Abstract
Cleavage and polyadenylation (C/P) of nascent transcripts is essential for maturation of the 3′ ends of most eukaryotic mRNAs. Over the past three decades, biochemical studies have elucidated the machinery responsible for the seemingly simple C/P reaction. Recent genomic analyses have indicated that most eukaryotic genes have multiple cleavage and polyadenylation sites (pAs), leading to transcript isoforms with different coding potentials and/or variable 3′ untranslated regions (UTRs). As such, alternative cleavage and polyadenylation (APA) is an important layer of gene regulation impacting mRNA metabolism. Here, we review our current understanding of APA and recent progress in this field.
APA is widespread in eukaryotes
The mechanism of 3′ end processing of nascent transcripts in eukaryotic cells is determined by the type of RNA polymerase used for transcription and specific signals embedded in the transcript (reviewed in [1]). Except for most replication-dependent histone mRNAs in metazoans, and some protozoans (reviewed in [2]), all pre-mRNAs are processed by C/P, which involves endonucleolytic cleavage of the nascent RNA and synthesis of a poly(A) tail (reviewed in [3]), and is a necessary prelude to subsequent transcription termination. The site of C/P, also known as the poly(A) site or simply pA, is defined by surrounding cis elements (Box 1). Mutations affecting pA usage have been implicated in several human diseases (reviewed in [4]), such as thrombophilia and some thalassemias, underscoring the importance of 3′ end processing in gene expression and its relevance to human health.
Box 1. Core cis elements for cleavage and polyadenylation.
Cleavage and polyadenylation is controlled by cis elements located upstream and downstream near the pA. In metazoans, upstream elements include the hexamers AAUAAA/AUUAAA or other close variants, commonly referred to as the polyadenylation signal (PAS), U-rich elements, and UGUA elements. Downstream elements include U-rich elements and GU-rich elements. In addition, distal downstream G-rich elements can be frequently found for mammalian pAs. Although variants of A[A/U]UAAA have significantly lower activities than A[A/U]UAAA in C/P [94], they are prevalent. In the human genome, about half of the pAs have AAUAAA, ~16% have AUUAAA, ~20% have single nucleotide variants of A[A/U]UAAA, and ~10% of pAs do not have a recognizable AAUAAA-like sequence (at least in the −40 to −1 nt region relative to the pA). Importantly, the type of PAS varies with pA location, with the 3′-most pA having the highest frequency of AAUAAA [50]. This difference in PAS supports the notion that the 3′-most pAs are typically strong, ensuring proper termination of transcription, and upstream pAs are weak, allowing regulation. Variation in placement and occurrence also applies to other cis elements around the pA [89]. Statistical analysis suggests that the pA strength is defined by surrounding cis elements in a combinatorial manner [95]. Consistent with this, a functional pA requires only an A-rich upstream sequence and strong U-rich elements, and pAs of this type are more regulated across tissues [96].
Recent genomic studies have uncovered widespread occurrences of APA in metazoan protein-coding transcripts (Box 2): 70–79% of mammalian genes [5,6] and about half of the genes in flies [7], worms [8], and zebrafish [9] have been reported to display APA. Interestingly, large fractions of yeast and plant genes have also been reported to show APA (Box 2). Although the exact statistics can vary widely due to differences in experimental conditions and bioinformatic methods used to identify pAs (Box 2), it is clear that eukaryotic genes express isoforms that have significant differences in the 3′ end portion of the transcript. Here, we review our current understanding of the biochemistry and genomics of APA. We focus mostly on metazoans, and offer perspectives on future directions. Some aspects of APA have also been discussed in several recent reviews [10–12], to which readers are referred for full appreciation of this rapidly growing subject.
Box 2. Mapping pAs in genomes.
pAs are mapped by aligning cDNA/EST sequences to the genome. An unaligned poly(A) sequence immediately downstream of the last aligned position is considered to be derived from the poly(A) tail. Recent advances in deep sequencing technologies have enabled systematic mapping of pAs and quantitative analysis of APA. Several methods focusing on 3′ end sequencing have been developed in the past 2 years, which largely fall into three major categories: (i) oligo(dT)-based cDNA synthesis and sequencing, such as PAS-seq, PASAS, and PolyA-seq [5,42,49], which is essentially the same as traditional cDNA/EST sequencing; (ii) capture of 3′ end fragments by RNA ligation followed by cDNA sequencing, such as 3P-seq and 3′ READS [6,8]; and (iii) direct RNA sequencing (DRS) of the 3′ end region of transcript [97]. Like traditional cDNAs/ESTs, approach (i) is affected by internal priming; the oligo(dT) can prime at internal A-rich sequences during cDNA synthesis, despite the ease of the protocol. Approach (ii) eliminates the internal priming issue because oligo(dT) is not used for cDNA synthesis. However, the method can be complex and RNA ligation bias can affect quantitative comparison of different pA isoforms, if not properly controlled. Approach (iii) is the simplest but requires the Helicos sequencer, which is generally not accessible. In principle, it can still have the internal priming issue, but it has been reported to have few false positives [97,98].
Using deep sequencing methods, studies have found a large fraction of mRNA genes in metazoans have APA: 70–79% in mammals [5,6]; 55% in zebrafish [9]; 54% in Drosophila[7]; 30–40% in Caenorhabditis elegans[8,99,100]. Interestingly, 70–75% of mRNA genes in A. thaliana have been found to have multiple pAs [98,101], but the occurrence of intronic pAs is very low [98]. Surprisingly, a large fraction of yeast mRNA genes have also been found to have alternative pAs, although the statistics differ widely (43% [102] vs 70% [97]). One source of variation in statistics is clustering of closely spaced pAs. The cleavage reaction is typically not precise, presumably due to imprecise and overlapping placement of cis elements for C/P (Box 1), which leads to multiple pAs within a short window; for example, 24 nt for mammalian pAs [50]. These heterogeneous cleavage sites need to be clustered for analysis of APA.
