Review on how NF-kB member RelB regulates leukocyte biology by reprograming bioenergetics, metabolism and immune phenotypes, thereby influencing both homeostasis and inflammatory diseases.
Keywords: NF-κB, inflammation, endotoxin tolerance, bioenergy
Abstract
RelB is one of the more unusual members of the NF-κB family. This family, arguably the best known group of transcription regulators, regulates an astonishing array of cell types and biological processes. This includes regulation of cell growth, differentiation and death by apoptosis, and the development and function of the innate and adaptive-immune system. RelB is best known for its roles in lymphoid development, DC biology, and noncanonical signaling. Within the last few years, however, surprising functions of RelB have emerged. The N-terminal leucine zipper motif of RelB, a motif unique among the NF-κB family, may associate with more diverse DNA sequences than other NF-κB members. RelB is capable of direct binding to the AhR that supports the xenobiotic-detoxifying pathway. RelB can regulate the circadian rhythm by directly binding to the BMAL partner of CLOCK. Finally, RelB also couples with bioenergy NAD+ sensor SIRT1 to integrate acute inflammation with changes in metabolism and mitochondrial bioenergetics. In this review, we will explore these unique aspects of RelB, specifically with regard to its role in immunity.
Introduction
On its 25th birthday in 2012, biomedical sciences aptly honored the discovery of the highly conserved NF-κB family of master transcription regulators [1]. This remarkable family, arguably the best known of transcription factors, is active in an astonishing array of cell types and biological processes. These processes include cell growth, differentiation, apoptosis, and the development and function of the innate and adaptive-immune systems. NF-κB member RelA (p65), the original and still star member of the family, has been studied intensely and is the focus of numerous reviews. Whreas NF-κB member RelB has received less attention, it has been linked to a surprising array of overlapping and distinct processes that lie outside of many properties of other NF-κB members.
Statistically, outliers arise as a result of changes in system behavior and/or simply through natural deviations in populations. Malcolm Gladwell popularly applied this observation in his acclaimed book, Outliers, where he tried to understand the distinct success of some individuals within similarly intelligent and ambitious peer groups. In this review, we apply this “outlier” principle to understand the emerging story of RelB, an outlier in the NF-κB family of proteins with special attributes.
What features of this review merit the outlier distinction? Whereas RelB is best known for its roles in lymphoid development, DC biology, and noncanonical signaling [2], surprisingly distinct functions of RelB have emerged. RelB is a major contributor to chromatin biology, frequently functioning as a dual transcription factor that silences sets of genes by generating silent facultative heterochromatin and activates euchromatin of others [3]. RelB is required to repress immediate-response proinflammatory genes during endotoxin tolerance [4] and couples with the bioenergy NAD+ sensor SIRT1 to integrate acute inflammation with changes in metabolism and mitochondrial bioenergetics [5]. The N-terminal leucine zipper motif of RelB, a motif unique among the NF-κB family, may associate with more diverse DNA sequences than other NF-κB members [6]. Moreover, RelB is capable of direct binding to the AhR that supports the xenobiotic, detoxifying pathway [7], as well as suppressing circadian rhythm by directly binding to the BMAL partner of CLOCK [8].
GENE PRODUCT, GENE STRUCTURE, AND EXPRESSION
Gene product
When RelB was discovered in mice just over 20 years ago, it was identified as a family member by the RHD shared by all of the members of the NF-κB family [9]. In this first report, RelB associated with NF-κB p50 at its C-terminus to activate transcription, during which time, RelB-p50 dimers bound to the same NF-κB sites recognized by p50 homodimers. Not long after, a transcription repressor protein in Jurkat lymphoid cells, referred to as I-rel, was described [10]. I-rel associated with p50 or RelA to inhibit NF-κB transactivation but seemingly without direct DNA binding; because of this, I-rel appeared different from RelB. However, before long, it was discovered that RelB could, in fact, bind to DNA but only after forming a heterodimer with NF-κB p50 or p52 [11]. Investigators then concluded that there was no significant difference between RelB and I-rel, causing RelB to become the preferred name.
Like all NF-κB members, RelB contains an ∼300-residue region known as the RHD (Fig. 1). This region supports many of the NF-κB essential functions, such as DNA binding, dimerization, and nuclear localization [12]. RelB, unlike other NF-κB members, has an N-terminal leucine zipper motif [9], a domain that can typically interact with many proteins [13]. Whereas the exact function of this motif in RelB is unknown, it is highly conserved between mice and humans, and point mutations that affect the leucine zipper decrease the ability of RelB to activate transcription of target genes [14]. Studies of the crystal structure of RelB-p50 heterodimers, bound to DNA, indicate that whereas RelB contains four DNA-interacting residues that are highly conserved among NF-κB members, it can bind a more diverse set of NF-κB consensus sequences than other family members [6]. It is plausible but unproven that the RelB leucine zipper region contributes to RelB binding in a number of physiologic responses discussed subsequently, such as metabolism, bioenergetics, xenobiosis, and circadian rhythm.
Figure 1. Two-dimensional structure of NF-κB family members.
LZ, leucine zipper region; RHR, Rel homology region; TAD, transcription-activating domain; ANK, ankyrin repeat.
Gene structure and expression
The RelB gene is located on chromosome 19q13.32 and encodes an mRNA from 11 exons, making a protein of 579 aa [15]. The 5′ RelB promoter has two κB sites but no TATA region. As analyzed by the SABiosciences (Qiagen, Valencia, CA, USA) site list of DNA domain-binding sites, RelB DNA potentially can be regulated by many different transcription factors at its promoter, introns, and exons; among these putative regulatory sites are domains that bind glucocorticoid receptors, c-Jun, c-Fos, sp-1, and B-zip chromatin repressor and insulator CCCTC-binding factor (CTFC) that defines the boundary between euchromatin/heterochromatin and IFN response factor 1 (IRF1). As analyzed by the Santa Cruz gene browser (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and data entered from the Encyclopedia of DNA Elements (ENCODE) project [16], RelB has an enhancer region near the beginning of its reference sequence, as identified by binding of histone H3 lysine 4 methyl-1, H3 lysine 27 acetylation, and PU.1. However, functional studies of this potential regulatory region have not been reported.
RelB is often constitutively expressed in lymphocytes and DCs [17, 18] but not in most innate-immune cells (e.g., monocytes, macrophages, and neutrophils) where the basal expression level of RelB mRNA and protein is minimal and requires a stimulus for the gene to be induced [19]. The best-studied transactivator of induced RelB mRNA in immune cells is RelA, which can bind proximal promoter NF-κB sites [19]; this report also showed that RelB can autoregulate mRNA levels by binding its own promoter NF-κB sites. CD40L, an activator of the noncanonical NF-κB pathway, increases RelB expression in B cells, likely by activating RelA [20]. AP-1 regulates RelB expression by binding a site 1503 bp upstream of the start site when cytomegalovirus protein IE1 is present [21]. In contrast, the steroid hormone 1α,25-dihydroxyvitamin D3 inhibits RelB mRNA expression [22] by unknown promoter interactions; this process blocks DC differentiation. In response to proinflammatory signals, such as LPS binding to TLR4, RelA up-regulates RelB mRNA expression significantly in innate-immune cells [4, 23]. Finally, the cellular bioenergy sensor SIRT1 promotes RelB mRNA expression in human THP1 promonocytes by an unknown mechanism [5], thereby linking cellular metabolism and bioenergetics with immune function.
RelB mRNA levels and protein synthesis are regulated further at the translational level. The miR-146a directly binds to RelB mRNA to limit mRNA and block translation in a specific population of monocytic cells [24]. In mice, Ly6Chigh monocytes represent a proinflammatory population, roughly comparable with M1 macrophages [25]. In these Ly6Chigh monocytes, miR-146 is induced by RelA. Thus, activation of RelA can induce RelB expression and generate miR-146 to repress the expression of RelA itself via disrupting IL-1R-associated kinase 1 and TRAF6 [26, 27] and by directly limiting RelB protein translation. This suppression of RelB by miR-146a can enhance the immune response of the proinflammatory Ly6Chigh monocyte subpopulation [24]. The miR146 functions are complicated, and it is not known how the differential effects of miR-146 on RelA and RelB are coordinated temporally during inflammatory and immune reactions.
Protein stabilization
Once expressed, the RelB protein is very labile while by itself [28]. Early work recognized this and identified NF-κB2 as an important RelB stabilizer [29, 30]. NF-κB2 has two forms: p100 and p52. The p100 form is more common in the unstimulated state, and it prevents RelB DNA binding. This association with p100 requires phosphorylation of RelB at serine 368 [29]. The processing-inhibitory domain of p100 prevents the proteosome from converting p100 to p52 until the former has been ubiquitinated [31]. This process is slower and less efficient than the conversion of p105 to p50 [32]. Interestingly, the association of p100 with RelB provides a cooperative stabilizing state for RelB and p100, limiting the processing of p100 to p52 [29].
As p100, the C-terminus of NF-κB2 acts as a RelB inhibitor [14], where it sequesters RelB in the cytosol to repress its activity (Fig. 2). The factors that typically repress RelA through a similar sequestration method, namely IkBα, IκBβ, or p105, do not have the same effect on RelB [33]. The C-terminus of NF-κB2/p100, however, is capable of sequestering RelA-p50 dimers [34]. In fact, the ankyrin repeat domain of p100 shows sufficient homology to the IκB proteins to classify it as a fourth IκB protein, IκBδ. Degradation of IκBδ allows for the release of RelA-p50 and up-regulation of RelB and NF-κB2 [35]. In contrast, the p52 form of NF-κB2 has the opposite effect on RelB as intact p100, forming a functional heterodimer rather than acting as a repressor [36] after proteosomal processing of p100 to p52 [37]. Interestingly, RelB appears to affect the rate of this proteosomal processing. In RelB−/− cells, there is less p100 and more p52 present. This phenotype is reversed if WT RelB is reintroduced [29]. Thus, in apparent circular logic, RelB manages to inhibit itself by stabilizing its own inhibitor.
