Abstract
Most psychiatric disorders are moderately to highly heritable. The degree to which genetic variation is unique to individual disorders or shared across disorders is unclear. To examine shared genetic etiology, we use genome-wide genotype data from the Psychiatric Genomics Consortium (PGC) for cases and controls in schizophrenia, bipolar disorder, major depressive disorder, autism spectrum disorders (ASD) and attention-deficit/hyperactivity disorder (ADHD). We apply univariate and bivariate methods for the estimation of genetic variation within and covariation between disorders. SNPs explained 17–29% of the variance in liability. The genetic correlation calculated using common SNPs was high between schizophrenia and bipolar disorder (0.68 ± 0.04 s.e.), moderate between schizophrenia and major depressive disorder (0.43 ± 0.06 s.e.), bipolar disorder and major depressive disorder (0.47 ± 0.06 s.e.), and ADHD and major depressive disorder (0.32 ± 0.07 s.e.), low between schizophrenia and ASD (0.16 ± 0.06 s.e.) and non-significant for other pairs of disorders as well as between psychiatric disorders and the negative control of Crohn’s disease. This empirical evidence of shared genetic etiology for psychiatric disorders can inform nosology and encourages the investigation of common pathophysiologies for related disorders.
The current classification of psychiatric disorders reflects clinical syndromes with largely unknown etiology and is based on historical descriptions provided by prominent clinicians over the last 125 years. Family (including twin and adoption) studies provide consistent evidence that genetic factors are involved in these syndromes1. In principle, family studies allow quantification of the shared genetic etiology of disorders, through the estimation of heritability (the proportion of variance in liability attributable to additive genetic factors), and the genetic correlation between them. However, difficulties in ascertaining samples of sufficient size mean that there are few estimates of genetic correlations. Nonetheless, family studies suggest correlated familial genetic liabilities to bipolar disorder and schizophrenia2,3, bipolar disorder and major depressive disorder2,3, and ASD and ADHD4–6 (Supplementary Table 1). Phenotypic and genetic overlap has also been suggested for ASD and schizophrenia7–11, ASD and bipolar disorder9, bipolar disorder and ADHD12, and major depressive disorder and ADHD13. Some of these relationships have been supported by recent evidence of shared molecular risk factors14–16, but the extent of these relationships remains unclear, given the small proportion of risk associated with individually identified variants.
The genomics era provides new opportunities to explore the shared genetic etiology of disorders. Genome-wide association studies (GWAS) assess common genetic polymorphisms (for example, SNPs) at several hundred thousand positions in the genome. The experimental paradigm of GWAS involves the identification of individual variants associated with case-control status17. However, these data can also be used to estimate the total variance in liability explained by SNPs (SNP heritability, ) through the estimation of genetic similarities (relationships) between cases and controls using SNP genotypes18,19. The pairwise genetic relationships that contribute to the estimate are very small, but the large number of pairwise relationships in a case-control sample generates estimates with reasonable precision. The value is an estimate of the total variance in liability to disease explained by SNPs together. Genetic variation is estimated when case-case pairs and control-control pairs are, on average, more similar across the genome than case-control pairs. The value is a lower bound for total narrow-sense heritability, as the former cannot include contributions from causal variants not tagged by the measured SNPs, mostly less common and rare causal variants. A bivariate extension20 of these genome-wide methods estimates the genetic correlation (rg SNP) explained by SNPs between case-control samples collected independently for two disorders (Online Methods). The correlation is positive when the cases of one disorder show higher genetic similarity to the cases of the other disorder than they do to their own controls. A negative correlation is possible if the cases of one disorder are less similar across the genome to the cases of another disorder than they are to controls of the other disorder. A genetic correlation of zero is estimated if the genome-wide relationship between cases of one disorder is the same with the cases as with the controls of another disorder. As a correlation, a high rg SNP value is achieved when the covariance term between the traits is similar in magnitude to the variance terms. Therefore, we also report the SNP-based coheritability of pairs of disorders, which is the covariance between disorders on the liability scale and allows comparison of the shared liability attributable to SNPs on the same scale as . Here we apply univariate and bivariate methods to the five disorders of the PGC—schizophrenia21, bipolar disorder22, major depressive disorder23, ASD24,25 and ADHD26—analyzed in the PGC Cross-Disorder Group association study25, together with additional ADHD data sets27–30 (Table 1).
Table 1.
Univariate analyses: sample description, SNP-based heritabilities and recurrence risk to first-degree relatives
| Schizophrenia | Bipolar disorder | Major depressive disorder | ASD | ADHD | ||
|---|---|---|---|---|---|---|
| SNPs (imputed) | 915,354 | 995,971 | 962,093 | 982,100 | 917,066 | |
| Cases | 9,087 | 6,704 | 9,041 | 3,303 | 4,163 | |
| Controls | 12,171 | 9,031 | 9,381 | 3,428a | 12,040a | |
| N cohorts | 17 | 11 | 9 | 8 | 8 | |
| Primary reference | 21 | 22 | 23 | 24,25 | 26–30 | |
| CC (s.e.) | 0.41 (0.015) | 0.44 (0.021) | 0.18 (0.017) | 0.31 (0.046) | 0.25 (0.020) | |
|
| ||||||
| Disorder risk for the study-based population (disorder risk, K)b | ||||||
| K | 0.01 | 0.01 | 0.15 | 0.01 | 0.05 | |
|
|
0.23 (0.008) | 0.25 (0.012) | 0.21 (0.021) | 0.17 (0.025) | 0.28 (0.023) | |
| λ1st-SNP (s.e) | 2.10 (0.05) | 2.23 (0.08) | 1.27 (0.03) | 1.75 (0.14) | 1.71 (0.07) | |
| λ1st | 8.8 | 9.6 | 1.5 | 8.7 | 3.5 | |
|
| ||||||
| Lower bound for disorder risk (K) | ||||||
| K | 0.004 | 0.007 | 0.1 | 0.001 | 0.03 | |
|
|
0.19 (0.007) | 0.23 (0.010) | 0.19 (0.018) | 0.11 (0.017) | 0.24 (0.020) | |
| λ1st-SNP (s.e) | 2.14 (0.06) | 2.25 (0.08) | 1.31 (0.03) | 1.79 (0.15) | 1.77 (0.07) | |
| λ1st | 14.4 | 11.7 | 1.7 | 29.4 | 4.5 | |
|
| ||||||
| Upper bound for disorder risk (K) | ||||||
| K | 0.012 | 0.015 | 0.2 | 0.015 | 0.08 | |
|
|
0.24 (0.009) | 0.27 (0.013) | 0.23 (0.023) | 0.19 (0.028) | 0.32 (0.026) | |
| λ1st-SNP (s.e) | 2.10 (0.05) | 2.20 (0.07) | 1.24 (0.02) | 1.74 (0.13) | 1.65 (0.06) | |
| λ1st | 8.0 | 7.7 | 1.4 | 7.0 | 2.8 | |
|
| ||||||
| Heritability estimated from twin/family studies61 | ||||||
| h2 | 0.81 | 0.75 | 0.37 | 0.80 | 0.75 | |
CC is the SNP-based heritability estimated on case-control scale. is the SNP-based heritability on liability scale, given assumed K. All estimates of are highly significantly different from zero. λ1st-SNP is the recurrence risk to first-degree relatives calculated from and K. λ1st is the recurrence risk to first-degree relatives calculated from h2 from twin and/or family studies and K.
RESULTS
SNP heritabilities for the five disorders
In our linear mixed model, we estimate the variance in case-control status explained by SNPs18 (heritability on the observed scale; CC estimates in Table 1). Cases in case-control samples are highly ascertained compared to in the population, and, because the cohorts for different disorders had different proportions of cases, CC estimates were difficult to interpret and compare. For this reason, we report values on the liability scale, in which a linear transformation18 is applied based on a user-specified estimate of the risk of the disorder in the study base population (disorder risk, K). For each disorder, we considered three values of K (Table 1), and we converted values to predicted risk to first-degree relatives (λ1st SNP) given K. We benchmarked the λ1st SNP risk values to risk to first-degree relatives (λ1st), consistent with estimates of heritability reported from family studies given K. Our estimates of λ1st SNP values were robust, and our estimates of values were reasonably robust, to the likely range of K values and show that a key part of the heritabilities or familial risk estimated from family studies is associated with common SNPs. Twice the standard error of estimates approximates the magnitude of the parameter that is possible to detect as being significantly different from zero, given the available sample sizes31.
