Abstract
Biodegradable polymeric nanoparticles are widely recognized as efficacious drug delivery vehicles, yet the rational engineering of nanoparticle surfaces in order to improve biodistribution, reduce clearance, and/or improve targeting remains a significant challenge. We have previously demonstrated that an amphiphilic conjugate of avidin and palmitic acid can be used to modify poly(lactic-co-glycolic acid) (PLGA) particle surfaces to display functional avidin groups, allowing for the facile attachment of biotinylated ligands for targeting or steric stabilization. Here, we hypothesized that the incorporation, density, and stability of surface-presented avidin could be modulated through varying the lipophilicity of its fatty acid conjugate partner. We tested this hypothesis by generating a set of novel conjugates incorporating avidin and common fatty acids. We found that conjugation to linoleic acid resulted in a ∼60% increase in the incorporation of avidin on the nanoparticle surface compared to avidin–palmitic acid, which exhibited the highest avidin incorporation in previous studies. Further, the linoleic acid–avidin conjugate yielded nanoparticles with enhanced ability to bind biotinylated ligands compared to the previous method; nanoparticles modified with avidin–linoleic acid bound ∼170% more biotin–HRP than those made with avidin–palmitic acid and ∼1300% more than particles made without conjugated avidin. Most critically, increased ligand density on anti-CD4-targeted nanoparticles formulated with the linoleic acid–avidin conjugate resulted in a 5% increase in binding of CD4+ T cells. Thus we conclude that the novel avidin–linoleic acid conjugate facilitates enhanced ligand density on PLGA nanoparticles, resulting in functional enhancement of cellular targeting.
Keywords: PLGA, Nanoparticle, Modification, Targeted, Drug delivery, T cells
1. Introduction
Biodegradable polymeric nanoparticles (NPs) have long been investigated as drug delivery vehicles. These particles can be used to solubilize concentrated drug payloads, improve drug stability and bioavailability, and extend drug effect through sustained delivery [1,2]. Among the most commonly used and extensively investigated biodegradable polymers are poly(lactic-co-glycolic acid) (PLGA) and its constituent polymers, polylactic acid (PLA) and polyglycolic acid (PGA) [1]. These polymers have a long history of safe use in humans and their degradation under physiologic conditions releases lactic and/or glycolic acid monomers that are easily metabolized or eliminated [3]. Copolymers of PLGA are of particular interest for drug delivery as the degradation rate of the polymer and subsequent drug release rate can be modulated by varying the ratio of the PLA to PGA segments; higher ratios of the more hydrophobic PLA decrease the penetration of water and overall degradation rate of the polymer while higher ratios of the more hydrophilic PGA have the opposite effect [1,3].
The biodistribution and pharmacokinetic properties of nanoscale drug delivery vehicles are largely dependent on their size, material composition, and surface properties [4–6]. There has been tremendous interest in the development of biodegradable nanoparticles that display targeting ligands in order to improve the biodistribution, safety, and efficacy of encapsulated agents. Antibody, aptamer and even small molecule-targeted PLGA nanoparticles have been shown to be preferentially bound to or internalized by target cells, compared to cells lacking the targeted receptor or ligand [7–10]. In vivo, targeting has been demonstrated to increase dose accumulation and persistence at sites of disease, such as tumor beds, where the target ligand is either uniquely or highly expressed [11,12]. Interestingly, localization or internalization of nanoparticles can enhance the potency of encapsulated agents, as measured by, for example, lowered IC50 values of chemotherapeutic drugs [7,10,12,13]. However, as PLGA lacks functional chemical groups on the aliphatic polyester backbone, a significant challenge has been the development of methods that enable facile surface modification of nanoparticles made from this polymer [1,14]. One popular method has been to utilize co-block polymers, such as those based on PLGA and the hydrophilic polymer polyethylene glycol (PEG), that contain functionalized endgroups which enable covalent conjugation of ligands. These polymers have long been used to fabricate nanoparticles in which the hydrophilic properties of PEG and relatively hydrophobic properties of PLGA determine the formation of core–shell structures [15,16]. Covalent conjugation of ligands to functionalized PEG, either before [10] or after [11] particle manufacture therefore results in their preferential display on the particle surface. However, one downside of this approach is the potential exposure of the drug-loaded NP to hydrolysis and the potential for ligand denaturation. Significant loss of the surface ligand can occur, likely due to hydrolysis of the PLGA [15], and a comparison of covalent conjugation versus adsorption has shown that conjugation can compromise the binding ability of targeting antibodies [7]. Likewise, fine control over the number or density of ligands has not been well discussed in the literature.
We have previously demonstrated that an amphiphilic avidin–palmitic acid conjugate can be utilized to present functional avidin groups on the surface of drug-loaded PLGA scaffolds and microparticles [17]. The avidin molecules are then available to bind biotinylated ligands at any point after particle manufacture and storage. Importantly, this methodology enables titration of targeting ligands and thereby precise control over the surface properties. For example, avidin-coated polystyrene particles, although ineffective as in vivo drug delivery vehicles, have been useful in investigating the effect of NP targeting due to the wide availability of biotinylated ligands [18]. Compared to other surface functionalization techniques, this novel methodology for modifying PLGA NP surfaces spares potentially labile ligands from harsh manufacturing processes and does not require modification of either the encapsulant or the polymer. We have demonstrated the versatility of this linker system by modifying PLGA nanoparticles with polyethylene glycol (PEG) for improved biodistribution of doxorubicin [19] and enhanced transport across mucosal barriers [20], lipoglycans for enhancement of encapsulated vaccine efficacy [21], ligand modification to improve cell uptake [22], and targeting antibodies for enhanced T-cell stimulation [23] and improved cytokine delivery [24].
As the preferential surface presentation of the avidin–fatty acid conjugate is thought to be driven by its amphiphilic nature [17], we hypothesized that varying the fatty acid lipophilicity would influence the density and stability of avidin–lipid incorporation in PLGA nanoparticles. Ligand density is a critical factor in the efficacy of targeted drug delivery systems; higher density is a particularly useful feature for ligands that, in their monomeric form, have a weak affinity for their target receptors [4,6,25–27], such as single-chain variable fragments (SCvF) and peptide/major histocompatibility complexes (peptide/MHCs), which have weak affinity to target T cell receptors [27–29]. Thus, the results of this study suggest new opportunities in the design of a high avidity nanoparticle platform for targeted drug delivery in a number of therapeutic scenarios.
