Abstract
Cytochrome P450 2E1 (CYP2E1) plays an important role in alcohol-induced toxicity and oxidative stress. Recently, we showed that this predominantly microsomal protein is also localized in rat hepatic mitochondria. In this report, we show that the N-terminal 30 amino acids of CYP2E1 contain a chimeric signal for bimodal targeting of the apoprotein to endoplasmic reticulum (ER) and mitochondria. We demonstrate that the cryptic mitochondrial targeting signal at sequence 21–31 of the protein is activated by cAMP-dependent phosphorylation at Ser-129. S129A mutation resulted in lower affinity for binding to cytoplasmic Hsp70, mitochondrial translocases (TOM40 and TIM44) and reduced mitochondrial import. S129A mutation, however, did not affect the extent of binding to the signal recognition particle and association with ER membrane translocator protein Sec61. Addition of saturating levels of signal recognition particle caused only a partial inhibition of CYP2E1 translation under in vitro conditions, and saturating levels of ER resulted only in partial membrane integration. cAMP enhanced the mitochondrial CYP2E1 (referred to as P450MT5) level but did not affect its level in the ER. Our results provide new insights on the mechanism of cAMP-mediated activation of a cryptic mitochondrial targeting signal and regulation of P450MT5 targeting to mitochondria.
Accurate targeting of proteins to their designated subcellular compartments is critical for maintaining the distinctive structural and functional characteristics of individual cellular components. At least three major types of import/transport systems have been described for targeting proteins translated in the cytoplasm to different organelles in the eukaryotic cells. 1) Proteins destined for the ER,1 Golgi, plasma membrane, and also those secreted out of cells, are targeted to the ER through a signal recognition particle (SRP)-dependent mechanism. This pathway is mostly co-translational and involves the delivery of nascent chains by SRP to the translocon complex on the ER membrane (1, 2). 2) Protein targeting to mitochondria occurs mostly by a post-translational mechanism, although exceptions to this generality have been reported (3, 4). As part of the mitochondrial targeting pathway, an unfolded polypeptide is brought in contact with the outer and inner membrane translocase complexes (TOM and TIM, respectively) (5). The protein is unidirectionally translocated through transmembrane protein channels formed of TOM40 and TIM23/TIM17 subunits, and its entry into the matrix space is finally facilitated by an ATP-dependent pull exerted by the mitochondrial Hsp70 chaperone protein (6). 3) The peroxisomal protein targeting, although it occurs post-translationally, involves a distinct set of cytosolic receptors. These proteins not only guide the precursor proteins to the peroxisomal membrane receptor Pex, they also lead the polypeptide into the matrix compartment and eventually recycle back to the cytosol for reutilization (7, 8). As predicted by the signal hypothesis, the targeting specificity of proteins is dictated by the signal sequences that function as specific “address codes” (2). The signal sequences required for protein targeting to the ER, mitochondria and peroxisomes are quite different, and it is generally believed that a protein may carry only one type of signal, thus ensuring its targeting to the correct membrane compartment.
In contrast to this general view, recent studies from our laboratory showed that xenobiotic inducible CYP1A1 and CYP2B1 contain “atypical” signals at the N termini, which we named “chimeric signals,” that were capable of targeting the apoprotein chains to both the ER and mitochondria under in vitro and in vivo conditions (9, 10). In the case of CYP1A1, the protein chains that escaped ER targeting (~25% of the total pool) were cleaved past N-terminal residues 4 and 32, thereby activating a cryptic mitochondrial targeting signal (9). In the case of CYP2B1, an intact but Ser-128-phosphorylated protein was transported to mitochondria with high efficiency (10). Ser-128-phosphorylated CYP2B1 nascent chains showed vastly reduced efficiency for binding to SRP, but increased mitochondrial transport. These latter results suggested that protein kinase A (PKA)-dependent phosphorylation at Ser-128 activates the cryptic mitochondrial targeting signal. Dual localization of yeast fumarase with identical N terminus in the cytosol and mitochondria has been reported to involve a novel mitochondrial processing of the primary translation product for subsequent export to the cytosol (11).
Recently we studied the nature of hepatic mitochondrial CYP2E1 (alternatively referred to as P450MT5), which exhibits immunological and molecular properties similar to microsomal CYP2E1 (12). The ethanol- and pyrazole-inducible CYP2E1 is thought to play important roles in the metabolism of ethanol, acetone, and induction of oxidative stress (13, 14). Our results with combinatorial approaches showed that mitochondrial P450MT5 has identical primary structure to that of microsomal CYP2E1, although the former exhibited a much higher level of Ser-129 phosphorylation, and significantly different helical and β-sheet contents (12). In the present study we demonstrate that CYP2E1 contains a cryptic mitochondrial targeting signal at amino acid residues 21–31, in addition to the previously demonstrated ER targeting signal within the first 30 residues of the protein (15, 16). A comparison of chimeric signals of CYP1A1, CYP2B1, and CYP2E1 shows subtle differences in terms of affinity for binding to SRP and mitochondrial translocases, but all of them show a distinct resistance to translational arrest in response to saturating levels of SRP, and propensity for translation in a membrane-free form. Our results also show for the first time that Ser-129 phosphorylation increases the affinity of the protein for binding to cytoplasmic Hsp70 (CyHsp70) family chaperones as a possible basis for increased mitochondrial targeting of P450MT5 but does not affect the binding of nascent chains to SRP or the major ER translocator protein Sec61.
EXPERIMENTAL PROCEDURES
Construction of Expression Plasmids
Full-length CYP2E1 cDNA (17) was generated by reverse transcriptase-based PCR. N-terminal and point mutations containing a start codon preceded by a Kozac consensus sequence were constructed essentially as described before (9, 10), and cloned in the pCMV4 and pGEM7zf (Promega Biotech, Madison, WI) vectors. Various fusion cDNA constructs were generated as described before (10) and cloned in pGEM7zf vector.
In Vitro Import of Labeled Proteins into Mitochondria
Rat liver or yeast mitochondria were isolated and used for in vitro import using a system described before (9, 10). In vitro translation products were generated in the TNT rabbit reticulocyte lysate system (RRL, Promega, Madison, WI) in presence of added [35S]Met (40 µCi/50-µl reaction, PerkinElmer Life Sciences), according to the recommended protocol of the manufacturer. In some experiments the translation products were phosphorylated for 45 min at 30 °C by supplementing the translation mix with 10 units of PKA (Sigma) and 100 µm ATP as described (18). The import assays were carried out in a 200-µl final volume as described before (9) and treated with 0–300 µg/ml trypsin for 20 min on ice. Trypsin-treated samples were mixed with 10 m excess of trypsin inhibitor, and mitochondria were recovered by sedimentation through 1 m sucrose as described (9). Mitochondrial proteins were solubilized in 2× Laemmli sample buffer for 10 min at 75 °C and analyzed by SDS-PAGE (19) and fluorography.
Expression of CYP2El cDNAs in COS Cells
Various cDNAs cloned in pCMV4 vector were transfected in COS-7 cells using FuGENE 6, a non-liposomal transfection reagent (Roche Molecular Biochemicals), according to the recommended protocol of the manufacturer. Cells from 6–8 plates (100 mm), after 24 h of transfection, were homogenized in sucrose-mannitol buffer containing protease inhibitor mixture with Teflon fitted glass homogenizer (~25 strokes). Isolation of mitochondria by differential centrifugation and sucrose density banding, and isolation of microsomes were described before (9).
SDS-PAGE and Immunoblot Analysis
Proteins were resolved by SDS-PAGE (19) and transferred to nitrocellulose membranes for immunoblot analysis as described (20). Polyclonal antibodies against CYP2E1 (Oxford Biomedical Research, Oxford, MI), mitochondrial transcription factor A, calreticulin (ABR, Golden, CO), and monoclonal antibody against CyHsp70 (Sigma) were used. Immunoblots were developed with chemiluminescence Super Signal Ultra kit (Pierce), and the blots were imaged and quantitated using a Fluor-S imaging system (Bio-Rad).
Chemical Cross-linking of CYP2E1 with Mitochondrial Translocases, SRP, and ER Translocator Protein Sec61
For cross-linking with mitochondrial translocator proteins, fusion proteins 1–160 CYP2El/dihy-drofolate reductase (DHFR) and 1–160 S129A/DHFR, subjected to phosphorylation in presence of added PKA and used for in vitro transport in isolated yeast mitochondria, and translocation arrest was initiated by adding 1 µm methotrexate as described before (6, 10). Translocation intermediates were cross-linked with 500 µm m-maleim-idobenzoyl-N-hydroxysuccinimidate ester (MBS), a membrane-permeable cross-linker (Pierce) for 20 min at room temperature (6). Termination of reaction with 80 mm glycine and 5 mm β-mercaptoethanoL, and immunoprecipitation with anti-CYP2El antibody (1:50) or antibodies to mitochondrial translocase proteins were essentially as described before (10, 21). Immunoprecipitated proteins were subjected to SDS-PAGE, and the gels were imaged in a Bio-Rad GS525 Molecular Imager.
Cross-linking of 35S-labeled CYP2E1 or CYP2B1 proteins with SRP was carried out co-translationally (21) as modified recently (10). In vitro translation was carried out in the wheat germ lysate (WGL) in the presence of increasing amounts of SRP (0.125–2 units). When phosphorylation was required, PKA (10 units/reaction) was added to the reaction mixture. After 30 min at 30 °C, reactions were stopped by adding 1 mm cycloheximide and the incubation was continued for 10 min to allow chain completion. Cross-linking was performed as described above using the water-soluble m-maleimidobenzoyl-N-hydroxysulfosuccinimi-date ester (S-MBS) cross-linker. Immunoprecipitation was performed using antibodies against CYP2E1, CYP2B1, or 54–kDa subunit of SRP. Immunoprecipitates were subjected to electrophoresis, and gels were imaged and quantified using a Bio-Rad GS525 Molecular Imager.
Cross-linking of CYP2E1 and CYP2B1 with ER membrane translocator protein Sec61 was carried out following the procedure of Greenfield and High (22) with some modifications. COS cells were co-transfected with both CYP2E1 and CYP2B1 cDNA constructs. Cells were harvested after 8 h, suspended in Hepes-sucrose (2 mm HEPES, pH 7.4, 70 mm sucrose, 210 mm Mannitol, 2 mm EDTA) at ~5 × 106 cells/ml, subjected to cross-linking with 500 µm MBS for 1 h at room temperature, and used for isolating microsomes and cytosol fractions. Cross-linked CYP2E1, CYP2B1, and calreticulin proteins were first immunoprecipitated with respective antibodies followed by immunoblot analysis with antibody to Sec61α or CyHsp70.
Membrane Integration Assay
Proteins were translated in WGL with or without added PKA (10 units), in the presence of 2 units/25 µl of unwashed or KCl-washed canine reticular endoplasmic membranes (10). Reactions were stopped by adding 1 mm cycloheximide, membranes were extracted with 0.1 m Na2CO3 (pH 11) for 1 h on ice and fractionated in to soluble (200,000 × g supernatant) and insoluble (pellet fractions) as described before (23). Both the membrane pellet and the protein extract were resuspended in 40 µl of 10 mm Tris-HCl (pH 7) containing 2% SDS and subjected to SDS-PAGE.