APA impacts the cellular transcriptome and proteome
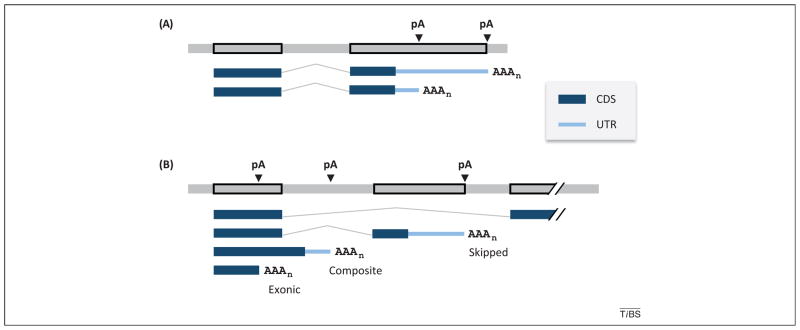
A given gene can encode transcripts with multiple pAs located in different regions (Figure 1). With respect to the impact on protein-coding, alternative pAs in the 3′-most exon typically leads to variable 3′ UTRs, whereas pAs in upstream introns and exons cause both coding sequence (CDS) and 3′ UTR changes. Alternative pAs in introns can be further divided into two subtypes depending on the configuration of the terminal exon in which they reside: namely, skipped terminal exons (a whole exon is skipped due to splicing), and composite terminal exons (partially internal and partially terminal) (Figure 1). A recent study of the mouse transcriptome indicated that about 68% of mRNA transcripts have APA in the 3′-most exon and 42% have pAs in upstream regions (31% have both types of pA) [6]. The diverse APA configurations have been shown to impact gene expression at different levels.
Figure 1.
Alternative cleavage and polyadenylation sites (pAs) in a gene. (A) Alternative cleavage and polyadenylation (APA) in 3′-most exon. A hypothetical gene is shown, with two pAs located in the 3′-most exon. The top gray line is genomic DNA with exons boxed, and bottom lines are mRNAs. Coding sequence (CDS) and 3′ untranslated region (UTR) are shown as thick and thin blue lines, respectively, as indicated in the graph, splicing as bent line, and pAs as arrowheads. AAAn indicates the poly(A) tail. (B) APA in upstream regions. The type of terminal exon is indicated. The top mRNA shows only splicing. Skipped, skipped terminal exon; composite, composite (internal/terminal) exon; exonic, upstream exon.
Regulation of mRNA metabolism via alternative 3′ UTRs
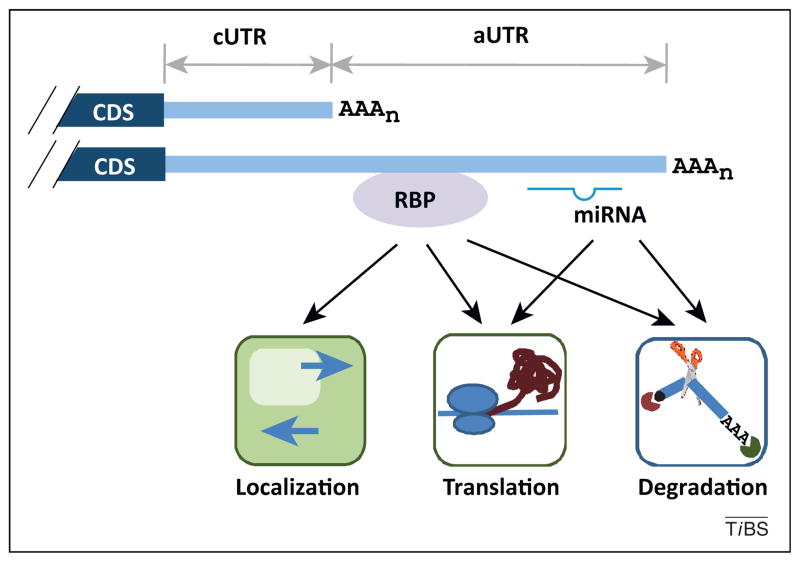
For mouse genes with APA, the shortest and longest 3′ UTRs of a given gene can differ significantly in length, with the median length being 249 and 1,773 nucleotides (nt), respectively [6]. For simplicity, we refer to the region in the 3′ UTR before the proximal pA the constitutive UTR or cUTR, and the region after that, the alternative UTR or aUTR (Figure 2). The 3′ UTR, via its embedded cis elements, plays a key role in regulating mRNA metabolism including its localization, stability, and translation; therefore, APA isoforms with different 3′ UTR lengths have distinct properties.
Figure 2.
Regulation of cis elements in 3′ untranslated regions (UTRs) by alternative cleavage and polyadenylation (APA). Two mRNA isoforms are shown. The 3′ UTR region upstream of the proximal cleavage and polyadenylation site (pA) is called the constitutive UTR (cUTR), and the downstream region is called the alternative UTR (aUTR). RNA-binding protein (RBP) and miRNA targeting to the aUTR are shown. Impacts on mRNA localization, translation, and degradation are indicated. CDS, coding sequence.
Metazoan 3′ UTRs are frequently targeted by miRNAs; a type of small RNA (~22 nt) that modulates mRNA stability and/or translation of target transcripts [13,14]. Bioinformatic surveys have indicated that, for mammalian genes expressing alternative 3′ UTR isoforms, more than half of conserved miRNA target sites are located in aUTRs [15,16]. Evasion of miRNA targeting by short mRNAs, that is, those without aUTRs, can have significant biological implications, particularly for genes having regulatory roles. For example, cells that express short mRNA isoforms of the proto-oncogenes cyclin D2 (CCND2) and insulin-like growth factor 2 mRNA binding protein 1 (IGF2BP1) that lack let-7 and/or miR-15/16 target sites are more proliferative and oncogenic, respectively, than those that express long mRNA isoforms [17]. Another important example is that differential expression of short isoforms of the myogenic regulator Pax3 (paired box 3) that lack miR-206 target sites leads to variation of muscle stem cells in different muscle types [18].
Box 3. Core protein factors for C/P.
The C/P machinery is composed of 15–20 core polypeptides, including four protein complexes and several single proteins. The protein complexes include CPSF, containing CPSF-160, CPSF-100, CPSF-73, CPSF-30, and hFip1 (human Fip1); CstF, containing CstF-77, CstF-64, and CstF-50; CFI, containing CFI-68/CFI-59 and CFI-25; and CFII, containing Pcf11 and Clp1. Single proteins include Symplekin, PAP, PABPN, and RNAP II. A proteomic study of the human C/P machinery has identified homologs of several yeast C/P factors, such as WDR33, RBBP6 (retinoblastoma binding protein 6), PP1 (protein phosphatase 1) α and β [103]. WDR33 has been shown to be a subunit of CPSF. In addition, mammals have a paralog of CstF-64, τCstF-64, which is also found in the C/P machinery, suggesting its general role in C/P. Extensive protein–protein interactions have been found between C/P factors (reviewed in [104]). Both CFI and CstF are dimers in the complex. Many C/P factors have RNA-binding activities. CPSF-73 is the endonuclease that cleaves pre-mRNA; CPSF-160 interacts with the PAS sequence, such as A[A/U]UAAA; Fip1 and CPSF-30 bind preferentially to U-rich sequences; CFI binds the UGUA element; and CstF-64/τCstF-64 interact with U/GU-rich elements. Some proteins that do not bind RNA have scaffolding functions, such as Symplekin and CstF-77. The carboxy terminal domain (CTD) of RNAP II, with a large size and dynamic phosphorylation patterns, is believed to provide a platform with which other factors can associate with before or during C/P. The C/P machinery purified from human cells has a sedimentation coefficient of ~50S and shows a distinct ‘kidney’ shape as imaged by electron microscopy [103].