2. Materials and methods
2.1. Preparation of avidin–fatty acid bioconjugates and avidin-functionalized nanoparticles
Stable avidin–lipid conjugates were formed using a zero-length crosslinking agent to create a covalent bond between the lipid carboxyl end groups and free amines on the avidin protein. Lipids (butyric, caprylic, palmitic, stearic, or linoleic acid; all from Sigma) were first reacted in 0.1× PBS with 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide (EDC) and N-hydroxylsulfosuccinimide (sulfo-NHS) (Invitrogen) to convert the terminal carboxyl group to an amine-reactive sulfo-NHS ester. Avidin (Sigma) at 5 mg/ml was then reacted with 10-fold molar excess of the NHS-functionalized fatty acid in 0.1× PBS and the solution was gently mixed at 37 °C for 2 h. Reactants were then dialyzed against 1.0× PBS at 37 °C for 24 h to remove excess reactants and/or hydrolyzed esters.
PLGA nanoparticles (NPs) were manufactured using an oil-in-water emulsion method. One hundred (100) mg of PLGA with molecular weight of 92–112 kDa and 50/50 lactide:glycolide ratio (Durect Corporation) were dissolved overnight in 2 ml dichloromethane. To make BSA-FITC loaded NPs, 100 μl of BSA-FITC (Fisher Scientific) (10 mg/ml in 1× PBS) were added to the polymer solution with vortexing. This solution was then added dropwise with vortexing to a 4 ml aqueous solution consisting of 2 ml (5 mg/ml) avidin–lipid bioconjugate and 2 ml 5% PVA (MW 30-70 kDa, Sigma) to make surface-functionalized, “avidin+” NPs. To make “blank” or “unconjugated” NPs controls, 2 ml of 1× PBS or 2 ml of freshly dissolved avidin (5 mg/ml in PBS) were used, respectively, instead of the avidin-lipid conjugate solution. The organic polymer/aqueous surfactant emulsion was then sonicated on ice 3× at 10 s intervals using a 600 watt Misonix 3000 sonicator with a 3/16” microtip. Solvent was removed and particles hardened by magnetic stirring for 3 h in 120 ml of 0.3% PVA aqueous solution. Nanoparticles were collected by centrifugation for 10 min at 10,000 rpm and resuspended/washed in sterile DI water. Particles were washed a total of 3 times to remove excess surfactant, conjugate, and encapsulant prior to lyophilization and storage at −20 °C Nanoparticles of polylactide (PLA) (100/0 lactide:glycolide ratio, Durect Corporation) were made in identical fashion.
2.2. Characterization of avidin-lipid conjugates
Avidin-lipid conjugates were previously characterized by HPLC [17]. We also examined the biotin-binding ability of conjugates using 4′-hydroxyazobenzene-2-carboxylic acid (HABA) (Sigma). HABA binds to avidin to yield a yellow-orange complex that absorbs at 500 nm. As the dye binds with weaker affinity to avidin (Kd = 5.8 × 10−6 M) than biotin (Kd = 1 × 10−15 M), traditionally the HABA assay is utilized to quantify biotin concentrations as the addition of free biotin displaces the HABA dye with an associated decrease in absorbance [30]. Here, the linear relationship between avidin concentration and absorbance was used to calculate avidin concentration.
2.3. Characterization of nanoparticles
Nanoparticle morphology was characterized by scanning electron microscopy (SEM). Samples were sputter-coated with gold using a Dynavac Mini Coater and imaged with a Philips XL30 SEM using a LaB electron gun with an accelerating voltage of 5–10 kV. Particle size and distribution were determined using ImageJ image analysis software (available from the NIH). Mean particle diameter was calculated by analysis of >250 counts per sample and statistical difference between groups assessed by two-tailed Student's t tests. The hydrodynamic mean effective diameter and polydispersity were measured in 1× PBS using a ZetaPALS particle sizing instrument (Brookhaven Instruments Corporation, Holtsville, NY). Particle counts (number of particles per milligram of sample) were obtained using a Nanosight instrument (NanoSight, Ltd., Wiltshire, UK).
2.4. Quantification of avidin incorporation, stability, and effect on release profile
Avidin incorporation in nanoparticles was quantified using a micro-BCA assay (Fisher Scientific). Unloaded, surface-modified nanoparticles were suspended in dilutions starting at 2 mg/ml in 1× PBS and 150 μl of sample or standard added to a 96 well u-bottom microplate. 150 μl of the micro-BCA working reagent was added to each well and the plate incubated for 2 h at 37 °C. The plate was then centrifuged to collect NPs and 100 μl of supernatant transferred to a new plate. Protein content was measured by absorbance at 562 nm and background measured at 650 nm. Mean avidin incorporation was calculated by performing experiments in triplicate and studies were repeated 2-3 times; statistical significant between groups was calculated using two-tailed Student's t tests. The stability of the avidin modification was assessed under physiologically relevant conditions. Briefly, 10 mg of unloaded nanoparticles (avidin-lipid modified nanoparticles, as well as “blank” and “unconjugated” nanoparticles) was incubated in triplicate in 1× PBS. To determine if avidin was simply adsorbed to the particle surface, nanoparticles were pipetted vigorously and vortexed for 60 s, centrifuged for 10 min at 13.2 krpm, and supernatant collected. Avidin content in the supernatant was quantified by micro-BCA assay. Additional time points were collected at 1, 2, 12, and 168 h to assess long-term stability.
The effect of surface modification on potential release of encapsulated agents was measured using BSA-FITC (Fisher Scientific) as a model encapsulated protein. Briefly, 10 mg of “blank,” “unconjugated,” and avidin–lipid modified nanoparticles was incubated in triplicate in 1× PBS on a rotary shaker at 37 °C. At designated time points, the samples were centrifuged and supernatant collected. BSA-FITC concentration was detected spectrofluorimetrically at excitation 490 nm/emission 525 nm. Statistical significant in the differences in cumulative release of avidin or BSA-FITC between groups was calculated at each time point by Student's t test.
2.5. Quantification of ligand capture by surface-modified nanoparticles
Horseradish peroxidase (HRP) is a 44 kDa enzyme commonly used for signal amplification in the detection of target molecules; biotin or avidin-conjugated versions are often used in secondary steps for ELISA and immunohistochemistry. Biotin–HRP (Accurate Chemicals & Scientific Corp.) was used here to quantify surface-bound avidin on NPs after particle formulation. 100 μl of NPs (1 mg/ml in 1× PBS) were added to u-bottom 96 well microplates and incubated at room temperature for 15 min with 100 μl biotin–HRP at varying dilutions. The plate was then centrifuged, supernatant discarded, and NPs resuspended in fresh PBS. This wash step was repeated 3× to remove unbound or weakly adsorbed enzyme. After the final wash, NPs were resuspended in 100 μl PBS and transferred to a new plate along with standards comprised of serial dilutions of soluble biotin–HRP. 100 μl of TMB substrate solution (Fisher) was added and the reaction stopped with 50 μl of 1 N HCl. The plate was centrifuged again and supernatant transferred to a new plate for absorbance readings at 450 and 570 nm. Experiments were conducted in triplicate, studies were repeated 2−3 times, and statistical significance between individual groups was calculated using two-tailed Student's t tests.