SNP coheritabilities and SNP correlations (rg SNP)
The relationships between disorders were expressed as SNP-based coheritabilities (Fig. 1). The rg SNP value was high between schizophrenia and bipolar disorder at 0.68 (0.04 standard error (s.e.)), moderate between schizophrenia and major depressive disorder at 0.43 (0.06 s.e.), bipolar disorder and major depressive disorder at 0.47 (0.06 s.e.), and ADHD and major depressive disorder at 0.32 (0.07 s.e.), low between schizophrenia and ASD at 0.16 (0.06 s.e.) and non-significant for other pairs of disorders (Supplementary Table 1). The rg SNP value for correlation is expected to be equal to the rg value from family studies only if genetic correlation is the same across the allelic frequency spectrum and if the linkage disequilibrium (LD) between genotyped and causal variants is similar for both disorders. The sample size for ASD was the smallest but still could detect correlations of >|0.18| different from zero in bivariate analyses with all other disorders.
Figure 1.
Evidence for genome-wide pleiotropy between psychiatric disorders. Proportion of variance in liability (SNP-based heritability) and proportion of covariance in liability between disorder (SNP-based coheritability) for five major psychiatric disorders. The 95% error bars represent the estimates ± 1.96 s.e. SCZ, schizophrenia; MDD, major depressive disorder; BPD, bipolar disorder.
Our results provide empirical evidence that schizophrenia, bipolar disorder and major depressive disorder have shared genetic etiology. Because some schizophrenia and bipolar disorder cohorts were collected in the same clinical environments, we investigated the possible impact of the non-independent collection of schizophrenia and bipolar disorder samples sets but found no significant change in the estimates related to this (Supplementary Table 2). The correlation between schizophrenia and ASD was significant but small (0.16, 0.06 s.e.; P = 0.0071). In general, our analyses suggested that, whereas common genetic variants contribute to both childhood-onset disorders (ASD and ADHD) and disorders usually diagnosed after childhood (schizophrenia, bipolar disorder and major depressive disorder), the sharing of common variants between these groups is modest.
The pattern of our results (in which pairs of disorders demonstrated genetic overlap) was consistent with polygenic profile score32 results from PGC cross-disorder analyses25. The profile score method uses SNP associations from one disorder to construct a linear predictor in another disorder. The profile scores explained small but significant proportions of the variance25, expressed as Nagelkerke’s R2 (maximum of 2.5% between schizophrenia and bipolar disorder). To achieve high R2 values requires accurate estimation of the effect sizes of individual SNPs and depends on the size of the discovery sample. In contrast, our approach uses SNPs to estimate genome-wide similarities between pairs of individuals, resulting in unbiased estimates of the relationships between disorders, with larger sample sizes generating smaller standard errors for the estimates. Our estimates were on the liability scale, allowing direct comparison to genetic parameters estimated in family studies, whereas a genetic interpretation of Nagelkerke’s R2 values is less straightforward33.
Genomic partitioning of SNP heritabilities and coheritabilities
The heritabilities explained by SNPs can be partitioned according to SNP annotation by the estimation of genetic similarity matrices from multiple, non-overlapping SNP sets. For the five disorders and the five disorder pairs showing significant SNP correlation, we partitioned the and SNP-based coheritabilities explained by functional annotation, allocating SNPs to one of three sets: (i) SNPs in genes preferentially expressed in the central nervous system (CNS+)34,35, (ii) SNPs in other genes and (iii) SNPs not in genes, with genes defined by 50-kb boundaries extending from their start and stop positions. The SNPs in the CNS+ gene set represented 0.20 of the total set, both in number and megabases of DNA. However, the proportion of the variance explained by SNPs attributable to this SNP set was significantly greater than 0.20 for schizophrenia (0.30; P = 7.6 × 10−8) and bipolar disorder (0.32; P = 5.4 × 10−6) and for schizophrenia and bipolar disorder coheritability (0.37; P = 8.5 × 10−8) (Fig. 2 and Supplementary Table 3). For other disorders or pairs of disorders, the estimates explained by CNS+ SNPs did not differ from the values expected by chance (Supplementary Table 3), although their large standard errors suggest that we cannot address this question with precision. For data from the schizophrenia and bipolar disorder pair, we also partitioned the heritabilities explained by SNPs by minor allele frequency (MAF) (Supplementary Table 4) and by chromosome (Supplementary Fig. 1). The high standard errors on estimates limited interpretation, but the results are consistent with a polygenic architecture comprising many common variants of small effect dispersed throughout the genome. The MAF partitioning suggests that a key part of the variance explained by SNPs is attributable to common causal variants (this was investigated in detail for schizophrenia35), but the low contribution to the total variance explained by SNPs with MAF of <0.1 reflects, at least in part, under-representation of SNPs with low MAFs in the analysis (minimum MAF = 0.01) relative to those present in the genome.
Figure 2.
Genomic partitioning of SNP-based heritability and SNP-based coheritability by annotation. Shown is the proportion of SNPs attributable to genes in the CNS+ set (red), the proportion of SNP-based heritability attributable to SNPs in the CNS+ set (dark green), the proportion of SNP-based coheritability attributable to SNPs in the CNS+ set (light green) and the proportion of SNP-based heritability for Crohn’s disease attributed to SNPs in the CNS+ set (orange). The 95% error bars represent the estimates ± 1.96 s.e. ***P < 1 × 10−5 in a test of whether the proportion of heritability explained by SNPs was equal to the proportion of SNP for the CNS+ set.
Within-disorder heterogeneity
To benchmark the estimates of genetic sharing across disorders, we estimated sharing between data subsets for the same disorder. We split the data for each disorder into two or three independent sets and estimated values for each subset and the SNP-based coher-itability between each pair of subsets within a disorder (Fig. 3a and Supplementary Table 5). The estimates of from the data subsets were typically higher than the estimate from the combined sample; we note that published estimates from individual cohorts of bipolar disorder18, major depressive disorder36 and ASD37 were also higher. Because both traits in these data subset bivariate analyses are for the same disorder, the SNP-based coheritability is also an estimate of for the disorder, but these estimates were generally lower than the estimates of SNP-based heritability from individual data subsets. These results generated SNP-based correlations that were less than 1, sometimes significantly so (Supplementary Table 5). The SNP-based correlation between schizophrenia and bipolar disorder (0.68, 0.04 s.e.) was of comparable magnitude to the SNP-based correlations between bipolar disorder data sets (0.63, 0.11 s.e.; 0.88, 0.09 s.e.; and 0.55, 0.10 s.e.; Fig. 3a,b, SNP-based coherit-abilities), adding further weight to the conclusion that schizophrenia and bipolar disorder may be part of the same etiological spectrum.
Figure 3.
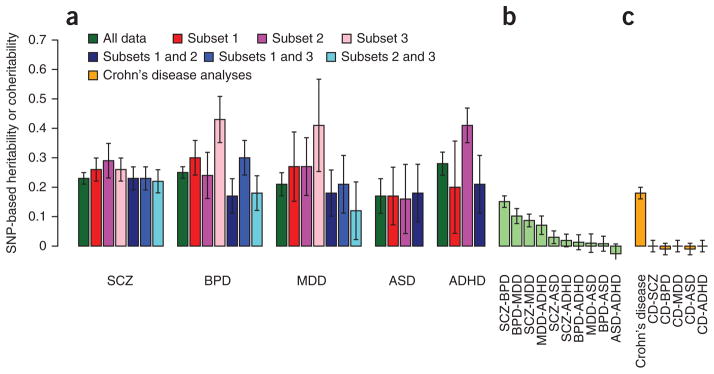
SNP-based heritabilities and coheritabilities. (a) For each disorder, SNP-based heritabilities are estimated from univariate analyses of the full data set (dark green) or of sample subsets (red and pink bars). These heritabilities are also estimated from bivariate analyses in which different subsets of the same disorder comprise the two traits (blue). Test of the heterogeneity of estimates, P value for Cochran’s Q: schizophrenia, 0.3; bipolar disorder, 1 × 10−6; major depressive disorder, 4 × 10−3; ADHD, 9 × 10−6; ASD, 0.99; Higgins’ I2: schizophrenia, 21%; bipolar disorder, 86%; major depressive disorder, 71%; ADHD, 91%; ASD, 0%). (b) For comparison, the coheritabilities using the full data sets reported in Figure 1 are shown. (c) As a negative control, estimates of coheritabilities with Crohn’s disease, a disease not expected to be genetically related to psychiatric disorders, are shown. We estimated 95% error bars using ± 1.96 s.e.