The 3′ UTRs also contain binding sites for a large group of RNA-binding proteins (RBPs) that regulate mRNA stability, including AU-rich elements (AREs), GU-rich elements (GREs), Puf (Pumilio and FBF) protein binding elements (reviewed in [19]), and structural elements [20]. Accordingly, APA isoforms having different RBP-binding sites can have variable stabilities. A case in point is the recent finding that a genetic polymorphism leading to differential expression of two APA isoforms of human interferon regulatory factor (IRF)5 mRNA is linked to risk of systemic lupus erythematosus [21]. Because of the presence of an ARE in the aUTR, the two isoforms have different decay rates, leading to different amounts of protein products [21]. It is notable that aUTRs are generally more AU-rich than cUTRs [16], making it possible that differences in mRNA stability between APA isoforms is a general phenomenon.
APA isoforms with different 3′ UTRs have also been shown to have different localization potentials. For example, whereas the brain-derived neurotropic factor (BDNF) short mRNA form is restricted to the neuronal soma, the long form is capable of localizing to the dendrites, where it is locally translated [22]; similarly, long and short isoforms of both myo-inositol monophosphatase-1 (IMPA1) [23] and RAN binding protein 1 (RANBP1) [24] mRNAs are localized to the axon and soma, respectively. Notably, genes in general tend to be expressed as long 3′ UTR isoforms in neurons [25], making it plausible that differential localization of APA isoforms is a widespread mechanism in these cells. Interestingly, a recent study of mRNA localization mediated by muscleblind-like (Mbnl) RNA-binding proteins in C2C12 mouse myoblasts indicated that APA isoforms in these nonpolarized cells can be differentially localized[26]. Knockdown of Mbnl causes a change in the distribution of mRNAs between cytosolic and insoluble fractions, and mRNAs with Mbnl binding sites in aUTRs are more affected by the knockdown, suggesting at least some differential localization is mediated by Mbnl. Similarly, differences in nuclear retention between APA isoforms have also been reported for transcripts containing RNA structures formed by inverted Alu sequences in aUTRs [27]. Interestingly, this difference in mRNA localization exists in HeLa cells but not in embryonic stem (ES) cells.
Effect on protein isoforms
Most intronic pAs are situated between coding exons, leading to isoforms encoding different proteins [6]. One classic case is the genes encoding immunoglobulin heavy chain M (IgM) proteins [28]. IgM mRNAs switch from using a distal pA to a proximal pA during activation of B cells, which results in a shift in protein production from a membrane-bound form to a secreted form. A genomic survey has found that the membrane-anchoring potential of 376 mouse genes can change through alternative usage of 3′ terminal exons [29], similar to IgM mRNAs. Interestingly, a promising therapeutic strategy is to switch from expression of mRNA isoforms encoding membrane receptors to expression of isoforms encoding soluble proteins that function as dominant negatives. For example, switching isoforms by antisense-based attenuation of splicing for the vascular endothelial growth factor receptor-2 (VEGFR2/KDR) inhibits angiogenesis [30].
To a much lesser extent, pAs can be located in 5′ UTRs or CDSs, although the significance of such sites is largely unexplored. Presumably, pAs in CDSs lead to transcripts without an in-frame stop codon, which are subject to rapid degradation via the nonstop decay mechanism [31]. However, in some cases, an in-frame stop codon is created using the poly(A) tail sequence, as demonstrated recently in the case of the gene encoding glutamylprolyl tRNA synthetase (EPRS) [32]. Full-length EPRS is part of the GAIT (γ-interferon-activated inhibitor of translation) complex, which inhibits translation of mRNAs containing the GAIT complex-interacting element. The truncated protein, resulting from a pA in its CDS, has a dominant-negative function. The UAA stop codon of the truncated EPRS is derived partly from a tyrosine codon (UAC) and partly from the poly(A) tail. Genome-wide analysis has revealed that at least seven more genes can have the same conversion of a tyrosine codon to a stop codon due to pA in the CDS [32].
Long noncoding RNAs (lncRNAs)
lncRNAs are an emerging class of transcripts typically defined as having >200 nt and no coding potential (reviewed in [33]). Most if not all of these RNAs also use C/P for 3′ end processing. A recent study has indicated that at least 66% of annotated mouse lncRNA genes (2600 in total) display APA, and lncRNA genes are more likely than mRNA genes to have alternative pAs in upstream regions [6], suggesting differences in APA evolution between mRNA and lncRNA genes. A study of NEAT1 (nuclear paraspeckle assembly transcript 1), a lncRNA that plays a role in forming paraspeckles (ribonucleoprotein bodies in the interchromatin space of the nucleus), indicates that it is subject to APA [34]. The long 23-kb isoform NEAT1_2 but not the short 3.7-kb NEAT1_1 isoform is required for paraspeckle formation, and heterogeneous nuclear ribonucleoprotein (hnRNP) K, a paraspeckle protein, negatively regulates 3′ processing of NEAT1_1, thereby increasing the ratio of NEAT1_2 to NEAT1_1 and thus enhancing paraspeckle formation.
APA is dynamic under different biological conditions
Tissue specificity
Studies of individual genes over the past two decades have reported hundreds of cases in which APA isoforms are differentially expressed under different cellular conditions (some early studies are reviewed in [35]). Global analysis of APA isoforms using expressed sequence tag (EST) libraries has indicated variation of expression in different tissues [25,36], suggesting that APA isoform expression is not stochastic. Some human tissues have been found to have a global tendency favoring certain isoform types [25]. For example, promoter-distal pAs are generally preferred in neuronal tissues, and cells in the blood have the opposite trend [25]. In addition, distinct APA isoform expression patterns have been reported in testes and different stages of spermatogenesis [25,37]. By surveying the usage of 2590 orthologous pAs in brain, kidney, liver, muscle, and testis, Derti et al. [5] have recently reported that pA usage is more similar among the same tissues across different species than among different tissues within the same species; particularly for brain and liver samples, suggesting conservation of tissue-specific regulation of APA. A similar tissue bias in APA isoform expression has also been reported in flies [7] where, as in humans, brain and testis tissues have been found to express the longest and shortest 3′ UTRs, respectively [7].
