nodal specimens for EGFR mutation; for KRAS mutation and for ALK FISH (Table 2).
We then compared CP-EBUS-derived nodal tissue to lung biopsies, either using image-guided percutaneous transthoracic core-needle biopsies or lung biopsies obtained through bronchoscopy (Table 2). The success rates were higher when CP-EBUS-derived nodal tissue was compared to image-guided percutaneous transthoracic core-needle biopsies for EGFR mutation, for KRAS mutation and for ALK FISH (Table 2). The success rates for CP-EBUS-derived nodal tissue were similar to those of bronchoscopy-guided lung biopsies (Table 2).
We compared image-guided percutaneous transthoracic core-needle and bronchoscopy-guided lung biopsies in relation to estimated tumor cellularity, tumor area, use of touch preparation for rapid on-site evaluation, presence of extensive desmoplastic stromal response, number of slides cut from the paraffin block used for immunohistochemical/ancillary studies, number of biopsy passes, location site within the lung and mass size. The only two major differences were in the use of a touch preparation (image-guided percutaneous transthoracic core-needle biopsies 8/8 cases versus 0/13 cases for bronchoscopy-guided lung biopsies, p<0.0001) and the number of passes of needle for tissue acquisition (image-guided percutaneous transthoracic core-needle biopsies 8/11 cases with ≤ 2 passes versus 0/14 cases for bronchoscopy-guided lung transbronchial biopsies, p=0.072). Complications, including pneumothorax, were also more common in image-guided percutaneous transthoracic core-needle biopsies (6/11 cases) when compared to bronchoscopy-guided lung biopsies (0/14 cases, p=0.009).
Failed specimens using CP-EBUS-derived nodal tissue
We attempted to determine factors specifically associated with molecular testing failure in specimens originating from CP-EBUS. In most of the samples that failed at least one of the tumor genotyping techniques used, we observed that there were insufficient tumor cells in the cell block specimen (<100 tumor cells per slide and inferior dimension of tumor specimen). In CP-EBUS-derived nodal tissues that were successful 17/19 (89.4%) had ≥ 100 cells while in failure cases only 2/5 (40%) had ≤ 100 cells (p=0.042). We also evaluated as other possible characteristics the size of the nodal tissue biopsied, the location of the node, the number of passes per lymph node, use of touch preparation for rapid on-site evaluation, presence of extensive desmoplastic stromal response, and number of slides cut from the paraffin block used for immunohistochemical and ancillary studies. None of these characteristics were significantly different between success and failure CP-EBUS-derived nodal samples.
DISCUSSION
In this study, we provide evidence that CP-EBUS-TBNA is able to procure adequate lymph node tissue to carry out genetic analyses of EGFR, KRAS, and ALK using commercially-available techniques that are employed for everyday patient care. More than 20% of tumor samples submitted for tumor genotype at our academic medical center originated from CP-EBUS-TBNA of hilar or mediastinal nodes. In these metastases, the success rate for EGFR mutation analysis was 95.2%, for KRAS mutation 90.5%, and for ALK FISH 90.5%. More importantly, these CP-EBUS-TBNA samples were not inferior to surgical thoracic nodal samples and or to image-guided percutaneous transthoracic core-needle biopsies in our single institution series; results that may not reflect practice patterns at other centers.
The clinician (a medical oncologist, thoracic surgeon, or pulmonologist) is frequently faced with the choice of diagnostic sampling technique. Traditionally, image-guided percutaneous transthoracic core-needle biopsies have been used by most practices to obtain tissue from peripheral lung tumors even in the presence of hilar and/or mediastinal thoracic lymphadenopathy. However, our study shows that when compared to CP-EBUS-guided biopsies, image-guided percutaneous transthoracic core-needle biopsies of the lung had a significantly higher failure rate of tumor genotype techniques. The high failure rate of image-guided percutaneous transthoracic core-needle biopsies to yield adequate material for molecular studies has been reported by other academic medical centers (6). It is possible to speculate that tumor tissue loss during the aforementioned procedure and subsequent preparation explains these results. Indeed in our series, image-guided percutaneous transthoracic core-needle biopsies more frequently had tissue loss (in the form of cytological adequacy evaluation of core biopsy touch-preparation slides at time of biopsy) than bronchoscopic-derived lung biopsies.
