Abstract
Epigenetic mechanisms that maintain neurogenesis throughout adult life remain poorly understood1. Trithorax group (trxG) and Polycomb group (PcG) gene products are part of an evolutionarily conserved chromatin remodelling system that activate or silence gene expression, respectively2. Although PcG member Bmi1 has been shown to be required for postnatal neural stem cell self-renewal3,4, the role of trxG genes remains unknown. Here we show that the trxG member Mll1 (mixed-lineage leukaemia 1) is required for neurogenesis in the mouse postnatal brain. Mll1-deficient subventricular zone neural stem cells survive, proliferate and efficiently differentiate into glial lineages; however, neuronal differentiation is severely impaired. In Mll1-deficient cells, early proneural Mash1 (also known as Ascl1) and gliogenic Olig2 expression are preserved, but Dlx2, a key downstream regulator of subventricular zone neurogenesis, is not expressed. Over-expression of Dlx2 can rescue neurogenesis in Mll1-deficient cells. Chromatin immunoprecipitation demonstrates that Dlx2 is a direct target of MLL in subventricular zone cells. In differentiating wild-type subventricular zone cells, Mash1, Olig2 and Dlx2 loci have high levels of histone 3 trimethylated at lysine 4 (H3K4me3), consistent with their transcription. In contrast, in Mll1-deficient subventricular zone cells, chromatin at Dlx2 is bivalently marked by both H3K4me3 and histone 3 trimethylated at lysine 27 (H3K27me3), and the Dlx2 gene fails to properly activate. These data support a model in which Mll1 is required to resolve key silenced bivalent loci in postnatal neural precursors to the actively transcribed state for the induction of neurogenesis, but not for gliogenesis.
Neural stem cells (NSCs) and neurogenesis persist throughout life in the subventricular zone (SVZ) and dentate gyrus of the hippocampus. The Mll1 histone methyltransferase is expressed in adult SVZ cells5 as well as embryonic SVZ and ventricular zone (Supplementary Fig. 1). In development, Mll1 regulates the epigenetic maintenance of homeotic gene expression patterns2. Human MLL is a proto-oncogene, with chromosomal translocations resulting in MLL fusion proteins that produce human leukaemias with mixed lineage identity6. Mouse Mll1 is required for embryonic haematopoiesis, and most Mll1-null mice die between embryonic day (E)10.5 and E12 (ref. 7). We therefore used a conditional knockout ‘floxed’ allele of Mll1 (Mll1F/F)8.
To delete Mll1 from a subset of NSCs, we used the transgenic mouse hGFAP-Cre9, which exhibits excision of floxed alleles in precursors of the hippocampal dentate gyrus, cerebellar granular cells, and SVZ NSCs at E13.5 (ref. 10). hGFAP-Cre;Mll1F/F mice were born at the expected Mendelian ratios and were indistinguishable from their wild-type and hGFAP-Cre;Mll1F/+ littermates (hereafter referred to as controls). However, by postnatal day (P)15, hGFAP-Cre;Mll1F/F mice developed progressive growth retardation and ataxia, and the mice usually died between P25 and P30. We therefore initially analysed hGFAP-Cre;Mll1F/F mice and controls at P23–P25.
All brain regions that undergo considerable postnatal neurogenesis—including the cerebellar granule cell layer, hippocampal dentate gyrus (Supplementary Fig. 2) and the olfactory bulb (Fig. 1b)—showed a marked reduction in the size and the number of neurons in hGFAP-Cre;Mll1F/F mice. This suggested a common requirement for Mll1 in neurogenesis. To investigate the role of Mll1 in a NSC population, we focused on the SVZ-olfactory-bulb system. Throughout life, NSCs in the SVZ generate large numbers of neuroblasts that migrate to the olfactory bulb11–13 where they differentiate into interneurons (Fig. 1a, schematic). To evaluate the rate of SVZ neurogenesis, we injected animals with the cell-cycle marker 5-bromo-2-deoxyuridine (BrdU) 1 h before being culled and then co-immunostained sections for BrdU and the neuroblast marker doublecortin (DCX). hGFAP-Cre;Mll1F/F mice had 3–4-fold fewer BrdU+ DCX+ SVZ cells than the controls (Fig. 1c, d). Despite this decreased rate of BrdU incorporation, hGFAP-Cre;Mll1F/F mice had an expanded SVZ (Fig. 1c, right, yellow doubleheaded arrow) containing DCX+ cells; this accumulation developed after P7 (Supplementary Fig. 3) and could be explained if Mll1-deficient progenitor cells have impaired and/or abnormal migration, resulting in cell accumulation. Although Mll1-deficient progenitors retained expression of neuroblast markers such as DCX and Tuj1 (Supplementary Fig. 3), they also possessed ultrastructural characteristics of intermediate, transit-amplifying cells (Supplementary Fig. 5k, l). Indeed, neuroblast chain migration in hGFAP-Cre;Mll1F/F mice and from SVZ explants14 was severely disorganized, resulting in a 40% reduction in the migration distance in vitro (Supplementary Fig. 4). Thus, postnatal neuronal addition in the olfactory bulb was abrogated by a decreased rate of neurogenesis as well as by impaired neuroblast migration.