Interaction of CYP2E1 with CyHsp70
Interaction of CYP2E1 with CyHsp70 under in vitro and in vivo conditions was tested by co-immunoprecipitation. For studying in vitro interaction, CYP2E1 protein was synthesized and phosphorylated in RRL in the presence or the absence of PKA inhibitors (H89 or a myristoylated peptide, PKI, Calbiochem-Novabiochem Corp., San Diego, CA). Interaction of the CYP proteins with CyHsp70 present in the RRL under these conditions was estimated by co-immunoprecipitation. Reaction mixtures were immunoprecipitated with a polyclonal antibody against CYP2E1 raised in goat, and resolved by SDS-PAGE on a 10% gel. The bottom half of the gel containing the 52-kDa CYP proteins was subjected to autoradiography, and the top portion of the gel was transferred to a nitrocellulose membrane and probed with a mouse monoclonal antibody for Hsp70.
In vivo interaction was studied using post-mitochondrial supernatant fraction from cells transfected with CYP2E1 WT, CYP2E1 S129A and CYP2E1 KKmut cDNAs, in the presence or absence of dibutyryl cAMP (db-cAMP). CyHsp70 was co-immunoprecipitated with a polyclonal antibody against CYP2E1 raised in goat. The immunoprecipitated proteins were resolved by SDS-PAGE on a 12% gel and transferred to nitrocellulose, and the membrane was probed concomitantly with mouse monoclonal antibody directed against CyHsp70 and monoclonal antibody to CYP2E1.
Binding of CYP2El to Reconstituted TOM40
Yeast TOM40 protein expressed in Escherichia coli BL21 cells (24) was purified from the inclusion bodies to near homogeneity and used for reconstitution in liposomes essentially as described (24). Liposomes were prepared from azolectin (Sigma, type IVS) in MOPS-Tris (pH 6.9) buffer using a Branson sonifier as described before (25). Liposomes were freeze-thawed three times and solubilized by adding n-octyl glucopyranoside (6% v/v) on ice for 30 min. Solubilized liposomes (10 mg) were mixed with 0.1 mg of TOM40 protein and diluted 2-fold with 10 mm MOPS-Tris, pH 6.9, containing 0.5 mm PMSF and 0.5 mm DTT. Reconstitution was performed by dialysis of the mixture at 4 °C for 24 h against the same buffer. 35S-Labeled and phosphorylated RRL translation products (80,000–160,000 cpm) were incubated for 1 h at 30 °C with TOM40-containing or TOM40-free liposomes (250 µg) in a buffer containing 20 mm KC1 and 2% bovine serum albumin, followed by addition of 150 mm KCl. Liposomes were isolated by centrifugation at 200,000 × g, washed once with 150 mm KC1 containing buffer (26), resuspended in Laemmli buffer, and subjected to SDS-PAGE on a 12% gel. The amounts of radioactive protein binding were measured by imaging through the GS-525 imager (Bio-Rad).
Depletion of CyHsp70 from RRL
CYP2E1 WT and 1–160 CYP2E1/DHFR were translated in RRL (100-µl reactions) and incubated for 30 min on ice with 8 m urea (v/v). After dialysis for 6 h against three changes of 50 mm Tris-HCl buffer, pH 7.4, containing 1 mm MgCl2, 50 mm KC1, and 0.1 mm DTT, the mixture was incubated overnight with 10 µg of monoclonal anti-CyHsp70 antibody. CyHsp70 was depleted by adding protein A-agarose, followed by pelleting the immune complex. In vitro import with isolated rat liver mitochondria was carried out using translation products formed in control RRL or in RRL depleted of CyHsp70.
Results
Characteristics of the N-terminal Signal Sequence of CYP2E1
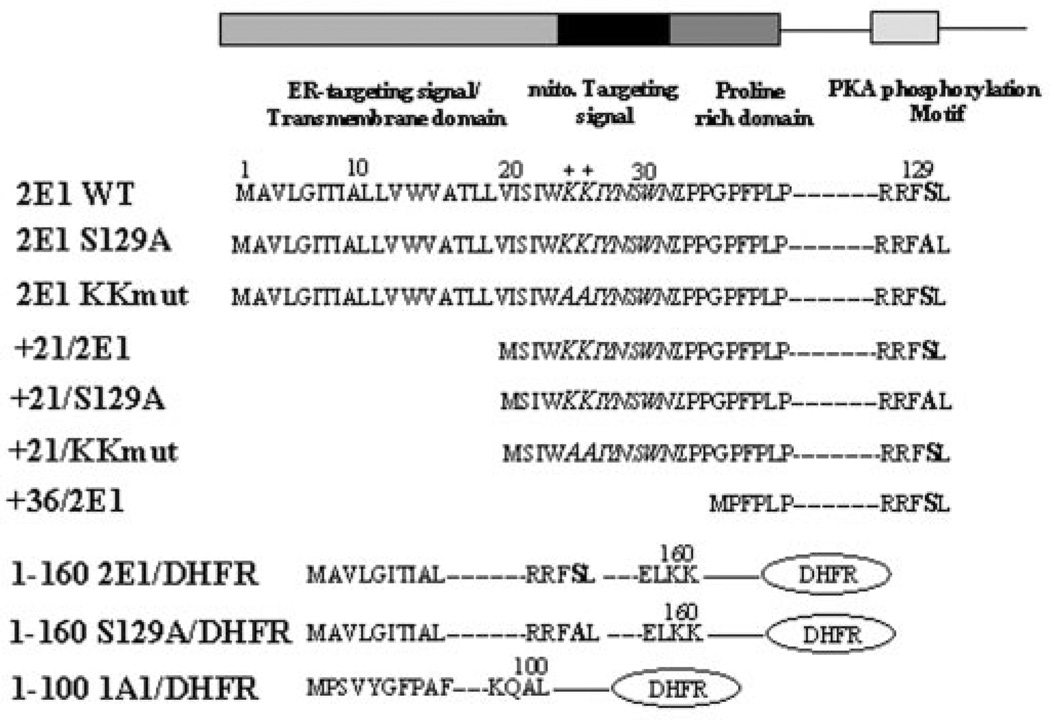
Fig. 1 shows the known and predicted microdomains at the N terminus of CYP2E1. The N-terminal 29-amino acid region of CYP2E1 contains an ER targeting and a transmembrane helical domain, but it lacks an efficient ER retention signal compared with other ER-targeted CYPs (27). The +33 to + 39 region of the protein contains the conserved proline-rich domain, which is suggested to help fold the protein facing the cytoplasmic side of the membrane. No canonical mitochondrial targeting signal preceding the proline-rich domain, similar to those in CYP1A1 and CYP2B1 (9, 10) is seen in the N-terminal 39-amino acid region of CYP2E1. However, sequence 21–31, which is part of the transmembrane anchor domain, contains two positively charged amino acids (at positions 24 and 25), which may potentially function as part of a cryptic mitochondrial targeting signal. As observed with other members of CYP family 2, CYP2E1 contains a unique PKA target phosphorylation site at Ser-129 (28, 29). Based on this general background, we generated a series of N-terminal deletions and point mutations targeted to the positive residues at positions 24 and 25 and to the phosphorylation site at position 129 (see Fig. 1). We also generated fusion constructs of sequence 1–160 of CYP2E1 (wild-type and phosphorylation site mutant S129A) fused to DHFR reporter protein as shown. A construct of sequence 1–100 of CYP1A1 fused to DHFR was also generated as shown (Fig. 1).
Fig. 1. Chimeric signal properties of the N-terminal end of CYP2E1.
The predicted signal properties of the N-terminal 35 amino acid sequences of CYP2E1 and various mutations targeted to this region (in bold) and fusion constructs used in this study are shown.
Localization of Mitochondrial Targeting Signal and Activation by cAMP-dependent Phosphorylation
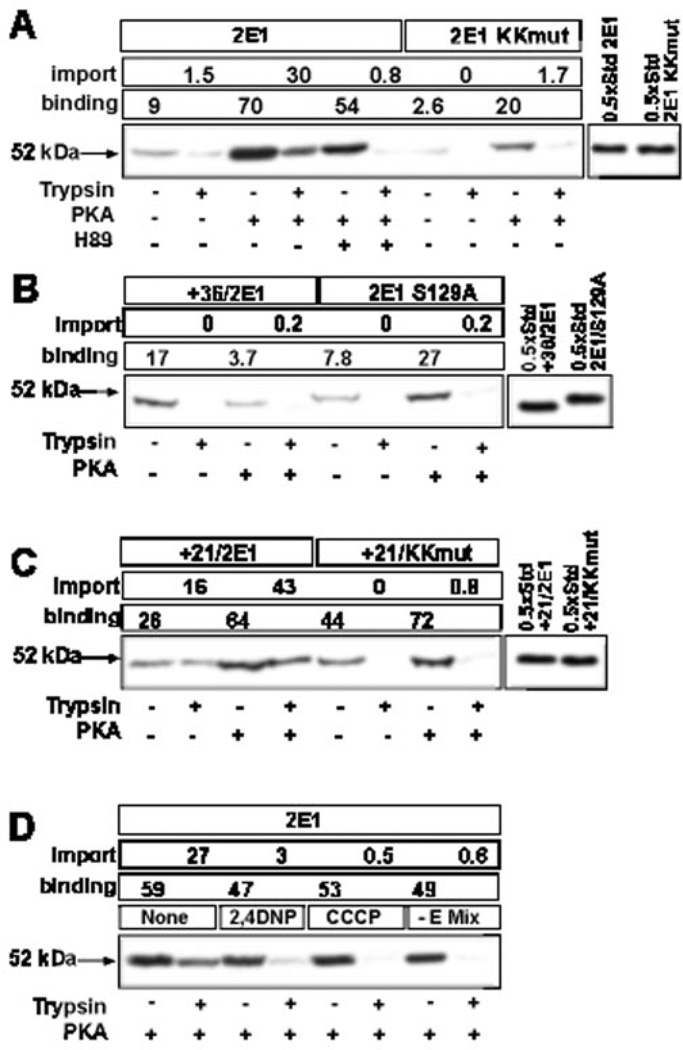
Mitochondrial targeting of CYP2E1 was studied using in vitro import of 35S-labeled proteins into isolated rat liver mitochondria using resistance to limited trypsin digestion as a criterion for protein import. ~9% of input wild-type CYP2E1 protein was found to be associated with mitochondria banded through 1 m sucrose, and only 1.5% of input protein was rendered resistant to trypsin digestion, suggesting inefficient binding and import (Fig. 2A). Phosphorylation of translation products by incubation with PKA and ATP strongly increased both protein binding and import to 70 and 30% of input counts, respectively (Fig. 2A). Addition of H89 (an inhibitor of PKA) during phosphorylation (Fig. 2A) or mutations targeted to the phosphorylation site Ser-129 (Fig. 2B, construct CYP2E1 S129A), reduced mitochondrial transport compared to the control unphosphorylated CYP2E1 level. Deletion of N-terminal 21 amino acid residues (+21/2E1) increased the transport by ~10-fold compared with intact and unphosphorylated CYP2E1 protein, and phosphorylation caused an additional increase in import to ~45% of input protein (Fig. 2C). Truncation to residue 36, on the other hand, nearly completely abolished the import, suggesting that the putative mitochondrial targeting signal resides between amino acid residues 21 and 36 (Fig. 2B). N-terminal deletion to residue 31 caused a similar impairment of transport, narrowing the potential mitochondrial targeting sequence from position 21 to 31 of the protein (data not shown). Mutations targeted to the two positively charged residues at 24 and 25 abolished both the basal transport and the PKA-dependent increase in import of wild-type and +21/2E1 (Fig. 2, A and C). Fig. 2D shows that import of phosphorylated CYP2E1 protein was dependent on transmembrane potential because 2,4-dinitrophenol and carbonyl cyanide-m-chlorophenyl hydrazone, which disrupt mitochondrial membrane potential, reduced the transport to 10 and 2% of control reactions, respectively. The transport was also dependent on ATP, because omission of energy mix reduced the transport. Activators (Mn2+ and Mg2+) and inhibitors (EDTA and O-phenanthroline) of matrix processing peptidase (MPP) had no significant effect on the efficiency of mitochondrial import of phosphorylated CYP2E1 protein or the size of the imported protein, further confirming that imported CYP2E1 is not proteolytically processed (results not shown).