Response to extracellular signals
Related to tissue specificity, the APA of a set of genes can be regulated by specific extracellular cues. For example, Flavell et al. [38] have found that promoter-proximal pA isoforms, which encode truncated proteins or short 3′ UTRs, are upregulated for a group of genes when neuronal cells are activated by depolarization agents. In the same study, additional analyses of microarray data sets from various cell types stimulated by extracellular factors, including epidermal growth factor (EGF) stimulation of astrocytes, interleukin-2 stimulation of T cells, and anti-IgM and CpG co-application to B cells, have revealed distinct APA patterns, suggesting that APA can be regulated in a pathway-specific manner in response to extra-cellular signals. Notably, regulation of 3′ end processing in some genes has been shown to be mediated by specific signaling pathways, such as modulation of pA usage of prothrombin pre-mRNA via the p38 kinase in response to the stress-inducing agent anisomycin [39] and that of BCL2-interacting killer (BIK) pre-mRNA via protein kinase C (PKC)δ in response to DNA damage [40].
Cell growth and development
APA also varies as a function of proliferation or differentiation status. For example, in a pioneering work on global APA changes, Sandberg et al. [15] have reported, based on microarray data, a general 3′ UTR shortening after T cells were activated to proliferate. In addition, they have found that the 3′ UTR length in a cell line is generally shorter than in its corresponding tissue, and that there is a general negative correlation between 3′ UTR length and expression of cell proliferation-related genes. This result has been echoed by Ji et al. [16], who have examined APA regulation in mouse embryonic development using ESTs, data from microarrays, and serial analysis of gene expression (SAGE). Despite variations among tissues, during development, 3′ UTR length has been found to correlate negatively with expression of genes involved in proliferation and positively with expression of genes involved in differentiation and morphogenesis [16]. 3′ UTRs are globally lengthened when proliferating myoblast cells are differentiated into quiescent myotubes [16] and shortened when differentiated cells are reprogrammed to ES cell-like induced pluripotent stem (iPS) cells [41]. A notable exception, however, has been observed with spermatogonial germ cells, whose reprogramming to ES cells involves 3′ UTR lengthening [41]. Notably, this is in line with the fact that germ cells are more proliferative than ES cells. Similar trends of 3′ UTR length regulation have been reported for comparisons of ES cells versus neural stem/progenitor (NSP) cells or neurons [42]. Although these studies have all pointed to a connection between 3′ UTR length and cell proliferation, cardiac hypertrophy, in which myocytes grow in size rather than in number, has also been found to involve 3′ UTR shortening [43]. Thus, a general rule may be that APA regulation is correlated with cell growth.
Cancer
Cancer cells are of course highly proliferative. In keeping with this, and consistent with the above, cancer cells have been found to express, in general, mRNAs with shortened 3′ UTRs, as first shown in transformed cell lines [44] and in mouse B-cell leukemia/lymphoma models [45], and more recently in human colorectal carcinomas [46] and breast and lung cancers [47]. In the study by Singh et al. [45], the APA profile was found to be informative in separating tumor subtypes with different survival consequences, indicating its relevance to cancer development and utility as a diagnostic marker. One key question concerning APA regulation in cancer is whether proliferation or transformation is the major driver of APA. Meta-analysis of microarray data from transformed and nontransformed cells with similar predicted proliferation rates has led to the conclusion that cell transformation has a significant role in 3′ UTR regulation [44]. However, a recent study has shown that, by comparing the same cells (BJ primary fibroblast and mammary epithelial cell line MCF10A) in proliferating, arrested, and transformed states, proliferation is a more important determinant of 3′ UTR length [48]. Adding to the complexity of 3′ UTR regulation in cancer, Fu et al. [49] have reported that, compared to MCF10A, breast cancer cell lines MCF7 and MB231 show shortened and lengthened 3′ UTRs, respectively. Notably, it has also been reported that, contrary to the general trend, some gene groups, such as cell–cell adhesion genes, tend to express mRNAs with lengthened 3′ UTRs in cancer cells [45,46]. Therefore, it remains to be fully delineated how APA of different transcripts is regulated in different cancer types and at different stages.
APA is modulated by multiple mechanisms
Regulation of core C/P factor expression
The core components of the mammalian C/P machinery include ~15 polypeptides, most of which exist in multi-subunit subcomplexes (Box 3 and Figure 3). Regulation of APA by modulation of core factor expression was first demonstrated for cleavage stimulation factor (CstF)-64, one of the subunits of the CstF complex (Box 3), which is strongly upregulated during B cell maturation, resulting in higher usage of the upstream intronic pA in the IgM pre-mRNA [28] (discussed previously). This has been shown to reflect the higher affinity of CstF-64 for the distal pA site, such that it is preferentially utilized when CstF-64 levels are low. Consistent with the dynamic regulation of APA in development and differentiation, expression of a substantial fraction of the core factors is highly regulated during embryonic development, reprogramming of differentiated cells, and differentiation of myoblasts [16,41]. Interestingly, the aggregated expression changes of C/P factors inversely correlate with global 3′ UTR length changes [41], suggesting that downregulation of 3′ end processing activity leads to lengthening of 3′ UTRs and vice versa. This view is consistent with reporter assay results showing decreased alternative pA usage when C2C12 cells are differentiated [16]. Additionally, isoforms using weak pAs are less expressed during differentiation and development, regardless of the pA location [6], even though upstream pA sites tend to be weaker than downstream sites [50]. Although the mechanisms controlling expression of C/P factors are not well understood, genes encoding C/P factors tend to have proliferation-related cis elements in their promoters, such as E2F binding sites [41,48]. Indeed, inhibition of expression of E2F factors 1 and 2 leads to 3′ UTR lengthening of a set of transcripts shown to have shortened 3′ UTRs in proliferating cells [48].