Figure 1. Mll1 is required for normal SVZ-olfactory bulb neurogenesis.
a, Schematics of SVZ-olfactory-bulb neurogenesis. Left, coronal section of the olfactory bulb (OB) indicating the region where newly born neuroblasts (red dots) initially arrive from the SVZ. GCL, granule cell layer. Middle, sagittal section showing paths of neuroblast migration from the SVZ to the olfactory bulb. Right, coronal section indicating the germinal SVZ (red dots); the blue box indicates regions shown in c. b, Haematoxylin and eosin (H&E)-stained coronal sections through the P25 olfactory bulb of control (left) and hGFAP-Cre;Mll1F/F (right) mice. The black box indicates the olfactory bulb core comprised of recently born neuroblasts. c, DCX (red) and BrdU (green) immunohistochemistry of the SVZ is shown. The size of the SVZ is indicated by a yellow doubleheaded arrow. LV, lateral ventricle; St, striatum. d, Quantification of BrdU+ DCX+ SVZ cells. hpf, high power field. Error bars, s.e.m.; three mice per group; *P = 0.025. Scale bars, 200 μm (b) and 20 μm (c).
In addition to producing neurons, SVZ NSCs generate astrocytes and oligodendrocytes15–17. Glial development in hGFAP-Cre;Mll1F/F mice was not impaired. In fact, expression of the astrocyte marker GFAP was increased in the SVZ of hGFAP-Cre;Mll1F/F mice (Fig. 2a) as well as in other brain regions (Supplementary Fig. 5e–h). The SVZ ependymal layer develops postnatally18, and these cells formed normally in hGFAP-Cre;Mll1F/F mice (Fig. 2b and Supplementary Fig. 5i, j). OLIG2 is expressed in developing and mature oligodendrocytes19,20, and OLIG2 expression in white matter was similar in hGFAP-Cre;Mll1F/F mice and controls (Fig. 2c, green). Furthermore, oligodendrocyte myelination of major axon tracts in hGFAP-Cre;Mll1F/F mice appeared normal as assessed by FluoroMyelin staining (Supplementary Fig. 5a–d) and myelin basic protein (MBP) immunohistochemistry (Fig. 2c, red). However, it was possible that local, non-SVZ glial progenitors had compensated for a defective SVZ stem cell population.
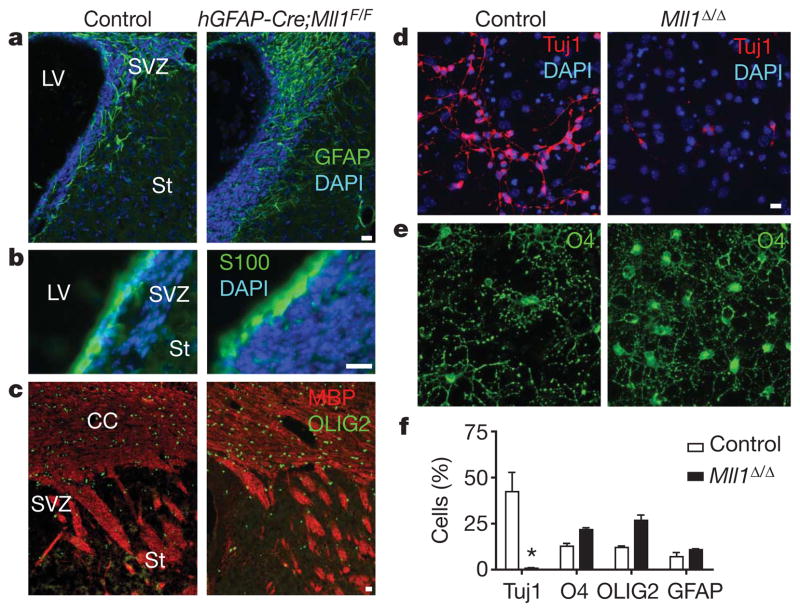
Figure 2. Mll1-deletion impairs postnatal SVZ-olfactory-bulb neurogenesis but not gliogenesis.