The estimates of heritability from both univariate (Fig. 3a, red and pink bars) and bivariate (Fig. 3a, blue bars) analyses are more heterogeneous for bipolar disorder, major depressive disorder and ADHD than they are for schizophrenia and ASD. Several factors could explain why SNP-based heritabilities from univariate analyses of a single data set could generate higher estimates than bivariate analyses of independent data sets35, including loss of real signal or dilution of artifacts. Loss of real signal might occur because individual cohorts are more homogeneous, both phenotypically (for example, owing to use of the same assessment protocols) and genetically (for example, because LD between causal variants and analyzed SNPs might be higher within than between cohorts). Artifacts could also generate consistent differences in case genotypes relative to control genotypes within case-control data sets. In the derivation of our methodology18, we emphasized that any factors making SNP genotypes of cases more similar to those of other cases and making the genotypes of controls more similar to those of other controls would produce SNP-based heritability. The fitting as covariates of principal components derived from the SNP data corrects both for population stratification and for genotyping artifacts, but residual population stratification could remain, although this bias should be small38. Partitioning SNP-based heritability by chromosome in analyses where each chromosome was fitted individually compared to analyses where all chromosomes were fitted jointly is an empirical strategy to assess residual stratification35,39, and we found no evidence of this type of stratification here (Supplementary Fig. 1). Stringent quality control (as applied here) helps to remove artifacts, but artifactual differences between cases and controls might remain, particularly for data sets in which cases and controls have been genotyped independently40. As more data sets accumulate, the contributions from artifacts are diluted because the random directional effects of artifacts (including population stratification) are not consistent across data sets. For this reason, significant SNP-based coheritabilities between subsets of the same disorder are unlikely to reflect artifacts and provide a lower bound for SNP-based heritability.
Pseudocontrols
One strategy adopted in GWAS to guard against artifacts from population stratification is to genotype family trio samples (cases and their parents) and then analyze the data as a case-control sample, with controls generated as genomic complements of the cases (pseudo-controls). ADHD subset 1 and most of the ASD sample comprised case-pseudocontrol samples and, consistent with this strategy limiting the impact of artifacts from population stratification or genotyping, it is noted that the lowest SNP-based heritability for the five psychiatric disorders was for ASD and that the estimate of SNP-based heritability was lower for ADHD subset 1 than for ADHD subset 2. However, under a polygenic model, assortative mating41 or preferential ascertainment of multiplex families could diminish the expected mean difference in liability between pseudocontrols and cases37, which would result in an underestimation of SNP-based heritability from case-pseudocontrol compared to case-control analyses and would also result in nonzero estimates of SNP-based heritability from pseudocontrol-control analyses, as shown in analysis of ASD data37.
SNP-based coheritabilities with Crohn’s disease
As a negative control analysis, we conducted bivariate analyses between each of the PGC data sets and Crohn’s disease samples from the International IBD Genetics Consortium (IIBDGC)42. Although onset of major depressive disorder is not uncommon after diagnosis with Crohn’s disease43 and although gastrointestinal pathology is a common comorbidity with ASD44, there is no strong evidence of a familial relationship between psychiatric disorders and Crohn’s disease. Despite substantial hSNP 2 values for Crohn’s disease (0.19, 0.01 s.e.), none of the SNP-based coheritabilities with the psychiatric disorders differed significantly from zero (Fig. 3c, Supplementary Table 6 and Supplementary Note). Lastly, genomic partitioning by annotation of the variance in Crohn’s disease explained by SNPs showed, as expected, no excess of variance attributable to SNPs in the CNS+ gene set (Fig. 2). Our results provide no evidence of common genetic pleiotropy in Crohn’s disease and ASD, consistent with a non-genetic, for example, microbial45, explanation for the comorbidity of gastrointestinal symptoms in ASD.
Potential impact of misclassification of disorders
Misclassification among disorders could inflate estimates of genetic correlation and/or coheritability46. Indeed, some level of misclas-sification in psychiatric disorders is expected. For example, longitudinal studies47,48 of first admissions with psychosis showed that, with long-term follow-up, ~15% of subjects initially diagnosed with bipolar disorder were rediagnosed with schizophrenia, whereas ~4% of schizophrenia diagnoses were reclassified as bipolar disorder. Cases selected for GWAS contributing to PGC are more likely to have achieved a stable diagnosis compared to first-admission cases. However, assuming these levels of misclassification, the genetic correlation between bipolar disorder and schizophrenia for true diagnoses is still high, estimated46 to be 0.55. Likewise, because a modest proportion of cases diagnosed with major depressive disorder, when followed over time, ultimately meet criteria for bipolar disorder49, our estimated genetic correlation between these two disorders may be modestly inflated by misclassification. However, if moderate-to-high genetic correlations between the major adult disorders are true, then overlapping symptoms and misdiagnosis among these disorders might be expected. The rg SNP value between schizophrenia and major depressive disorder is also unlikely to reflect misdiagnosis because misclassification between these disorders is rare49. Excluding 5 of the 18 PGC schizophrenia cohorts containing schizoaffective disorder cases21 (Supplementary Table 7) or major depressive disorder cohorts ascertained from community rather than clinical settings (Supplementary Table 8) had little impact on rg SNP estimates.
DISCUSSION
Our results show direct, empirical, quantified molecular evidence for an important genetic contribution to the five major psychiatric disorders. The estimates for each disorder—schizophrenia, 0.23 (0.01 s.e.), bipolar disorder, 0.25 (0.01 s.e.), major depressive disorder, 0.21 (0.02), ASD, 0.17 (0.02 s.e.) and ADHD, 0.28 (0.02 s.e.)—are considerably less than the heritabilities estimated from family studies (Table 1). Yet, they show that common SNPs make an important contribution to the overall variance, implying that additional individual, common SNP associations can be discovered as sample size increases50. values are a lower bound for narrow-sense heritability because they exclude contributions from some causal variants (mostly rare variants) not associated with common SNPs. Although SNP-based heritability estimates are similar for major depressive disorder and other disorders, much larger sample sizes will be needed, as high risk for a disorder implies lower power for equal sample size51. The values are all lower than those reported for height (0.45, 0.03 s.e.)39, but the estimates are in the same ballpark as those reported for other complex traits and diseases using the same quality control pipeline, such as for body mass index (BMI) (0.17, 0.03 s.e.)39, Alzheimer’s disease (0.24, 0.03 s.e.), multiple sclerosis (0.30, 0.03 s.e.) and endometriosis (0.26, 0.04 s.e.)40.
Our results show molecular evidence of the sharing of genetic risk factors across key psychiatric disorders. Traditionally, quantification of the genetic relationship between disorders has been thwarted by the need for cohorts of families or twins assessed for multiple disorders. Problems of achieving genetically informative samples of sufficient size and without associated ascertainment biases for the rarer psychiatric disorders have meant that few studies have produced meaningful estimates of genetic correlations. Notably, our estimates of heritability and genetic correlation are made using very distant genetic relationships between individuals, both within and between disorders, so that shared environmental factors are unlikely to contaminate our estimates. Likewise, our estimates are unlikely to be confounded by non-additive genetic effects, as the coefficients of non-additive genetic variance between very distant relatives are negligible52.
The estimates of SNP-based genetic correlation (rg SNP) between disorders reflect the genome-wide pleiotropy of variants tagged by common SNPs, and whether these are the same as correlations across the allelic frequency spectrum may differ between pairs of disorders. For example, a high rg SNP value but a low genetic correlation estimated from family studies (rg) could indicate that the same common variants contribute to genetic susceptibility for both disorders, although the diagnostic-specific variants are less common variants. For this reason, the comparison of rg SNP with rg estimated from family studies is not straightforward. Nonetheless, we benchmark our estimates in this way, calculating the increased risk of disorder B in first-degree relatives of probands with disorder A (λA,B) from the rg SNP value to allow comparison with literature values (Supplementary Table 1). A meta-analysis53 reported increased risk of bipolar disorder in first-degree relatives of probands with schizophrenia compared to first-degree relatives of control probands (λSCZ,BPD) of 2.1, which implies a maximum genetic correlation between the disorders of 0.3 (assuming that the disorder risks for schizophrenia and bipolar disorder are both 1% and their heritabilities are 81% and 75%, respectively; Table 1). However, a large-scale Swedish family and adoption study54 estimated the genetic correlation between schizophrenia and bipolar disorder to be +0.60, similar to that found here. Profiling scoring analysis using genome-wide SNPs32 was the first method to clearly demonstrate a genetic relationship based on molecular data, but quantification as a genetic correlation was not reported. The evidence of shared genetic risk factors for schizophrenia and bipolar disorder was strengthened by our analyses of the CNS+ gene set in which we saw a clear enrichment in variants shared by these two disorders.