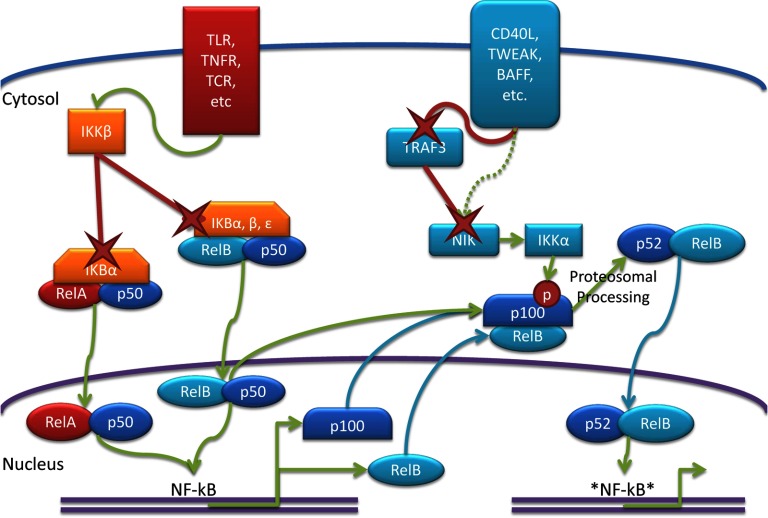
Figure 2. Schematic of the canonical and noncanonical NF-κB pathway.
Canonical signaling causes activation of IKKβ, which degrades (X) IκBα, releasing RelA-p50 heterodimers and allowing them to bind NF-κB sites. Among the genes up-regulated by this are RelB and p100. Canonical signaling also releases RelB-p50 heterodimers; however, these dimers are limited by p100. Noncanonical signaling degrades TRAF3, stabilizing NIK. IKKα is then activated, causing the phosphorylation of p100 and triggering its proteosomal processing into p52. The structure of RelB allows for binding to more diverse sites, as indicated by *NF-kB*.
Post-translational changes
To date, very little is known about specific post-translational changes that support or repress RelB biological activities; there are only three known post-translational modifications to RelB, all of them phosphorylations that regulate its stability. RelB is phosphorylated rapidly at threonine 84 and serine 552, causing cleavage at the N-terminus and its degradation by the proteosome [28]. These phosphorylations are mediated by glycogen synthase kinase-3β, specific inhibition of which blocks RelB phosphorylation and degradation [38]. This degradation does not occur when RelB is phosphorylated at serine 368, allowing its association with and stabilization by p100 [29]. Recent data from our lab show that activated NAD+-dependent deacetylase SIRT1 is required to load RelB on the TNF-α promoter in THP1 promonocytes [5]. Undoubtedly, other functional, post-translational modification will be discovered (e.g., deacetylation, methylation).
PROTEIN ACTIVATION PATHWAYS
Canonical NF-κB pathway
The NF-κB family of transcription factors is poised in an inactive state in cells and can be activated quickly by processes that are independent of transcription. There are two major pathways for activating the sequestered RelA, c-rel, and RelB transcription factors, leading to their subsequent binding to cognate DNA: the canonical and noncanonical NF-κB pathways (Fig. 2). In the canonical pathway, a surface receptor, such as a TLR, TNF-αR, or TCR, serves as the initial signaling molecule [2]. This activates a series of kinases that ultimately phosphorylates IκBα, IκBβ, and IκBϵ, leading to their proteasomal degradation and release of RelA and p50 heterodimers [2, 34]. Once the heterodimers have been released, they translocate into the nucleus, where they can bind DNA and regulate the immune response.
The degradation of IκB not only activates RelA-p50 heterodimers but also RelB-p50 heterodimers. Recent mathematical modeling supports that given the expression of RelB and p100 in DCs, there is insufficent p100 to prevent RelB nuclear localization [39]. Despite this, RelB remains in the cytosol, where RelB-p50 dimers are sequestered by IκBα, IκBβ, and IκBϵ, allowing them to be activated concurrently with RelA-p50 dimers. Active RelB-p50 dimers are limited, however, by p100, whose expression is up-regulated by canonical signaling [40]. RelB has a stronger affinity for p100 than for p50 [33]. Once p100 is present, it associates with the RelB-p50 dimers and represses their DNA-binding ability [41]. Although the frequency of this form of RelB protein control and activation is unknown, it explains conflicting reports of canonical and noncanonical cross-signaling [1].
Noncanonical NF-κB pathway
Whereas RelB-p50 dimers have been observed in other cell types, the noncanonical path is typically favored to release functional RelB activation [41]. This pathway, which functions in innate and adaptive immunity, uses a series of kinases and ubiquitin ligases to control the conversion of p100 to p52 and thus, regulates the activity of RelB. The noncanonical pathway is activated by extracellular signals, such as LTβR, CD40L, BAFF, and TWEAK, which cause TRAF3 degradation in the proteosome [42, 43]. In unstimulated cells, TRAF3 forms a complex with TRAF2, cIAP1/2 NIK [2, 42]. In this complex, NIK is ubiquitin-tagged by cIAP1/2 and subsequently degraded. The interaction between TRAF3 and NIK requires TRAF2, which links TRAF3 to the cIAP1/2 ubiquitin ligase complex [44]. Once TRAF3 is degraded by noncanonical signaling, NIK becomes stabilized and activates IKKα, and the two kinases phosphorylate p100 at serines 99, 108, 115, 123, and 872 [31, 45]. The phosphorylation of these residues creates a binding site for β-TrCP, a receptor for the β-TrCPSKp1–Cullin–F-box ubiquitin ligase [46]. This results in the partial proteosomal processing of p100, converting it to p52, releasing RelB from its inhibitor, and providing its functional partner. In mice lacking TRAF3, NIK stability is greater, and more p100 is processed into p52. This overactive processing of p100 results in an autoimmune phenotype, which can be rescued by knocking out NF-κB2 entirely [47].
Not surprisingly, CD40L, an activator of the noncanonical NF-κB pathway, increases RelB expression in B cells, likely via RelA activation [20]. RelB expression via RelA also occurs in mouse embryonic fibroblast cells treated with a LTβR-stimulating antibody, a noncanonical signal [40]. These cells showed degradation of IκBα and RelA activation prior to increased RelB expression. Proteosomal processing of p100 or IκBδ also causes an up-regulation in RelB by halting p100 sequestration of RelA-p50 dimers [35]. Pathways involving RelB are depicted in Fig. 2.
RelB, IMMUNE DEVELOPMENT, AND IMMUNOPATHOLOGY
Genetic deletions of RelB emphasize the importance of RelB in balancing adaptive and innate immunity. RelB whole animal −/− mice exhibit a complex phenotype with developmental defects and autoimmunity (Table 1). RelB −/− mice, as well as NF-κB2 −/−, lack Peyer's patch development [48]. RelB −/− also lack germinal centers and follicular DC networks and are also deficient in marginal sinus formation [49]. The spleens of RelB −/− mice have a decreased white pulp and an increased red pulp [50].
Table 1. Mice Exhibit a Complex Phenotype with Developmental Defects and Autoimmunity.
| Tissue type | RelB−/− phenotype | Immune cells affected | Immune cells responsible |
|---|---|---|---|
| Thymus | Disorganized medulla; lack of mature DCs; lack of UEA-1+ medullary epithelial cells; lack of negative selection in T cells | CD4+ T cells, CD8+ T cells | DCs, medullary epithelial cells |
| Spleen | Splenomegaly; lack of marginal zones; decreased white pulp; increased red pulp | B cells | Follicular DCs |
| LNs | Lack of germinal centers; impaired B cell proliferation | B cells | Follicular DCs |
| Peyer's patches | Lack of Peyer's patch formation | N/A | N/A |
| Periphery | Multiorgan inflammatory cell infiltration; skin lesions | Granulocytes, monocytes | T cells |
Basal RelB expression in lymphoid organs is generally low, and DC populations are mostly responsible for constitutive levels of RelB observed in the thymus, spleen, and lymph nodes [18]. It comes as no surprise, therefore, that many of the immune-system irregularities in RelB−/− mice can be traced back to DCs, which represent the primary junction of innate and adaptive immunity. These knockout mice lack many splenic and thymic DC structures, including the germinal centers, marginal zones, and thymic medulla [51], and show a severe multisystem inflammatory syndrome with T cell and monocytic infiltrates in multiple organs [52]. A combined knockout of RelB and p50, which would affect innate and adaptive immunity [53], has multiorgan damage, simulating the systemic inflammatory syndrome associated with sepsis. Taken together, the gene-deletion studies support the critical role of RelB in balancing pro- and anti-inflammatory immune responses, as suggested when I-rel was discovered [10].
RelB is essential for the proper development of the thymus and the proper “education” of T cells. In RelB knockout mice, the thymic medulla is highly disorganized, resulting in a population of T cells that has not undergone negative selection [54]. In addition to being disorganized, the mature DCs and UEA-1+ medullary epithelial cells are missing in RelB knockout mice [52]. Such mice develop T cell-dependent skin lesions resembling atopic dermatitis [55]. Interestingly, however, T cells in RelB knockout mice do develop to maturity [56] and express the normal surface markers [50].