Fig. 2. Sequence requirements for the in vitro transport of CYP2E1 protein.
35S-Labeled translation products generated in the presence or absence of added PKA were used for the import into rat liver mitochondria. Mitochondria from a companion set of reactions were treated with trypsin (300 µg/ml), and the proteins were resolved by SDS-PAGE on a 12% gel and subjected to fluorography. A–C, wild-type and various mutant CYP2E1 proteins were used as indicated. In D, mitochondria were pre-incubated for 20 min on ice with or without added 2,4-dinitrophenol (2,4DNP, 25 µm), carbonyl cyanide-m-chlorophenyl hydrazone (CCCP, 25 µm), or in the absence of added energy mix (E mix) before initiating import reaction with phosphorylated wild-type CYP2E1 translation product. The percentage binding and import are presented.
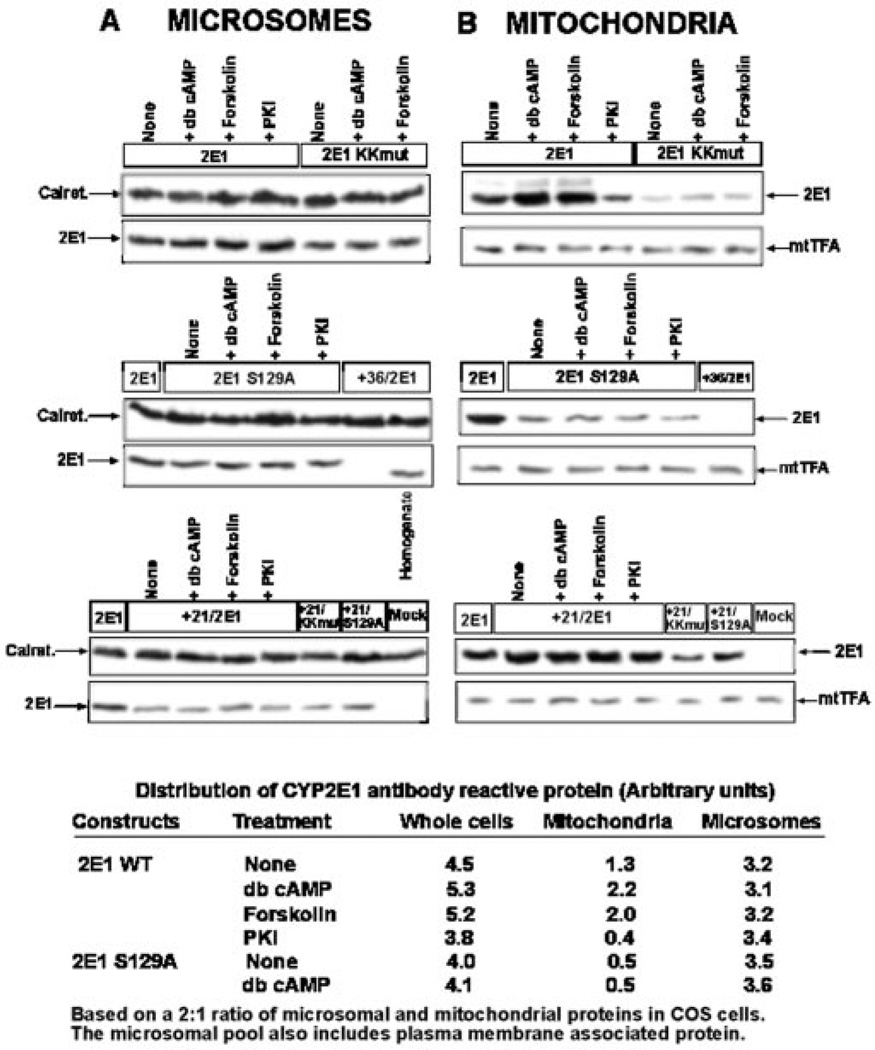
The role of PKA-dependent phosphorylation on the in vivo targeting of CYP2E1 to mitochondria was investigated using COS cells transfected with various wild-type and mutant cDNA constructs shown in Fig. 1. The effects of cAMP analog db-cAMP, cAMP inducer forskolin, and PKA-specific inhibitor myristoylated peptide (PKI) on subcellular distribution of CYP2E1 were examined. After 24 h of transfection, mitochondria and microsomes were isolated and analyzed by immunoblot analysis. As previously described, the cell fractionation procedure yielded mitochondrial preparations containing less than 1% microsomal contamination as tested by marker enzyme assays (9, 10).
Immunoblot analysis in Fig. 3 was carried out with equal amount (20 µg) of mitochondrial and microsomal proteins, and the levels of antibody reactive proteins in different subcellular compartments (Fig. 3, bottom) were calculated based on a recovery of 2:1 ratio of microsomal and mitochondrial proteins from COS cell homogenates. Results together show that, under normal culture conditions, the microsomal CYP2E1 content was ~2–3-fold higher than the mitochondrial content. The immunoblots in topmost panels show that addition of db-cAMP or forskolin resulted in a 50–70% increase in the mitochondrial CYP2E1 level but did not affect the level of microsomal CYP2E1. PKI caused a 70% inhibition of mitochondrial CYP2E1 but did not affect the level of microsome-associated CYP2E1 (see Fig. 3, top panels). Mitochondrial targeting of CYP2E1 was drastically reduced by K24A and K25A substitutions (2ElKKmut), but these mutations only marginally affected the microsomal targeting of the protein. Forskolin and db-cAMP did not affect the targeting of mutant protein to either of subcellular compartments (Fig. 3, top panels). Mutation at the Ser-129 phosphorylation site (2E1S129A) drastically reduced mitochondrial targeting while leaving ER targeting unaffected (Fig. 3, middle panels). As expected, db-cAMP, forskolin, or PKI did not affect the level of targeting of mutant protein either to mitochondria or to the ER. Furthermore, +36/2E1, lacking the transmembrane domain and also the putative mitochondrial targeting signal, was detected neither in the mitochondrial nor the microsomal fractions, although it was detected in the total homogenate (middle panel). Deletion to residue 21 (+21/2E1) reduced the microsomal targeting by ~70% but increased the mitochondrial targeting by nearly 2-fold (bottom panel). Mutations targeted to the positive residues at positions 24 and 25 (+21/KKmut) and the phosphorylation site (S129A) reduced the level of mitochondrial targeting of +21/2E1 by 70–80% without any significant effect on the microsomal targeting. These results are consistent with the in vitro results presented in Fig. 2, and show that Lys-24 and Lys-25 are critical for mitochondrial targeting of CYP2E1.
Fig. 3. Effects of phosphorylation on in vivo targeting of CYP2E1 protein to mitochondria and microsomes.
COS cells were transfected with indicated cDNA constructs or empty vector (Mock), and subcellular fractions were isolated as described under “Experimental Procedures.” Microsomal (A) and mitochondrial (B) proteins (20 µg each) or total homogenate fraction (100 µg) were resolved by SDS-PAGE on a 12% gel and subjected to immunoblot analysis. Blots were sequentially probed with a goat polyclonal antibody for CYP2E1 (1:2000 dilution) and with an anti-calreticulin (1:1000 dilution) antibody for microsomal proteins (A) or with an anti-mitochondrial transcription factor A antibody (1:3000 dilution) for mitochondrial proteins (B) as loading controls. db-cAMP (10 µm) and forskolin (10 µm) were added 3 h after transfection. PKI was added 1 h before transfection at a final concentration of 400 nm. The blots were quantitated by imaging through a Bio-Rad Fluor S imager, and the relative distribution of CYP2E1 in the microsomal and mitochondrial fractions was calculated based on a 2:1 recovery of the microsomal and mitochondrial protein from the total cell homogenate.
The quantitation of immunoblot data based on a 2:1 recovery of microsomal and mitochondrial proteins from transfected COS cells shows that the total cellular CYP2E1 level was increased by ~25% in cells treated with db-cAMP and forskolin and reduced by ~25% in cells treated with PKI. Results also show that only the mitochondrial CYP2E1 content changed significantly under these treatment conditions, whereas the microsomal content remained relatively unaltered. Mutations targeted to Ser-129 did not affect the total cellular level of CYP2E1 protein, although the mitochondrial level was drastically reduced with a marginal increase in the microsomal protein level.