2.6. Analysis of ligand targeting
CD4-targeted NPs were formulated by incubating rhodamine-loaded, avidin-modified NPs with 1000-fold molar excess of biotinylated rat anti-mouse CD4 antibody (Fisher) for 15 min at room temperature. NPs were washed 3× in 1× PBS in order to remove excess and unbound antibody and stored on ice in 1× PBS containing 1% fetal bovine serum (FBS). Splenocytes were collected from 6 to 8 week old C57/BL6 mice (Jackson Labs): briefly, mice were euthanized and spleens collected in RPMI supplemented with 5% FBS; splenocytes were collected by passing spleens through an 8 μm cell strainer. Cells were stored on ice in 1× PBS containing 1% FBS and used within 3 h of collection. 1×106 splenocytes were first stained with a non-blocking, FITC-labeled antibody to label CD4+ T cells. Cells were then incubated for 15 min at 37 °C with a 100-fold excess of NPs or controls. Number and percentage of cells with bound NPs was measured via flow cytometry (FACScan, BD Biosciences) by gating on cells with increased side scatter and positive for rhodamine fluorescence. Experiments were conducted in triplicate, repeated once, and statistical significant between individual groups was calculated using two-tailed Student's t tests.
3. Results
3.1. Development and characterization of avidin–lipid conjugates
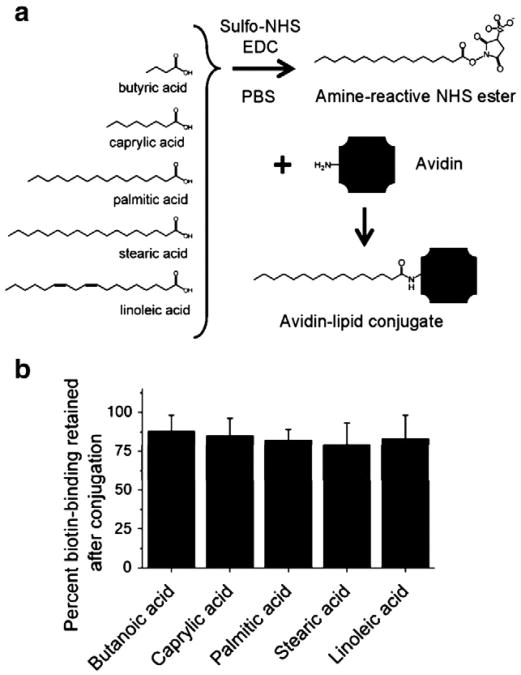
Avidin–lipid conjugates were formed using common fatty acids of varying chain length (Fig. 1a). The terminal carboxyl group on the lipid was first converted to an amine-reactive sulfo-NHS ester, and then reacted with the amines on avidin molecules to form covalent amide bonds (Fig. 1a). The biotin-binding potential of avidin after lipid conjugation was examined using the HABA assay. HABA binds to avidin in a concentration-dependent, reversible manner; the resulting linear increase in absorbance (Supplemental Fig. 1) allows for quantification of available biotin-binding sites. It was found that lipid conjugation via NHS/EDC chemistry did not significantly diminish the biotin-binding capacity of avidin. All conjugates retained greater than 80% of biotin-binding capacity compared to a control of unconjugated, fresh avidin in buffer alone (Fig. 1b).
Fig. 1.
Avidin–lipid formation and retention of biotin-binding capacity. (a) The terminal carboxyl on common fatty acids was converted into an amine-reactive sulfo-NHS ester and reacted with primary amines to form stable amide bonds to avidin. The subsequent bioconjugate was collected and purified by dialysis. (b) All conjugates retained more than 80% capacity to bind 4 -hydroxyazobenzene-2-carboxylic acid (HABA), indicating preservation of biotin-binding ability. Data represent mean±1 standard deviation across 3 or more samples.
3.2. Development and characterization of avidin–fatty acid modified nanoparticles
PLGA nanoparticles formed with avidin conjugates were found to be discrete, smooth, and spherical by SEM and no significant differences in appearance were observed between groups (Fig. 2a). SEM images were analyzed using ImageJ software by counting N250 particles per image file. Size distributions were relatively narrow and consistent among groups — a representative histogram is presented in (Fig. 2b). The mean particle diameter across all groups was found to be 220 nm±50 nm (±1 standard deviation) and there were no statistically significant differences between individual groups (Fig. 2c). To confirm the potential presence of aggregates, the hydrodynamic diameter of particles was assessed in 1× PBS by dynamic light scattering. The diameter of suspended NPs was found to be consistent with the SEM measurements and samples were found to have a polydispersity index under 0.2 (Fig. 2d). Treatment of avidin-modified NPs with biotinylated PEG did not significantly alter NP size or polydispersity (Fig. 2d), which was not surprising as the relatively low polydispersity index of non-PEGylated, avidin-modified NPs suggests a low degree of particle aggregation. We note that similar avidin-modified NPs were found to non-specifically adsorb 1 μg bovine serum albumin (BSA) per mg NPs after in vitro incubation with serum-containing PBS [19]. In those studies, PEGylation of NPs was shown to result in a 4-fold reduction of BSA adsorption [19].
Fig. 2.
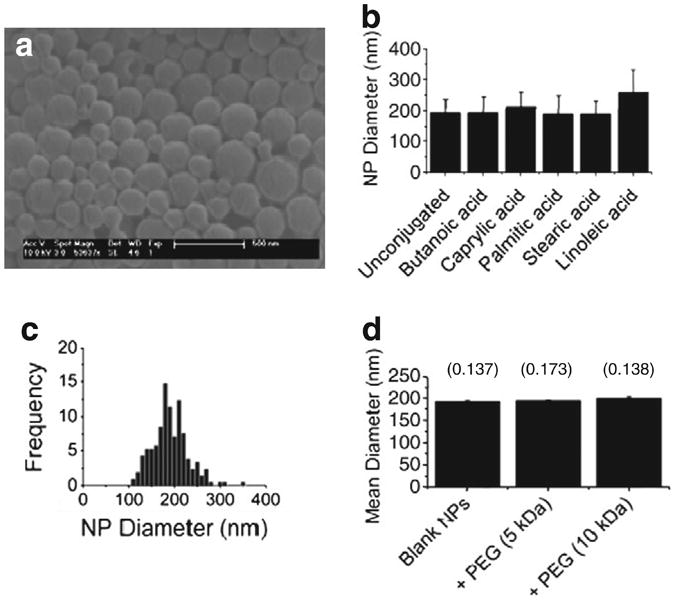
Size distribution of avidin–modified nanoparticles. (a) Representative scanning electron micrograph (SEM) of PLGA nanoparticles (NPs) formulated with avidin–lipid. (b) Mean diameter of NPs obtained by image analysis. X-axis denotes specific lipid conjugated to avidin or unconjugated avidin control. Data represent mean ± 1 standard deviation (n>250 counts per sample). (c) Representative size distribution of avidin–palmitic acid modified NPs. (d) Hydrodynamic diameter of avidin–palmitic acid modified NPs in 1× PBS was measured by dynamic light scattering. Addition of biotinylated PEG resulted in modest, statistically insignificant increase in apparent mean diameter with no effect on sample polydispersity (reported above columns). Each data point represents 10 measurements of the same sample before and after addition of biotin–PEG to the sample. Biotin–PEG alone did not alter background measurements.