Given the limited negative selection, one would expect severe widespread inflammation in RelB knockout mice. In fact, it appears that eliminating RelB or the noncanonical signaling pathway can decrease the T cell response. T cells in RelB knockout mice show defects in production of IFN-γ, IL-2, and IL-4 in response to multiple stimuli, including LPS, anti-CD40 antibodies, and Toxoplasma gondii infection [57]. This suppression of the T cell response appears to be a result of an increase in CD25−forkhead p3− memory CD4+ T cells [58]. The decrease in T cell response is supported further in mice with mutated NIK, where T cells showed impaired IL-2 production and thus, reduced proliferation after CD3 stimulation [59]. Thus, in RelB−/− or NIK−/− mice, a subset of memory T cells suppresses the proliferation and IL-2 response of the T cells. When these suppressor memory cells are removed, T cells actually exhibit a hyper-responsive phenotype with higher expression of IL-2 and increased T cell proliferation [58].
RelB also coordinates an inducible proliferative response in B cells. In RelB knockout mice, stimulated B cells mature normally and undergo Ig class-switching [60]. The germinal centers and marginal zones, however, are absent from the spleen [49], where these zones are the primary sites of B cell proliferation [61]. As one might expect, without them, mice show a decrease in B cell proliferation and impaired production of antigen-specific antibodies [60]. Analogous to spleen, RelB knockout mice lack MZB; if RelB-negative regulators, TRAF2 or TRAF3, are knocked out, MZB cells have been shown to accumulate [62] with potential negative consequences. In mice overexpressing BAFF and a hyperactive noncanonical pathway, a B cell-associated autoimmune pathology develops [63]. Removal of the spleen and, with it, the MZB cells reverses this pathology.
Taken together, data support that the absence of RelB results in T cell autoimmunity, the excess of RelB results in B cell autoimmunity, and RelB is required for supporting differentiated DCs. Published data also support that RelB enhances development of innate immunity through its effects on bone marrow stroma and its interaction with hematopoietic progenitor cells [64, 65]. Through its role in DC development, RelB acts as a key balancing factor at the cusp of the adaptive and innate-immune responses.
Nonimmune cell functions of RelB
Whereas RelB is found primarily acting in immune cells and tissue, RelB is known to respond to immune signals and environmental stressors in nonimmune cell types. RelB regulates cytokine production in lung epithelial cells in response to immune agonists [66]. In these cells, RelB was seen acting as a dual regulator, where RelB knockdowns showed increased IL-6 but decreased CCL-20 and RANTES production. Emerging data indicate that RelB is also involved in the xenobiotic response in the lung (discussed subsequently) [67]. In kidney epithelial cells, RelB responds to a combination of inflammatory and xenobiotic signals. When renal proximal tubule epithelial cells are treated with TNF-α and the chemotherapeutic agent cisplatin, there is a disruption of actin stress fibers that leads to apoptosis [68]. Knockdown of RelB through RNA interference prevents this morphological change and rescues these cells.
RelB is also important in muscle, where it regulates growth, differentiation, and possibly apoptosis. In skeletal muscle, IKKα is required for IGF-II-mediated myoblast differentiation, and NIK overexpression is sufficient for increased myogenin expression and myoblast fusion [69]. Additionally, regenerating muscle fibers in mdx mice show increased RelB nuclear localization and decreased RelA nuclear localization [70]. RelA has the opposite effect on myogenesis. Whereas RelA−/− mice are embryonically lethal, RelA−/− TNF-α−/− double-knockout mice are viable and show significantly more myotubes and increased expression of myofibrillar proteins when compared with TNF-α−/− single knockouts [71]. In cardiomyocytes, hyperosmotic stress can cause RelA and RelB but neither p50 nor p52 to translocate to the nucleus [72]. This translocation increases proapoptotic cleavage of caspase-3 and -9; however, it remains unclear whether this is a result of RelA homodimers or RelA-RelB heterodimers. As discussed subsequently, RelB also promotes mitochondrial biogenesis in myocytes [73].
Finally, RelB plays a role in regulation of antiapoptotic genes in prostate and mammary tissue, as evidenced by the presence of a noncanonical-activating mutation in these cancers and reviewed in detail elsewhere [74]. RelB has been found to be activated in prostate cancers, where it up-regulates manganese-SOD by directly binding the promoter [75]. This up-regulation increases the cancer's resistance to ionizing radiation, a resistance that can be reversed through inhibition of RelB. In mouse mammary tumors, RelB bound the promoters of cyclin D1 and c-myc, up-regulating their expression and promoting tumor growth [76]. In ERα-negative mammary tumors, there is increased RelB-mediated expression of the prosurvival gene Bcl-2 [77].
EMERGING CONCEPTS
RelB functions as a modifier of chromatin structure to support phenotypic transitions during the acute inflammatory response and explains a NF-κB paradox
NF-κB-mediated inflammation temporally progresses in overlapping but distinct phenotypic phases. Robust activation of the canonical NF-κB pathway releases RelA and p50 to translocate rapidly to cognate DNA and up-regulate expression of many proinflammatory genes [78]. The products of these genes, such as IL-1β and TNF-α, can generate uncontrolled inflammation if left unchecked [79]. Within several hours, however, transcription of a set of NF-κB-responsive genes is repressed by cytosolic and nuclear-located, negative-feedback processes; other sets of genes are up-regulated reciprocally [80]. These signal- and gene-set-specific, multiple adaptive processes support the classically described [81] phenotype of endotoxin tolerance if exposed to a second TLR4 stimulus [82, 83]. In fact, this sustained but reversible tolerance is so pronounced that many animals, given a tolerizing dose of endotoxin, can survive a second, otherwise lethal dose [84]. During human and animal sepsis, this property is displayed by many cells that regulate immune responses, including innate-immune phagocytes (monocyte, macrophages, neutrophils, and NK cells) [85–87] and adaptive-immune T cells, B cells, and DCs [88, 89]. Thus, during acute-immune and inflammatory responses that are excessive and uncontrolled properly, a global shift from active to repressed immunity occurs, where RelA and RelB play a critical role in the switch from host-offensive and -defensive responses to acute infection.
As discussed previously, the initial, canonical signaling pathway from TLR receptors activates RelA rapidly, whereas RelB activation occurs at a slower rate [19, 90]. With sufficient nuclear accumulation of RelB, RelA is replaced and transcription blunted. The early cellular studies of RelB function in fibroblasts with RelB deletion showed increased RelA, p50, and IκBα expression and that transfecting RelB cDNA into these fibroblasts reversed this process [91]. Similarly, RelB-overexpressing cells limited activation of RelA-dependent genes in fibroblasts and lymphoid cells [92]. The repression of RelA activation and nuclear mRNA transactivation function occurs despite an open TLR4 pathway to IκB degradation [93], RelA release, and nuclear translocation [90]. These observations generated a confusing NF-κB paradox of activation of the canonical NF-κB pathway with nuclear translocation without binding of RelA to cognate DNA of immediate response genes [3, 94]. Chan et al. [95] resolved this paradox by showing that cognate-binding sites became inaccessible to RelA because of epigenetic silencing of proinflammatory genes such as IL-1β. Later, Yoza et al. [4] showed in cultures of human promonocytes and in human sepsis that RelB expression was required to limit expression of proinflammatory genes during the adaptive endotoxintolerant phase that follows early TLR4 responses and occurs during human sepsis. Importantly, these studies showed that the facultative heterochromatin that depends on RelB induction is reversible [96]. As discussed subsequently, bioenergy shifts and SIRT1 activation are proximal to RelB, and both proteins are required to initiate chromatin structural changes that silence acute proinflammatory genes.
What is the precise molecular mechanism by which RelB limits acute inflammation and represses innate immunity? After its delayed induction following TLR4 responses, RelB represses RelA by blocking its transfer to DNA or by replacing RelA bound to DNA and then modifying promoter chromatin structure. First, RelB forms heterodimers with RelA at its prebound nuclear site to limit its availability to exposed and cognate DNA [97]. These heterodimers occur in endotoxin-tolerant cells, where tolerance is reversed by knocking down RelB [4]. Secondly, RelB displaces RelA bound to p50; such a dimer shift was observed first on the IL-12p40 promoter in DCs, wherein an increase in RelB binding after LPS correlated with a decrease in RelA and RNA PolII binding to the promoter [98]. Similarly, RelB replaces RelA on the TNF-α and IL-1β promoters (and likely many other immediate-response genes) of monocytes and neutrophils to generate a repressive state by epigenetically promoting a switch from active euchromatin to facultative heterochromatin [99]; this process occurs during the immunosuppressive phase of human sepsis. To generate silent facultative heterochromatin, RelB supports accumulation of critical histone and DNA modifiers, resulting in nucleosome positional changes [100, 101]. During silencing, H3 lysine 9 is dimethylated, concomitant with reduced H3 serine 10 phosphorylation [95]. To modify chromatin structure further, RelB directly binds to the H3 lysine methyltransferase G9a [100], allowing histone linker protein H1 and heterochromatin-binding protein-1 to accumulate and support CpG DNA methylation by DNA (cytosine-5-)-methyltransferase 3a/b [100]. Importantly, forcing RelB expression in nontolerant cells with low RelB expression recapitulates formation of silent heterochromatin, but this process uses unknown, post-translational events, as the phenotype requires stimulation of TLR4 [101]. These RelB-dependent chromatin modifications, perhaps combined with the sequestration of RelA and its displacement from NF-κB-binding sites, effectively halt further inflammatory signaling and suppress the innate-immune response until RelB promoter binding diminishes.