Effects of Phosphorylation on the Binding of CYP2El Apoprotein to SRP
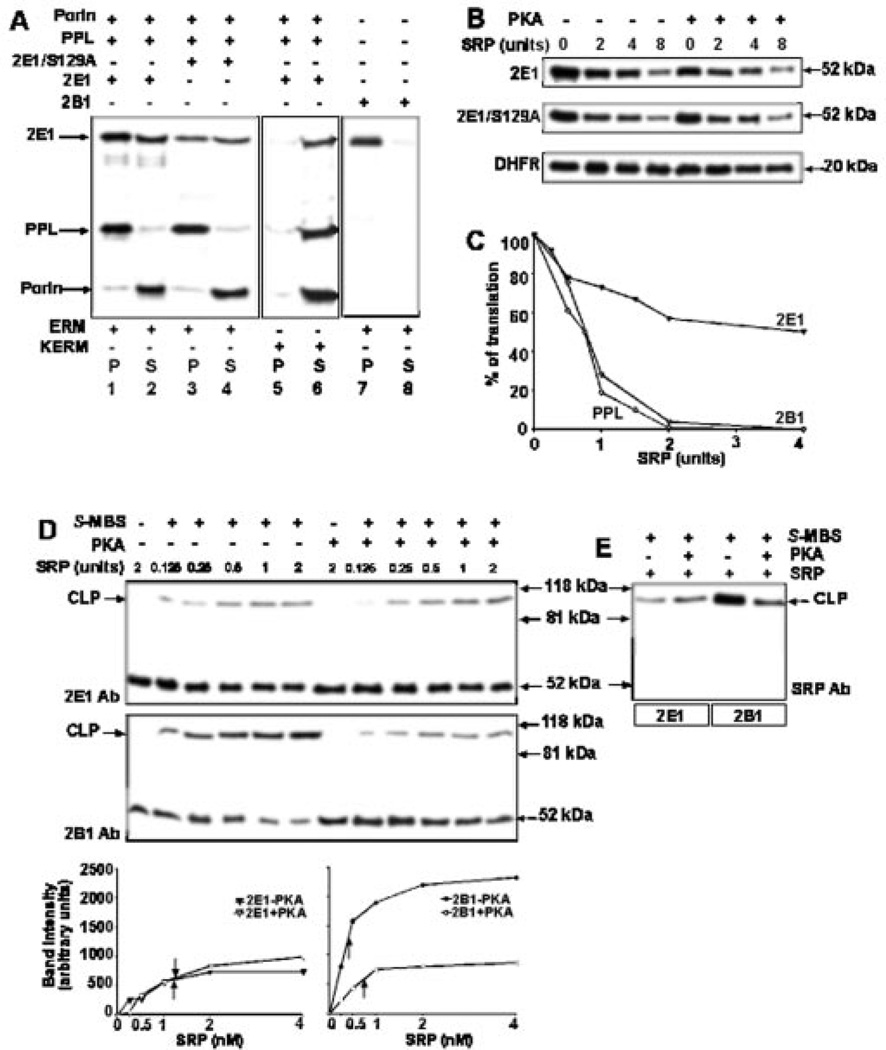
In Fig. 4A (lanes 1–6), we compared the extent of ER membrane integration of wild-type and S129A mutant proteins that were translated in the WGL in the presence of [35S]Met, with or without added PKA (10 units), and 5 units of unwashed canine rough ER or KCl-washed ER membranes. Preprolactin (PPL) as a marker protein containing a classical ER targeting signal, and porin as a marker for a non-ER-targeted protein were also co-translated. The extent of membrane association was determined by extraction with alkaline Na2CO3. In the absence of added PKA, both wild-type and CYP2E1/S129A mutant proteins partitioned almost equally in the insoluble (P) and soluble (S) fractions, indicating that ~50% of the CYP2E1 was not efficiently inserted in the ER membranes and instead remained in the soluble fraction (Fig. 4A, lanes 1 and 2). Although not shown, in vitro translated CYP2E1 in RRL showed a similar distribution. Addition of PKA did not alter the distribution of CYP2E1 and CYP2E1/S129A mutant between insoluble and soluble compartments (data not shown). Notably, the S129A mutant protein also partitioned in the membrane-bound and -free phases in a manner similar to the wild-type phosphorylated protein, suggesting that phosphorylation has no effect on the efficiency of CYP2E1 targeting to the ER. As expected, more than 90% of ER-targeted PPL was recovered in the insoluble fraction and more than 90% of the mitochondrial outer membrane protein porin was recovered in the soluble fraction (Fig. 4A, lanes 1–4). When translation was performed in the presence of KCl-washed ER membranes that are deficient in SRP, almost 100% of the CYP2E1 protein partitioned in the soluble fraction, indicating that insertion of CYP2E1 in the ER membranes was SRP-de-pendent (Fig. 4A, lanes 5 and 6). Results also show that CYP2B1, another similarly oriented transmembrane CYP, was almost completely recovered in the membrane fraction under similar in vitro translation conditions (Fig. 4A, lanes 7 and 8), suggesting that CYP2B1 is targeted to the ER more efficiently than CYP2E1.
Fig. 4. Effect of phosphorylation of CYP2E1 on ER membrane association and SRP binding.
A, extent of integration of wild-type and S129A mutant CYP2E1 proteins into ER membrane. Proteins were translated in WGL in the presence of [35S]Met and 5 units of unwashed endoplasmic reticular membranes (ERM, lanes 1–4) or KCl-washed membranes (KERM, lanes 5 and 6). PPL and porin were co-translated. CYP2B1, another N-terminally anchored membrane protein, was used also as a control (lanes 7 and 8). B, wild-type CYP2E1, CYP2E1 S129A mutant and DHFR proteins were translated in WGL in presence of [35S]Met with or without added PKA (10 units) and indicated amounts of SRP. Aliquots of translation reaction (20 µl) were resolved by SDS-PAGE on 12% gels. C, rates of SRP-mediated translation inhibition of CYP2E1, CYP2B1, and PPL from reactions run as in B. The gels were imaged and quantitated in a Bio-Rad Fluor S molecular imager. The activity in the absence of added SRP for each protein was regarded as 100%. D and E, levels of SRP binding to phosphorylated and unphosphorylated CYP2E1 by chemical cross-linking. Translation of wild-type CYP2E1 (top panel of D and also first two lanes in E) or CYP2B1 (bottom panel of D and last two lanes of E) was carried out in WGL in presence of [35S]Met, increasing amounts of SRP, with or without added PKA (10 units). Cross-linking with S-MBS was performed as described under “Experimental Procedures.” Reaction products with or without added cross-linker were immunoprecipitated with polyclonal antibody to CYP2E1 or CYP2B1 as indicated in D. Cross-linked products (CLP) are indicated. The extent of cross-linking as a function of SRP concentration is presented underneath panel D. In E, the immunoprecipitation was carried out with antibody to the 54-kDa subunit of SRP.
Under in vitro conditions, SRP binds to nascent chains containing an ER targeting signal, causing an arrest of chain elongation and thus, translation inhibition (30). Elongation arrest is thought to be a physiologically important function of SRP (31). We therefore used this property as an indicator of the affinity of SRP for phosphorylated or unphosphorylated CYP2E1 nascent chains (Fig. 4B). Cytosolic protein DHFR was used as an internal control. In the first approach, CYP2E1, CYP2E1/S129A, or DHFR were translated in the WGL system in presence of increasing concentrations of SRP. The rate of translation of both wild-type and S129A mutant CYP2E1 was inhibited with increasing amounts of SRP with a maximum inhibition of 70–80% at the highest SRP concentration (8 units) used. Addition of PKA had no significant effect on the rate of inhibition by SRP, further confirming that phosphorylation does not alter the extent of CYP2E1 binding to SRP. The in vitro translation of DHFR was not inhibited under these conditions. A comparison of the rate of SRP-mediated inhibition of CYP2E1, CYP2B1, and PPL is presented in Fig. 4C. It is noteworthy that, at the highest SRP concentration of 4 units, the translation of both CYP2B1 and PPL was inhibited to near 100%. Under these conditions, CYP2E1 was inhibited only by ~40–45%. These results show that the chimeric signals of the two CYP proteins respond differently to added SRP suggesting possible difference in binding affinities.
In a more direct approach, we studied the extent of association of phosphorylated and unphosphorylated CYP2E1 with SRP and compared with that of CYP2B1 by chemical cross-linking (Fig. 4, D and E). Proteins were translated in WGL in the presence of [35S]Met and increasing amounts of SRP and subjected to cross-linking with S-MBS. The cross-linked products were immunoprecipitated with antibody to CYP2E1 (Fig. 4D, top panel) and CYP2B1 (Fig. 4D, bottom panel), respectively. Immunoprecipitates with both antibodies showed slow migrating cross-linked products of ~95 kDa, in addition to 52-kDa CYP proteins. As expected, the immunoprecipitates showed increasing levels of cross-linked product with increasing SRP in both cases reaching saturation levels at ~4 units of SRP (see the quantitation at the bottom of Fig. 4D). In the case of CYP2E1, phosphorylated (+PKA) as well as unphosphorylated (−PKA) translation products yielded similar levels of cross-linking. However, in support of our recent results on extent of ER targeting (10), the unphosphorylated CYP2B1 yielded 3–4-fold higher level of cross-linking as compared with unphosphorylated CYP2B1. The data on cross-linking (Fig. 4D and the quantitation underneath) show that the association constant for CYP2B1 (0.41 nm) nearly doubled after phosphorylation (0.76 nm). However, the association constant for CYP2E1 (1.23 nm) was not significantly affected by phosphorylation (1.34 nm). The results of SRP mediated translation inhibition (Fig. 4C) and cross-linking (Fig. 4D) together show the following. 1) In marked variation from that observed with CYP2B1, phosphorylation has no effect on the extent of CYP2E1 binding to SRP. 2) The chimeric N-terminal signals of CYP2E1 and CYP2B1 have different affinities for SRP, with unphosphorylated CYP2E1 showing a significantly higher association constant as compared with unphosphorylated CYP2B1.
In Fig. 4E, the reaction mixtures with 2 units of SRP, and with or without added PKA, were also immunoprecipitated with antibody to the 54-kDa subunit of SRP, confirming that the cross-linked product indeed contains a component of SRP in addition to the CYP proteins.
Association of Phosphorylated CYP2E1-DHFR Fusion Protein with Mitochondrial Translocases
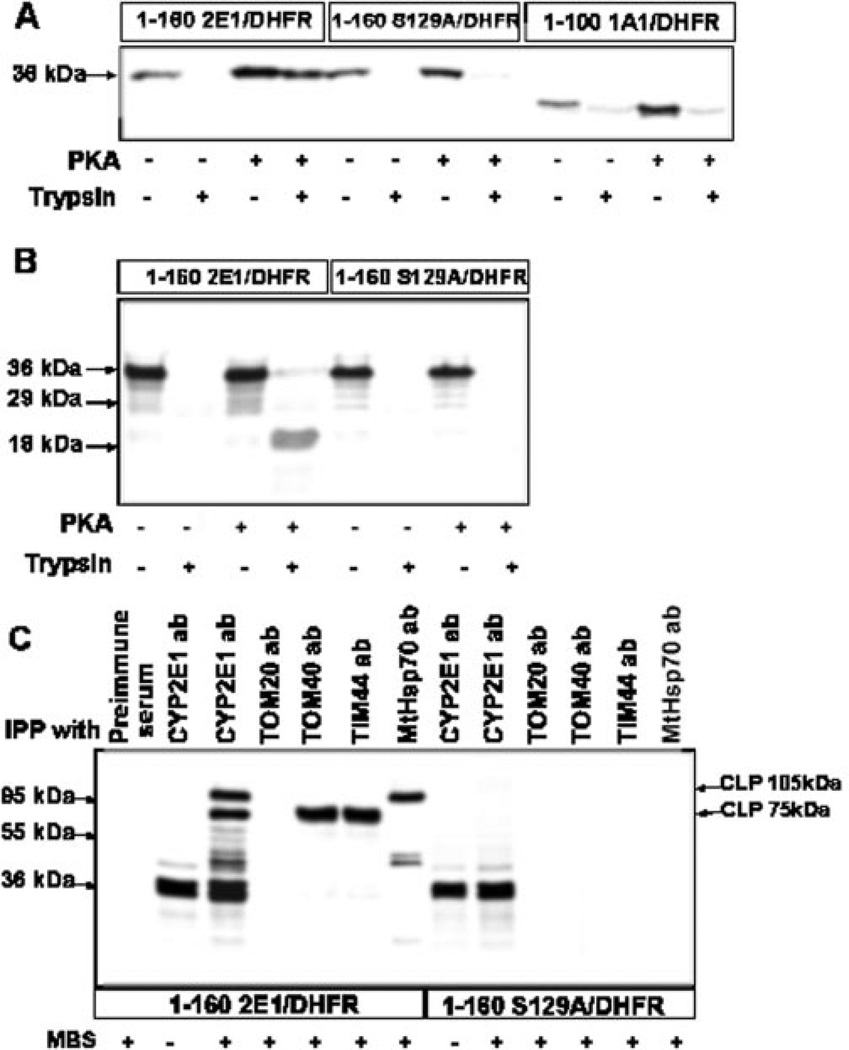
Chemical cross-linking was carried out to determine the interaction of CYP2E1 signal sequence with components of mitochondrial TOM and TIM complexes. We used fusion proteins containing the first 160 amino acids of wild-type or S129A mutant CYP2E1 proteins fused to DHFR (6). Fusion protein composed of the first 100 amino acids of CYP1A1 fused to DHFR served as a negative control, because the mitochondrial import of CYP1A1 is not regulated by phosphorylation. The translation products with or without added PKA were used for import into yeast mitochondria, translocation was arrested by adding methotrexate and translocation intermediates were generated (6, 10). As expected, only phosphorylated 1–160 CYP2E1/DHFR was imported into mitochondria and rendered resistant to trypsin (Fig. 5A). The unphosphorylated fusion protein was not imported at a significant level, although it bound to mitochondria at a significant level. The S129A mutant protein, on the other hand, was only marginally imported even when it was translated in the presence of added PKA. Finally, the 1–100: CYP1A1/DHFR fusion protein was imported inefficiently, whether it was translated in the presence or absence of added PKA. Fig. 5B shows the formation of a translocation-arrested intermediate in the presence of added methotrexate. No detectable unphosphorylated 1–160 CYP2E1/DHFR fusion protein was resistant to trypsin digestion, possibly because of inefficient binding of the protein to mitochondrial translocases. The phosphorylated 1–160 CYP2E1/DHFR fusion protein, however, yielded an 18-kDa trypsin-resistant product representing the translocation-arrested component. Fusion protein with mutated phosphorylation site, 1–160 S129A/DHFR, failed to yield any trypsin-resistant product, further confirming inefficient import of the protein.