a, Immunohistochemistry for the astrocyte marker GFAP (green) in P25 coronal brain sections of control (left) and hGFAP-Cre;Mll1F/F (right) mice. Nuclei are counterstained with 4,6-diamidino-2-phenylindole (DAPI; blue). b, Immunohistochemistry for the S100+ (green) ependymal cells. c, Immunohistochemistry for markers of oligodendrocytes, OLIG2 (green) and MBP (red). CC, corpus callosum. d, SVZ cultures after 4 days of differentiation from control (left) and Mll1Δ/Δ (right) mice immunostained for the neuronal marker Tuj1 (red). e, O4+ (green) oligodendrocytes in the same fields of view as those in d. f, Quantification of cell differentiation. Error bars, s.e.m. of triplicate cultures; *P = 0.016. Scale bars, 20 μm.
Therefore, to specifically examine the developmental potential of SVZ stem cells, we used SVZ monolayer NSC cultures21. MLL1 was expressed in control SVZ cultures, whereas cultures from hGFAP-Cre;Mll1F/F mice were 95–96% MLL1-deleted (Supplementary Fig. 6a–e). Mll1Δ/Δ and control SVZ cultures had similar proliferation rates (27.5% ± 0.7% BrdU+ cells in Mll1Δ/Δ cultures versus 26.7% ± 2.4% in controls; mean ± s.e.m.). In differentiation conditions, Mll1Δ/Δ cultures produced >20-fold fewer Tuj1+ cells compared to controls (Fig. 2d, f). Mll1Δ/Δ cultures had comparable numbers of pycnotic cells under both proliferative (79% of control, 31% s.d.) and differentiation (68% of control, 15% s.d.) conditions, suggesting that Mll1 was not required for SVZ cell survival. Similar to our observations in vivo, Mll1Δ/Δ cultures efficiently produced GFAP+ astrocytes and O4+ and OLIG2+ oligodendrocytes (Fig. 2e, f and Supplementary Fig. 6i, j). Thus, Mll1Δ/Δ SVZ NSC cultures had a similar proliferation rate and comparable cell death rate, but they produced fewer neuronal cells while remaining gliogenic.
Because Mll1 was deleted in radial glial precursors at E13.5, it was possible that many transcriptional changes had ‘accumulated’ before the genesis of SVZ NSCs. Therefore, we next studied the effect of Mll1 deletion after postnatal SVZ NSC genesis. To accomplish this, we derived SVZ NSC cultures from P6–P7 Mll1F/F or Mll1+/+ mice that also carried the ZEG reporter transgene; ZEG expresses green fluorescent protein (GFP) in cells that have undergone Cre-mediated recombination22. Cultures were then infected with an adenovirus expressing Cre, and 48–72 h later GFP+ cells were isolated by fluorescent activated cell sorting (FACS) (Supplementary Fig. 7a). Ninety-five to ninety-eight per cent of GFP+ Mll1F/F;ZEG cells (hereafter referred to as ZEG;Mll1Δ/Δ cells) did not express MLL1; conversely, 100% of GFP+ Mll1+/+;ZEG cells (hereafter referred to as ZEG; control cells) expressed MLL1 protein (Supplementary Fig. 7b–e). There was no significant difference in cell death between proliferating ZEG;Mll1Δ/Δ and ZEG;control stem cell cultures as assessed by propidium iodide and annexin A5 staining (Supplementary Fig. 7f). After FACS isolation, ZEG;Mll1Δ/Δ and ZEG;control cells also had similar BrdU incorporation rates (Supplementary Fig. 7g). Under differentiation conditions, ZEG;Mll1Δ/Δ SVZ cells produced threefold fewer Tuj1+ cells than ZEG;control cells (Fig. 3a–c).Differentiating ZEG;Mll1Δ/Δ cells had a trend towards fewer activated caspase 3+ cells (Fig. 3c), indicating that cell death did not account for the decreased neurogenesis. Furthermore, ZEG;Mll1Δ/Δ cells produced glial populations efficiently; in fact, there was approximately twofold more O4+ oligodendrocytes and 50% more GFAP+ astrocytes (Fig. 3d, e and Supplementary Fig. 7h, i) in ZEG;Mll1Δ/Δ SVZ cultures. Thus, Mll1 was required after the genesis of SVZ stem cells specifically for the neuronal lineage.