Our finding of a substantial rg SNP of +0.43 between schizophrenia and major depressive disorder is notable and contrary to conventional wisdom about the independence of familial risk for these disorders. However, because major depressive disorder is common, even a high genetic correlation implies only modest incremental risk. Assuming the disorder risks and heritabilities for schizophrenia and major depressive disorder given in Table 1, then the genetic correlation between them of 0.43 predicts increased risk of major depressive disorder in first-degree relatives of probands with schizophrenia compared to first-degree relatives of control probands (λSCZ,MDD) of 1.6. In fact, meta-analysis of five interview-based research studies of families are broadly consistent with our results (λSCZ,MDD = 1.5, 95% confidence interval (CI) = 1.2–1.8; Supplementary Table 9), suggesting that familial coaggregation of major depressive disorder and schizophrenia reflects genetic effects rather than resulting from living in a family environment that includes a severely ill family member. If replicated by future work, our empirical molecular genetic evidence of a partly shared genetic etiology for schizophrenia and major depressive disorder would have key nosological and research implications, incorporating major depressive disorder as part of a broad psychiatric genetic spectrum. A shared genetic etiology for bipolar disorder and major depressive disorder has been shown in family studies2,3, but the rg SNP value of 0.47 was lower than the estimate of 0.65 from a twin study55.
Our results show a small but significant rg SNP value between schizophrenia and ASD. A lower genetic correlation between schizophrenia and ASD than between schizophrenia and bipolar disorder is consistent with Swedish national epidemiological studies, which reported higher odds ratios in siblings for schizophrenia and bipolar disorder54 than for schizophrenia and ASD9. These results imply a modest overlap of common genetic etiological processes in these two disorders, consistent with emerging evidence from the discovery of copy number variants, in which both shared variants (for example, 15q13.3, 1q2.1 and 17q12 deletions56,57) and mutations in the same genes although with different variants (deletions associated with schizophrenia and duplications associated with autism and vice-versa10). The small ASD sample size thwarted attempts at further explorative partitioning of the SNP-based coheritability for schizophrenia and ASD.
The lack of overlap between ADHD and ASD is unexpected and is not consistent with family and data linkage studies, which indicate that the two disorders share genetic risk factors5,6,58,59. Some rare copy number variants are seen in both disorders16. As noted above, the use of pseudocontrols for many of the ASD and ADHD cohorts may affect all results for these disorders. Ideally, we would investigate the impact of pseudocontrols, given the hierarchical diagnostic system (autism but not autism spectrum is an exclusion criterion for most ADHD data sets), on estimates of SNP-based coheritability, but the small ASD sample size prohibits such analyses. We also found no overlap between ADHD and bipolar disorder, despite support from meta-analysis results of an increased risk for ADHD in relatives of individuals with bipolar disorder I (a subtype of bipolar disorder with more extreme manic symptoms than the other major bipolar disorder subtype) and an increased risk for bipolar disorder I in relatives of individuals with ADHD12. These findings could mean that the familial link between the two disorders is mediated by environmental risk factors or that shared genetic factors are not part of the common allelic spectrum. Alternatively, the etiological link between ADHD and bipolar disorder might be limited to bipolar disorder I or early-onset bipolar disorder12, which, therefore, is difficult for us to detect. Our finding of genetic overlap between ADHD and major depressive disorder is consistent with evidence from studies showing increased rates of ADHD in the families of depressed probands and increased rates of depression in families of probands with ADHD12,13.
Our results should be interpreted in the context of four potentially important methodological limitations. First, any artifacts that make SNP genotypes more similar between cases than between cases and controls could inflate estimates of SNP-based heritability18, but to a much lesser extent for SNP-based coheritability. Second, the sample sizes varied considerably across the five disorders. Although values are expected to be unbiased, estimates from smaller samples are accompanied by larger standard errors, blurring their interpretation. Third, although applying similar diagnostic criteria, the clinical methods of ascertainment and the specific study protocols, including which specific interview instruments were employed, varied across sites. We cannot now determine the degree to which our results might have been influenced by between-site differences in the kinds of patients seen or in their assessments. Fourth, by combining samples from geographic regions, contributions from less common associated variants specific to particular populations are diluted compared to what would have been achieved if the same sample size had been ascertained from a single homogeneous population.
In summary, we report SNP-based heritabilities that are significantly greater than zero for all five disorders studied. We have used the largest psychiatric GWAS data sets currently available, and our results provide key pointers for future studies. Our results demonstrate that the dearth of significant associations from psychiatric GWAS so far, particularly for major depressive disorder, ASD and ADHD, reflects lack of power to detect common associated variants of small effect rather than the absence of such variants. Hence, as sample sizes increase, the success afforded to other complex genetic diseases50 in increasing the understanding of their etiologies is achievable for psychiatric disorders, as is already being shown for schizophrenia60. We also provide evidence of substantial sharing of the genetic risk variants tagged by SNPs between schizophrenia and bipolar disorder, bipolar disorder and major depressive disorder, schizophrenia and major depressive disorder, ADHD and major depressive disorder, and, to a lesser extent, between schizophrenia and ASD. Our results will likely contribute to the efforts now under way to base psychiatric nosology on a firmer empirical footing. Furthermore, they will encourage investigations into shared pathophysiologies across disorders, including potential clarification of common therapeutic mechanisms.
ONLINE METHODS
Data and quality control
A summary of the data available for analysis is listed in Table 1 and comprise data used in the PGC–Cross-Disorder Group analysis25 together with newly available ADHD samples27–30. Data upload to the PGC central server follows strict guidelines to ensure local ethics committee approval for all contributed data (PGC; see URLs). Data from all study cohorts were processed through the stringent PGC pipeline25. Imputation of autosomal SNPs used CEU (Utah residents of Northern and Western European ancestry) and TSI (Toscani in Italia) HapMap Phase 3 data as the reference panel21. For each analysis (univariate or bivariate), we retained only SNPs that had MAF of >0.01 and imputation R2 of >0.6 in all contributing cohort subsamples (imputation cohorts). Different quality control strategies were investigated in detail for the raw and PGC imputed genotyped data of the International Schizophrenia Consortium, a subset of the PGC schizophrenia sample35. The Crohn’s disease samples from IIBDGC42 were processed through the same quality control and imputation pipeline as the PGC data, generating a data set of 5,054 cases and 11,496 controls from 6 imputation cohorts.
In each analysis, individuals were excluded to ensure that all cases and controls were completely unrelated in the classical sense, so that no pairs of individuals had a genome-wide similarity relationship greater than 0.05 (equivalent to about second cousins). This procedure removed ancestry outliers (over and above those already removed in the PGC quality control pipeline; Supplementary Fig. 2) and ensured that overlapping control sets were allocated randomly between disorders in the bivariate analyses. Exact numbers of cases and controls used in each analysis are listed in Supplementary Tables 1–8.
Linear mixed model for estimation of SNP-based heritability and coherit-ability
We used the methods presented in Lee et al.18,35. Briefly, we estimated the variance in case-control status explained by all SNPs using a linear mixed model
where y is a vector of case (y = 1) or control (y = 0) status (the observed scale), β is a vector for fixed effects of the overall mean (intercept), sex, sample cohort and 20 ancestry principal components, g is the vector of random additive genetic effects based on aggregate SNP information and e is a vector of random error effects. X is an incidence matrix for the fixed effects relating these effects to individuals. The variance structure of phenotypic observations is
where is additive genetic variance tagged by the SNPs, is error variance, A is the realized similarity relationship matrix estimated from SNP data19 and I is an identity matrix. All variances were estimated on the observed case-control scale and were transformed to the liability scale, which requires specification of the disorder risk K to estimate . Risk to first-degree relatives was calculated from K and on the basis of the liability threshold model62.
The bivariate analyses used a bivariate extension of equation (1) (ref. 20). The two traits were measured in different individuals, but the equations were related through the genome-wide similarities estimated from SNPs. Genetic and residual variances for the traits were estimated as well as the genetic covariance σg12. The genetic correlation coefficient (rg) was calculated by (σg12/(σg1σg2)) and is approximately the same on the observed case-control scale as on the liability scale20 and so does not depend on specifications of K. The covariance σg12 can be transformed to the liability scale, accounting for assumed disorder risks and proportions of cases and controls in the samples of each disorder20, and it equals the coheritability52 rgh1h2. We used the approximated χ2 test statistic (estimate/s.e.)2 to test whether estimates were significantly different from zero. We checked that this simple approximation agreed well with the more formal and computer-intensive likelihood ratio test for several examples. Heterogeneity of SNP-based heritabilities was tested using Cochran’s Q (ref. 63) and Higgins’ I2 (ref. 64) values, acknowledging potential non-independence of the six estimates (three subsets plus three subset pairs).