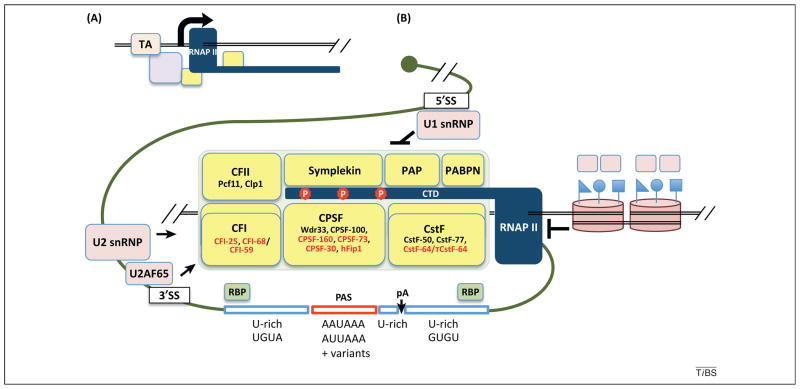
Figure 3.
Regulation of the cleavage and polyadenylation machinery in 3′ end processing. (A) Some cleavage and polyadenylation (C/P) factors (yellow boxes) are recruited at the promoter by transcription activators (TAs) or other unknown mechanisms. (B) The pA is surrounded by several core cis elements, as indicated. The components of the C/P complex are indicated, and those binding to core elements are shown in red. Cleavage factor (CF)I and cleavage stimulation factor (CstF) are dimers, shown as two overlapping boxes. Phosphorylation of the RNA polymerase (RNAP) II carboxy terminal domain (CTD) is indicated. Splicing factors, such as U1 small nuclear ribonucleoprotein (snRNP), U2 snRNP, and U2AF65, interact with the C/P complex. Arrow indicates positive regulation of C/P and T-ended line indicates negative regulation. Two nucleosomes are shown (right), as well as attached symbols representing histone marks, such as H3K36me3. Other RNA-binding proteins (RBPs) can bind to sequences near the pA, thereby enhancing or inhibiting C/P. CPSF, cleavage and polyadenylation specificity factor; hFip1, human Fip1; PABPN, nuclear poly(A)-binding protein; PAP, poly(A) polymerase; PAS, polyadenylation signal; Wdr33, WD repeat domain 33.
Although the global downregulation of C/P factors correlates with 3′ UTR lengthening, perturbations of individual C/P factors have presented a more complex picture. First, knockdown of cleavage factor I (CFI)-68, a subunit of the CFI complex, has been found to cause a general shortening, not lengthening, of 3′ UTRs [51]. Second, CstF-64 knockdown in the same study did not induce dramatic changes in pA choice. This result can be explained, however, because downregulation of CstF-64 triggers a compensatory upregulation of the highly homologous τCstF-64, likely offsetting the effect of CstF-64 knockdown [52]. Third, knockdown of the nuclear poly(A)-binding protein PABPN1 has been found to induce global 3′ UTR shortening [53]. Importantly, ectopic expression of trePABPN1(A17), a short triplet repeat expansion mutant that is found in autosomal dominant oculopharyngeal muscular dystrophy (POMD) patients, leads to a similartrendof3′ UTR shortening in both cultured cells and mouse tissues, suggesting a connection between 3′ UTR regulation and etiology of POMD. Although the mechanistic details remain elusive, the apparent importance of a factor known to be involved in the polyadenylation step of 3′ end processing for pA choice highlights the interplay between cleavage and polyadenlyation.
RBPs
As with alternative splicing, RBPs can affect APA. Indeed, since the early work on the hnRNP protein polypyrimidine tract-binding protein (PTB) [54], a growing number of RBPs have been found to regulate C/P (reviewed in [55]). Some RBPs inhibit pA usage by occluding the binding of core C/P factors, and some enhance pA usage by recruiting core factors. Both modes of action can be elicited by the same protein in different contexts, as shown for the neuronal RBP Nova: its binding near the pA is inhibitory, whereas binding distant from the pA can be enhancing [56]. Given the tissue-specific expression of some RBPs, such proteins may play roles in defining tissue-specific APA [57]. For example, Hilgers et al. [58] have shown that the RBP ELAV (embryonic lethal abnormal vision) in flies mediates neural-specific 3′ UTR lengthening by suppressing proximal pA usage. Similarly, its mammalian homologs, the Hu proteins, have been shown to inhibit pAs with U-rich elements in HeLa cells [59]. Interestingly, the mRNA for HuR, a ubiquitously expressed Hu family member, is also subject to APA regulation, either by HuR itself [60] or by other neuron-specific Hu proteins (HuB/C/D) [61]. Although the former serves as a negative feedback mechanism to control its own expression, the latter is implicated in balancing the prodifferentiation activity of HuB/C/D and the pro-proliferation activity of HuR. It is also worth noting that regulation of APA by RBP is not limited to metazoans. In fact, one of the classic examples of RBP-mediated APA has come from a plant study, which revealed that binding of FCA, an RBP involved in control of flowering, to FY, the plant homolog of C/P factor WDR33 (WD repeat domain 33), regulates APA of the FCA pre-mRNA itself [62].
Splicing and U1 small nuclear ribonucleoprotein (snRNP)
Splicing and C/P are frequently interconnected. This is indicated, for example, by the interactions between key factors involved in these two processes such as cleavage and polyadenylation specificity factor (CPSF)-160 and U1 snRNP [63]; U2 snRNP and CPSF [64]; and U2AF65 and CFI [65]. Coupling of C/P and splicing was initially suggested to help define the 3′ terminal exon [66]. However, U1 snRNP, via its inhibition of poly(A) polymerase (PAP)α [67], can exert a negative role in pA usage. As such, weak 5′ splice sites and large introns allow increased usage of composite terminal exon pAs [68,69]. Significantly, inhibition of U1 snRNP activity by antisense morpholino oligo-nucleotides against U1 snRNA leads to activation of cryptic pAs near transcription start sites [70], a phenomenon not observed when splicing is generally attenuated by spliceostatin A, a U2 snRNP inhibitor. Additionally, mild attenuation of U1 snRNP function that does not inhibit splicing has been found to increase usage of proximal pAs in 3′ UTRs and in upstream alternative terminal exons [71]. This mechanism, dubbed telescripting, has been shown to cause pre-mRNA shortening via APA during rapid and transient transcriptional upregulation upon activation of neurons.