In addition to its direct recruitment of a repressive histone methyltransferase, RelB can associate with an ATP-dependent SWI/SNF nucleosome-remodeling complex [102]; RelB-dependent changes in SWI/SNF exist in endotoxin-tolerant monocytes with remodeled nucleosomes, and nucleosome repositioning is reversed by knockdown of RelB [101]. The SWI/SNF complex is a classic chromatin-remodeling enzyme, capable of activating or repressing genes, and affects development and differentiation in lymphocytes and stem cells [103]; interestingly, RelB also affects these two processes. Subunits of the SWI/SNF complex are recruited to proinflammatory gene promoters following MyD88 signaling, indicating that the complex is needed for optimal TLR4 immune responses [104]. Recently, it was shown, using Requiem protein, that RelB associates with the SWI/SNF complex in multiple human cell lines to induce production of CXCL13 adherence protein in response to noncanonical signaling in human tumor cell lines [102].
Taken together, an emerging paradigm emphasizes a distinct role for RelB as a direct contributor to dynamic changes in chromatin structure during inflammatory and innate-immune responses. The role of RelB in generating repressor cells of adaptive immunity during acute-immune responses has not been examined to our knowledge. Importantly, these data indicate the dual function of RelB as an epigenetic chromatin modifier.
RelB lies at the interface between SIRT1-dependent bioenergy sensing and control of metabolism and mitochondrial function
Another developing theme, indicating distinct attributes of RelB, is its connection to metabolism, in metabolic responsiveness and regulation; this unexpected pathway requires partnership with the NAD+-sensing sirtuin family member, SIRT1 (one of seven SIRTS in animals [105]). After TLR4 signaling, the RelB-mediated effects on gene regulation support a metabolic shift toward Warburg-type glycolysis, which diminishes glucose as a source for mitochondrial respiration [106]. This shift occurs concomitant with an increase in the NAD+/NADH ratio [107], providing NAD+ to activate the deacetylase function of SIRT1.
Nuclear-located SIRT1 initiates a two-step shift to modify inflammation and metabolism. First, SIRT1 binds to nuclear RelA and deactivates it by deacetylating lysine 310. Second, SIRT1 induces RelB transcription and promotes RelB binding to the TNF-α and IL-1β proximal promoters [5]. As discussed previously, SIRT1 is required to support chromatin modifications that require RelB. It is not known whether SIRT1 deacetylates RelB directly but it does deacetylate lysine 16 of histone H4 [5]. Thus, RelB expression and activity are supported by SIRT1 after it senses changes in cell nutrition and bioenergy. Coincident with a shift from the proinflammatory state to adaptation/endotoxin tolerance and an immunosuppressive anti-inflammatory phenotype is a switch to increased fatty acid flux and transport into mitochondria as a fuel source for an oxygen metabolism alternative to glucose (unpublished data). Taken together, data indicate that RelB is a dual chromatin regulator, repressing euchromatin by generating heterochromatin and activating the euchromatin of specific sets of nonrepressed genes, which combine to change inflammation and metabolic phenotypes.
Whether RelB is regulating metabolism directly is still being investigated, but there are some early signs that it may have that capacity. In mouse myocytes, deleting RelB decreases expression of many genes that enhance mitochondrial biogenesis, whereas activating RelB and IKKα by the noncanonical pathway increased mitochondrial mass [73]. Interestingly, this RelB-dependent path is mediated by RelB regulation of PGC-1β, which similar to PGC-1α, is a master switch, supporting mitochondrial biogenesis [108]. Furthermore, RelB can localize to mitochondria, as determined by histochemical analysis [109]. Whereas it remains unclear what, if any, functional ramifications that RelB mitochondrial localization has, it is known that RelA, IKKβ, and IκBα localize to mitochondrial and decrease mitochondrial respiration [110, 111]. We have observed that RelB supports expression of SIRT3, which localizes to mitochondria and enhances many mitochondrial functions (unpublished data). SIRT3 deacetylates and activates mitochondrial proteins, including the NADH dehydrogenase (ubiquinone)-1α-subcomplex subunit 9 subunit of complex I, succinate dehydrogenase, glutamate dehydrogenase, isocitrate dehydrogenase, AMPK, long-chain acyl-CoA dehydrogenase, and other regulators of oxidative metabolism [112]. The net result of SIRT3 activity is a nearly 100% increase of ATP levels compared with SIRT3−/− mice; thus, any regulation of SIRT3 by RelB has major implications for oxidative metabolism. Taken together, the data support that the SIRT1 and RelB axis branches to reprogram chromatin that directs the inflammatory process and chromatin that directs intermediary metabolism and mitochondrial biogenesis and bioenergetics. Both branches likely require the dual effects of RelB as an activator of euchromatin and an inducer of silent heterochromatin. Chromatin immunoprecipitation-seqencing studies of RelB DNA binding across the genome are needed to better understand this integrated network of gene-specific chromatin topology.
RelB binds to the AhR that supports xenobiotic responses
An emerging line of research indicates that RelB binds to many proteins not necessarily aligned with the NF-κB pathway. One of these is AhR, a transcription factor involved in the xenobiotic response responsible for detoxifying polychlorinated biphenyls and polycyclic aromatic hydrocarbons via increased expression of the cytochrome P450 family [113]. RelB–AhR interactions and downstream effects promote detoxification of tobacco smoke. The smoke induces the AhR pathway, which induces RelB expression and acts with RelB to limit toxicity in fibroblasts and lung [67, 114]. This increase in RelB causes a decrease in COX-2 and PG-mediated inflammation. AhR can promote the generation of proinflammatory Th17 cells or anti-inflammatory regulatory T cells, depending on the stimulus [115]; these researchers also found that RelB and AhR are capable of forming a dimer [7] that can recognize NF-κB sites normally bound by RelB-p52 dimers, as well as newly characterized RelB/AhR-responsive elements, such as one on the IL-8 promoter. Thus, it appears that RelB links to cellular defenses for toxins as well as viable microbes.
RelB regulates circadian rhythm
A surprising, new role for RelB is its direct-participation, circadian-cycle pathway [8]. The CLOCK/BMAL1 complex binds to E-box elements to alter expression of clock-controlled genes that change approximately every 12 h. These clock regulators influence several metabolic pathways, including mitochondrial biogenesis factor PGC-1α [116]. PER and CRY are clock-controlled genes that once expressed, feed back to inhibit CLOCK/BMAL1 and act as the other side of the cycle. The circadian rhythm is controlled by the levels of NAD+/NADH and the expression of rate-limiting nicotinamide phosphoribosyltransferase, which generates NAD+ [117]. This supports changes in SIRT1 deacetylase activity, which supports the rhythmic circadian shifts. It was not surprising then that RelB–a direct partner of SIRT1 in inflammation and metabolism—modifies the circadian rhythm by directly binding to BMAL1 [8]. Strikingly, BMAL1 is a member of the aryl hydrocarbon nuclear transporter of AhR family [118]. So, the biologic clock, xenobiotic and immune pathways, and metabolism may converge with the help of RelB, the physiologic implications of which are substantial.
Proinflammatory signals, such as TNF-α, disrupt the circadian cycle by blocking CLOCK/BMAL1-mediated transcription of E-box regulatory elements [119]. These signals can decrease expression of PER, BMAL1, and D-site of albumin promoter-binding protein [120]. When the CLOCK/BMAL1-negative regulator CRY is knocked out, mice show an increase inflammatory signaling and RelA activation [121]. Similarly, when a dominant-negative form of CLOCK is expressed, inflammatory cytokines IL-6 and IL-1β are repressed [8]. Administration of endotoxin to humans is associated with a block in circadian rhythm in peripheral blood leukocytes [122] and during sepsis [123]. As RelB represses CLOCK/BMAL1-mediated transcription, independent of CRY [8], it appears that RelB has yet another distinct feature when it is expressed during immune and inflammatory responses. Figure 3 provides a schematic of emerging features of RelB in leukocyte biology.
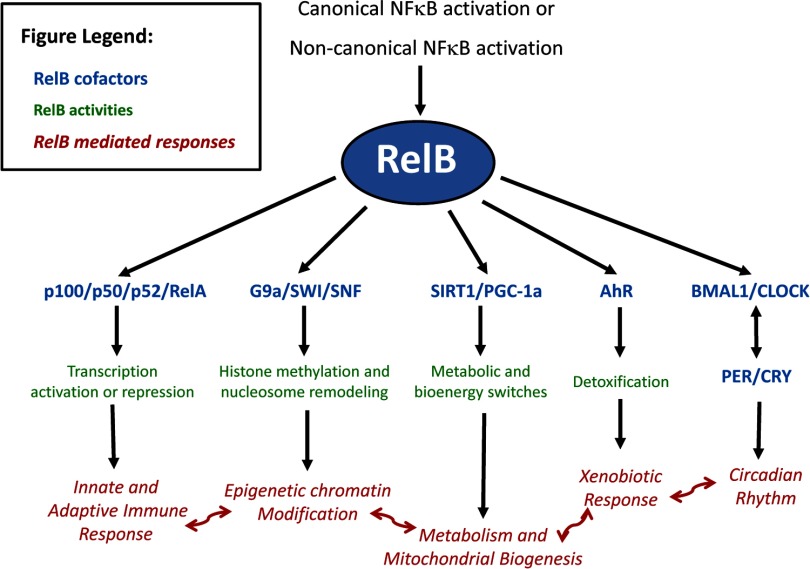
Figure 3. Overview of emerging fields of RelB biology.