Disorder risk for the study-based population (disorder risk, K)
Estimates of and SNP-based coheritability from the linear model are on the case-control scale and so depend partly on the proportion of cases and controls in the sample. Transformation to the liability scale allowed benchmarking of to estimates of heritability from family studies, and the transformation accounts for the proportion of cases in the sample and depends on the assumed disorder risk (K). The appropriate choice of K depends on the definitions of both the phenotype (including ascertainment strategy) and the population, which might differ between cohorts. We considered lower and upper bounds for K in Table 1 to cover the range of possible values. rg SNP estimates are independent of scale and hence are not dependent on the choice of K.
Genome-partitioning linear mixed model
We partitioned the variance explained by the SNPs in several ways. For example, for the univariate linear model
with
where n is the number of subsets from any non-overlapping partitioning of SNPs; n = 22 for the joint analysis by chromosome, n = 5 for the analysis by MAF bin and n = 3 for the analysis of SNP by gene annotation in which SNPs were classed as CNS+ genes (2,725 genes representing 547 Mb), SNPs in other genes (14,804 genes representing 1,069 Mb) and the remaining SNPs not in genes. Gene boundaries were set at ± 50 kb from the 5′ and 3′ UTRs of each gene, and CNS+ genes were the four sets identified by Raychaudhuri et al.34 (one set comprised genes expressed preferentially in the brain compared to other tissues, and the other three sets comprised genes annotated to be involved in neuronal activity, learning and synapses). The CNS+ set was found to explain more of the SNP-based heritability than expected by chance for schizophrenia35. All methods have been implemented into the freely available GCTA software65.
aSome cohorts include cases and pseudocontrols, where pseudocontrols are the genomic complements of the cases derived from genotyping of proband-parent trios.
bUsed in Figures 1 and 3 Supplementary Tables 1–8.
Supplementary Material
Acknowledgments
This research was directly supported by the Australian Research Council (FT0991360 and DE130100614) and the Australian National Health and Medical Research Council (613608, 1011506 and 1047956). The PGC Cross-Disorder Group is supported by National Institute of Mental Health (NIMH) grant U01 MH085520. Statistical analyses were carried out on the Genetic Cluster Computer (see URLs), which is financially supported by the Netherlands Scientific Organization (NOW; 480-05-003; principal investigator D.P.) along with a supplement from the Dutch Brain Foundation and VU University. Numerous (>100) grants from government agencies along with substantial private and foundation support worldwide enabled the collection of phenotype and genotype data, without which this research would not be possible; grant numbers are listed in primary PGC publications or in the Supplementary Note.
S Hong Lee1, Stephan Ripke2,3, Benjamin M Neale2,3, Stephen V Faraone4,5, Shaun M Purcell2,3,6, Roy H Perlis3,7, Bryan J Mowry1,8, Anita Thapar9,10, Michael E Goddard11,12, John S Witte13, Devin Absher14, Ingrid Agartz15,16, Huda Akil17, Farooq Amin18, Ole A Andreassen15,19, Adebayo Anjorin20, Richard Anney21, Verneri Anttila2, Dan E Arking22, Philip Asherson23, Maria H Azevedo24, Lena Backlund25, Judith A Badner26, Anthony J Bailey27, Tobias Banaschewski28, Jack D Barchas29, Michael R Barnes30, Thomas B Barrett31, Nicholas Bass20, Agatino Battaglia32, Michael Bauer33, Mònica Bayés34, Frank Bellivier35–38, Sarah E Bergen3,7,39, Wade Berrettini40, Catalina Betancur41–43, Thomas Bettecken44, Joseph Biederman45, Elisabeth B Binder44, Donald W Black46, Douglas H R Blackwood47, Cinnamon S Bloss48,49, Michael Boehnke50,51, Dorret I Boomsma52–54, Gerome Breen23,55, René Breuer56, Richard Bruggeman57, Paul Cormican21, Nancy G Buccola58, Jan K Buitelaar59, William E Bunney60, Joseph D Buxbaum61, William F Byerley62,63, Enda M Byrne1, Sian Caesar64, Wiepke Cahn65, Rita M Cantor66, Miguel Casas67,68, Aravinda Chakravarti22, Kimberly Chambert3, Khalid Choudhury20, Sven Cichon69–72, C Robert Cloninger73, David A Collier23, Edwin H Cook74, Hilary Coon75, Bru Cormand76–78, Aiden Corvin21, William H Coryell46, David W Craig79, Ian W Craig23, Jennifer Crosbie80, Michael L Cuccaro81, David Curtis82, Darina Czamara44,83, Susmita Datta84, Geraldine Dawson85–87, Richard Day88, Eco J De Geus52–54, Franziska Degenhardt69,71, Srdjan Djurovic15,89, Gary J Donohoe21, Alysa E Doyle90, Jubao Duan91, Frank Dudbridge92, Eftichia Duketis93, Richard P Ebstein94, Howard J Edenberg95,96, Josephine Elia40,97, Sean Ennis98, Bruno Etain35,38,99,100, Ayman Fanous101,102, Anne E Farmer23, I Nicol Ferrier103, Matthew Flickinger50,51, Eric Fombonne104,105, Tatiana Foroud96, Josef Frank56, Barbara Franke59, Christine Fraser9,10, Robert Freedman106, Nelson B Freimer107, Christine M Freitag93, Marion Friedl108, Louise Frisén25, Louise Gallagher21, Pablo V Gejman91, Lyudmila Georgieva9,10, Elliot S Gershon26, Daniel H Geschwind109,110, Ina Giegling108, Michael Gill21, Scott D Gordon111, Katherine Gordon-Smith9,64, Elaine K Green112, Tiffany A Greenwood113, Dorothy E Grice114,115, Magdalena Gross116, Detelina Grozeva9, Weihua Guan50,51,117, Hugh Gurling20, Lieuwe De Haan118, Jonathan L Haines119, Hakon Hakonarson120,121, Joachim Hallmayer122, Steven P Hamilton62, Marian L Hamshere9,123, Thomas F Hansen124,125, Annette M Hartmann108, Martin Hautzinger126, Andrew C Heath73, Anjali K Henders111, Stefan Herms69,72, Ian B Hickie127, Maria Hipolito128, Susanne Hoefels116, Peter A Holmans9,123, Florian Holsboer44, Witte J Hoogendijk129, Jouke-Jan Hottenga52,54, Christina M Hultman39, Vanessa Hus130, Andrés Ingason124,125, Marcus Ising44, Stéphane Jamain35,38,99,100, Edward G Jones131,256, Ian Jones9,10, Lisa Jones64, Jung-Ying Tzeng132, Anna K Kähler39, René S Kahn65, Radhika Kandaswamy20, Matthew C Keller133, James L Kennedy134, Elaine Kenny21, Lindsey Kent135, Yunjung Kim136, George K Kirov9,10, Sabine M Klauck137, Lambertus Klei138, James A Knowles139, Martin A Kohli44, Daniel L Koller96, Bettina Konte108, Ania Korszun140, Lydia Krabbendam141, Robert Krasucki20, Jonna Kuntsi23, Phoenix Kwan50,51, Mikael Landén39,142, Niklas Långström39, Mark Lathrop143, Jacob Lawrence20, William B Lawson128, Marion Leboyer35,38,99,100, David H Ledbetter144, Phil H Lee7, Todd Lencz145–147, Klaus-Peter Lesch148,149, Douglas F Levinson150, Cathryn M Lewis23, Jun Li151, Paul Lichtenstein39, Jeffrey A Lieberman152, Dan-Yu Lin153, Don H Linszen154, Chunyu Liu155, Falk W Lohoff40, Sandra K Loo107,156, Catherine Lord157, Jennifer K Lowe109,110, Susanne Lucae44, Donald J MacIntyre47, Pamela A F Madden73, Elena Maestrini158, Patrik K E Magnusson39, Pamela B Mahon159, Wolfgang Maier116, Anil K Malhotra145–147, Shrikant M Mane160, Christa L Martin144, Nicholas G Martin111, Manuel Mattheisen71,125,161,162, Keith Matthews88, Morten Mattingsdal15,163, Steven A McCarroll3, Kevin A McGhee47, James J McGough164, Patrick J McGrath152, Peter McGuffin23, Melvin G McInnis165, Andrew McIntosh47,166, Rebecca McKinney113, Alan W McLean47,166, Francis