3.3. Quantification of surface-bound avidin
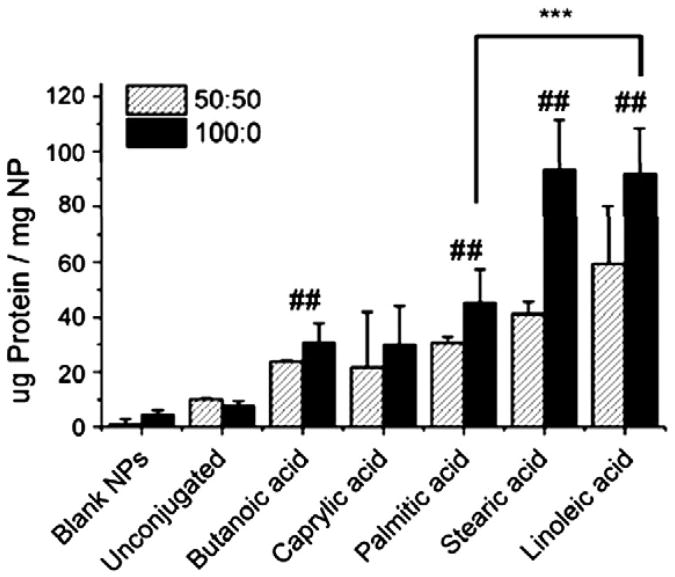
The avidin content on PLGA nanoparticles was assessed via the micro-BCA assay. Total protein content ranged from 10 ± 1 to 60 ± 21 μg of avidin per milligram of NPs and increased with increasing chain length of the lipid (Fig. 3, diagonal fill). Blank PLGA NPs (no avidin used in NP formulation) were used as a negative control. “Unconjugated” PLGA NPs (non-lipid conjugated avidin used in formulation) were included to examine non-specific avidin adsorption that might occur during NP manufacture. For all but one group (caprylic acid), avidin–lipid conjugates demonstrated significantly higher avidin incorporation in NPs compared to unmodified avidin alone (##, p<0.05 by Student's t test) (Fig. 3).
Fig. 3.
Lipid and polymer hydrophobicity influence avidin incorporation. Total protein content in NPs was measured using the colorimetric micro bicinchoninic assay (micro-BCA) kit. X-axis denotes lipid group conjugated to avidin, or non avidin–modified “blank” control, NPs. Nanoparticles were made with PLGA polymer containing 50:50 ratio of lactide:glycolide monomer (diagonal fill) or PLA polymer (100:0 ratio of lactide:glycolide monomer, solid fill). Conjugation to butanoic, palmitic, stearic, or linoleic acid resulted in a statistically significant increase in avidin incorporation in both PLGA and PLA NPs when compared to blank NPs and NPs made with unconjugated avidin (##, p<0.05 by Student 's t test). Linoleic acid was found to be a better conjugation partner than palmitic acid, resulting in a 100% increase in avidin incorporation in PLA NPs (***, p<0.01 by Student's t test).
Increasing lipophilicity of the fatty acid–avidin conjugate (i.e. chain length of the lipid) used in nanoparticle formulation resulted in increased incorporation and surface display of avidin; we tested whether increasing the lipohilicity of the constituent polymer would enhance this effect. To assess the effect of polymer hydrophobicity on avidin–lipid incorporation, nanoparticles were made using polymer of the same molecular weight but consisting entirely of lactide repeat units (100:0 lactide:glycolide ratio, or “PLA”) (Fig. 3, solid fill). Use of the more hydrophobic PLA polymer resulted in increases in the incorporation into nanoparticles made with avidin-stearic acid (from 41 ± 4 to 94 ± 18 μg avidin/mg NP) or avidin–linoleic acid (from 59 ± 21 to 92 ± 16 μg avidin/mg NP) compared to analogous PLGA nanoparticles (Fig. 3). Maximum avidin incorporation across all formulations was achieved by the use of the avidin–linoleic acid or avidin–stearic acid conjugates; compared to avidin–palmitic acid, utilization of linoleic acid as a conjugate partner increased avidin incorporation in both PLGA and PLA NPs by approximately 100% (Fig. 3). This difference was highly significant in the PLA NPs (***, p<0.01 by Student's t test) (Fig. 3).
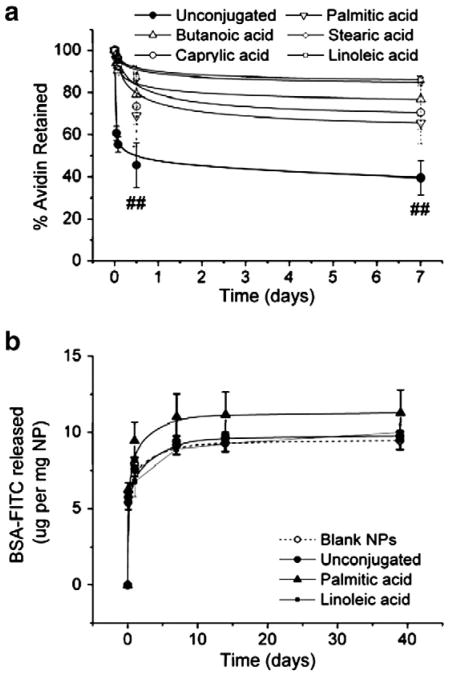
We next tested for the preservation of protein functionality under physiologically relevant conditions. Freeze-dried PLGA NPs were suspended in 1× PBS at 37 °C on a rotating shaker. At fixed times after re-suspension (1 h, 12 h, and 7d), PLGA NPs were subjected to vortexing and vigorous pipetting in 1× PBS for 15 min. The particles were then centrifuged and the supernatant collected for analysis of free avidin. The protein content in the supernatant was measured to determine the percentage of total incorporated avidin that was lost during these washes. Significantly, we observed that up to 40% of the avidin measured in particles made with unconjugated avidin was washed off in the first hour while only 2–6% of incorporated avidin was lost when conjugated to any lipid (Fig. 4a). After 7 d of incubation, approximately 60% of initially associated avidin was lost from NPs made without a fatty acid conjugate (Fig. 4a). Among the conjugated avidin groups, a maximum loss of 34% of initial avidin was observed in NPs modified with avidin–palmitic acid, while only ∼12% loss was observed in particles made with linoleic acid–avidin (Fig. 4a). The total amount of avidin released from lipid-modified NPs ranged from 6 ± 2 to 13 ± 2 μg, with NPs made with linoleic acid–avidin losing less avidin from their surfaces than those made with palmitic acid–avidin despite a much larger total amount of avidin incorporation in the linoleic acid–avidin NPs (Supplemental Fig. 2).