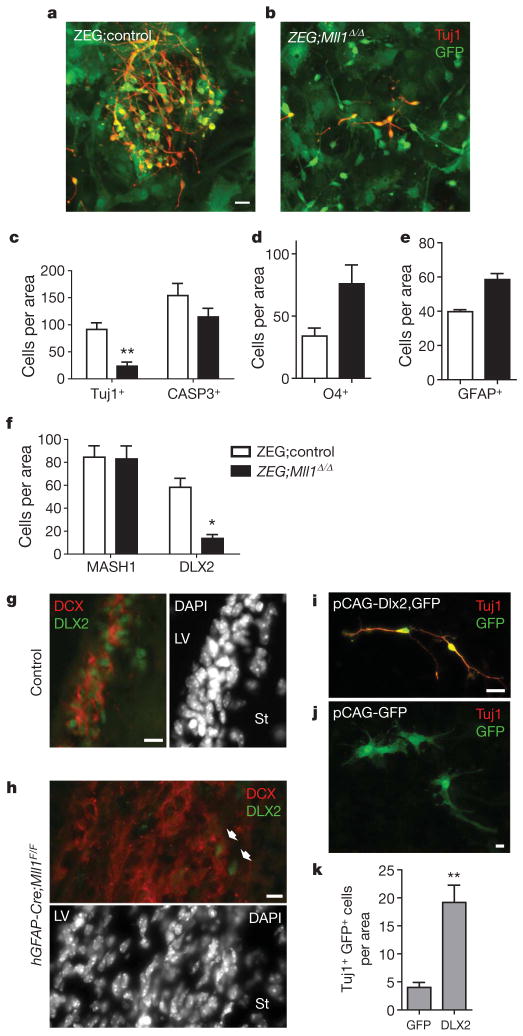
Figure 3. Mll1-dependent DLX2 expression is required for postnatal SVZ neurogenesis.
a, Immunocytochemistry for Tuj1+ (red) neuroblasts in FACS-isolated GFP+ ZEG;control SVZ cells after differentiation. b, ZEG;Mll1Δ/Δ cultures stained for Tuj1. c, Quantification of Tuj1+ neuronal differentiation and activated caspase 3+ cells after 4 days of differentiation. **P = 0.002. d, e, Quantification of glial differentiation. The number of O4+oligodendrocytes (d) and GFAP+astrocytes (e) was counted after 4–5 days of differentiation. f, Quantification of MASH1+ and DLX2+ cells after 2 days of differentiation. *P = 0.02. g, h, Immunohistochemistry for DLX2 (green) and DCX (red) in SVZ coronal brain sections of control (g) and hGFAP-Cre;Mll1F/F mice (h). i–k, Enforced Dlx2 expression can rescue neurogenesis in Mll1Δ/Δ SVZ cultures. Immunocytochemistry for Tuj1 (red) and GFP (green) after transfection of both pCAG-Dlx2 and pCAG-GFP plasmids (i) and pCAG-GFP plasmid alone (j). k, Quantification of neuronal lineage rescue by Dlx2 transfection. **P = 0.004; error bars, s.e.m.; 3–6 replicates per group. Scale bars, 10 μm (g, h) and 20 μm (a, b, i, j).
Because Mll1 is important for the epigenetic regulation of specific gene expression, we next examined the expression of genes important for SVZ neurogenesis. NSC markers SOX2 (ref. 23) and Nestin were present in 100% of cells in both ZEG;Mll1Δ/Δ and ZEG;control cells under proliferation conditions (Supplementary Fig. 7j, k). During early differentiation, MASH1 expression, a bHLH factor important for SVZ neurogenesis and oligodendrogliogenesis24, was also not altered in ZEG;Mll1Δ/Δ cultures (Fig. 3f and Supplementary Fig. 8a, b). However, DLX2, a homeodomain-containing transcription factor important for olfactory bulb interneuron development and migration in the embryo25, was decreased ~fourfold in ZEG;Mll1Δ/Δ cultures (Fig. 3f and Supplementary Fig. 8c, d). In vivo immunostaining showed a similar impairment of DLX2 expression in the SVZ of hGFAP-Cre;Mll1F/F mice (Fig. 3g, h). Normally, Dlx2 is expressed in transit-amplifying cells and is then maintained in migratory neuroblasts. hGFAP-Cre;Mll1F/F mice still had DLX2 in a few cells at the base of the mass of DCX+ cells, however, most these cells did not express this transcription factor.