J McMahon167, William M McMahon75, Andrew McQuillin20, Helena Medeiros139, Sarah E Medland111, Sandra Meier56, Ingrid Melle15,19, Fan Meng17, Jobst Meyer168, Christel M Middeldorp52,54, Lefkos Middleton169, Vihra Milanova170, Ana Miranda171, Anthony P Monaco172,173, Grant W Montgomery111, Jennifer L Moran3, Daniel Moreno-De-Luca174, Gunnar Morken175,176, Derek W Morris21, Eric M Morrow177,178, Valentina Moskvina9,123, Pierandrea Muglia179, Thomas W Mühleisen69,71,180, Walter J Muir47,166,256, Bertram Müller-Myhsok44,83, Michael Murtha181, Richard M Myers14, Inez Myin-Germeys141, Michael C Neale102, Stan F Nelson107, Caroline M Nievergelt113, Ivan Nikolov9,10, Vishwajit Nimgaonkar182,183, Willem A Nolen184, Markus M Nöthen69,71, John I Nurnberger96,185, Evaristus A Nwulia128, Dale R Nyholt111, Colm O’Dushlaine3, Robert D Oades186, Ann Olincy106, Guiomar Oliveira24,187, Line Olsen124,125, Roel A Ophoff 107,188,189, Urban Osby25, Michael J Owen9,10, Aarno Palotie190, Jeremy R Parr103, Andrew D Paterson191,192, Carlos N Pato139, Michele T Pato139, Brenda W Penninx53,54,193, Michele L Pergadia73, Margaret A Pericak-Vance81, Benjamin S Pickard47,166, Jonathan Pimm20, Joseph Piven87, Danielle Posthuma194–196, James B Potash46, Fritz Poustka93, Peter Propping71, Vinay Puri20, Digby J Quested197, Emma M Quinn21, Josep Antoni Ramos-Quiroga67,68, Henrik B Rasmussen124,125, Soumya Raychaudhuri2,3, Karola Rehnström190, Andreas Reif 198, Marta Ribasés67,199, John P Rice200, Marcella Rietschel56, Kathryn Roeder201, Herbert Roeyers202, Lizzy Rossin3, Aribert Rothenberger203, Guy Rouleau204, Douglas Ruderfer6, Dan Rujescu108, Alan R Sanders91, Stephan J Sanders174,181,205,206, Susan L Santangelo207,208, Joseph A Sergeant209, Russell Schachar80, Martin Schalling25, Alan F Schatzberg210, William A Scheftner211, Gerard D Schellenberg212, Stephen W Scherer213, Nicholas J Schork48,214, Thomas G Schulze159,215, Johannes Schumacher71, Markus Schwarz216, Edward Scolnick3, Laura J Scott50,51, Jianxin Shi217, Paul D Shilling113, Stanley I Shyn218, Jeremy M Silverman115, Susan L Slager219, Susan L Smalley107,156, Johannes H Smit53,193, Erin N Smith48,214, Edmund J S Sonuga-Barke202,220, David St. Clair221, Matthew State174,181,205, Michael Steffens222, Hans-Christoph Steinhausen223–225, John S Strauss226, Jana Strohmaier56, T Scott Stroup227, James S Sutcliffe228, Peter Szatmari229–231, Szabocls Szelinger79, Srinivasa Thirumalai232, Robert C Thompson17, Alexandre A Todorov73, Federica Tozzi179, Jens Treutlein56, Manfred Uhr44, Edwin J C G van den Oord233, Gerard Van Grootheest53,193, Jim Van Os141, Astrid M Vicente234–236, Veronica J Vieland237, John B Vincent226, Peter M Visscher1,238, Christopher A Walsh239–242, Thomas H Wassink46, Stanley J Watson17, Myrna M Weissman243, Thomas Werge124,125,244, Thomas F Wienker245, Ellen M Wijsman246,247, Gonneke Willemsen52,53, Nigel Williams9,10, A Jeremy Willsey181,205, Stephanie H Witt56, Wei Xu192, Allan H Young103,248, Timothy W Yu249, Stanley Zammit9,10, Peter P Zandi250, Peng Zhang50,51,165, Frans G Zitman251, Sebastian Zöllner50,51,165, International Inflammatory Bowel Disease Genetics Consortium (IIBDGC)252, Bernie Devlin138, John R Kelsoe113,253, Pamela Sklar6, Mark J Daly2,3, Michael C O’Donovan9,10, Nicholas Craddock9,10, Patrick F Sullivan136, Jordan W Smoller3,7, Kenneth S Kendler102,254,255,257 & Naomi R Wray1,257
Footnotes
The University of Queensland, Queensland Brain Institute, Brisbane, Queensland, Australia.
Analytic and Translational Genetics Unit, Massachusetts General Hospital and Harvard Medical School, Boston, Massachusetts, USA.
Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, Massachusetts, USA.
Department of Psychiatry, State University of New York (SUNY) Upstate Medical University, Syracuse, New York, USA.
Department of Neuroscience and Physiology, SUNY Upstate Medical University, Syracuse, New York, USA.
Division of Psychiatric Genomics, Department of Psychiatry, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Psychiatric and Neurodevelopmental Genetics Unit, Massachusetts General Hospital, Boston, Massachusetts, USA.
Queensland Centre for Mental Health Research, Wacol, Queensland, Australia.
Medical Research Council (MRC) Centre for Neuropsychiatric Genetics and Genomics, Cardiff University School of Medicine, Cardiff, UK.
Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University School of Medicine, Cardiff, UK.
Biosciences Research Division, Department of Environment and Primary Industries Victoria, Melbourne, Victoria, Australia.
Faculty of Land and Environment, University of Melbourne, Melbourne, Victoria, Australia.
Department of Epidemiology and Biostatistics, University of California, San Francisco, San Francisco, California, USA.
HudsonAlpha Institute of Biotechnology, Huntsville, Alabama, USA.
KG Jebsen Centre for Psychosis Research, Institute of Clinical Medicine, University of Oslo, Oslo, Norway.
Department of Research, Diakonhjemmet Hospital, Oslo, Norway.
Molecular Psychiatry Laboratory, Molecular and Behavioral Neuroscience Institute, University of Michigan, Ann Arbor, Michigan, USA.
Department of Psychiatry and Behavioral Sciences, Atlanta Veterans Affairs Medical Center, Emory University, Atlanta, Georgia, USA.
Division of Mental Health and Addiction, Oslo University Hospital, Oslo, Norway.
Mental Health Sciences Unit, University College London, London, UK.
Department of Psychiatry, Trinity College Dublin, Dublin, Ireland.
McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
MRC Social, Genetic and Developmental Psychiatry (SGDP) Centre, The Institute of Psychiatry, King’s College London, London, UK.
Faculty of Medicine, University of Coimbra, Coimbra, Portugal.
Department of Molecular Medicine and Surgery, Center for Molecular Medicine, Karolinska Institutet, Stockholm, Sweden.
Department of Psychiatry, University of Chicago, Chicago, Illinois, USA.
Department of Psychiatry, University of British Columbia, Vancouver, British Columbia, Canada.
Department of Child and Adolescent Psychiatry and Psychotherapy, Central Institute of Mental Health, Medical Faculty Mannheim, University of Heidelberg, Mannheim, Germany.
Department of Psychiatry, Weill Medical College, Cornell University, New York, New York, USA.
GlaxoSmithKline, London, UK.
Portland Veterans Affairs Medical Center, Portland, Oregon, USA.
Stella Maris Institute for Child and Adolescent Neuropsychiatry, Calambrone, Pisa, Italy.
Department of Psychiatry and Psychotherapy, Carl Gustav Carus University Hospital, Dresden, Germany.
Centro Nacional de Análisis Genómico (CNAG), Parc Científic de Barcelona (PCB), Barcelona, Spain.
Institut National de la Santé et de la Recherche Médicale (INSERM) U955, Psychiatrie Génétique, Créteil, France.
Université Denis Diderot, Paris, France.
Assistance Publique–Hôpitaux de Paris (AP-HP), Groupe Hospitalier Saint-Louis, Lariboisiere, F Widal, Departement de Psychiatrie, Paris, France.
ENBREC (European Network of Bipolar Research Expert Centres) Group, Fondation FondaMental, Créteil, France.
Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm, Sweden.
Department of Psychiatry, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
INSERM U952, Paris, France.