Fig. 4.
Lipid conjugation stabilizes avidin incorporation but does not impede release of encapsulated bovine serum albumin. (a) Avidin-modified NPs were incubated in 1× PBS at 37 °C with rotation to determine the release of avidin under physiologic conditions. Avidin content in supernatant was measured by BCA and compared to Fig. 3a to determine percent loss of initial avidin. Avidin retention among all lipid-modified groups ranged from 66 to 86% of total incorporated avidin and was significantly higher than retention in particles made with the unconjugated protein (##, p<0.05 by Student's t test). Data represent mean±1 standard deviation (n=3 samples). (b) Release of model protein was measured in 1× PBS at 37 °C. BSA-FITC was encapsulated in nanoparticles made without avidin, with avidin, and with avidin–fatty acid conjugates as noted. Modification with avidin–lipid did not significantly alter the release profile.
We have previously demonstrated the controlled delivery of bioactive proteins such as IL-2 [23] and leukemia inhibitory factor [24,31], as well as plasmid DNA [22], from avidin-coated PLGA particles of varying sizes. To investigate whether the avidin surface modification had any effects on the encapsulant release profile, we encapsulated BSA-FITC as a model protein. BSA-FITC-loaded nanoparticles were formulated with avidin–palmitic acid, avidin–linoleic acid, unconjugated avidin control, or no avidin (“blank”) control. No statistically significant differences were observed among any of the groups at each time point (Fig. 4b) and release profiles were consistent with previous reports [32,33].
3.4. Quantification of capture of biotinylated ligands
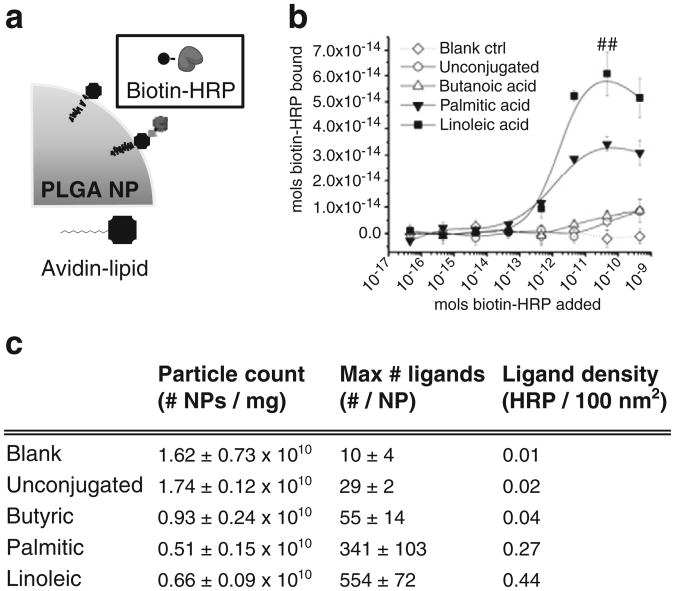
To quantify the biotin-binding capability of the avidin surface modification, avidin–modified PLGA NPs were incubated with increasing doses of biotin–HRP (Fig. 5a). NPs were subjected to three washes in PBS to remove excess/unbound ligands and blank NPs were used as an additional control for nonspecific biotin–HRP binding. Conjugation of avidin to linoleic acid resulted in a 13-fold increase in maximum ligand binding compared to unconjugated avidin NPs (##, p<0.05) with a maximum of 6.1 ± 1.0 × 10−14 mol of biotin–HRP bound per mg NPs (Fig. 5b). Binding was saturated under these conditions (i.e. it did not increase when a 1000-fold excess (10−11 mol) of biotin–HRP was added per milligram of NPs). Particle counts were measured using a Nanosight imaging system in order to determine the average number of NPs per mg of sample and the number of ligands per individual NP (Fig. 5c). These results suggest a high ligand density of approximately 1 ligand per 230 nm2 of surface area for nanoparticles made with avidin–linoleic acid (Fig. 5c), based on an average nanoparticle radius of 100 nm (determined as in Fig. 2b).
Fig. 5.
Quantification of biotinylated ligand capture. Utilization of the avidin–biotin linker system enables versatile and facile modification of NPs after the manufacturing process; schematic shown in (a). (b) Avidin-modified NPs were reconstitutedin1×PBS containing varying concentrations of biotin–HRP. A maximum of 6.1±1.0×10−14 mol biotin–HRP bound per milligram of NPs was achieved using avidin–linoleic acid, representing a 1300% increase over unconjugated avidin or a 170% increase over avidin–palmitic acid (##, p<0.05 by Student's t test). (c) Particle counts (Nanosight) and calculated ligand densities for PLGA NPs made with different avidin–fatty acid conjugates as noted. Ligand density was calculated by dividing the number of bound active ligands by total nanoparticle surface area (based on size measurements as determined in Fig. 2b).