RelB interacts with factors or complexes in blue. The green processes are those in which RelB is involved, whereas red represents the ultimate responses that RelB regulates.
PERSPECTIVE
Clearly, the RelB gene distinctly impacts many phenotypic features of immune and inflammatory responses. These crucial functions are cell type-specific and act spatially and temporally, as the protein modifies chromatin structure directly or indirectly and influences protein–protein interactions. A better understanding of RelB could have broad implications for treating immune and inflammatory diseases, as well as metabolic disorders. We suggest that the following areas for research and the attendant unanswered questions will yield important, new insights into RelB function and the integration of the immune response with other biological processes.
1. RelB expression
Aside from RelA, it remains unknown what factors may control RelB expression. What enhancer-promoter regions, transcription factors, and chromatin modifiers control basal and inducible expression of RelB? How does this influence immune cell-type specificity for RelB basal or induced expression? What noncoding RNAs and miRNAs regulate RelB expression, and how does RelB regulate these functional RNAs?
2. RelB effects across the genome
RelB may directly or indirectly bind to thousands of DNA sites, as well as other transcription factors and histone modifiers. How does RelB combine with SIRT1 to temporally regulate sets of genes across the genome? Is this network driven by bioenergy shifts?
3. RelB leucine zipper-mediated protein–protein interactions
Many proteins involved in inflammation, metabolism, and cellular bioenergetics contain leucine zippers. What is the role of the RelB leucine zipper in extending RelB spatial and temporal properties during inflammation and immune responses?
4. Xenobiotic responses and RelB
AhR not only up-regulates RelB in response to toxins, it also interacts directly with it to promote or inhibit the inflammatory and immune response. What is the role of RelB in regulating xenobiotic paths of detoxification, and how do these pathways integrate with immune defenses against microbes?
5. Integration of immune responses with metabolism and cellular bioenergetics
Cell sensing by sirtuins and their effects on bioenergy integrate with immunity, at least in part, via RelB. How do these pathways affect acute and chronic innate and adaptive-immune responses at levels of inflammation, intermediary metabolism, and mitochondrial biogenesis and bioenergetics? What nuclear chromatin and non-nuclear protein–protein interactions are involved?
6. Circadian rhythm and immunity
RelB modifies circadian rhythm directly by binding BMAL/CLOCK, a process dependent on NAD+ availability and activation of SIRT1. How do these changes in circadian rhythm alter the immune responses and their links to metabolism and bioenergetics? What are the physiologic implications on RelB arresting the circadian rhythm?
7. The polarity of the SIRT1:RelB axis
SIRT1's control of RelB can repress immune and inflammatory responses when increased and augment them when reduced. Can this new paradigm be used to improve therapies for acute or chronic inflammatory and immune diseases?
CONCLUDING REMARKS
We have presented our perspective on RelB as an outlier within the NF-κB family. We described that it applies its unique attributes to play diverse roles in leukocyte biology. As a modifier of chromatin, RelB can mediate a transient response, such as acute inflammation, or participate in cell differentiation, such as maturation of DCs. Beyond this, it forms an unusual partnership with the bioenergy regulator, SIRT1, to coordinate a shift in the clinical phases of acute inflammation from offensive attack during infection to defensive host protection during resolution. It can participate in the response to xenobiotic toxins as well. RelB supports the Randle cycle of fatty-acid metabolism and mitochondrial oxidation, and it modifies the circadian rhythm—a known partner with intermediary metabolism and with inflammation—directly. This remarkably successful protein deserves thoughtful study; understanding its outlier character further may reveal novel approaches to treat acute and chronic inflammatory and immune disorders.
ACKNOWLEDGMENTS
This lab is funded by U.S. National Institutes of Health grants RO1 AI065791 and RO1 AI079144.
Footnotes
- −/−
- deficient
- β-TrCP
- β-transducin repeat-containing protein
- AhR
- aryl hydrocarbon receptor
- BAFF
- B cell-activating factor of the TNF ligand superfamily, member 13b
- BMAL
- aryl hydrocarbon receptor nuclear translocator-like
- CD40L
- CD40 ligand
- cIAP
- cellular inhibitor of apoptosis
- CLOCK
- clock circadian regulator
- CRY
- cryptochrome
- I-rel
- inhibitor-v-rel reticuloendotheliosis viral oncogene homolog
- miR
- microRNA
- MZB
- marginal zone B cell
- NIK
- NF-κB-inducing kinase
- PER
- period circadian clock 1
- PGC-1α
- peroxisome proliferator-activated receptor γ coactivator-1 α
- Rel
- v-rel reticuloendotheliosis viral oncogene homolog
- RHD
- v-rel reticuloendotheliosis viral oncogene homolog homology domain
- SIRT
- sirtuin NAD+-dependent deacetylase
- SWI/SNF
- switch/sucrose nonfermentable
- TWEAK
- TNF-like weak inducer of apoptosis, member 12
- UEA-1
- ulex europeaus agglutinin-1
AUTHORSHIP
P.M. researched and wrote this review and generated Figs. 1 and 2 and Table 1. C.M. edited extensively and helped to outline this review. B.Y. edited this review and generated Fig. 3.
DISCLOSURES
The authors declare no conflict of interest.
REFERENCES
- 1. Hayden M. S. (2012) A less-canonical, canonical NF-κB pathway in DCs. Nat. Immunol. 13, 1139–1141 [DOI] [PubMed] [Google Scholar]
- 2. Vallabhapurapu S., Karin M. (2009) Regulation and function of NF-κB transcription factors in the immune system. Annu. Rev. Immunol. 27, 693–733 [DOI] [PubMed] [Google Scholar]
- 3. McCall C. E., Yoza B. K. (2007) Gene silencing in severe systemic inflammation. Am. J. Respir. Crit. Care Med. 175, 763–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yoza B. K., Hu J. Y., Cousart S. L., Forrest L. M., McCall C. E. (2006) Induction of RelB participates in endotoxin tolerance. J. Immunol. 177, 4080–4085 [DOI] [PubMed] [Google Scholar]
- 5. Liu T. F., Yoza B. K., El Gazzar M., Vachharajani V. T., McCall C. E. (2011) NAD+-dependent SIRT1 deacetylase participates in epigenetic reprogramming during endotoxin tolerance. J. Biol. Chem. 286, 9856–9864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Moorthy A. K., Huang D. B., Wang V. Y., Vu D., Ghosh G. (2007) X-Ray structure of a NF-κB p50/RelB/DNA complex reveals assembly of multiple dimers on tandem κB sites. J. Mol. Biol. 373, 723–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vogel C. F., Sciullo E., Li W., Wong P., Lazennec G., Matsumura F. (2007) RelB, a new partner of aryl hydrocarbon receptor-mediated transcription. Mol. Endocrinol. 21, 2941–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bellet M. M., Zocchi L., Sassone-Corsi P. (2012) The RelB subunit of NFκB acts as a negative regulator of circadian gene expression. Cell Cycle 11, 3304–3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ryseck R. P., Bull P., Takamiya M., Bours V., Siebenlist U., Dobrzanski P., Bravo R. (1992) RelB, a new Rel family transcription activator that can interact with p50-NF-κ B. Mol. Cell. Biol. 12, 674–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ruben S. M., Klement J. F., Coleman T. A., Maher M., Chen C. H., Rosen C. A. (1992) I-Rel: a novel rel-related protein that inhibits NF-κ B transcriptional activity. Genes Dev. 6, 745–760 [DOI] [PubMed] [Google Scholar]
- 11. Bours V., Azarenko V., Dejardin E., Siebenlist U. (1994) Human RelB (I-Rel) functions as a κ B site-dependent transactivating member of the family of Rel-related proteins. Oncogene 9, 1699–1702 [PubMed] [Google Scholar]
- 12. Baldwin A. S., Jr., (1996) The NF-κ B and IκB proteins: new discoveries and insights. Annu. Rev. Immunol. 14, 649–683 [DOI] [PubMed] [Google Scholar]
- 13. Alber T. (1992) Structure of the leucine zipper. Curr. Opin. Genet. Dev. 2, 205–210 [DOI] [PubMed] [Google Scholar]
- 14. Dobrzanski P., Ryseck R. P., Bravo R. (1993) Both N- and C-terminal domains of RelB are required for full transactivation: role of the N-terminal leucine zipper-like motif. Mol. Cell. Biol. 13, 1572–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen F. (2002, Jan.) RELB (v-rel reticuloendotheliosis viral oncogene homolog B). Atlas Genet. Cytogenet. Oncol. Haematol. Retrieved from http://AtlasGeneticsOncology.org/Genes/RELBID324.html