Centre National de la Recherche Scientifique (CNRS) Unité Mixte de Recherche (UMR) 7224, Paris, France.
Université Pierre et Marie Curie, Paris, France.
Max Planck Institute of Psychiatry, Munich, Germany.
Clinical and Research Programs in Pediatric Psychopharmacology and Adult ADHD, Massachusetts General Hospital and Harvard Medical School, Boston, Massachusetts, USA.
Department of Psychiatry, University of Iowa, Iowa City, Iowa, USA.
Division of Psychiatry, University of Edinburgh, Royal Edinburgh Hospital, Edinburgh, UK.
The Scripps Translational Science Institute, La Jolla, California, USA.
Scripps Health, La Jolla, California, USA.
Department of Biostatistics, School of Public Health, University of Michigan, Ann Arbor, Michigan, USA.
Center for Statistical Genetics, School of Public Health, University of Michigan, Ann Arbor, Michigan, USA.
Department of Biological Psychology, VU University, Amsterdam, The Netherlands.
EMGO+ (ExtraMuraalGeneeskundig Onderzoek) Institute for Health and Care Research, Amsterdam, The Netherlands.
Neuroscience Campus Amsterdam, Amsterdam, The Netherlands.
National Institute of Heath Research (NIHR) Biomedical Research Centre for Mental Health, South London, London, UK and Maudsley National Health Service (NHS) Trust and Institute of Psychiatry, London, UK.
Department of Genetic Epidemiology in Psychiatry, Central Institute of Mental Health, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany.
Department of Psychiatry, University Medical Center Groningen, University of Groningen, Groningen, The Netherlands.
School of Nursing, Louisiana State University Health Sciences Center, New Orleans, Louisiana, USA.
Department of Cognitive Neuroscience, Donders Institute for Brain, Cognition and Behavior, Radboud University Medical Centre, Nijmegen, The Netherlands.
Department of Psychiatry and Human Behavior, University of California–Irvine, Irvine, California, USA.
Seaver Autism Center for Research and Treatment, Department of Psychiatry, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Department of Psychiatry, University of California, San Francisco, San Francisco, California, USA.
NCIRE (Northern California Institute of Q Research and Education), San Francisco, California, USA.
Department of Psychiatry, Birmingham University, Birmingham, UK.
Department of Psychiatry, Rudolf Magnus Institute of Neuroscience, University Medical Center, Utrecht, The Netherlands.
David Geffen School of Medicine, University of California, Los Angeles, Los Angeles, California, USA.
Department of Psychiatry, Hospital Universitari Vall d’Hebron, CIBERSAM (Centro de Investigación Biomédica en el Area de Salud Mental), Barcelona, Spain.
Department of Psychiatry and Legal Medicine, Universitat Autònoma de Barcelona, Barcelona, Spain.
Department of Genomics, Life & Brain Center, University of Bonn, Bonn, Germany.
Institute of Neuroscience and Medicine (INM-1), Research Center Jülich, Jülich, Germany.
Institute of Human Genetics, University of Bonn, Bonn, Germany.
Division of Medical Genetics, Department of Biomedicine, University of Basel, Basel, Switzerland.
Department of Psychiatry, Washington University School of Medicine, St. Louis, Missouri, USA.
Department of Psychiatry, Institute for Juvenile Research, University of Illinois, Chicago, Illinois, USA.
Department of Psychiatry, University of Utah, Salt Lake City, Utah, USA.
Departament de Genètica, Facultat de Biologia, Universitat de Barcelona, Barcelona, Spain.
Biomedical Network Research Centre on Rare Diseases (CIBERER), Barcelona, Spain.
Institut de Biomedicina de la Universitat de Barcelona (IBUB), Barcelona, Spain.
The Translational Genomics Research Institute, Phoenix, Arizona, USA.
Neurosciences and Mental Health Program, The Hospital for Sick Children, University of Toronto, Toronto, Ontario, Canada.
John P. Hussman Institute for Human Genomics, University of Miami, Miami, Florida, USA.
East London NHS Foundation Trust, Queen Mary, University of London, London, UK.
Munich Cluster for Systems Neurology (SyNergy), Munich, Germany.
Genetics Institute, University College London, London, UK.
Autism Speaks, New York, New York, USA.
Department of Psychiatry, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA.
Carolina Institute for Developmental Disabilities, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA.
Division of Neuroscience, Medical Research Institute, University of Dundee, Ninewells Hospital & Medical School, Dundee, UK.
Department of Medical Genetics, Oslo University Hospital, Oslo, Norway.
Psychiatric and Neurodevelopmental Genetics Unit, Massachusetts General Hospital and Harvard Medical School, Boston, Massachusetts, USA.
Department of Psychiatry and Behavioral Sciences, NorthShore University Health System and University of Chicago, Evanston, Illinois, USA.
Department of Non-Communicable Disease Epidemiology, London School of Hygiene and Tropical Medicine, London, UK.
Department of Child and Adolescent Psychiatry, Psychosomatics and Psychotherapy, JW Goethe University Frankfurt, Frankfurt, Germany.
Psychology Department, National University of Singapore, Singapore.
Department of Biochemistry and Molecular Biology, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Department of Medical and Molecular Genetics, Indiana University School of Medicine, Indianapolis, Indiana, USA.
AI Dupont Hospital for Children, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
School of Medicine, Medical Science University College, Dublin, Ireland.
Université Paris Est, Faculté de Médecine, Créteil, France.
AP-HP, Hôpital H Mondor–A Chenevier, Département de Psychiatrie, Créteil, France.
Department of Psychiatry, Georgetown University School of Medicine, Washington, DC, USA.
Virginia Institute of Psychiatric and Behavioral Genetics, Virginia Commonwealth University, Richmond, Virginia, USA.
Institute of Neuroscience, Newcastle University, Newcastle upon Tyne, UK.
Department of Psychiatry, Oregon Health & Science University, Portland, Oregon, USA.
Institute for Development & Disability, Oregon Health & Science University, Portland, Oregon, USA.
Department of Psychiatry, University of Colorado Denver, Aurora, Colorado, USA.
Center for Neurobehavioral Genetics, University of California, Los Angeles, Los Angeles, California, USA.
Department of Psychiatry, University of Halle, Halle, Germany.
Department of Neurology, David Geffen School of Medicine, University of California, Los Angeles, Los Angeles, California, USA.
Center for Autism Research and Treatment, Semel Institute, David Geffen School of Medicine, University of California, Los Angeles, Los Angeles, California, USA.
Queensland Institute of Medical Research, Brisbane, Queensland, Australia.
Department of Biomedical and Biological Sciences, Plymouth University, Plymouth, UK.
Department of Psychiatry, University of California, San Diego, La Jolla, California, USA.
Division of Tics, OCD and Related Disorders, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Department of Psychiatry, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
Department of Psychiatry, University of Bonn, Bonn, Germany.
Division of Biostatistics, University of Minnesota, Minneapolis, Minnesota, USA.
Department of Psychiatry, Academic Medical Centre, University of Amsterdam The Netherlands.
Center for Human Genetics Research, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
The Center for Applied Genomics, Division of Human Genetics, The Children’s Hospital of Philadelphia, Philadelphia, Pennsylvania, USA.
Department of Pediatrics, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania, USA.
Department of Psychiatry, School of Medicine, Stanford University, Stanford, California, USA.
Biostatistics and Bioinformatics Unit, Cardiff University, Cardiff, UK.
Institute of Biological Psychiatry, Copenhagen University Hospital, Roskilde, Denmark.
The Lundbeck Initiative for Integrative Psychiatric Research, iPSYCH, Roskilde, Denmark.
Department of Clinical and Developmental Psychology, Eberhard Karls University of Tübingen, Tübingen, Germany.
Brain and Mind Research Institute, University of Sydney, Sydney, New South Wales, Australia.
Department of Psychiatry and Behavioral Sciences, Howard University College of Medicine, Washington, DC, USA.
Department of Psychiatry, Erasmus Medical Center, Rotterdam, The Netherlands.
Department of Psychology, University of Michigan, Ann Arbor, Michigan, USA.
Center for Neuroscience, University of California, Davis, Davis, California, USA.
Bioinformatics Research Center, North Carolina State University, Raleigh, North Carolina, USA.
Department of Psychology, University of Colorado, Boulder, Colorado, USA.
Psychiatric Neurogenetics Section, Centre for Addiction and Mental Health, Toronto, Ontario, Canada.
School of Medicine, University of St Andrews, St Andrews, UK.
Department of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA.