3.5. Effect of ligand density on targeting of CD4+ T lymphocytes
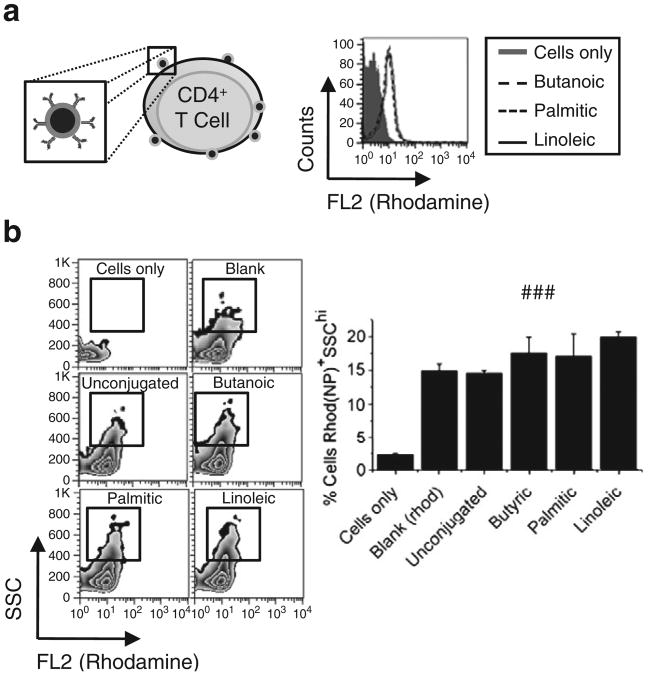
Formulation of PLGA NPs with the avidin–linoleic acid conjugate resulted in a higher avidin surface density than achieved with the palmitic acid–avidin conjugate without any detriment to protein encapsulation or delivery; we examined the functional advantages of this enhanced density on cellular targeting. Rhodamine-loaded NPs with surface-presented avidin were targeted against CD4+ T cells (Supplemental Fig. 3) via capture of biotinylated anti-CD4. Incubation of cells with CD4-targeted, rhodamine-loaded NPs resulted in a statistically significant shift in the population of cells positive for rhodamine (Rhod+): no significant differences in mean channel fluorescence were observed between any NP groups (Fig. 6a). NPs alone are identifiable by low forward scatter (FSC) and high side scatter (SSC) (Supplemental Fig. 3); therefore, the CD4+Rhod+ cell population was further examined for the presence of cells with high side scatter (SSChi); representative FACS plots are shown in (Fig. 6b). A statistically significant increase in the CD4+Rhod+SSChi population was observed when cells were treated with NPs formulated with the linoleic acid–avidin conjugate compared to blank (###, p<0.01 by Student's t test) or unconjugated avidin control NPs (###, p<0.01 by Student's t test), while no statistically significant difference was noted with any other group (Fig. 6b). Using fluorescent microscopy and cryo-EM, we have previously shown that CD4-targeted, cytokine-loaded NPs bind to the exterior of CD4+ T cells and are not internalized after in vitro incubation [24]. The results of the current investigation appear to be consistent with the previous finding; here, the side scatter and FL2 signal of cells is increased by cell surface-bound NPs due to, respectively, the opacity and encapsulated rhodamine of the NPs. We hypothesize that the relatively modest size of the increase in particle binding may be due in part to the relatively high number (∼98,000) of CD4 molecules per CD4+ T cell [34]; interaction between solid nanoparticles and CD4+ T cells has previously been demonstrated with a density of merely 2–3 antibodies per nanoparticle [35]. Enhanced ligand density may prove to be even more potent in targeting applications that involve low target-receptor avidity or low target antigen density [27].
Fig. 6.
Effect of ligand density on cell targeting. Increased density of surface ligands enhances targeting effect. (a) Rhodamine-loaded, avidin-modified NPs were modified with biotinylated antibodies targeting the CD4 T lymphocyte surface ligand. (b) NP binding to CD4+ T cells was evaluated by flow cytometry via increase in side scatter and fluorescence. (c) Increased ligand density increases the number of cells with bound NPs, as shown in FACS plots. (d) Quantification of the percentage of cells with bound NPs revealed an increase associated with incorporation of avidin via the linoleic acid–avidin conjugate (linoleic) compared to blank NPs (Blank (rhod)) and NPs formulated with unconjugated avidin (unconjugated) (###, p<0.01 by Student's t test).
4. Discussion
We previously demonstrated that avidin–palmitic acid conjugates are versatile tools for surface modification of PLGA scaffolds and microparticles [17]. Here, we examined the effects of fatty acid lipophilicity on the incorporation and stability of avidin–lipid conjugates in PLGA nanoparticles. Our results indicate that, among the fatty acids tested, linoleic acid provides the highest density of avidin displayed on the nanoparticle surface. Importantly, improvements in avidin density resulted in an enhancement of ligand binding capacity and a functional increase in nanoparticle targeting to T cells in vitro. Thus, the results of this study may be relevant to nanoparticle targeting and localization to cells in a number of therapeutic settings.
The total amount and stability of avidin bound to the NP surface depended on both lipid and polymer hydrophobicity. Conjugation to lipid significantly increased the amount of incorporated avidin on the NP surface; an approximately 10-fold difference was observed between unconjugated avidin (7–10 μg of avidin per mg NPs) and avidin conjugated to linoleic acid (60–92 μg per mg NP) (Fig. 3). Increasing fatty acid lipophilicity increased avidin incorporation into NP; a conjugation to linoleic acid (chain length C18:2) afforded a 3-fold increase in avidin incorporation compared to conjugation to butyric acid (C4:0) and a 2-fold increase in incorporation compared to palmitic acid (C16:0) (***p<0.01, Fig. 3). Utilization of the more hydrophobic PLA polymer resulted in a modest increase in avidin incorporation (Fig. 3).
Conjugation to lipid increased the stability of avidin incorporation: of the 7–10 μg of unconjugated avidin non-specifically incorporated in NPs, more than 60% was released after one week of incubation in PBS (Fig. 4a). Conversely, conjugation to lipid reduced the loss to 14–34% of total incorporated avidin conjugate (Fig. 4a). Interestingly, while we observed significant differences in the total amount of initially incorporated avidin among lipid-modified groups, the comparative differences between these groups in total amount of avidin lost over one week were relatively small (Supplemental Fig. 2). Moreover, the total amount of avidin added to NP formulation was constant across all groups. Thus, we conclude that the incorporation of avidin and stability of modification is indeed dependent on the presence of the lipid conjugate, as opposed to any inherent adsorption or encapsulation of avidin.
Use of fatty acid–avidin conjugates in PLGA NP formulation did not impact either the rate of release or the encapsulation efficiency of an incorporated protein (Fig. 4b). A prior study indicated that the incorporation of surface avidin on NPs via fatty acid conjugates resulted in reduced release of DNA caused by impaired release [22]. The use of a protein drug payload in this study, as opposed to DNA, likely explains the difference in results; nevertheless, the data presented here demonstrate that NPs modified to display avidin on their surfaces via fatty acid conjugates retain their full potential with regard to protein delivery (Fig. 4b). We do not anticipate that incorporation of more surface-presented avidin via the linoleic acid–avidin conjugate will diminish the potential for encapsulation and sustained delivery of small molecule drugs, which has already been validated in surface-modified PLGA NPs made with the palmitic acid–avidin conjugate [19].