Coupling of pA usage and transcription
Similar to the coupling of transcription and alternative splicing, in which the promoter sequence can affect splicing of specific exons (reviewed in [72]), accumulating evidence indicates that transcriptional activity impacts pA choice. Two types of coupling are conceivable: kinetic coupling, in which the elongation rate of transcription regulates pA choice, and recruitment coupling, in which factor recruitment at the promoter influences downstream pA usage. In support of the kinetic coupling model, it has long been known that cis elements that cause pausing of RNA polymerase (RNAP) II, such as G-rich elements, facilitate pA usage [73]; presumably this is because of the slow assembly of the C/P complex relative to transcription. Consistent with this, defects in the transcriptional elongation factors RPB2, TFIIS, and SPT5 enhance usage of upstream pAs in yeast [74], and the elongation factor ELL2 (eleven-nineteen lysine-rich leukemia gene 2) has been shown to modulate the usage of the proximal pA of IgH pre-mRNAs [75]. More direct evidence comes from a recent study of a mutant Drosophila strain expressing an RNAP II with a slower elongation rate [76]. In this strain, expression of the proximal pA isoform of the polo gene is increased. Importantly, APA of polo is physiologically relevant because transgenic flies lacking the distal pA die at the pupa stage due to perturbed proliferation of precursor cells of the abdomen.
Studies showing that transcriptional activators (TAs) can bring about more efficient 3′ end processing provide support for the recruitment coupling model. Such enhancement has been observed both in transfection assays in vivo [77] and in a transcription-coupled 3′ end processing assay in vitro [78]. Globally, inducible genes tend to generate isoforms with shorter 3′ UTRs when expressed at high levels, and isoforms with longer 3′ UTRs when expressed at low levels [79]. In addition, C/P factors have been found at the promoter region of genes by chromatin immunoprecipitation (ChIP) analysis [80–83]. Med23, one of the Mediator subunits, can also directly interact with several RNA processing proteins, including CFI-59, and its knockdown leads to many changes of pA choice [84]. Furthermore, TAs can recruit C/P factors either directly [85] and/or via the RNAP II-associated factor (PAF) elongation complex [78], leading to enhanced 3′ end processing.
Chromatin structure and histone modifications?
Considerable evidence now indicates that alternative splicing can be affected by chromatin structure and modifications [86], but is APA? Several studies have suggested this may be the case. For example, studies from yeast and humans have shown that the region around the pA is depleted of nucleosomes [87]. Interestingly, highly used alternative pAs tend to have a higher downstream affinity for nucleosomes [88], suggesting chromatin organization might potentially impact APA. However, because highly used pAs have different downstream sequences than less used sites [89], and sequence composition is an important determinant of nucleosome levels, it is not clear to what extent the variation in nucleosome density is due to sequence alone versus active remodeling. Nevertheless, a correlation between nucleosome levels and accumulation of RNAP II after the pA has been observed [90], suggesting that nucleosomes in this region can cause RNAP II pausing. Additionally, variations in nucleosome density and levels of H3K36me3, a type of histone modification enriched at gene 3′ ends, between genes expressed at different levels, have been found to be much greater at proximal pAs than at distal pAs [79], suggesting an interplay between pA usage and nucleosome configuration and histone modification. Supporting this idea, the PAF complex, which associates with RNAP II and plays roles in histone methylation, has been shown to interact with the C/P machinery and influence pA usage [82].
Concluding remarks
Building upon biochemical studies of 3′ end processing in the past three decades, recent advances at the molecular and systems levels concerning APA have stimulated interest in comprehending this dynamic process and its widespread implications for regulation of gene expression and cell growth control. Many questions remain to be addressed in the coming years. For instance, with the availability of powerful deep sequencing methods to study APA isoforms, thorough understanding of APA in different species, tissue types, developmental stages, and disease states is within reach. Whether APA can be used for cancer diagnosis/prognosis should be rigorously explored. Additionally, the impact of APA on RNA metabolism needs to be fully studied, particularly at the systems level. For example, how different isoforms are sorted in polarized versus nonpolarized cells is largely unknown. Similarly, to what extent APA impacts protein output in different cell types, and in cell growth and differentiation, during which APA is globally regulated, needs to be addressed. Whether the presence or absence of miRNA target sites through APA can engage mRNAs in competition for miRNAs, a hypothetical scenario known as ‘competing endogenous RNA (ceRNA)’ [91], needs to be explored.
Furthermore, given the diverse consequences of perturbation of core C/P factors on APA, and the role of other factors including tissue-specific RBPs, pA usage appears to be more complex than previously thought. Regulation of pA is reminiscent of transcriptional control at the 5′ end of genes, involving factors binding to both core promoters and enhancers. Here, RBPs and core factors appear to interact with different cis elements around the pA, and pA usage seems to be determined in a combinatorial manner. How regulation of these factors, including post-translational modifications, leads to APA needs to be explored. Along these lines, it is worth noting that depletion of yeast mRNA export adaptor protein Yra1, which interacts with C/P factor Pcf11, leads to widespread APA [92]. Thus, how other proteins interacting with the C/P machinery can change APA needs to be established. Finally, a clearer picture of APA regulation by chromatin organization, histone modifications, and DNA methylation [93], is expected to emerge in the coming years. How APA patterns are regulated, reinforced, or memorized at the epigenetic level will be unraveled.
Acknowledgments
We thank our laboratory members for helpful discussions. We apologize for not citing many important papers related to the topic owing to space limitation. This work was funded by grants GM 84089 (B.T.) and GM 28983 (J.L.M.) from National Institutes of Health.