Fig. 5. Interaction of phosphorylated CYP2E1 with mitochondrial protein translocases.
A, 1–160 CYP2E1/DHFR, 1–160 S129A/DHFR, and 1–100 CYP1A1/DHFR fusion proteins translated in the RRL were used for in vitro import. B, effect of methotrexate-mediated translocation arrest of wild-type (1–160 CYP2E1/DHFR) and mutant (1-160 S129A/DHFR) fusion proteins. Reaction mixtures were preincubated on ice for 20 min with 1 µm methotrexate before initiating the import. C, chemical cross-linking of translocation-arrested 1–160 CYP2E1/DHFR and 1-160 S129A/DHFR fusion proteins with mitochondrial translocases and MtHsp70. Both fusion proteins were translated in presence of PKA (10 units), and translocation arrest was caused by adding 1 µm methotrexate as in B. IPP, immunoprecipitation; ab, antibody. Cross-linking with MBS cross-linker and immunoprecipitation were carried out as described under “Experimental Procedures.” In A and B, proteins were resolved by SDS-PAGE on 14% gels and in C on 10% gel. Cross-linked products (CLP) are indicated.
In the next series of experiments, methotrexate-arrested intermediates of wild-type and S129A mutant fusion proteins (with added PKA) were subjected to chemical cross-linking with membrane-permeable cross-linker MBS, and the cross-linked products were immunoprecipitated with antibodies against subunits of translocase complexes or CYP2E1 antibody. Results in Fig. 5C show that pre-immune IgG did not interact with 35S-labeled 1–160 CYP2E1/DHFR fusion protein. In reaction mixture without added cross-linker, only the input 36-kDa fusion protein was immunoprecipitated by antibody to CYP2E1. A companion reaction with added cross-linker yielded two major cross-linked products of ~105 and ~75 kDa, and a number of minor products in addition to the 36-kDa input fusion protein. Immunoprecipitation with specific antibodies showed that the 75-kDa band in lane 3 represents two similarly migrating cross-linked products with TOM40 and TIM44, and the 105-kDa band represents cross-linked product with mitochondrial Hsp70 (MtHsp70) from the matrix side. Results also showed that the fusion protein did not cross-link significantly with TOM20. This observation is consistent with unpublished results in our laboratory that chimeric signals of both CYP2B1 and CYP2E1 bypass interaction with TOM20 and directly associate with TOM22.2 As expected, the mutant protein 1–160 S129A/DHFR failed to cross-link significantly with any of the translocase subunits tested. Both the wild-type and S129A mutant fusion proteins showed double bands, indicating alternate translation at an immediate downstream AUG codon. These results suggest that Ser-129 phosphorylation is important for CYP2E1 interaction with the mitochondrial translocase complexes and for the mitochondrial import.
Phosphorylated CYP2E1 Binds to CyHsp70 and Mitochondrial TOM40 Proteins with Increased Affinity
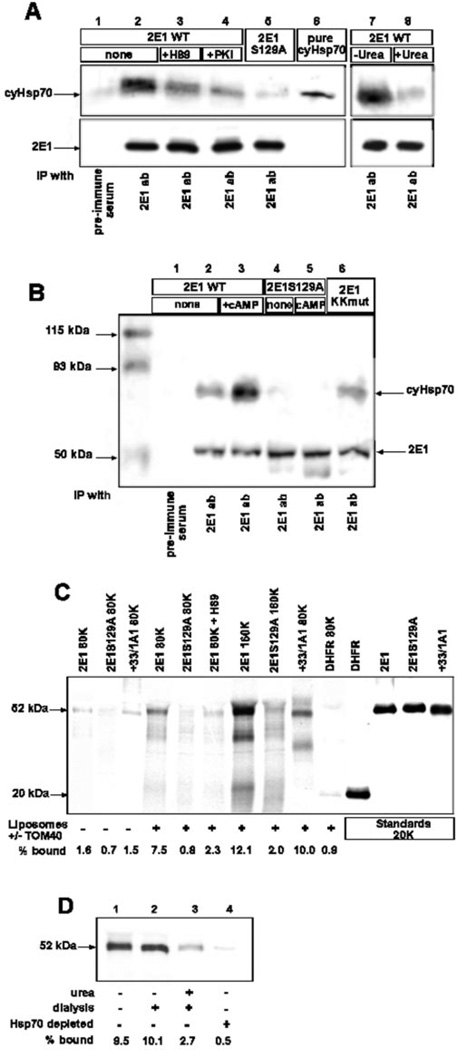
With a view to understand the mechanism of cAMP-mediated increase in mitochondrial targeting of CYP2E1, we studied two different extramitochondrial steps of protein targeting, namely binding of nascent protein to CyHsp70 and its subsequent binding to liposome reconstituted TOM40. Association with CyHsp70 was tested by co-immunoprecipitation of in vitro translated CYP2E1 with endogenous CyHsp70 present in the RRL (Fig. 6A). Interaction of CYP2E1 with CyHsp70 in vivo in transfected COS cells was tested by immunoprecipitation of post-mitochondrial protein fraction (Fig. 6B). The in vitro translation products were immunoprecipitated with CYP2E1 antibody, and subjected to immunoblot analysis with CyHsp70 antibody. As seen from Fig. 6A (lane 2), CYP2E1 antibody co-immunoprecipitated the highest amount of CyHsp70 from reaction with phosphorylated wild-type CYP2E1 (without added inhibitors). The pre-immune antibody, on the other hand, did not precipitate either of the two proteins to a significant level (lane 1). The inhibitors of PKA, H89 and PKI, added during translation markedly reduced the co-precipitation of CyHsp70 protein (lanes 3 and 4). Additionally, the S129A mutant protein also drastically reduced the level of CyHsp70 co-precipitation, suggesting that phosphorylation increases the CYP2E1 binding to CyHsp70 (lane 5). As expected, treatment with 4 m urea followed by dialysis (see lane 8) drastically reduced the binding of nascent protein to CyHsp70 as compared with control samples not treated with urea but dialyzed (lane 7). The levels of CYP2E1 proteins in these reactions were not limiting as indicated by the intensity of bands in the lower panel of Fig. 6A.
Fig. 6. Ser-129 phosphorylation increases the affinity of CYP2E1 for binding to CyHsp70 and reconstituted TOM40 proteins.
In A, wild-type and S129A mutant CYP2E1 proteins were translated and phosphorylated in RRL in the presence or absence of H89 (100 nm) or PKI (1 µm). Treatment with 4 m urea was carried out at 30 °C for 25 min, followed by dialysis over two changes of 20 mm Tris-Cl, pH 7.5, 1 mm MgCl2, 1 mm DTT, 50 mm KC1. Control samples without added urea were also incubated and dialyzed similarly, but with added 100 µm ATP. Translation products (25 µl) were immunoprecipitated (IP) either with pre-immune serum or with polyclonal antibody (ab) against CYP2E1, and the immunoprecipitates were resolved by SDS-PAGE on a 10% gel. Purified CyHsp70 (1 µg) was run alongside. The upper part of the gel (containing Hsp70) was transferred to nitrocellulose and probed with monoclonal antibody to CyHsp70. The bottom part of the gel (containing CYP2E1) was subjected to autoradiography. In B, post-mitochondrial supernatants were isolated from COS cells transfected with WT/CYP2E1, CYP2E1/129A, or CYP2El/KKmut in the absence or presence of db-cAMP (10 µm). The supernatants (1 ml) were immunoprecipitated either with pre-immune serum or with polyclonal antibody against CYP2E1 and separated by SDS-PAGE on a 12% gel. After transfer to nitrocellulose, the membrane was incubated with monoclonal antibodies against Hsp70 and CYP2E1. In C, 35S-labeled +33/CYP1A1 and DHFR proteins (80,000 cpm each) or phosphorylated CYP2E1 and CYP2E1/S129A proteins (80,000 and 160,000 cpm as indicated) were incubated with liposomes containing reconstituted TOM40 or with an equal amount of liposomes lacking TOM40. The amount of bound proteins was quantified by autoradiography. In D, binding experiments were carried out as in C using 160,000 cpm each of translation product in RRL: control untreated translation product (lane 1), translation product incubated without added urea followed by dialysis (lane 2), translation product incubated with urea followed by dialysis (lane 3), and translation mix depleted of CyHsp70 by immunoadsorption (lane 4) as described under “Experimental Procedures.” Treatment with urea and dialysis were carried out as in A.
Fig. 6B shows the pattern of co-immunoprecipitation using post-mitochondrial protein fractions of COS cells transfected with WT CYP2E1, CYP2E1/S129A, or CYP2El/KKmut in the presence or absence of added db-cAMP. Results show that CyHsp70 was co-immunoprecipitated with antibody to CYP2E1 only in cells transfected with WT CYP2E1 but not with S129A mutant CYP2E1, suggesting the importance of phosphorylation for in vivo interaction. Mutations at the N-terminal positively charged residues (KKmut) had no effect on CYP2E1 interaction with CyHsp70 protein. As expected, addition of db-cAMP markedly increased the binding further confirming the role of PKA-mediated phosphorylation for CYP2E1 binding to CyHsp70. Results in Fig. 6B also show that the levels of CYP2E1 were nearly similar in different cell extracts, ensuring that varying levels of co-immunoprecipitation of CyHsp70 were not the result of limiting CYP2E1 levels. Additionally, pre-immune antibody did not immunoprecipitate significant CYP2E1 or CyHsp70 proteins (Fig. 6B, lane 1).