To demonstrate that Dlx2 is a key developmental regulator for Mll1-dependent neurogenesis, we co-transfected Dlx2 and GFP expression plasmids into Mll1Δ/Δ SVZ cultures and induced differentiation. Transfection of the GFP plasmid alone was performed as a control. Concordance of GFP and DLX2 expression in co-transfected cultures was ~85% (Supplementary Fig. 9a, b). Mll1Δ/Δ SVZ cells co-transfected with Dlx2 and GFP had a fourfold increase in the number of Tuj1+GFP+cells (Fig. 3i, quantification in panel k) after 4 days of differentiation as compared to the GFP-transfected controls (Fig. 3j).
To determine whether Dlx2 is a direct target of MLL1, we performed chromatin immunoprecipitation (ChIP) from differentiating SVZ NSC cultures with anti-MLL1 antibodies. We found that MLL1 specifically bound to the Dlx2 promoter region and the region immediately downstream from the transcriptional start site, but not to the chromatin 1 kb upstream (Fig. 4a).
Figure 4. Dlx2 is trimethylated at both H3K4 and H3K27 in Mll1Δ/Δ cells.
At the top is a schematic indicating the location of the primer sets used for qChIP. a, MLL1 qChIP of the Dlx2 locus. b, qRT–PCR analysis of Dlx2, Mash1 and Olig2 in wild-type (grey bars) and Mll1Δ/Δ (black bars) cells during early differentiation. c, qChIP analysis of H3K4me3 levels at Dlx2, Mash1 and Olig2 loci. d, qChIP for H3K27me3 levels. e, Model of Mll1 function in the specification of the neuronal lineage from NSCs. NSCs have bivalent chromatin domains at key neurogenic genes (for example, Dlx2). In this state, precursors can form astrocytes and oligodendrocytes (blue arrows). In order for neurogenesis to proceed (red arrow), MLL1 is required for the resolution of specific bivalent loci, possibly by recruiting H3K27-specific demethylases (K27DM). Error bars, s.d.; n = 3.
Methylation of histone lysine residues is a critical determinant of active and silent gene expression states. H3K4me3 correlates strongly with active transcription whereas H3K27me3 is associated with gene silencing. Chromatin regions containing high levels of both H3K4me3 and H3K27me3 have been termed ‘bivalent domains’ and are silenced but thought to be ‘poised’ for activation26. MLL contains a catalytic SET domain that can methylate histone H3 at K4 (ref. 27). MLL family members can also recruit H3K27-specific histone demethylases28,29. Thus MLL proteins possess two non-mutually exclusive mechanisms for promoting transitions between transcriptionally restrictive and permissive chromatin states. We therefore investigated H3 methylation patterns at the promoter regions of Dlx2, Mash1 and Olig2. In differentiating SVZ cells, Dlx2 expression was Mll1-dependent whereas Mash1 and Olig2 expression were not (Fig. 4b). ChIP analysis of wild-type cells demonstrated that there were high levels of H3K4me3 and low levels of H3K27me3 at all three loci, coherent with their transcriptionally active state (Fig. 4c, d, yellow bars). Surprisingly, loss of MLL1 did not affect H3K4me3 at any of the analysed loci, suggesting that other H3K4 methyltransferases are important for maintenance of H3K4me3 at the Dlx2 locus. In Mll1Δ/Δ cells, H3K27me3 levels were strongly increased at Dlx2, but not at Mash1 or Olig2 loci (Fig. 4d, brown bars). Moreover, in the absence of MLL1, we observed H3K27me3 spreading 1 kb upstream of the transcriptional start site (Fig. 4d), consistent with previous descriptions of bivalent loci30. Thus, in the absence of MLL1, the Dlx2 locus is bivalent in differentiating SVZ NSCs.
Bivalent domains mark developmentally important loci in pluripotent and multipotent cells26. Upon differentiation into lineage-specific precursors, many bivalent domains are resolved into H3K4me3 or H3K27me3 monovalent domains, However, some loci remain bivalent30, possibly reflecting remaining gene expression plasticity. The presence of bivalent domains in tissue-specific stem cell populations suggests that there is a requirement for H3K27me3 demethylase activity at specific loci throughout development, and, in the case of SVZ neurogenesis, into adulthood. Our data are consistent with a model in which MLL1 function, perhaps mediated by H3K27me3 demethylase recruitment, is essential for bivalent domain resolution at Dlx2 during neurogenesis (Fig. 4e). In the absence of MLL1, the Dlx2 locus remains bivalent and therefore silenced. Deletion of Mll1 in broader and/or earlier populations of NSCs in development may reveal a similar requirement of Mll1 for embryonic neurogenesis.