References
- 1.Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Genes Dev. 2009;23:1247–1269. doi: 10.1101/gad.1792809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marzluff WF, et al. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 4.Danckwardt S, et al. 3′ end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J. 2008;27:482–498. doi: 10.1038/sj.emboj.7601932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derti A, et al. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012;22:1173–1183. doi: 10.1101/gr.132563.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoque M, et al. Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat Methods. 2013;10:133–139. doi: 10.1038/nmeth.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smibert P, et al. Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell Rep. 2012;1:277–289. doi: 10.1016/j.celrep.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jan CH, et al. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature. 2011;469:97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ulitsky I, et al. Extensive alternative polyadenylation during zebrafish development. Genome Res. 2012;22:2054–2066. doi: 10.1101/gr.139733.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Giammartino DC, et al. Mechanisms and consequences of alternative polyadenylation. Mol Cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi Y. Alternative polyadenylation: new insights from global analyses. RNA. 2012;18:2105–2117. doi: 10.1261/rna.035899.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol. 2012;19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 15.Sandberg R, et al. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji Z, et al. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci USA. 2009;106:7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayr C, Bartel DP. Widespread shortening of 3′ UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boutet SC, et al. Alternative polyadenylation mediates microRNA regulation of muscle stem cell function. Cell Stem Cell. 2012;10:327–336. doi: 10.1016/j.stem.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garneau NL, et al. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 20.Kim YK, et al. Staufen1 regulates diverse classes of mammalian transcripts. EMBO J. 2007;26:2670–2681. doi: 10.1038/sj.emboj.7601712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graham RR, et al. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc Natl Acad Sci USA. 2007;104:6758–6763. doi: 10.1073/pnas.0701266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.An JJ, et al. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134:175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andreassi C, et al. An NGF-responsive element targets myo-inositol monophosphatase-1 mRNA to sympathetic neuron axons. Nat Neurosci. 2010;13:291–301. doi: 10.1038/nn.2486. [DOI] [PubMed] [Google Scholar]
- 24.Yudin D, et al. Localized regulation of axonal RanGTPase controls retrograde injury signaling in peripheral nerve. Neuron. 2008;59:241–252. doi: 10.1016/j.neuron.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H, et al. Biased alternative polyadenylation in human tissues. Genome Biol. 2005;6:R100. doi: 10.1186/gb-2005-6-12-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang ET, et al. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012;150:710–724. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takagaki Y, et al. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87:941–952. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- 29.Davis MJ, et al. Differential use of signal peptides and membrane domains is a common occurrence in the protein output of transcriptional units. PLoS Genet. 2006;2:e46. doi: 10.1371/journal.pgen.0020046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vorlova S, et al. Induction of antagonistic soluble decoy receptor tyrosine kinases by intronic polyA activation. Mol Cell. 2011;43:927–939. doi: 10.1016/j.molcel.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vasudevan S, et al. Non-stop decay – a new mRNA surveillance pathway. Bioessays. 2002;24:785–788. doi: 10.1002/bies.10153. [DOI] [PubMed] [Google Scholar]
- 32.Yao P, et al. Coding region polyadenylation generates a truncated tRNA synthetase that counters translation repression. Cell. 2012;149:88–100. doi: 10.1016/j.cell.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naganuma T, et al. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 2012;31:4020–4034. doi: 10.1038/emboj.2012.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edwalds-Gilbert G, et al. Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 1997;25:2547–2561. doi: 10.1093/nar/25.13.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beaudoing E, Gautheret D. Identification of alternate polyadenylation sites and analysis of their tissue distribution using EST data. Genome Res. 2001;11:1520–1526. doi: 10.1101/gr.190501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu D, et al. Systematic variation in mRNA 3′-processing signals during mouse spermatogenesis. Nucleic Acids Res. 2007;35:234–246. doi: 10.1093/nar/gkl919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flavell SW, et al. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Danckwardt S, et al. p38 MAPK controls prothrombin expression by regulated RNA 3′ end processing. Mol Cell. 2011;41:298–310. doi: 10.1016/j.molcel.2010.12.032. [DOI] [PubMed] [Google Scholar]
- 40.Li W, et al. Star-PAP control of BIK expression and apoptosis is regulated by nuclear PIPKIalpha and PKCdelta signaling. Mol Cell. 2012;45:25–37. doi: 10.1016/j.molcel.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ji Z, Tian B. Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS ONE. 2009;4:e8419. doi: 10.1371/journal.pone.0008419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shepard PJ, et al. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA. 2011;17:761–772. doi: 10.1261/rna.2581711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park JY, et al. Comparative analysis of mRNA isoform expression in cardiac hypertrophy and development reveals multiple post-transcriptional regulatory modules. PLoS ONE. 2011;6:e22391. doi: 10.1371/journal.pone.0022391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh P, et al. Global changes in processing of mRNA 3′ untranslated regions characterize clinically distinct cancer subtypes. Cancer Res. 2009;69:9422–9430. doi: 10.1158/0008-5472.CAN-09-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morris AR, et al. Alternative cleavage and polyadenylation during colorectal cancer development. Clin Cancer Res. 2012;18:5256–5266. doi: 10.1158/1078-0432.CCR-12-0543. [DOI] [PubMed] [Google Scholar]
- 47.Lembo A, et al. Shortening of 3′UTRs correlates with poor prognosis in breast and lung cancer. PLoS ONE. 2012;7:e31129. doi: 10.1371/journal.pone.0031129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elkon R, et al. E2F mediates enhanced alternative polyadenylation in proliferation. Genome Biol. 2012;13:R59. doi: 10.1186/gb-2012-13-7-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fu Y, et al. Differential genome-wide profiling of tandem 3′ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res. 2011;21:741–747. doi: 10.1101/gr.115295.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tian B, et al. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martin G, et al. Genome-wide analysis of pre-mRNA 3′ end processing reveals a decisive role of human cleavage factor I in the regulation of 3′ UTR length. Cell Rep. 2012;1:753–763. doi: 10.1016/j.celrep.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 52.Yao C, et al. Transcriptome-wide analyses of CstF64-RNA interactions in global regulation of mRNA alternative polyadenylation. Proc Natl Acad Sci USA. 2012;109:18773–18778. doi: 10.1073/pnas.1211101109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jenal M, et al. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell. 2012;149:538–553. doi: 10.1016/j.cell.2012.03.022. [DOI] [PubMed] [Google Scholar]
- 54.Moreira A, et al. The upstream sequence element of the C2 complement poly(A) signal activates mRNA 3′ end formation by two distinct mechanisms. Genes Dev. 1998;12:2522–2534. doi: 10.1101/gad.12.16.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 2010;38:2757–2774. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Licatalosi DD, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hilgers V, et al. ELAV mediates 3′ UTR extension in the Drosophila nervous system. Genes Dev. 2012;26:2259–2264. doi: 10.1101/gad.199653.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu H, et al. Hu proteins regulate polyadenylation by blocking sites containing U-rich sequences. J Biol Chem. 2007;282:2203–2210. doi: 10.1074/jbc.M609349200. [DOI] [PubMed] [Google Scholar]