Next, we assessed the extent of binding of phosphorylated and unphosphorylated CYP2E1 to liposome-reconstituted TOM40 protein. We used +33/1A1, whose mitochondrial targeting does not require phosphorylation, S129A mutant CYP2E1, and cytosolic protein DHFR as controls. As shown in Fig. 6C, the wild-type CYP2E1, S129A mutant, and +33/1A1 proteins did not bind significantly to liposomes without added TOM40 protein. The phosphorylated wild-type CYP2E1 protein bound to reconstituted TOM40 protein in a concentration-dependent manner (7.5–12%). The S129A mutant protein under these conditions bound at 0.8–2% level. The +33/1A1 protein also bound efficiently (10%) to reconstituted TOM40. As expected, DHFR, a cytosolic protein, bound at a very low level. Although these results suggest that phosphorylation increases the affinity of CYP2E1 for TOM40, use of total translation mix containing CyHsp70 leaves open the possibility that increased binding is because of conformational changes induced by the latter. This question was addressed using reaction mixture treated with urea (followed by dialysis) to disrupt the complex or translation mix depleted of CyHsp70 by immunoadsorption following urea treatment. As shown in Fig. 6D, control dialyzed reaction mixture without urea treatment (lane 2) bound to reconstituted TOM40 at approximately the same level (9–10%) as non-dialyzed and untreated control (lane 1). The translation product from urea-treated and dialyzed sample, however, showed a markedly reduced binding of —2.7% (lane 3). Furthermore, the translation mix depleted of CyHsp70 (see Fig. 7) showed no detectable binding (lane 4). These results suggest that CyHsp70 binding directly or indirectly facilitates the binding of nascent CYP2E1 to reconstituted TOM40.
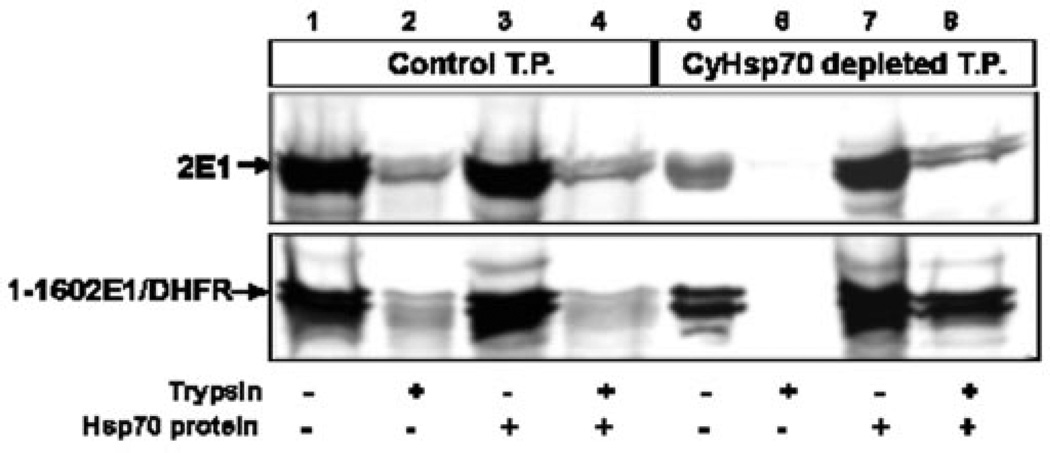
Fig. 7. Role of CyHsp70 on mitochondrial import of CYP2E1.
35S-Labeled translation products (CYP2E1 WT and 1–160 CYP2E1/DHFR) were generated in the presence of added PKA. Untreated (lanes 1–4) translation products (T.P.) or CyHsp70-depleted translation products (lanes 5–8) were used for mitochondrial import. For each sample, a companion set of mitochondria was treated with 300 µg/ml trypsin. In some cases, purified CyHsp70 was added before the import reaction (lanes 3, 4, 7, and 8).
Requirement for CyHsp70 in Mitochondrial Import of CYP2E1
The role of CyHsp70 in the mitochondrial import of CYP2E1 in vitro was directly tested by selective depletion of, and supplementation with, purified CyHsp70. Initial experiments (not presented) showed that immunodepletion of CyHsp70 drastically inhibited in vitro translation of CYP2E1 nascent protein. We therefore translated CYP2E1 and 1–160 CYP2E1/DHFR in RRL in presence of added PKA, and the RRL was then depleted of CyHsp70 by immunoadsorption. —20% of labeled CYP2E1 chains were lost during the depletion steps, which include treatment with urea, followed by dialysis and immunoadsorption. As seen in Fig. 7, in vitro translation products (CYP2E1, top panel; CYP2E1-DHFR fusion protein, bottom panel) in control and depleted-RRL were used for in vitro mitochondrial import assay. Mitochondrial binding was reduced by 50–80% (lane 5) with in vitro translation products depleted of CyHsp70, and the mitochondrial import was nearly completely abolished (lane 6). However, both binding to mitochondria (lane 7) and import (lane 8) were nearly completely restored by supplementing the reaction mixture with purified CyHsp70. However, addition of purified CyHsp70 to control RRL did not have any effect on either the binding or import of CYP2E1 and CYP2E1-DHFR proteins (lanes 3 and 4). The translation products as well as imported products in CyHsp70-deleted extracts appear as doubles, which may represent translation products with alternate start sites or minor degradation. These results further support the view that CYP2E1 nascent chain binding to CyHsp70 is important for its mitochondrial transport.
Role of Phosphorylation on the Extent of CYP2El Interaction with ER Translocator Protein Sec61 in Transfected COS Cells
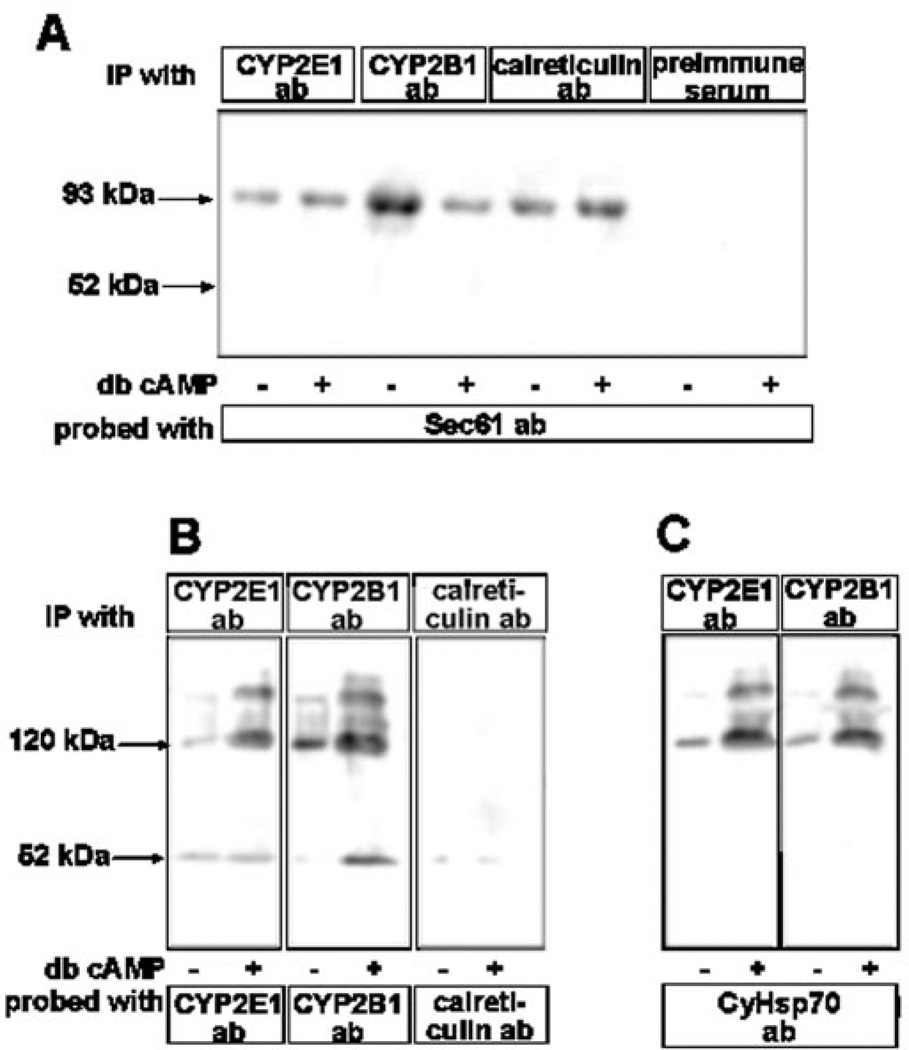
The significance of in vitro data on the binding of phosphorylated and unphosphorylated CYP2E1 nascent protein to SRP and CyHsp70 (Figs. 4 and 6) was further assessed in a whole cell setting by using chemical cross-linking. COS cells co-transfected with CYP2E1 and CYP2B1 cDNA constructs (for 8 h), and treated with or without db-cAMP, were subjected to cross-linking with membrane-permeable cross-linker MBS. The CYP proteins in association with the ER-specific protein Sec61 or the cytosolic fraction were detected by immunoprecipitation. As an initial first step of protein targeting to ER, the SRP-bound nascent chains associate with the ER translocator protein Sec61, and remain bound to this protein until polypeptide chains are completed (32). We have therefore used association with Sec61 as a criterion for ER association. As shown in Fig. 6A, the microsomal proteins from cells incubated with cross-linker were subjected to immunoprecipitation with various antibodies and the immunoprecipitates were further probed with Sec61 antibody by immunoblot analysis. Results show that a significant amounts of CYP2E1, CYP2B1, and calreticulin exist in association with Sec61, as indicated by the level of ~93-kDa cross-linked product. Although not shown, immunoblot analysis failed to detect 52-kDa CYP2E1 and CYP2B1 proteins in this fraction. Results also show that cAMP did not affect the level of CYP2E1 cross-linking with Sec61, whereas this treatment markedly reduced the level of CYP2B1 cross-linking with Sec61. The latter results are consistent with our previous observation that PKA-mediated phosphorylation reduces the level of microsomal targeting by way of reduced affinity of the apoprotein for SRP (10). Finally, treatment with cAMP marginally increased the level of cross-linked product with calreticulin, which is likely the result of increased translation rates. As expected, pre-immune serum did not yield any antibody reactive protein species.
The steady state levels of proteins in the soluble fraction of cAMP-treated and untreated control cells were assessed by immunoprecipitation followed by immunoblot analysis (Fig. 6B). In the case of CYP2E1 and CYP2B1 proteins, most of the antibody reactive proteins were detected in higher molecular forms of~ 120–210 kDa. Low levels of 52-kDa species were also detected in both cases. As expected of a true ER-targeted protein, no significant calreticulin was detected in the soluble cytoplasmic fraction. Results in Fig. 6B also show that the level of both CYP2E1 and CYP2B1 in the putative cross-linked species (120–210 kDa) markedly increased in cells treated with cAMP. Because the in vitro studies in Fig. 6 showed that phosphorylated CYP2E1 binds to CyHsp70 with increased efficiency, we reprobed the blots with Hsp70 antibody. Immunoblot in Fig. 6C shows that the slow migrating complexes indeed represent CYP2E1 and CYP2B1 cross-linked to CyHsp70 protein. The nature of the ~210-kDa species remains unknown, although it might represent a dimer of CYP2El-CyHsp70 cross-linked product. These results provide confirmatory evidence that phosphorylation does not alter the rate of CYP2E1 targeting to the ER, but significantly increases the CyHsp70-bound pool in the cytosol.