- 16. ENCODE Project Consortium, Dunham I., Kundaje A., Aldred S. F., Collins P. J., Davis C. A., Doyle F., Epstein C. B., Frietze S., Harrow J., et al. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ammon C., Mondal K., Andreesen R., Krause S. W. (2000) Differential expression of the transcription factor NF-κB during human mononuclear phagocyte differentiation to macrophages and dendritic cells. Biochem. Biophys. Res. Commun. 268, 99–105 [DOI] [PubMed] [Google Scholar]
- 18. Carrasco D., Ryseck R. P., Bravo R. (1993) Expression of relB transcripts during lymphoid organ development: specific expression in dendritic antigen-presenting cells. Development 118, 1221–1231 [DOI] [PubMed] [Google Scholar]
- 19. Bren G. D., Solan N. J., Miyoshi H., Pennington K. N., Pobst L. J., Paya C. V. (2001) Transcription of the RelB gene is regulated by NF-κB. Oncogene 20, 7722–7733 [DOI] [PubMed] [Google Scholar]
- 20. Mineva N. D., Rothstein T. L., Meyers J. A., Lerner A., Sonenshein G. E. (2007) CD40 ligand-mediated activation of the de novo RelB NF-κB synthesis pathway in transformed B cells promotes rescue from apoptosis. J. Biol. Chem. 282, 17475–17485 [DOI] [PubMed] [Google Scholar]
- 21. Wang X., Sonenshein G. E. (2005) Induction of the RelB NF-κB subunit by the cytomegalovirus IE1 protein is mediated via Jun kinase and c-Jun/Fra-2 AP-1 complexes. J. Virol. 79, 95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dong X., Craig T., Xing N., Bachman L. A., Paya C. V., Weih F., McKean D. J., Kumar R., Griffin M. D. (2003) Direct transcriptional regulation of RelB by 1α,25-dihydroxyvitamin D3 and its analogs: physiologic and therapeutic implications for dendritic cell function. J. Biol. Chem. 278, 49378–49385 [DOI] [PubMed] [Google Scholar]
- 23. Li T., Morgan M. J., Choksi S., Zhang Y., Kim Y. S., Liu Z. G. (2010) MicroRNAs modulate the noncanonical transcription factor NF-κB pathway by regulating expression of the kinase IKKα during macrophage differentiation. Nat. Immunol. 11, 799–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Etzrodt M., Cortez-Retamozo V., Newton A., Zhao J., Ng A., Wildgruber M., Romero P., Wurdinger T., Xavier R., Geissmann F., Meylan E., Nahrendorf M., Swirski F. K., Baltimore D., Weissleder R., Pittet M. J. (2012) Regulation of monocyte functional heterogeneity by miR-146a and Relb. Cell Rep. 1, 317–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ley K., Miller Y. I., Hedrick C. C. (2011) Monocyte and macrophage dynamics during atherogenesis. Arterioscler. Thromb. Vasc. Biol. 31, 1506–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhao J. L., Rao D. S., Boldin M. P., Taganov K. D., O'Connell R. M., Baltimore D. (2011) NF-κB dysregulation in microRNA-146a-deficient mice drives the development of myeloid malignancies. Proc. Natl. Acad. Sci. USA 108, 9184–9189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. El Gazzar M., Church A., Liu T., McCall C. E. (2011) MicroRNA-146a regulates both transcription silencing and translation disruption of TNF-α during TLR4-induced gene reprogramming. J. Leukoc. Biol. 90, 509–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Marienfeld R., Berberich-Siebelt F., Berberich I., Denk A., Serfling E., Neumann M. (2001) Signal-specific and phosphorylation-dependent RelB degradation: a potential mechanism of NF-κB control. Oncogene 20, 8142–8147 [DOI] [PubMed] [Google Scholar]
- 29. Maier H. J., Marienfeld R., Wirth T., Baumann B. (2003) Critical role of RelB serine 368 for dimerization and p100 stabilization J. Biol. Chem. 278, 39242–39250 [DOI] [PubMed] [Google Scholar]
- 30. Fusco A. J., Savinova O. V., Talwar R., Kearns J. D., Hoffmann A., Ghosh G. (2008) Stabilization of RelB requires multidomain interactions with p100/p52. J. Biol. Chem. 283, 12324–12332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xiao G., Harhaj E. W., Sun S. C. (2001) NF-κB-inducing kinase regulates the processing of NF-κB2 p100. Mol. Cell 7, 401–409 [DOI] [PubMed] [Google Scholar]
- 32. Heusch M., Lin L., Geleziunas R., Greene W. C. (1999) The generation of NF-κB2 p52: mechanism and efficiency. Oncogene 18, 6201–6208 [DOI] [PubMed] [Google Scholar]
- 33. Solan N. J., Miyoshi H., Carmona E. M., Bren G. D., Paya C. V. (2002) RelB cellular regulation and transcriptional activity are regulated by p100. J. Biol. Chem. 277, 1405–1418 [DOI] [PubMed] [Google Scholar]
- 34. Basak S., Kim H., Kearns J. D., Tergaonkar V., O'Dea E., Werner S. L., Benedict C. A., Ware C. F., Ghosh G., Verma I. M., Hoffmann A. (2007) A fourth IκB protein within the NF-κB signaling module. Cell 128, 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shih V. F., Tsui R., Caldwell A., Hoffmann A. (2011) A single NFκB system for both canonical and non-canonical signaling. Cell Res. 21, 86–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wietek C., O'Neill L. A. (2007) Diversity and regulation in the NF-κB system. Trends Biochem. Sci. 32, 311–319 [DOI] [PubMed] [Google Scholar]
- 37. Beinke S., Ley S. C. (2004) Functions of NF-κB1 and NF-κB2 in immune cell biology. Biochem. J. 382, 393–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Neumann M., Klar S., Wilisch-Neumann A., Hollenbach E., Kavuri S., Leverkus M., Kandolf R., Brunner-Weinzierl M. C., Klingel K. (2011) Glycogen synthase kinase-3β is a crucial mediator of signal-induced RelB degradation. Oncogene 30, 2485–2492 [DOI] [PubMed] [Google Scholar]
- 39. Shih V. F., Davis-Turak J., Macal M., Huang J. Q., Ponomarenko J., Kearns J. D., Yu T., Fagerlund R., Asagiri M., Zuniga E. I., Hoffmann A. (2012) Control of RelB during dendritic cell activation integrates canonical and noncanonical NF-κB pathways. Nat. Immunol. 13, 1162–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Müller J. R., Siebenlist U. (2003) Lymphotoxin β receptor induces sequential activation of distinct NF-κ B factors via separate signaling pathways. J. Biol. Chem. 278, 12006–12012 [DOI] [PubMed] [Google Scholar]
- 41. Derudder E., Dejardin E., Pritchard L. L., Green D. R., Korner M., Baud V. (2003) RelB/p50 dimers are differentially regulated by tumor necrosis factor-α and lymphotoxin-β receptor activation: critical roles for p100. J. Biol. Chem. 278, 23278–23284 [DOI] [PubMed] [Google Scholar]
- 42. Liao G., Zhang M., Harhaj E. W., Sun S. C. (2004) Regulation of the NF-κB-inducing kinase by tumor necrosis factor receptor-associated factor 3-induced degradation. J. Biol. Chem. 279, 26243–26250 [DOI] [PubMed] [Google Scholar]
- 43. Sun S. C. (2011) Non-canonical NF-κB signaling pathway. Cell Res. 21, 71–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zarnegar B. J., Wang Y., Mahoney D. J., Dempsey P. W., Cheung H. H., He J., Shiba T., Yang X., Yeh W. C., Mak T. W., Korneluk R. G., Cheng G. (2008) Noncanonical NF-κB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat. Immunol. 9, 1371–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xiao G., Fong A., Sun S. C. (2004) Induction of p100 processing by NF-κB-inducing kinase involves docking IκB kinase α (IKKα) to p100 and IKKα-mediated phosphorylation. J. Biol. Chem. 279, 30099–30105 [DOI] [PubMed] [Google Scholar]
- 46. Liang C., Zhang M., Sun S. C. (2006) β-TrCP binding and processing of NF-κB2/p100 involve its phosphorylation at serines 866 and 870. Cell. Signal. 18, 1309–1317 [DOI] [PubMed] [Google Scholar]
- 47. He J. Q., Zarnegar B., Oganesyan G., Saha S. K., Yamazaki S., Doyle S. E., Dempsey P. W., Cheng G. (2006) Rescue of TRAF3-null mice by p100 NF-κ B deficiency. J. Exp. Med. 203, 2413–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yilmaz Z. B., Weih D. S., Sivakumar V., Weih F. (2003) RelB is required for Peyer's patch development: differential regulation of p52-RelB by lymphotoxin and TNF. EMBO J. 22, 121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Weih D. S., Yilmaz Z. B., Weih F. (2001) Essential role of RelB in germinal center and marginal zone formation and proper expression of homing chemokines. J. Immunol. 167, 1909–1919 [DOI] [PubMed] [Google Scholar]
- 50. Weih F., Carrasco D., Durham S. K., Barton D. S., Rizzo C. A., Ryseck R. P., Lira S. A., Bravo R. (1995) Multiorgan inflammation and hematopoietic abnormalities in mice with a targeted disruption of RelB, a member of the NF-κ B/Rel family. Cell 80, 331–340 [DOI] [PubMed] [Google Scholar]
- 51. Gerondakis S., Grumont R., Gugasyan R., Wong L., Isomura I., Ho W., Banerjee A. (2006) Unravelling the complexities of the NF-κB signalling pathway using mouse knockout and transgenic models. Oncogene 25, 6781–6799 [DOI] [PubMed] [Google Scholar]