Interestingly, the absence of rapid on-site evaluation for CP-EBUS procedures did not seem to affect the success rate in our series and may have indeed led to a more thorough evaluation and repeated sampling of the lymph nodes. Also of note, in traditional image-guided percutaneous transthoracic core-needle biopsies of the lung there is an increased risk of complications (including pneumothorax) when doing multiple needle passes; a concern that is not significant with CP-EBUS-TBNA (7). However, the importance of extensive training in CP-EBUS-TBNA, particularly in complete and systematic staging for lung cancer, cannot be overemphasized (8).
The use of CP-EBUS-TBNA for tumor genotype in NSCLC has been reported by a few other groups, with successful analysis of at least one genotype (such as EGFR or KRAS mutations) in 72.2% to 98.4% of samples obtained (9-14). One of the largest published series described a cohort of 774 patients in 5 centers of the United Kingdom with known or suspected lung cancer who had CP-EBUS-TBNA done to determine if these samples were suitable for NSCLC subclassification and genotyping. The available tissue allowed subclassification of 77% of samples and genotyping for EGFR mutation in 90% of the cases (15). In our study, all samples were obtained using routine day-to-day procedures, underwent routine pathological subtyping with immunohistochemical markers, and then were subsequently sent for multiple tumor genotype analyses with a success rate that exceeded 90%. The aforementioned studies confirm that CP-EBUS-TBNA samples are adequate as a starting material for routine tumor genotype studies.
In summary, commercially-available tumor genotype techniques for EGFR mutation analysis, KRAS mutation analysis and ALK FISH are feasible in most EBUS-TBNA NSCLC samples. These encouraging findings may indicate that further expansion of routine tumor genotype into the care of patients with NSCLC may be possible using targeted sample acquisition through EBUS.
ACKNOWLEDMENTS
We would like to thank all current and former members of the Thoracic Oncology Clinic at Beth Israel Deaconess Medical Center, and our patients.
Funding/Grant Support:
This work was funded in part through fellowships from the American Society of Clinical Oncology Conquer Cancer Foundation (DBC), an American Cancer Society grant (RSG 11-186, DBC), a Lung Cancer Foundation of America - International Association for the Study of Lung Cancer grant (DBC), and National Institutes of Health grant (CA090578, DBC).
Footnotes
Conflict of interest:
DBC has received consulting fees from Pfizer, Roche and AstraZeneca. No other conflict of interest is stated.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- (1).Gaughan EM, Costa DB. Genotype-driven therapies for non-small cell lung cancer: focus on EGFR, KRAS and ALK gene abnormalities. Ther Adv Med Oncol. 2011;3:113–125. doi: 10.1177/1758834010397569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Lindeman NI, Cagle PT, Beasley MB, Chitale DA, Dacic S, Giaccone G, et al. Molecular Testing Guideline for Selection of Lung Cancer Patients for EGFR and ALK Tyrosine Kinase Inhibitors: Guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Thorac Oncol. 2013;8:823–859. doi: 10.1097/JTO.0b013e318290868f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Annema JT, van Meerbeeck JP, Rintoul RC, Dooms C, Deschepper E, Dekkers OM, et al. Mediastinoscopy vs endosonography for mediastinal nodal staging of lung cancer: a randomized trial. JAMA. 2010;304:2245–2252. doi: 10.1001/jama.2010.1705. [DOI] [PubMed] [Google Scholar]