DISCUSSION
A recent study from our laboratory showed that hepatic mitochondrial P450MT5 from pyrazole-treated rats exhibits primary sequence identical to that of similarly induced microsomal CYP2E1, except that it has a higher level of Ser-129 phosphorylation (12). In support of these results, here we show that Ser-129 phosphorylated CYP2E1 protein is efficiently targeted to mitochondrial compartment both in vitro and in vivo. It was recently shown that the N-terminal 29 amino acid residues, the putative transmembrane anchor domain of CYP2E1, contain the signal for ER targeting although it is inefficient for retaining the protein in the ER (26). This study demonstrates that the sequence region between residues 21 and 31 also contains a cryptic signal for mitochondrial targeting, which is activated by phosphorylation at Ser-129. In this study, we show that Ser-129-phosphorylated CYP2E1 with intact N terminus is efficiently targeted to mitochondrial compartment both in vitro and in vivo. We demonstrate that the sequence region between 21 and 31 also contains a cryptic mitochondrial targeting signal, which is activated by phosphorylation at Ser-129. Previously we showed that cAMP-mediated phosphorylation markedly increases the efficiency of CYP2B1 targeting to the mitochondrial compartment (10). In extension of these results, we now show that phosphorylation at Ser-129 markedly increases the extent of CYP2E1 protein binding to cytoplasmic chaperone Hsp70 under both in vitro and in vivo conditions, which in turn increases the affinity of the nascent protein for mitochondrial translocase TOM40. It is likely that CyHsp70 binding induces a conformational change in CYP2E1 protein, thus facilitating its interaction with TOM40 and other mitochondrial translocases. The phosphorylation-mediated signal activation, therefore, appears to be related to pre-translocation steps of chaperone binding and presentation to translocase complexes.
CYP2E1 is believed to play important roles in modulating oxidative stress-related cellular pathologies and toxicity. Its expression at both mRNA and protein levels is induced under various pathophysiological conditions, including diabetes, obesity, starvation (13), and alcohol ingestion (33). CYP2E1 metabolizes endogenous compounds of physiological importance, such as lipid hydroperoxides and ketone bodies (34, 35), in addition to various exogenous chemicals (36, 37). Recent studies show that CYP2E1 has a direct role in the production of reactive O2 species, induction of oxidative stress, and apoptosis (38, 14). Because of the expanding role of mitochondrial genetic and membrane systems in diverse metabolic processes, integration and execution of apoptotic signal (39), and generation of stress signaling that affects nuclear gene expression (40, 41), it is important to understand the mechanism of CYP2E1 targeting and its regulation.
The present study demonstrates that the two positively charged residues at positions 24 and 25 are critical for mitochondrial targeting. The chimeric N-terminal signal (residues 1–31) of CYP2E1 appears to resemble the bimodal targeting signal of CYP2B1 with subtle differences. A recent study using transient transfection of CYP2E1 lacking the N-terminal 1–29 residues of the protein concluded that the mitochondrially targeted component is a 40-kDa soluble protein, which is generated by the MPP-mediated cleavage at an undetermined N-terminal site (42, 43). Using a full-length protein, however, we were unable to demonstrate any detectable processing either under conditions when MPP activity was induced by Mg2+ and Mn2+ or by incubation with purified yeast MPP protein (results not presented). Furthermore, in our hands any deletion beyond the positively charged residue at position 25 abolished mitochondrial targeting under both in vivo and in vitro conditions. The physiological significance of the earlier study (43) using a truncated protein that lacked the critical components of the mitochondrial targeting signal remains unclear.
Protein phosphorylation plays a critical role in many cellular processes, including signal amplification, modulation of structure/function of proteins, and protein ubiquitination (44). In extension of our recent observation with CYP2B1, the present study demonstrates yet another function for protein phosphorylation in the activation of an otherwise inactive mitochondrial targeting signal. Specifically, our results show that phosphorylation at an internal site results in increased affinity of nascent CYP2E1 for binding to CyHsp70 in vitro as well as in vivo, and for a major mitochondrial outer membrane translocase, TOM40. In this respect, our present and previous studies (10) together provide an important insight into a novel cAMP-dependent regulation of mitochondrial biogenesis. A notable difference from the previously published results with CYP2B1 is that phosphorylation at Ser-129 did not alter the efficiency of nascent CYP2E1 protein binding to SRP. In contrast, the phosphorylated nascent CYP2B1 showed a markedly lower affinity for binding to SRP. We believe that phosphorylation of CYP2E1 translated on free ribosomes for mitochondrial targeting is a distinct event from the phosphorylation of microsome-associated CYP2E1, which has been implicated in the regulation of catalytic function of the microsomal CYP (45) and its ubiquitination (46).
Another major difference between the bimodal targeting of CYP2E1, CYP2B1, and CYP1A1 is their mode of translation. Our results show that in vitro translation of CYP2B1 is nearly completely inhibited by saturating levels of SRP, whereas the translation of CYP2E1 was inhibited by only ~40%. Inhibition of CYP1A1 followed a pattern intermediary to these two proteins (9).3 Extent of membrane insertion as tested by alkaline Na2CO3 extraction and binding to SRP by chemical cross-linking essentially follow this pattern. CYP2E1 showed a generally low affinity for binding to SRP, as indicated by cross-linking, and a large fraction (>50%) of CYP2E1 was not inserted into added ER membrane, further suggesting that part of the nascent chains is translated in membrane-free form. On the other hand, under low cAMP conditions, CYP2B1 was translated as a bona fide ER-targeted protein (10), with a markedly higher efficiency for binding to SRP and >90% of nascent chains associating with ER. CYP1A1 behaved in an intermediary manner in that ~25–30% of the nascent chains escaped ER targeting irrespective of the phosphorylation status (9). The contrasting efficiencies of phosphorylated CYP2E1 and CYP2B1 for binding to SRP is further supported by cross-linking studies under in vivo conditions (Fig. 8), which show no change in interaction of CYP2E1 with Sec61 under induced cAMP conditions, when interaction of CYP2B1 with Sec61 is markedly decreased. A comparison of the structural properties of sequence 1–36, representing the chimeric signals of those three CYP proteins is presented in Table I. The PROT-SCALE-based analysis shows that the chimeric signal of CYP2E1 is least hydrophobic and has least α-helical content, and is conformationally most unstable as compared with that of CYP2B1, which is at the other end of the spectrum with respect to these properties. CYP1A1 exhibits intermediary properties.
Fig. 8. Effects of cAMP on the extent of interaction of CYP2E1 with Sec61 and CyHsp70 proteins in intact cells.
COS cells were co-transfected with CYP2E1 and CYP2B1 cDNA constructs and incubated for 8 h in the absence or presence of added db-cAMP (10 µm). Cells were then incubated for 1 h with 500 µm membrane-permeable cross-linker; MBS and subcellular fractions were isolated as described under “Experimental Procedures.” In A, microsomal proteins (500 µg each) were immunoprecipitated (IP) with indicated antibodies (ab) and the immunoprecipitates were probed with antibody to Sec61 protein by immunoblot analysis. In B, cytosolic proteins (500 µg each) were immunoprecipitated with antibodies to CYP2E1, CYP2B1, or calreticulin and the immunoprecipitates were probed the same antibody by immunoblot analysis. In C, immunoprecipitates, as in B, were probed with antibody to CyHsp70. Details of transfection, chemical cross-linking, cell fractionation, and immunoprecipitation were as described under “Experimental Procedures.”
Table I. Comparison of the N’-terminal regions of CYPs that contain chimeric signals for bimodal targeting.
The N-terminal amino acid 1–36 sequence regions were analyzed using ExPaSy (Expert Protein Analysis System) molecular biology server of the Swiss Institute of Bioinformatics. The number of + indicates the degree of α-helicity or hydrophobicity.
| Signal parameters | CYP1A1 | CYP2B1 | CYP2E1 |
|---|---|---|---|
| α-Helicity | + + | + + + | + |
| Hydrophobicity | + + + | + + + + + | + + |
| Stability | Stable | Very stable | Unstable |
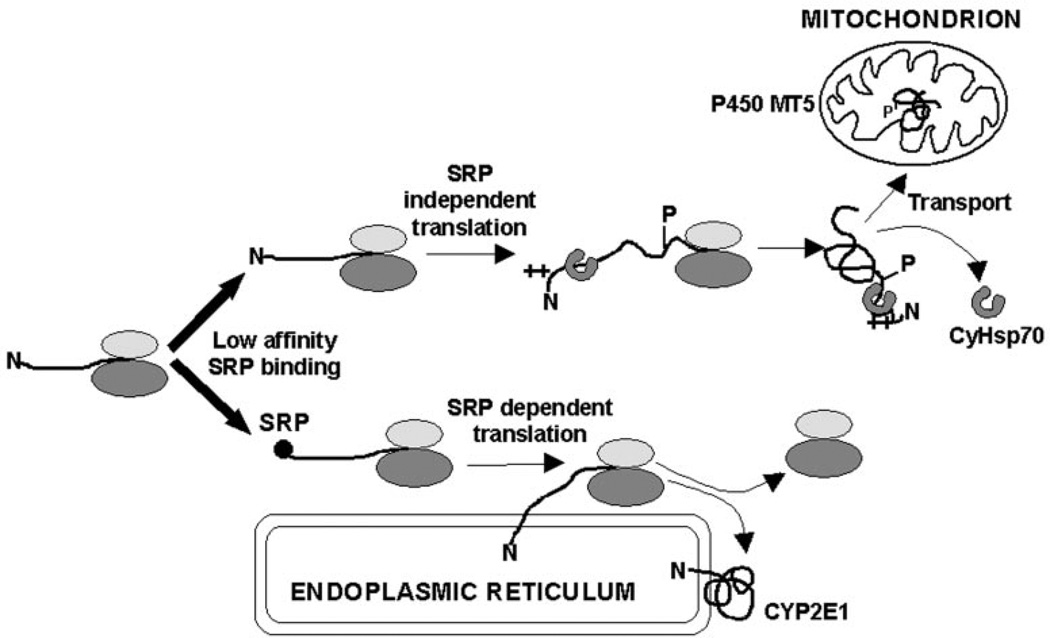
A common property among the three CYPs (2E1, 2B1, and 1A1) with chimeric signals is their inability to undergo elongation arrest during in vitro translation in the presence of added SRP (10, 47). Elongation arrest is thought to increase the translocation efficiency of nascent chains possibly by extending the time of its interaction with the translocation machinery (2, 29). In the case of CYP2B1, a high affinity of the ER targeting signal for SRP under non-phosphorylated state probably enables its efficient targeting to the ER. In the case of CYP1A1 and CYP2E1, the relatively low efficiency of the N-terminal signal for SRP binding, coupled with inability to undergo elongation arrest may be the reason why ~25–50% of the nascent chains escape ER targeting and undergo translation in a membrane-free state. Phosphorylated CYP2B1, on the other hand, behaves similarly to the chimeric signal of CYP2E1, in that it shows lower affinity for SRP binding, thus showing higher propensity for translation in a membrane-free form. Consistent with this, results of in vitro and in vivo studies together show that phosphorylated CYP2E1 binds to CyHsp70 with increased efficiency, which in turn increases the efficiency with which the nascent protein binds to TOM40. Based on these observations, we propose a model for mitochondrial CYP2E1 targeting (Fig. 9), which implies that lower affinity for SRP binding under both high cAMP or low cAMP conditions causes >50% of the nascent chains to escape ER targeting and undergo translation as membrane free protein. cAMP-dependent phosphorylation at Ser-129 of the protein results in the activation of cryptic mitochondrial signal, which includes more efficient association with cytoplasmic chaperones and more efficient binding to mitochondrial translocases. We postulate that these latter two events enable preferential targeting of complete, unprocessed, but phosphorylated apoprotein to mitochondria (Fig. 8). In variance from the mechanism described for CYP2B1, cAMP regulates only the mitochondrial targeting but not the ER targeting of CYP2E1.