- 60.Dai W, et al. RNA-binding protein HuR autoregulates its expression by promoting alternative polyadenylation site usage. Nucleic Acids Res. 2012;40:787–800. doi: 10.1093/nar/gkr783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mansfield KD, Keene JD. Neuron-specific ELAV/Hu proteins suppress HuR mRNA during neuronal differentiation by alternative polyadenylation. Nucleic Acids Res. 2012;40:2734–2746. doi: 10.1093/nar/gkr1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simpson GG, et al. FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell. 2003;113:777–787. doi: 10.1016/s0092-8674(03)00425-2. [DOI] [PubMed] [Google Scholar]
- 63.Lutz CS, et al. Interaction between the U1 snRNP-A protein and the 160-kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes Dev. 1996;10:325–337. doi: 10.1101/gad.10.3.325. [DOI] [PubMed] [Google Scholar]
- 64.Kyburz A, et al. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. Mol Cell. 2006;23:195–205. doi: 10.1016/j.molcel.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 65.Millevoi S, et al. An interaction between U2AF 65 and CF I(m) links the splicing and 3′ end processing machineries. EMBO J. 2006;25:4854–4864. doi: 10.1038/sj.emboj.7601331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Niwa M, et al. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev. 1990;4:1552–1559. doi: 10.1101/gad.4.9.1552. [DOI] [PubMed] [Google Scholar]
- 67.Gunderson SI, et al. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol Cell. 1998;1:255–264. doi: 10.1016/s1097-2765(00)80026-x. [DOI] [PubMed] [Google Scholar]
- 68.Tian B, et al. Widespread mRNA polyadenylation events in introns indicate dynamic interplay between polyadenylation and splicing. Genome Res. 2007;17:156–165. doi: 10.1101/gr.5532707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peterson ML, Perry RP. The regulated production of mu m and mu s mRNA is dependent on the relative efficiencies of mu s poly(A) site usage and the c mu 4-to-M1 splice. Mol Cell Biol. 1989;9:726–738. doi: 10.1128/mcb.9.2.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaida D, et al. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–668. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Berg MG, et al. U1 snRNP determines mRNA length and regulates isoform expression. Cell. 2012;150:53–64. doi: 10.1016/j.cell.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kornblihtt AR. Promoter usage and alternative splicing. Curr Opin Cell Biol. 2005;17:262–268. doi: 10.1016/j.ceb.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 73.Yonaha M, Proudfoot NJ. Specific transcriptional pausing activates polyadenylation in a coupled in vitro system. Mol Cell. 1999;3:593–600. doi: 10.1016/s1097-2765(00)80352-4. [DOI] [PubMed] [Google Scholar]
- 74.Cui Y, Denis CL. In vivo evidence that defects in the transcriptional elongation factors RPB2, TFIIS, and SPT5 enhance upstream poly(A) site utilization. Mol Cell Biol. 2003;23:7887–7901. doi: 10.1128/MCB.23.21.7887-7901.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martincic K, et al. Transcription elongation factor ELL2 directs immunoglobulin secretion in plasma cells by stimulating altered RNA processing. Nat Immunol. 2009;10:1102–1109. doi: 10.1038/ni.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pinto PA, et al. RNA polymerase II kinetics in polo polyadenylation signal selection. EMBO J. 2011;30:2431–2444. doi: 10.1038/emboj.2011.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rosonina E, et al. Transcriptional activators control splicing and 3′-end cleavage levels. J Biol Chem. 2003;278:43034–43040. doi: 10.1074/jbc.M307289200. [DOI] [PubMed] [Google Scholar]
- 78.Nagaike T, et al. Transcriptional activators enhance polyadenylation of mRNA precursors. Mol Cell. 2011;41:409–418. doi: 10.1016/j.molcel.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ji Z, et al. Transcriptional activity regulates alternative cleavage and polyadenylation. Mol Syst Biol. 2011;7:534. doi: 10.1038/msb.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Glover-Cutter K, et al. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Venkataraman K, et al. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 2005;19:1315–1327. doi: 10.1101/gad.1298605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rozenblatt-Rosen O, et al. The tumor suppressor Cdc73 functionally associates with CPSF and CstF 3′ mRNA processing factors. Proc Natl Acad Sci USA. 2009;106:755–760. doi: 10.1073/pnas.0812023106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Calvo O, Manley JL. Strange bedfellows: polyadenylation factors at the promoter. Genes Dev. 2003;17:1321–1327. doi: 10.1101/gad.1093603. [DOI] [PubMed] [Google Scholar]
- 84.Huang Y, et al. Mediator complex regulates alternative mRNA processing via the MED23 subunit. Mol Cell. 2012;45:459–469. doi: 10.1016/j.molcel.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Uhlmann T, et al. The VP16 activation domain establishes an active mediator lacking CDK8 in vivo. J Biol Chem. 2007;282:2163–2173. doi: 10.1074/jbc.M608451200. [DOI] [PubMed] [Google Scholar]
- 86.Luco RF, et al. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Spies N, et al. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell. 2009;36:245–254. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hu J, et al. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA. 2005;11:1485–1493. doi: 10.1261/rna.2107305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Grosso AR, et al. Dynamic transitions in RNA polymerase II density profiles during transcription termination. Genome Res. 2012;22:1447–1456. doi: 10.1101/gr.138057.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Salmena L, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Johnson SA, et al. The export factor Yra1 modulates mRNA 3′ end processing. Nat Struct Mol Biol. 2011;18:1164–1171. doi: 10.1038/nsmb.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wood AJ, et al. Regulation of alternative polyadenylation by genomic imprinting. Genes Dev. 2008;22:1141–1146. doi: 10.1101/gad.473408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sheets MD, et al. Point mutations in AAUAAA and the poly (A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 1990;18:5799–5805. doi: 10.1093/nar/18.19.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cheng Y, et al. Prediction of mRNA polyadenylation sites by support vector machine. Bioinformatics. 2006;22:2320–2325. doi: 10.1093/bioinformatics/btl394. [DOI] [PubMed] [Google Scholar]
- 96.Nunes NM, et al. A functional human poly(A) site requires only a potent DSE and an A-rich upstream sequence. EMBO J. 2010;29:1523–1536. doi: 10.1038/emboj.2010.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ozsolak F, et al. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell. 2010;143:1018–1029. doi: 10.1016/j.cell.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sherstnev A, et al. Direct sequencing of Arabidopsis thaliana RNA reveals patterns of cleavage and polyadenylation. Nat Struct Mol Biol. 2012;19:845–852. doi: 10.1038/nsmb.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mangone M, et al. The landscape of C. elegans 3′UTRs. Science. 2010;329:432–435. doi: 10.1126/science.1191244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Haenni S, et al. Analysis of C. elegans intestinal gene expression and polyadenylation by fluorescence-activated nuclei sorting and 3′-end-seq. Nucleic Acids Res. 2012;40:6304–6318. doi: 10.1093/nar/gks282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wu X, et al. Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. Proc Natl Acad Sci USA. 2011;108:12533–12538. doi: 10.1073/pnas.1019732108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wilkening S, et al. An efficient method for genome-wide polyadenylation site mapping and RNA quantification. Nucleic Acids Res. 2013;41:e65. doi: 10.1093/nar/gks1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shi Y, et al. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell. 2009;33:365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mandel CR, et al. Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci. 2008;65:1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]