- 52. Burkly L., Hession C., Ogata L., Reilly C., Marconi L. A., Olson D., Tizard R., Cate R., Lo D. (1995) Expression of relB is required for the development of thymic medulla and dendritic cells. Nature 373, 531–536 [DOI] [PubMed] [Google Scholar]
- 53. Weih F., Durham S. K., Barton D. S., Sha W. C., Baltimore D., Bravo R. (1997) p50-NF-κB complexes partially compensate for the absence of RelB: severely increased pathology in p50(−/−)relB(−/−) double-knockout mice. J. Exp. Med. 185, 1359–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Naspetti M., Aurrand-Lions M., DeKoning J., Malissen M., Galland F., Lo D., Naquet P. (1997) Thymocytes and RelB-dependent medullary epithelial cells provide growth-promoting and organization signals, respectively, to thymic medullary stromal cells. Eur. J. Immunol. 27, 1392–1397 [DOI] [PubMed] [Google Scholar]
- 55. Barton D., HogenEsch H., Weih F. (2000) Mice lacking the transcription factor RelB develop T cell-dependent skin lesions similar to human atopic dermatitis. Eur. J. Immunol. 30, 2323–2332 [DOI] [PubMed] [Google Scholar]
- 56. DeKoning J., DiMolfetto L., Reilly C., Wei Q., Havran W. L., Lo D. (1997) Thymic cortical epithelium is sufficient for the development of mature T cells in relB-deficient mice. J. Immunol. 158, 2558–2566 [PubMed] [Google Scholar]
- 57. Caamaño J., Alexander J., Craig L., Bravo R., Hunter C. A. (1999) The NF-κ B family member RelB is required for innate and adaptive immunity to Toxoplasma gondii. J. Immunol. 163, 4453–4461 [PubMed] [Google Scholar]
- 58. Ishimaru N., Kishimoto H., Hayashi Y., Sprent J. (2006) Regulation of naive T cell function by the NF-κB2 pathway. Nat. Immunol. 7, 763–772 [DOI] [PubMed] [Google Scholar]
- 59. Matsumoto M., Yamada T., Yoshinaga S. K., Boone T., Horan T., Fujita S., Li Y., Mitani T. (2002) Essential role of NF-κ B-inducing kinase in T cell activation through the TCR/CD3 pathway. J. Immunol. 169, 1151–1158 [DOI] [PubMed] [Google Scholar]
- 60. Attar R. M., Caamaño J., Carrasco D., Iotsova V., Ishikawa H., Ryseck R. P., Weih F., Bravo R. (1997) Genetic approaches to study Rel/NF-κ B/I κ B function in mice. Semin. Cancer Biol. 8, 93–101 [DOI] [PubMed] [Google Scholar]
- 61. Kosco-Vilbois M. H., Bonnefoy J. Y., Chvatchko Y. (1997) The physiology of murine germinal center reactions. Immunol. Rev. 156, 127–136 [DOI] [PubMed] [Google Scholar]
- 62. Gardam S., Sierro F., Basten A., Mackay F., Brink R. (2008) TRAF2 and TRAF3 signal adapters act cooperatively to control the maturation and survival signals delivered to B cells by the BAFF receptor. Immunity 28, 391–401 [DOI] [PubMed] [Google Scholar]
- 63. Thien M., Phan T. G., Gardam S., Amesbury M., Basten A., Mackay F., Brink R. (2004) Excess BAFF rescues self-reactive B cells from peripheral deletion and allows them to enter forbidden follicular and marginal zone niches. Immunity 20, 785–798 [DOI] [PubMed] [Google Scholar]
- 64. Tan J. K., Ni K., Le F., O'Neill H. C. (2007) Hematopoiesis of immature myeloid dendritic cells in stroma-dependent spleen long-term cultures occurs independently of NF-κB/RelB function. Exp. Hematol. 35, 1580–1593 [DOI] [PubMed] [Google Scholar]
- 65. Zhao C., Xiu Y., Ashton J., Xing L., Morita Y., Jordan C. T., Boyce B. F. (2012) Noncanonical NF-κB signaling regulates hematopoietic stem cell self-renewal and microenvironment interactions. Stem Cells 30, 709–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tully J. E., Nolin J. D., Guala A. S., Hoffman S. M., Roberson E. C., Lahue K. G., van der Velden J., Anathy V., Blackwell T. S., Janssen-Heininger Y. M. (2012) Cooperation between classical and alternative NF-κB pathways regulates proinflammatory responses in epithelial cells. Am. J. Respir. Cell. Mol. Biol. 47, 497–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Baglole C. J., Maggirwar S. B., Gasiewicz T. A., Thatcher T. H., Phipps R. P., Sime P. J. (2008) The aryl hydrocarbon receptor attenuates tobacco smoke-induced cyclooxygenase-2 and prostaglandin production in lung fibroblasts through regulation of the NF-κB family member RelB. J. Biol. Chem. 283, 28944–28957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Benedetti G., Fokkelman M., Yan K., Fredriksson L., Herpers B., Meerman J., van de Water B., de Graauw M. (2013) The nuclear factor κB family member RelB facilitates apoptosis of renal epithelial cells caused by cisplatin/tumor necrosis factor α synergy by suppressing an epithelial to mesenchymal transition-like phenotypic switch. Mol. Pharmacol. 84, 128–138 [DOI] [PubMed] [Google Scholar]
- 69. Canicio J., Ruiz-Lozano P., Carrasco M., Palacin M., Chien K., Zorzano A., Kaliman P. (2001) Nuclear factor κ B-inducing kinase and Iκ B kinase-α signal skeletal muscle cell differentiation. J. Biol. Chem. 276, 20228–20233 [DOI] [PubMed] [Google Scholar]
- 70. Charan R. A., Hanson R., Clemens P. R. (2012) Deubiquitinating enzyme A20 negatively regulates NF-κB signaling in skeletal muscle in mdx mice. FASEB J. 26, 587–595 [DOI] [PubMed] [Google Scholar]
- 71. Bakkar N., Wang J., Ladner K. J., Wang H., Dahlman J. M., Carathers M., Acharyya S., Rudnicki M. A., Hollenbach A. D., Guttridge D. C. (2008) IKK/NF-κB regulates skeletal myogenesis via a signaling switch to inhibit differentiation and promote mitochondrial biogenesis. J. Cell Biol. 180, 787–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Eisner V., Quiroga C., Criollo A., Eltit J. M., Chiong M., Parra V., Hidalgo K., Toro B., Díaz-Araya G., Lavandero S. (2006) Hyperosmotic stress activates p65/RelB NFκB in cultured cardiomyocytes with dichotomic actions on caspase activation and cell death. FEBS Lett. 580, 3469–3476 [DOI] [PubMed] [Google Scholar]
- 73. Bakkar N., Ladner K., Canan B. D., Liyanarachchi S., Bal N. C., Pant M., Periasamy M., Li Q., Janssen P. M., Guttridge D. C. (2012) IKKα and alternative NF-κB regulate PGC-1β to promote oxidative muscle metabolism. J. Cell Biol. 196, 497–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Baud V., Jacque E. (2008) The alternative NF-κB activation pathway and cancer: friend or foe? Med. Sci. (Paris) 24, 1083–1088 [DOI] [PubMed] [Google Scholar]
- 75. Josson S., Xu Y., Fang F., Dhar S. K., St Clair D. K., St Clair W. H. (2006) RelB regulates manganese superoxide dismutase gene and resistance to ionizing radiation of prostate cancer cells. Oncogene 25, 1554–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Demicco E. G., Kavanagh K. T., Romieu-Mourez R., Wang X., Shin S. R., Landesman-Bollag E., Seldin D. C., Sonenshein G. E. (2005) RelB/p52 NF-κB complexes rescue an early delay in mammary gland development in transgenic mice with targeted superrepressor IκB-α expression and promote carcinogenesis of the mammary gland. Mol. Cell. Biol. 25, 10136–10147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wang X., Belguise K., Kersual N., Kirsch K. H., Mineva N. D., Galtier F., Chalbos D., Sonenshein G. E. (2007) Oestrogen signalling inhibits invasive phenotype by repressing RelB and its target BCL2. Nat. Cell Biol. 9, 470–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bohrer H., Qiu F., Zimmermann T., Zhang Y., Jllmer T., Mannel D., Bottiger B. W., Stern D. M., Waldherr R., Saeger H. D., Ziegler R., Bierhaus A., Martin E., Nawroth P. P. (1997) Role of NFκB in the mortality of sepsis. J. Clin. Invest. 100, 972–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wang H., Ma S. (2008) The cytokine storm and factors determining the sequence and severity of organ dysfunction in multiple organ dysfunction syndrome. Am. J. Emerg. Med. 26, 711–715 [DOI] [PubMed] [Google Scholar]
- 80. McCall C. E., Yoza B., Liu T., El Gazzar M. (2010) Gene-specific epigenetic regulation in serious infections with systemic inflammation. J. Innate Immun. 2, 395–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Beeson P. (1947) Tolerance to bacterial pyrogens. J. Exp. Med. 86, 29–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Granowitz E. V., Porat R., Mier J. W., Orencole S. F., Kaplanski G., Lynch E. A., Ye K., Vannier E., Wolff S. M., Dinarello C. A. (1993) Intravenous endotoxin suppresses the cytokine response of peripheral blood mononuclear cells of healthy humans. J. Immunol. 151, 1637–1645 [PubMed] [Google Scholar]
- 83. Biswas S. K., Lopez-Collazo E. (2009) Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends Immunol. 30, 475–487 [DOI] [PubMed] [Google Scholar]
- 84. Sanchez-Cantu L., Rode H. N., Christou N. V. (1989) Endotoxin tolerance is associated with reduced secretion of tumor necrosis factor. Arch. Surg. 124, 1432–1436 [DOI] [PubMed] [Google Scholar]