Fig. 9. A model for the biogenesis of microsomal CYP2E1 and mitochondrial MT5.
In summary, we provide confirmatory evidence for a novel class of chimeric signals with dual targeting property. Results also provide valuable new insight as to how cellular cAMP levels regulate the biogenesis of mitochondrial enzymes associated with drug and alcohol toxicity by modulating the rate of mitochondrial targeting of CYP2E1 and also other CYPs.
Acknowledgments
We thank Drs. Reid Gilmore and David Andrews for generously providing some of the reagents and antibodies used in this study. We are thankful to Haider Raza and members of the Avadhani laboratory for comments and suggestions on the manuscript and to Molly Higgins for editorial help on the manuscript.
Footnotes
This work was supported in part by National Institutes of Health Grant GM-34883 (to N. G. A.) and GM-57067 (to D. P.).
The abbreviations used are: ER, endoplasmic reticulum; CYP, cytochrome P450; db-cAMP, dibutyryl cAMP; DHFR, dihydrofolate reductase; DTT, dithiothreitol; Hsp, heat shock protein; CyHsp70, cytoplasmic Hsp70; MtHsp70, mitochondrial Hsp70; MBS, m-maleimidobenzoyl-N-hydroxysuccinimidate ester; S-MBS, m-maleimidobenzoyl-N-hydroxysulfosuccinimidate ester; MOPS, 4-morpholinepropanesulfonic acid; MPP, matrix processing peptidase; PPL, preprolactin; PKA, protein kinase A; PKI, protein kinase A inhibitor myristoylated peptide; RRL, rabbit reticulocyte lysate; SRP, signal recognition particle; TIM, translocase of the inner membrane; TOM, translocase of the outer membrane; WGL, wheat germ lysate; WT, wild-type.
H. K. Anandatheerthavarada, M.-A. Robin, G. Biswas, N. B. V. Sepuri, D. M. Gordon, D. Pain, and N. G. Avadhani, unpublished results.
M.-A. Robin, H. K. Anandatheerthavarada, G. Biswas, N. B. V. Sepuri, D. M. Gordon, D. Pain, and N. G. Avadhani, unpublished results.
REFERENCES
- 1.Schatz G, Dobberstein B. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- 2.Keenan RJ, Freymann DM, Stroud RM, Walter P. Annu. Rev. Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- 3.Lithgow T. FEES Lett. 2000;476:22–26. doi: 10.1016/s0014-5793(00)01663-x. [DOI] [PubMed] [Google Scholar]
- 4.Fujiki M, Verner K. J. Biol. Chem. 1993;268:1914–1920. [PubMed] [Google Scholar]
- 5.Neupert W. Annu. Rev. Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- 6.Schneider HC, Berthold J, Bauer MF, Dietmeier K, Guiard B, Brunner M, Neupert W. Nature. 1994;371:768–777. doi: 10.1038/371768a0. [DOI] [PubMed] [Google Scholar]
- 7.Smith MD, Schnell DJ. Cell. 2001;105:293–296. doi: 10.1016/s0092-8674(01)00337-3. [DOI] [PubMed] [Google Scholar]
- 8.Subramani S, Roller A, Snyder WB. Annu. Rev. Biochem. 2000;69:399–418. doi: 10.1146/annurev.biochem.69.1.399. [DOI] [PubMed] [Google Scholar]
- 9.Addya S, Anandatheerthavarada HK, Biswas G, Bhagwhat SV, Mullick J, Avadhani NG. J. Cell Biol. 1997;139:589–599. doi: 10.1083/jcb.139.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anandatheerthavarada HK, Biswas G, Mullick J, Sepuri NBV, Otvos L, Pain D, Avadhani NG. EMBO J. 1999;18:5494–5504. doi: 10.1093/emboj/18.20.5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sass E, Blachinsky E, Rarniely S, Pines O. J. Biol. Chem. 2001;276:46111–46117. doi: 10.1074/jbc.M106061200. [DOI] [PubMed] [Google Scholar]
- 12.Robin M-A, Anandatheerthavarada HK, Fang JK, Cudic M, Otvos L, Avadhani NG. J. Biol. Chem. 2001;276:24680–24689. doi: 10.1074/jbc.M100363200. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka E, Terada M, Misawa S. J. Clin. Pharmacol. Ther. 2000;25:165–175. doi: 10.1046/j.1365-2710.2000.00282.x. [DOI] [PubMed] [Google Scholar]
- 14.Wu D, Cederbaum AI. Alcohol Clin. Exp. Res. 1999;23:67–76. [PubMed] [Google Scholar]
- 15.Monier S, Van Luc P, Kreibich G, Sabatini DD, Adesnik M. J. Cell Biol. 1988;107:457–470. doi: 10.1083/jcb.107.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szczesna-Skorupa E, Remper B. Methods Enzymol. 1991;206:64–75. doi: 10.1016/0076-6879(91)06077-g. [DOI] [PubMed] [Google Scholar]
- 17.Song B-J, Gelboin HV, Park S-S, Yang CS, Gonzalez FJ. J. Biol. Chem. 1986;261:16689–16697. [PubMed] [Google Scholar]
- 18.Roch JA, Waxman DJ. Methods Enzymol. 1991;206:305–315. doi: 10.1016/0076-6879(91)06100-h. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli UR. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Towbin H, Staehelin T, Gordon J. Proc. Natl. Acad. Sci. U. S. A. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laird V, High S. J. Biol. Chem. 1997;272:1983–1989. doi: 10.1074/jbc.272.3.1983. [DOI] [PubMed] [Google Scholar]
- 22.Greenfield JJA, High S. J. Cell Sci. 1999;112:1477–1486. doi: 10.1242/jcs.112.10.1477. [DOI] [PubMed] [Google Scholar]
- 23.Chuck SL, Lingappa VR. Cell. 1992;68:9–21. doi: 10.1016/0092-8674(92)90202-n. [DOI] [PubMed] [Google Scholar]
- 24.Gordon DM, Wang J, Amutha B, Pain D. Biochem. J. 2001;356:207–215. doi: 10.1042/0264-6021:3560207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hinnah SC, Hill K, Wagner R, Schlicher T, Soll J. EMBO J. 1997;16:7351–7360. doi: 10.1093/emboj/16.24.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hill R, Model K, Ryan MT, Dietmeier K, Martin F, Wagner R, Pfanner N. Nature. 1998;395:516–521. doi: 10.1038/26780. [DOI] [PubMed] [Google Scholar]
- 27.Szczesna-Skorupa E, Chen CD, Remper B. Arch. Biochem. Biophys. 2000;374:128–136. doi: 10.1006/abbi.1999.1628. [DOI] [PubMed] [Google Scholar]
- 28.Eliasson E, Mkrtchian S, Ingelman-Sundberg M. J. Biol. Chem. 1992;267:15765–15769. [PubMed] [Google Scholar]
- 29.Pyerin W, Tanaguchi H. EMBO J. 1989;8:3003–3010. doi: 10.1002/j.1460-2075.1989.tb08450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belin D, Bost S, Vassalli JD, Strub K. EMBO J. 1996;15:468–478. [PMC free article] [PubMed] [Google Scholar]
- 31.Mason N, Ciufo LF, Brown JD. EMBO J. 2000;19:4164–4174. doi: 10.1093/emboj/19.15.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song W, Raden D, Mandon EC, Gilmore R. Cell. 2000;100:333–343. doi: 10.1016/s0092-8674(00)80669-8. [DOI] [PubMed] [Google Scholar]
- 33.Dupont I, Bodenez P, Berthou F, Simon B, Bardou LG, Lucas D. Alcohol Alcoholism. 2000;35:98–103. doi: 10.1093/alcalc/35.1.98. [DOI] [PubMed] [Google Scholar]
- 34.Coons MJ, Vaz AD, Bestervelt LL. FASEB J. 1996;10:428–434. doi: 10.1096/fasebj.10.4.8647341. [DOI] [PubMed] [Google Scholar]
- 35.Roop DR, Coon MJ. Alcohol Clin. Exp. Res. 1986;10:S44–S49. doi: 10.1111/j.1530-0277.1986.tb05179.x. [DOI] [PubMed] [Google Scholar]
- 36.Guengerich FP, Shimada T. Chem. Res. Toxicol. 1991;4:391–407. doi: 10.1021/tx00022a001. [DOI] [PubMed] [Google Scholar]
- 37.Gonzalez FJ, Gelboin FJ. Drug Metab. Rev. 1994;26:165–183. doi: 10.3109/03602539409029789. [DOI] [PubMed] [Google Scholar]
- 38.Fataccioli V, Andraud E, Gentil M, French SM, Rouach H. Hepatology. 1999;29:14–20. doi: 10.1002/hep.510290106. [DOI] [PubMed] [Google Scholar]
- 39.Green DR, Reed JC. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 40.Biswas G, Adebanjo OA, Freedman BD, Anandatheerthavarada HK, Vijayasarathy C, Zaidi M, Rotlikoff M, Avadhani NG. EMBO J. 1999;18:522–533. doi: 10.1093/emboj/18.3.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amuthan G, Biswas G, Zhang SY, Rlein-Szanto A, Vijayasarathy C, Avadhani NG. EMBO J. 2001;20:1910–1920. doi: 10.1093/emboj/20.8.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neve EP, Ingelman-Sundberg M. FEBS Lett. 1999;460:309–314. doi: 10.1016/s0014-5793(99)01361-7. [DOI] [PubMed] [Google Scholar]
- 43.Neve EP, Ingelman-Sundberg M. J. Biol. Chem. 2001;276:11317–11322. doi: 10.1074/jbc.M008640200. [DOI] [PubMed] [Google Scholar]
- 44.Hunter T. Cell. 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- 45.Oesch-Bartlomowicz B, Padma PR, Becker R, Richter B, Hengstler JG, Freeman JE, Wolf CR, Oesch F. Exp. Cell Res. 1998;242:294–302. doi: 10.1006/excr.1998.4120. [DOI] [PubMed] [Google Scholar]
- 46.Banerjee A, Rocarek TA, Novak RF. Drug Metab. Disp. 2000;28:118–124. [PubMed] [Google Scholar]
- 47.Sakaguchi M, Mihara R, Sato R. Proc. Natl. Acad. Sci. U. S. A. 1984;81:3361–3364. doi: 10.1073/pnas.81.11.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]