Taken together, our results indicate that for lifelong neurogenesis, Mll1 is required by neural precursors to make the epigenetic transition to the neuronal lineage by mediating chromatin modifications at specific loci. Future analysis of the direct targets of MLL and bivalent loci in NSCs at different stages of development may lead to an epigenetic description of NSC lineage potential and a transcriptional program instructive for neurogenesis.
METHODS
Animals
Mll1F/F mice were maintained and genotyped as described8. hGFAP-Cre9, ZEG22 and R26R mice were obtained from Jackson Laboratories. To label proliferating cells, BrdU (10 mg ml−1 solution, 50 mg kg−1 body weight, Sigma) was injected intraperitoneally. Experiments were performed in accordance to protocols approved by Institutional Animal Care and Use Committee at UCSF.
Tissue preparation, immunohistochemistry and in situ hybridization
Brains were fixed by intracardiac perfusion as previously described13. Five-micrometre paraffin sections were used for H&E staining. For immunocytochemistry, 12-μm frozen sections were used with the following primary antibodies: mouse anti-GFAP (1:500, Chemicon), guinea pig anti-doublecortin (1:500, Chemicon), rat anti-BrdU (1:100, Abcam), anti-MASH (1:100 Pharmingen), rabbit anti-OLIG2 (1:10,000, gift from D. Rowitch), rabbit anti-DLX2 (1:1,000, gift from J. Rubenstein), rabbit anti-activated caspase 3 (1:500, Cell Signal Technology), anti-Nestin (1:500, Chemicon), rabbit anti-SOX2 (Abcam), mouse anti-S100 (1:500, Dako), mouse anti-MLL, N-terminal (1:500, Upstate), mouse anti-βIII-tubulin (Tuj1 clone, 1:250, Covance), mouse anti-myelin basic protein (1:2,000, Covance), mouse anti-O4 (1:50, Chemicon) and chicken anti-GFP (1:500, Aves). FluoroMyelin (Invitrogen) was used according to the manufacturer’s protocols. Sections were blocked with 10% goat serum plus 0.3% Triton X-100 (Sigma) in PBS for 30 min at 25 °C, before primary antibody incubation at 4 °C overnight. BrdU staining was performed as in ref. 13. Goat Alexa-Fluor secondary antibodies (Invitrogen) were used, and nuclei were counterstained with DAPI (Sigma). Mll1 in situ hybridization was as previously described5. X-Gal staining and electron microscopy was performed as previously described13.
Cell culture
Mouse SVZ monolayer cultures were produced essentially as previously described21. In brief, SVZ microdissections were dissociated with 0.25% trypsin and trituration. Cells were plated at ~30,000 cells cm−2 in 6-well plates (Corning) in proliferation medium (DMEM/F12/N2, 5% FCS, 20 ng ml−1 EGF, 20 ng ml−1 bFGF, 35 μg ml−1 bovine pituitary extract (media and N2 are from Invitrogen; growth factors are from Peprotech; FCS is from Hyclone). Non-attached cells were collected after 1 day and replated into 6-well plates. After ~7 days, the cells were confluent and these were routinely passaged 1:2 with 0.25% trypsin. Cells were passaged 4–6 times before use in experiments. Differentiation of cultures was induced by removing the EGF, FGF and FCS from the media21. For FACS, cells were dissociated with 0.25% trypsin and passed through a 40-μm mesh. A FACSAria (BD Biosciences) cell sorter with a 70-μm nozzle was used at the low pressure setting. Cells were collected into DMEM/F12 with 20% FCS. FACS isolated cells were centrifuged (500g, 15 min), resuspended in proliferation medium, and plated at ~100,000 cells cm−2. For cell proliferation analysis, BrdU was used at 10 μM. For immunostaining, cultures were fixed with 4% paraformaldehyde. Primary and secondary antibodies were used as indicated above, with the exception that O4 staining was performed without Triton X-100. For plasmid transfections, SVZ cells were plated at ~75,000 cells cm−2 into 8- or 16-well Lab-Tek CCR2 chamber slides (Nunc) in proliferation medium. The next morning, 5 μg cm−2 of pCAG-GFP or both pCAG-GFP and pCAG-Dlx2 was transfected with Lipofectamine (Invitrogen). Twenty-four hours after transfection, differentiation was induced. Annexin A5 staining with APC-conjugated antibodies (BD Biosciences) was performed as per the manufacturer protocols and quantified on a fluorescent flow cytometer.