- (4).Herth FJ, Annema JT, Eberhardt R, Yasufuku K, Ernst A, Krasnik M, et al. Endobronchial ultrasound with transbronchial needle aspiration for restaging the mediastinum in lung cancer. J Clin Oncol. 2008;26:3346–3350. doi: 10.1200/JCO.2007.14.9229. [DOI] [PubMed] [Google Scholar]
- (5).Yasufuku K, Nakajima T, Motoori K, Sekine Y, Shibuya K, Hiroshima K, et al. Comparison of endobronchial ultrasound, positron emission tomography, and CT for lymph node staging of lung cancer. Chest. 2006;130:710–718. doi: 10.1378/chest.130.3.710. [DOI] [PubMed] [Google Scholar]
- (6).Tam AL, Kim ES, Lee JJ, Ensor JE, Hicks ME, Tang X, et al. Feasibility of image-guided transthoracic core-needle biopsy in the BATTLE lung trial. J Thorac Oncol. 2013;8:436–442. doi: 10.1097/JTO.0b013e318287c91e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Varela-Lema L, Fernandez-Villar A, Ruano-Ravina A. Effectiveness and safety of endobronchial ultrasound-transbronchial needle aspiration: a systematic review. Eur Respir J. 2009;33:1156–1164. doi: 10.1183/09031936.00097908. [DOI] [PubMed] [Google Scholar]
- (8).Folch E, Majid A. Point: Are 50 supervised Procedures Required to Develop Competency in Performing Endobronchial Ultrasound-Guided Transbronchial Needle Aspiration for Mediastinal Staging? Yes. Chest. 2013;143:888–891. doi: 10.1378/chest.12-2462. [DOI] [PubMed] [Google Scholar]
- (9).Nakajima T, Yasufuku K, Suzuki M, Hiroshima K, Kubo R, Mohammed S, et al. Assessment of epidermal growth factor receptor mutation by endobronchial ultrasound-guided transbronchial needle aspiration. Chest. 2007;132:597–602. doi: 10.1378/chest.07-0095. [DOI] [PubMed] [Google Scholar]
- (10).Garcia-Olive I, Monso E, Andreo F, Sanz-Santos J, Taron M, Molina-Vila MA, et al. Endobronchial ultrasound-guided transbronchial needle aspiration for identifying EGFR mutations. Eur Respir J. 2010;35:391–395. doi: 10.1183/09031936.00028109. [DOI] [PubMed] [Google Scholar]
- (12).Schuurbiers OC, Looijen-Salamon MG, Ligtenberg MJ, van der Heijden HF. A brief retrospective report on the feasibility of epidermal growth factor receptor and KRAS mutation analysis in transesophageal ultrasound- and endobronchial ultrasound-guided fine needle cytological aspirates. J Thorac Oncol. 2010;5:1664–1667. doi: 10.1097/JTO.0b013e3181f0bd93. [DOI] [PubMed] [Google Scholar]
- (12).van Eijk R, Licht J, Schrumpf M, Talebian YM, Ruano D, Forte GI, et al. Rapid KRAS, EGFR, BRAF and PIK3CA mutation analysis of fine needle aspirates from non-small-cell lung cancer using allele-specific qPCR. PLoS One. 2011;6:e17791. doi: 10.1371/journal.pone.0017791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Santis G, Angell R, Nickless G, Quinn A, Herbert A, Cane P, et al. Screening for EGFR and KRAS mutations in endobronchial ultrasound derived transbronchial needle aspirates in non-small cell lung cancer using COLD-PCR. PLoS One. 2011;6:e25191. doi: 10.1371/journal.pone.0025191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Nakajima T, Yasufuku K, Nakagawara A, Kimura H, Yoshino I. Multigene mutation analysis of metastatic lymph nodes in non-small cell lung cancer diagnosed by endobronchial ultrasound-guided transbronchial needle aspiration. Chest. 2011;140:1319–1324. doi: 10.1378/chest.10-3186. [DOI] [PubMed] [Google Scholar]
- (15).Navani N, Brown JM, Nankivell M, Woolhouse I, Harrison RN, Jeebun V, et al. Suitability of endobronchial ultrasound-guided transbronchial needle aspiration specimens for subtyping and genotyping of non-small cell lung cancer: a multicenter study of 774 patients. Am J Respir Crit Care Med. 2012;185:1316–1322. doi: 10.1164/rccm.201202-0294OC. [DOI] [PMC free article] [PubMed] [Google Scholar]