- 85. Cavaillon J. M., Adib-Conquy M. (2005) Monocytes/macrophages and sepsis. Crit. Care Med. 33, S506–S509 [DOI] [PubMed] [Google Scholar]
- 86. McCall C. E., Grosso-Wilmoth L. M., LaRue K., Guzman R. N., Cousart S. L. (1993) Tolerance to endotoxin-induced expression of the interleukin-1 β gene in blood neutrophils of humans with the sepsis syndrome. J. Clin. Invest. 91, 853–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Souza-Fonseca-Guimaraes F., Adib-Conquy M., Cavaillon J. M. (2012) Natural killer (NK) cells in antibacterial innate immunity: angels or devils? Mol. Med. 18, 270–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Matsushita H., Ohta S., Shiraishi H., Suzuki S., Arima K., Toda S., Tanaka H., Nagai H., Kimoto M., Inokuchi A., Izuhara K. (2010) Endotoxin tolerance attenuates airway allergic inflammation in model mice by suppression of the T-cell stimulatory effect of dendritic cells. Int. Immunol. 22, 739–747 [DOI] [PubMed] [Google Scholar]
- 90. Yoza B. K., Hu J. Y., Cousart S. L., McCall C. E. (2000) Endotoxin inducible transcription is repressed in endotoxin tolerant cells. Shock 13, 236–243 [DOI] [PubMed] [Google Scholar]
- 91. Xia Y., Pauza M. E., Feng L., Lo D. (1997) RelB regulation of chemokine expression modulates local inflammation. Am. J. Pathol. 151, 375–387 [PMC free article] [PubMed] [Google Scholar]
- 92. Marienfeld R., May M. J., Berberich I., Serfling E., Ghosh S., Neumann M. (2003) RelB forms transcriptionally inactive complexes with RelA/p65. J. Biol. Chem. 278, 19852–19860 [DOI] [PubMed] [Google Scholar]
- 93. LaRue K. E., McCall C. E. (1994) A labile transcriptional repressor modulates endotoxin tolerance. J. Exp. Med. 180, 2269–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Madge L. A., May M. J. (2011) The NFκB paradox: RelB induces and inhibits gene expression. Cell Cycle 10, 6–7 [DOI] [PubMed] [Google Scholar]
- 95. Chan C., Li L., McCall C. E., Yoza B. K. (2005) Endotoxin tolerance disrupts chromatin remodeling and NF-κB transactivation at the IL-1promoter. J. Immunol. 175, 461–468 [DOI] [PubMed] [Google Scholar]
- 96. Yoza B. K., McCall C. E. (2011) Facultative heterochromatin formation at the IL-1 β promoter in LPS tolerance and sepsis. Cytokine 53, 145–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Jacque E., Tchenio T., Piton G., Romeo P. H., Baud V. (2005) RelA repression of RelB activity induces selective gene activation downstream of TNF receptors. Proc. Natl. Acad. Sci. USA 102, 14635–14640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Saccani S., Pantano S., Natoli G. (2003) Modulation of NF-κB activity by exchange of dimers. Mol. Cell. 11, 1563–1574 [DOI] [PubMed] [Google Scholar]
- 99. McCall C. E., El Gazzar M., Liu T., Vachharajani V., Yoza B. (2011) Epigenetics, bioenergetics, and microRNA coordinate gene-specific reprogramming during acute systemic inflammation. J. Leukoc. Biol. 90, 439–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Chen X., El Gazzar M., Yoza B. K., McCall C. E. (2009) The NF-κB factor RelB and histone H3 lysine methyltransferase G9a directly interact to generate epigenetic silencing in endotoxin tolerance. J. Biol. Chem. 284, 27857–27865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. El Gazzar M., Liu T., Yoza B. K., McCall C. E. (2010) Dynamic and selective nucleosome repositioning during endotoxin tolerance. J. Biol. Chem. 285, 1259–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Tando T., Ishizaka A., Watanabe H., Ito T., Iida S., Haraguchi T., Mizutani T., Izumi T., Isobe T., Akiyama T., Inoue J., Iba H. (2010) Requiem protein links RelB/p52 and the Brm-type SWI/SNF complex in a noncanonical NF-κB pathway. J. Biol. Chem. 285, 21951–21960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Wilson B. G., Roberts C. W. (2011) SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer 11, 481–492 [DOI] [PubMed] [Google Scholar]
- 104. Kayama H., Ramirez-Carrozzi V. R., Yamamoto M., Mizutani T., Kuwata H., Iba H., Matsumoto M., Honda K., Smale S. T., Takeda K. (2008) Class-specific regulation of pro-inflammatory genes by MyD88 pathways and IκBζ. J. Biol. Chem. 283, 12468–12477 [DOI] [PubMed] [Google Scholar]
- 105. Milne J. C., Denu J. M. (2008) The sirtuin family: therapeutic targets to treat diseases of aging. Curr. Opin. Chem. Biol. 12, 11–17 [DOI] [PubMed] [Google Scholar]
- 106. Liu T. F., Vachharajani V. T., Yoza B. K., McCall C. E. (2012) NAD+-dependent sirtuin 1 and 6 proteins coordinate a switch from glucose to fatty acid oxidation during the acute inflammatory response. J. Biol. Chem. 287, 25758–25769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Hart D. W., Gore D. C., Rinehart A. J., Asimakis G. K., Chinkes D. L. (2003) Sepsis-induced failure of hepatic energy metabolism. J. Surg. Res. 115, 139–147 [DOI] [PubMed] [Google Scholar]
- 108. Puigserver P., Wu Z., Park C. W., Graves R., Wright M., Spiegelman B. M. (1998) A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92, 829–839 [DOI] [PubMed] [Google Scholar]
- 109. Stasik I., Rapak A., Zioło E., Strzadała L. (2008) The mitochondrial localization of RelB and NFATx in immature T cells. Cell. Mol. Biol. Lett. 13, 493–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Cogswell P. C., Kashatus D. F., Keifer J. A., Guttridge D. C., Reuther J. Y., Bristow C., Roy S., Nicholson D. W., Baldwin A. S., Jr., (2003) NF-κ B and I κ B α are found in the mitochondria. Evidence for regulation of mitochondrial gene expression by NF-κ B. J. Biol. Chem. 278, 2963–2968 [DOI] [PubMed] [Google Scholar]
- 111. Johnson R. F., Witzel I. I., Perkins N. D. (2011) p53-Dependent regulation of mitochondrial energy production by the RelA subunit of NF-κB. Cancer Res. 71, 5588–5597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Giralt A., Villarroya F. (2012) SIRT3, a pivotal actor in mitochondrial functions: metabolism, cell death and aging. Biochem. J. 444, 1–10 [DOI] [PubMed] [Google Scholar]
- 113. Gonzalez F. J., Fernandez-Salguero P. (1998) The aryl hydrocarbon receptor: studies using the AHR-null mice. Drug Metab. Dispos. 26, 1194–1198 [PubMed] [Google Scholar]
- 114. McMillan D. H., Baglole C. J., Thatcher T. H., Maggirwar S., Sime P. J., Phipps R. P. (2011) Lung-targeted overexpression of the NF-κB member RelB inhibits cigarette smoke-induced inflammation. Am. J. Pathol. 179, 125–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Vogel C. F., Matsumura F. (2009) A new cross-talk between the aryl hydrocarbon receptor and RelB, a member of the NF-κB family. Biochem. Pharmacol. 77, 734–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Huang W., Ramsey K. M., Marcheva B., Bass J. (2011) Circadian rhythms, sleep, and metabolism. J. Clin. Invest. 121, 2133–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Imai S. (2009) Nicotinamide phosphoribosyltransferase (Nampt): a link between NAD biology, metabolism, and diseases. Curr. Pharm. Des. 15, 20–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Shimba S., Watabe Y. (2009) Crosstalk between the AHR signaling pathway and circadian rhythm. Biochem. Pharmacol. 77, 560–565 [DOI] [PubMed] [Google Scholar]
- 119. Cavadini G., Petrzilka S., Kohler P., Jud C., Tobler I., Birchler T., Fontana A. (2007) TNF-α suppresses the expression of clock genes by interfering with E-box-mediated transcription. Proc. Natl. Acad. Sci. USA 104, 12843–12848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Hashiramoto A., Yamane T., Tsumiyama K., Yoshida K., Komai K., Yamada H., Yamazaki F., Doi M., Okamura H., Shiozawa S. (2010) Mammalian clock gene cryptochrome regulates arthritis via proinflammatory cytokine TNF-α. J. Immunol. 184, 1560–1565 [DOI] [PubMed] [Google Scholar]
- 121. Narasimamurthy R., Hatori M., Nayak S. K., Liu F., Panda S., Verma I. M. (2012) Circadian clock protein cryptochrome regulates the expression of proinflammatory cytokines. Proc. Natl. Acad. Sci. USA 109, 12662–12667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Haimovich B., Calvano J., Haimovich A. D., Calvano S. E., Coyle S. M., Lowry S. F. (2010) In vivo endotoxin synchronizes and suppresses clock gene expression in human peripheral blood leukocytes. Crit. Care Med. 38, 751–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Verceles A. C., Silhan L., Terrin M., Netzer G., Shanholtz C., Scharf S. M. (2012) Circadian rhythm disruption in severe sepsis: the effect of ambient light on urinary 6-sulfatoxymelatonin secretion. Intensive Care Med. 38, 804–810 [DOI] [PubMed] [Google Scholar]