Microscopy and quantification
For quantification of cell cultures, at least five non-overlapping fields of view were analysed at the fluorescent microscope (Olympus AX70) with ×20 to ×60 objectives; in some cases, digital images were captured and immunostained cells were counted using Photoshop (Adobe Systems) or ImageJ (NIH) software. For in vivo SVZ cell quantification, we collected >3 non-overlapping confocal images from each tissue section using a Zeiss Axiovert 200M with a ×63-oil objective; from each animal, at least three separate tissue sections were analysed.
qRT–PCR
Total cellular RNA was isolated by Trizol method (Invitrogen) and quantified using the NanoDrop spectrophotometer. One microgram of total RNA was treated with Turbo DNase (Ambion) then reverse transcribed with VILO Superscript (Invitrogen). Complementary DNA corresponding to 5 ng of total RNA was used as a template in qPCR analysis performed on a Roche LC480 with SybrGreen (Roche). Relative expression for the studied genes was normalized to the mean signals of six mouse housekeeping genes: Atpaf2, Dhps, Gapdh, Nosip, Pdha1 and Tufm.
Quantitative chromatin immunoprecipitation
qChIP was performed as essentially as previously described31. In brief, 107 cells were fixed for 10–20 min in 1% formaldehyde and then washed with 125 mM glycine. Cells were pelleted by centrifugation at 1,000g. After freeze-thawing, cells were suspended in 1 ml swelling buffer and separated by 20 strokes in a Dounce homogenizer. The cell suspension was pelleted and resuspended in 250 μl of lysis buffer, then sonicated in a Diagenode Bioruptor with ten 30 s pulses over a 15 min period at the high-energy setting. The cell lysate was pelleted by centrifugation at 1.6 × 104 g for 15 min at 4 °C. The supernatant was diluted tenfold in ice-cold dilution buffer containing protease inhibitors (Roche) and subsequently cleared by centrifugation at 1.6 × 104 g for 15 min at 4 °C. An aliquot of supernatant was reserved as the input control. The remaining portion was incubated overnight with the appropriate antibodies (H3K4 tri-methyl, Abcam; H3K27 tri-methyl, Upstate; MLL rabbit polyclonal, gift from D. Allis). Immune complexes were collected by 30 min incubation with Protein A Dynabeads, washed once with dilution buffer, five times with RIPA buffer and once with TE buffer. Complexes were eluted with 1% SDS and 0.84% NaHCO3 solution. NaCl was added to 150 mM, and the eluates were decrosslinked by incubation at 65 °C overnight. The eluates were treated with RNase and proteinase K, extracted with phenol/chloroform and ethanol precipitated with glycogen as a carrier. An input control was processed in parallel. DNA was dissolved in ultrapure water and subjected to qPCR analysis with the Roche LC480 and SybrGreen. Serial dilutions of mouse genomic DNA was used for standardization. For qChIP and qRT–PCR, error estimates are standard deviations and were propagated by the least square formula. Recovery of genomic DNA as the percentage input was calculated as the ratio of copy numbers in the immunoprecipitate to the input control. Primer sequences are available on request.
Supplementary Material
Acknowledgments
We thank J. Rubenstein for anti-DLX2 antibodies and the pCAG-Dlx2 plasmid, D. Rowitch for anti-OLIG2 antibodies, and Y. Dou and R. Roeder for anti-MLL1 antibodies. This work was supported by the Neurosurgery Research and Education Foundation/American Association of Neurological Surgeons, Sandler Family Foundation, Northern California Institute for Research and Education, and the Clinical and Translational Research Institute at the University of California, San Francisco (D.A.L.), California Institute for Regenerative Medicine New Faculty Award and The Chicago Community Trust Searle Scholar Award (J.W.), and the Goldhirsch Foundation, J.G. Bowes Research Fund, and National Institutes of Health (NIH) 5R37-NS028478 (A.A.-B.).
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions D.A.L. conceived the project, designed and performed experiments, coordinated collaborations, and wrote the manuscript. Y.-C.H. worked on most experiments, quantified all in vivo data, and helped prepare the figures. T.S. and J.W. performed ChIP experiments, helped analyse data and contributed ideas. A.L.M and P.A.E. provided the Mll1F/F mouse, helped perform preliminary experiments in Mll1+/− mice and contributed ideas. J.M.G.V. provided electron microscopy data and histological interpretation. A.A.-B. contributed ideas, interpreted results and helped write the manuscript. All authors discussed the results and edited the manuscript.
Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Hsieh J, Gage FH. Epigenetic control of neural stem cell fate. Curr Opin Genet Dev. 2004;14:461–469. doi: 10.1016/j.gde.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 2.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Molofsky AV, et al. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fasano CA, et al. shRNA knockdown of Bmi-1 reveals a critical role for p21-Rb pathway in NSC self-renewal during development. Cell Stem Cell. 2007;1:87–99. doi: 10.1016/j.stem.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Lim DA, et al. In vivo transcriptional profile analysis reveals RNA splicing and chromatin remodeling as prominent processes for adult neurogenesis. Mol Cell Neurosci. 2006;31:131–148. doi: 10.1016/j.mcn.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Daser A, Rabbitts TH. Extending the repertoire of the mixed-lineage leukemia gene MLL in leukemogenesis. Genes Dev. 2004;18:965–974. doi: 10.1101/gad.1195504. [DOI] [PubMed] [Google Scholar]
- 7.Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 8.Jude CD, et al. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell. 2007;1:324–337. doi: 10.1016/j.stem.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhuo L, et al. hGFAP-cre transgenic mice for manipulation of glial and neuronal function in vivo. Genesis. 2001;31:85–94. doi: 10.1002/gene.10008. [DOI] [PubMed] [Google Scholar]
- 10.Han YG, et al. Hedgehog signaling and primary cilia are required for the formation of adult neural stem cells. Nature Neurosci. 2008;11:277–284. doi: 10.1038/nn2059. [DOI] [PubMed] [Google Scholar]
- 11.Luskin MB. Restricted proliferation and migration of postnatally generated neurons derived from the forebrain subventricular zone. Neuron. 1993;11:173–189. doi: 10.1016/0896-6273(93)90281-u. [DOI] [PubMed] [Google Scholar]
- 12.Lois C, Alvarez-Buylla A. Long-distance neuronal migration in the adult mammalian brain. Science. 1994;264:1145–1148. doi: 10.1126/science.8178174. [DOI] [PubMed] [Google Scholar]
- 13.Doetsch F, Caille I, Lim DA, Garcia-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- 14.Wichterle H, Garcia-Verdugo JM, Alvarez-Buylla A. Direct evidence for homotypic, glia-independent neuronal migration. Neuron. 1997;18:779–791. doi: 10.1016/s0896-6273(00)80317-7. [DOI] [PubMed] [Google Scholar]
- 15.Marshall CA, Suzuki SO, Goldman JE. Gliogenic and neurogenic progenitors of the subventricular zone: who are they, where did they come from, and where are they going? Glia. 2003;43:52–61. doi: 10.1002/glia.10213. [DOI] [PubMed] [Google Scholar]
- 16.Picard-Riera N, et al. Experimental autoimmune encephalomyelitis mobilizes neural progenitors from the subventricular zone to undergo oligodendrogenesis in adult mice. Proc Natl Acad Sci USA. 2002;99:13211–13216. doi: 10.1073/pnas.192314199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Menn B, et al. Origin of oligodendrocytes in the subventricular zone of the adult brain. J Neurosci. 2006;26:7907–7918. doi: 10.1523/JNEUROSCI.1299-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spassky N, et al. Adult ependymal cells are postmitotic and are derived from radial glial cells during embryogenesis. J Neurosci. 2005;25:10–18. doi: 10.1523/JNEUROSCI.1108-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu QR, et al. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- 21.Scheffler B, et al. Phenotypic and functional characterization of adult brain neuropoiesis. Proc Natl Acad Sci USA. 2005;102:9353–9358. doi: 10.1073/pnas.0503965102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- 23.Zappone MV, et al. Sox2 regulatory sequences direct expression of a β-geo transgene to telencephalic neural stem cells and precursors of the mouse embryo, revealing regionalization of gene expression in CNS stem cells. Development. 2000;127:2367–2382. doi: 10.1242/dev.127.11.2367. [DOI] [PubMed] [Google Scholar]
- 24.Parras CM, et al. Mash1 specifies neurons and oligodendrocytes in the postnatal brain. EMBO J. 2004;23:4495–4505. doi: 10.1038/sj.emboj.7600447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Long JE, et al. Dlx-dependent and -independent regulation of olfactory bulb interneuron differentiation. J Neurosci. 2007;27:3230–3243. doi: 10.1523/JNEUROSCI.5265-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 27.Milne TA, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 28.Lee MG, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 29.Swigut T, Wysocka J. H3K27 demethylases, at long last. Cell. 2007;131:29–32. doi: 10.1016/j.cell.2007.09.026. [DOI] [PubMed] [Google Scholar]
- 30.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wysocka J, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.