Critically, the functionality of the avidin protein was maintained after conjugation and formulation of nanoparticles (Fig. 5). Biotin–HRP was utilized as a model macromolecular ligand; thus, non-specific binding, avidin functionality, and ligand functionality could be simultaneously assessed. The total avidin incorporation correlated with ability to bind biotinylated ligand. Non-specific binding of biotin–HRP to blank NPs or unconjugated avidin NPs was minimal (Fig. 5). Therefore,we conclude that conjugation of avidin to lipid results in preferential surface presentation of the functional avidin groups. More specifically, the use of the lineoleic acid–avidin conjugate enables superior surface incorporation of avidin compared to the other conjugates assessed. While we measured a high ligand density, on the order of 1 active ligand per 200–300 nm2 of nanoparticle surface area (Fig. 5), we note that steric factors may reduce the availability of avidin binding sites. The results of this study suggest that direct conjugation of ligands to fatty acids may enable further increases in the density of surface ligands on PLGA nanoparticles.
Monoclonal antibodies have been effectively used to target pathological CD4+ T lymphocytes implicated in a variety of autoimmune disease processes in both mice and humans [36–38]. We have previously demonstrated the surface decoration of avidin–modified NPs with biotinylated anti-CD4 for the targeted delivery of cytokine therapeutics to CD4+ T cells, resulting in enhanced cytokine effect in vitro and in vivo [24,31]. Here, we demonstrate a method by which to control and optimize targeting ligand properties. Increased binding of NPs to CD4+ T cells was observed when NPs were manufactured using the avidin–linoleic acid conjugate and subsequently modified with biotinylated antibodies against the mouse CD4+ T cell surface ligand (Fig. 6). This improved methodology for attaching targeting ligands to PLGA nanoparticles holds great promise for several reasons: 1) the density of ligands can be easily manipulated through either particle manufacturing or titration of ligands; 2) this modification can take place after particle manufacture, sparing potentially labile ligands from harsh formulation conditions; 3) incorporation of targeting ligands does not diminish the utility of NPs for protein or small molecule drug delivery, and; 4) the effects of combinations of ligands can be easily investigated without altering the core nanoparticle properties. Thus, the novel surface modification technique described here represents a versatile methodology for the development of biodegradable nanoparticles with enhanced capacity for targeted drug delivery.
Supplementary Material
Acknowledgments
NIH grant EB000487 to W.M.S.
Footnotes
Appendix A. Supplementary data: Supplementary data to this article can be found online at doi: 10. 1016/j.jconrel.2011.06.025.
References
- 1.Saltzman WM. Drug Delivery. Oxford; New York: 2001. [Google Scholar]
- 2.Soppimath KS, Aminabhabi TM, Kulkarni AR, Rudzinski WE. Biodegradable polymeric nanoparticles as drug delivery devices. J Control Release. 2001;70:1–20. doi: 10.1016/s0168-3659(00)00339-4. [DOI] [PubMed] [Google Scholar]
- 3.Anderson JM, Shive MS. Biodegradation and biocompatibility of PLA and PLGA microspheres. Adv Drug Deliv Rev. 1997;28:5–24. doi: 10.1016/s0169-409x(97)00048-3. [DOI] [PubMed] [Google Scholar]
- 4.Moghimi SM, Hunter AC, Murray JC. Long-circulating and target-specific nanoparticles: theory to practice. Pharmacol Rev. 2001;53:283–318. [PubMed] [Google Scholar]
- 5.Davis ME, Chen Z, Shin DM. Nanoparticle therapeutics: an emerging treatment modality for cancer. Nat Rev Drug Discov. 2008;7:771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 6.Brigger I, Dubernet C, Couvreur P. Nanoparticles in cancer therapy and diagnosis. Adv Drug Deliv Rev. 2002;54:631–651. doi: 10.1016/s0169-409x(02)00044-3. [DOI] [PubMed] [Google Scholar]
- 7.Kocbek P, Obermajer N, Cegnar M, Kos J, Kristl J. Targeting cancer cells using PLGA nanoparticles surface modified with monoclonal antibody. J Control Release. 2007;120:18–26. doi: 10.1016/j.jconrel.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 8.Danhier F, Vroman B, Lecouturier N, Crokart N, Pourcelle V, Freichels H, Jerome C, Marchand-Brynaert J, Feron O, Preat V. Targeting of tumor endothelium by RGD-grafted PLGA-nanoparticles loaded with paclitaxel. J Control Release. 2009;140:166–173. doi: 10.1016/j.jconrel.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 9.Farokhzad OC, Cheng J, Teply BA, Sherifi I, Jon S, Kantoff PW, Richie JP, Langer R. Targeted nanoparticle–aptamer bioconjugates for cancer chemotherapy in vivo. Proc Natl Acad Sci U S A. 2006;103:6315–6320. doi: 10.1073/pnas.0601755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao H, Yung LY. Selectivity of folate conjugated polymer micelles against different tumor cells. Int J Pharm. 2008;349:256–268. doi: 10.1016/j.ijpharm.2007.07.040. [DOI] [PubMed] [Google Scholar]
- 11.Cheng J, Teply BA, Sherifi I, Sung J, Luther G, Gu FX, Levy-Nissenbaum E, Radovic-Moreno AF, Langer R, Farokhzad OC. Formulation of functionalized PLGA-PEG nanoparticles for in vivo targeted drug delivery. Biomaterials. 2007;28:869–876. doi: 10.1016/j.biomaterials.2006.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dhar S, Gu FX, Langer R, Farokhzad OC, Lippard SJ. Targeted delivery of cisplatin to prostate cancer cells by aptamer functionalized Pt(IV) prodrug-PLGAPEG nanoparticles. Proc Natl Acad Sci U S A. 2008;105:17356–17361. doi: 10.1073/pnas.0809154105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Acharya S, Dilnawaz F, Sahoo SK. Targeted epidermal growth factor receptor nanoparticle bioconjugates for breast cancer therapy. Biomaterials. 2009;30:5737–5750. doi: 10.1016/j.biomaterials.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 14.Keegan ME, Royce SM, Fahmy T, Saltzman WM. In vitro evaluation of biodegradable microspheres with surface-bound ligands. J Control Release. 2006;110:574–580. doi: 10.1016/j.jconrel.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Beletsi A, Panagi Z, Avgoustakis K. Biodistribution properties of nanoparticles based on mixtures of PLGA with PLGA-PEG diblock copolymers. Int J Pharm. 2005;298:233–241. doi: 10.1016/j.ijpharm.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 16.Yoo HS, Park TG. Biodegradable polymeric micelles composed of doxorubicin conjugated PLGA-PEG block copolymer. J Control Release. 2001;70:63–70. doi: 10.1016/s0168-3659(00)00340-0. [DOI] [PubMed] [Google Scholar]
- 17.Fahmy TM, Samstein RM, Harness CC, Saltzman WM. Surface modification of biodegradable polyesters with fatty acid conjugates for improved drug targeting. Biomaterials. 2005;26:5727–5736. doi: 10.1016/j.biomaterials.2005.02.025. [DOI] [PubMed] [Google Scholar]
- 18.Rajapaksa TE, Stover-Hamer M, Fernandez X, Eckelhoefer HA, Lo DD. Claudin 4-targeted protein incorporated into PLGA nanoparticles can mediate M cell targeted delivery. J Control Release. 2010;142:196–205. doi: 10.1016/j.jconrel.2009.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park J, Fong PM, Lu J, Russell KS, Booth CJ, Saltzman WM, Fahmy TM. PEGylated PLGA nanoparticles for the improved delivery of doxorubicin. Nanomedicine. 2009;5:410–418. doi: 10.1016/j.nano.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cu Y, Saltzman WM. Controlled surface modification with poly(ethylene)glycol enhances diffusion of PLGA nanoparticles in human cervical mucus. Mol Pharm. 2009;6:173–181. doi: 10.1021/mp8001254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demento SL, Eisenbarth SC, Foellmer HG, Platt C, Caplan MJ, Mark Saltzman W, Mellman I, Ledizet M, Fikrig E, Flavell RA, Fahmy TM. Inflammasome-activating nanoparticles as modular systems for optimizing vaccine efficacy. Vaccine. 2009;27:3013–3021. doi: 10.1016/j.vaccine.2009.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cu Y, LeMoellic C, Caplan MJ, Saltzman WM. Ligand-modified gene carriers increased uptake in target cells but reduced DNA release and transfection efficiency. Nanomedicine. 2010;6:334–343. doi: 10.1016/j.nano.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steenblock ER, Fahmy TM. A comprehensive platform for ex vivo T-cell expansion based on biodegradable polymeric artificial antigen-presenting cells. Mol Ther. 2008;16:765–772. doi: 10.1038/mt.2008.11. [DOI] [PubMed] [Google Scholar]
- 24.Park J, Gao W, Whiston R, Strom TB, Metcalfe S, Fahmy TM. Modulation of CD4+ T lymphocyte lineage outcomes with targeted, nanoparticle-mediated cytokine delivery. Mol Pharm. 2011;8:143–152. doi: 10.1021/mp100203a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bandyopadhyay A, Fine RL, Demento S, Bockenstedt LK, Fahmy TM. The impact of nanoparticle ligand density on dendritic-cell targeted vaccines. Biomaterials. 2011;32:3094–3105. doi: 10.1016/j.biomaterials.2010.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fahmy TM, Demento SL, Caplan MJ, Mellman I, Saltzman WM. Design opportunities for actively targeted nanoparticle vaccines. Nanomedicine (Lond) 2008;3:343–355. doi: 10.2217/17435889.3.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fahmy TM, Fong PM, Park J, Constable T, Saltzman WM. Nanosystems for simultaneous imaging and drug delivery to T cells. AAPS J. 2007;9:E171–E180. doi: 10.1208/aapsj0902019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corr M, Slanetz AE, Boyd LF, Jelonek MT, Khilko S, al-Ramadi BK, Kim YS, Maher SE, Bothwell AL, Margulies DH. T cell receptor-MHC class I peptide interactions: affinity, kinetics, and specificity. Science. 1994;265:946–949. doi: 10.1126/science.8052850. [DOI] [PubMed] [Google Scholar]
- 29.Sykulev Y, Brunmark A, Jackson M, Cohen RJ, Peterson PA, Eisen HN. Kinetics and affinity of reactions between an antigen-specific T cell receptor and peptide–MHC complexes. Immunity. 1994;1:15–22. doi: 10.1016/1074-7613(94)90005-1. [DOI] [PubMed] [Google Scholar]
- 30.Livnah O, Bayer EA, Wilchek M, Sussman JL. The structure of the complex between avidin and the dye, 2-(4′-hydroxyazobenzene) benzoic-acid (Haba) FEBS Lett. 1993;328:165–168. doi: 10.1016/0014-5793(93)80986-5. [DOI] [PubMed] [Google Scholar]
- 31.Gao W, Thompson L, Zhou Q, Putheti P, Fahmy TM, Strom TB, Metcalfe SM. Treg versus Th17 lymphocyte lineages are cross-regulated by LIF versus IL-6. Cell Cycle. 2009;8:1444–1450. doi: 10.4161/cc.8.9.8348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vila A, Sanchez A, Tobio M, Calvo P, Alonso MJ. Design of biodegradable particles for protein delivery. J Control Release. 2002;78:15–24. doi: 10.1016/s0168-3659(01)00486-2. [DOI] [PubMed] [Google Scholar]
- 33.Couvreur P, Puisieux F. Nanoparticles and microparticles for the delivery of polypeptides and proteins. Adv Drug Deliv Rev. 1993;10:141–162. [Google Scholar]
- 34.Davis KA, Abrams B, Iyer SB, Hoffman RA, Bishop JE. Determination of CD4 antigen density on cells: role of antibody valency, avidity, clones, and conjugation. Cytometry. 1998;33:197–205. doi: 10.1002/(sici)1097-0320(19981001)33:2<197::aid-cyto14>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 35.Velge-Roussel F, Breton P, Guillon X, Lescure F, Bru N, Bout D, Hoebeke J. Immunochemical characterization of antibody-coated nanoparticles. Experientia. 1996;52:803–806. doi: 10.1007/BF01923993. [DOI] [PubMed] [Google Scholar]
- 36.Horneff G, Burmester GR, Emmrich F, Kalden JR. Treatment of rheumatoid arthritis with an anti-CD4 monoclonal antibody. Arthritis Rheum. 1991;34:129–140. doi: 10.1002/art.1780340202. [DOI] [PubMed] [Google Scholar]
- 37.Onodera K, Lehmann M, Akalin E, Volk HD, Sayegh MH, Kupiec-Weglinski JW. Induction of “infectious” tolerance to MHC-incompatible cardiac allografts in CD4 monoclonal antibody-treated sensitized rat recipients. J Immunol. 1996;157:1944–1950. [PubMed] [Google Scholar]
- 38.Gavett SH, Chen X, Finkelman F, Wills-Karp M. Depletion of murine CD4+ T lymphocytes prevents antigen-induced airway hyperreactivity and pulmonary eosinophilia. Am J Respir Cell Mol Biol. 1994;10:587–593. doi: 10.1165/ajrcmb.10.6.8003337. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.