Division of Molecular Genome Analysis, German Cancer Research Center (DKFZ), Heidelberg, Germany.
Department of Psychiatry, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania, USA.
Department of Psychiatry, Zilkha Neurogenetic Institute, Keck School of Medicine, University of Southern California, Los Angeles, California, USA.
Wolfson Institute of Preventitive Medicine, Queen Mary University of London, London, UK.
Department of Psychiatry and Neuropsychology, Maastricht University Medical Centre, South Limburg Mental Health Research and Teaching Network, Maastricht, The Netherlands.
Institute of Neuroscience and Physiology, University of Gothenburg, Gothenburg, Sweden.
Centre National de Genotypage, Evry, France.
Geisinger Health System, Autism and Developmental Medicine Institute, Danville, Pennsylvania, USA.
Department of Psychiatry, Division of Research, The Zucker Hillside Hospital Division of the North Shore, Long Island Jewish Health System, Glen Oaks, New York, USA.
Center for Psychiatric Neuroscience, The Feinstein Institute of Medical Research, Manhasset, New York, USA.
Department of Psychiatry and Behavioral Science, Albert Einstein College of Medicine of Yeshiva University, Bronx, New York, USA.
Division of Molecular Psychiatry, ADHD Clinical Research Unit, Department of Psychiatry, Psychosomatics and Psychotherapy, University of Würzburg, Würzburg, Germany.
Department of Psychiatry and Psychology, School for Mental Health and Neuroscience (MHENS), Maastricht University, Maastricht, The Netherlands.
Department of Psychiatry and Behavioral Sciences, Stanford University, Stanford, California, USA.
Department of Human Genetics, University of Michigan, Ann Arbor, Michigan, USA.
New York State Psychiatric Institute, Columbia University, New York, New York, USA.
Department of Biostatistics, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA.
Department of Psychiatry, Academic Medical Centre University of Amsterdam, Amsterdam, The Netherlands.
Department of Psychiatry, Institute of Human Genetics, University of Illinois at Chicago, Chicago, Illinois, USA.
Department of Psychiatry and Biobehavioral Science, University of California, Los Angeles, Los Angeles, California, USA.
Center for Autism and the Developing Brain, Weill Cornell Medical College, White Plains, New York, USA.
Department of Pharmacy and Biotechnology, University of Bologna, Bologna, Italy.
Department of Psychiatry & Behavioral Sciences, Johns Hopkins University, Baltimore, Maryland, USA.
Yale Center for Genome Analysis, Orange, Connecticut, USA.
Department of Biomedicine, Aarhus University, Aarhus, Denmark.
Department of Genomic Mathematics, University of Bonn, Bonn, Germany.
Sørlandet Hospital, Kristiansand, Norway.
Child and Adolescent Psychiatry, Semel Institute, David Geffen School of Medicine, University of California, Los Angeles, Los Angeles, California, USA.
Department of Psychiatry, University of Michigan, Ann Arbor, Michigan, USA.
Molecular Medicine Centre, University of Edinburgh, Edinburgh, UK.
National Institute of Mental Health, US National Institutes of Health, Bethesda, Maryland, USA.
Department of Neurobehavioral Genetics, Trier University, Trier, Germany.
Neuroepidemiology and Ageing Research, School of Public Health, Imperial College London, London, UK.
Department of Psychiatry, First Psychiatric Clinic, Alexander University Hospital, Sofia, Bulgaria.
Department of Developmental and Educational Psychology, University of Valencia, Valencia, Spain.
Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, UK.
Office of the President, Tufts University, Medford, Massachusetts, USA.
Department of Psychiatry, Yale University, New Haven, Connecticut, USA.
Department of Psychiatry, St. Olavs Hospital, Trondheim, Norway.
Department of Neuroscience, Norwegian University of Science and Technology, Trondheim, Norway.
Department of Molecular Biology, Cell Biology and Biochemistry, Brown University, Providence, Rhode Island, USA.
Department of Psychiatry and Human Behavior, Brown University, Providence, Rhode Island, USA.
Neurosciences Centre of Excellence in Drug Discovery, GlaxoSmithKline Research and Development, Verona, Italy.
Life & Brain Center, University of Bonn, Bonn, Germany.
Child Study Center, Yale University, New Haven, Connecticut, USA.
Department of Psychiatry, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
Department of Human Genetics, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
Department of Psychiatry, Groningen University Medical Center, Groningen, The Netherlands.
Department of Psychiatry, Indiana University School of Medicine, Indianapolis, Indiana, USA.
Clinic for Child and Adolescent Psychiatry and Psychotherapy, University of Duisburg-Essen, Essen, Germany.
Research and Clinical Training Department, Pediatric Hospital, Centro Hospitalar e Universitário Coimbra, Coimbra, Portugal.
Department of Human Genetics, University of California, Los Angeles, Los Angeles, California, USA.
Department of Psychiatry, University Medical Center Utrecht, Utrecht, The Netherlands.
Sanger Institute, Hinxton, Cambridge, UK.
Program in Genetics and Genomic Biology, The Hospital for Sick Children, Toronto, Ontario, Canada.
Dalla Lana School of Public Health, University of Toronto, Toronto, Ontario, Canada.
Department of Psychiatry, VU University Medical Center, Amsterdam, The Netherlands.
Department of Functional Genomics, VU University, Amsterdam, The Netherlands.
Department of Clinical Genetics, VU Medical Center, Amsterdam, The Netherlands.
Department of Child and Adolescent Psychiatry, Erasmus University Medical Center, Rotterdam, The Netherlands.
Academic Department of Psychiatry, University of Oxford, Oxford, UK.
Department of Psychiatry, University of Würzburg, Würzburg, Germany.
Psychiatric Genetics Unit, Vall d’Hebron Research Institute, Barcelona, Spain.
Division of Biostatistics, Washington University School of Medicine, St. Louis, Missouri, USA.
Department of Statistics, Carnegie Mellon University, Pittsburgh, Pennsylvania, USA.
Department of Experimental Clinical & Health Psychology, Ghent University, Ghent, Belgium.
Child and Adolescent Psychiatry, University Medicine Göttingen, Göttingen, Germany.
Department of Neurology and Neurosurgery, McGill University, Montreal, Quebec, Canada.
Department of Genetics, Yale University, New Haven, Connecticut, USA.
Program on Neurogenetics, Yale University, New Haven, Connecticut, USA.
Department of Psychiatry, Maine Medical Center, Portland, Maine, USA.
Department of Psychiatry, Harvard Medical School, Boston, Massachusetts, USA.
Department of Clinical Neuropsychology, VU University, Amsterdam, The Netherlands.
Department of Psychiatry and Behavioral Science, Stanford University School of Medicine, Palo Alto, California, USA.
Rush Ambulatory Behavioral Health, Rush University Medical Center, Chicago, Illinois, USA.
Department of Pathology and Laboratory Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
The Centre for Applied Genomics, The Hospital for Sick Children, Toronto, Ontario, Canada.
The Scripps Research Institute, La Jolla, California, USA.
Department of Psychiatry & Psychotherapy, University of Göttingen, Göttingen, Germany.
Psychiatric Center Nordbaden, Wiesloch, Germany.
Division of Cancer Epidemiology and Genetics, National Cancer Institute, Bethesda, Maryland, USA.
Department of Psychiatry and Behavioral Sciences, University of Washington, Seattle, Washington, USA.
Mayo Clinic, Rochester, Minnesota, USA.
Developmental Brain & Behaviour Laboratory, Academic Unit of Psychology, University of Southampton, Southampton, UK.
Institute of Medical Sciences, University of Aberdeen, Foresterhill, Aberdeen, UK.
Research Department, Federal Institute for Drugs and Medical Devices (BfArM), Bonn, Germany.
Research Unit of Child and Adolescent Psychiatry, Aalborg University Hospital, Aalborg, Denmark.
Clinical Psychology and Epidemiology, University of Basel, Basel, Switzerland.
Department of Child and Adolescent Psychiatry, University of Zurich, Zurich, Switzerland.
Molecular Neuropsychiatry and Development Laboratory, Centre for Addiction and Mental Health, Toronto, Ontario, Canada.
Department of Psychiatry, Columbia University, New York, New York, USA.
Vanderbilt Brain Institute, Vanderbilt University, Nashville, Tennessee, USA.
Department of Psychiatry, University of Toronto, Toronto, Ontario Canada.
Neurosciences and Mental Health Program, Hospital for Sick Children, Toronto, Ontario Canada.
Centre for Addiction and Mental Health, Toronto, Ontario, Canada.
Oxford Health NHS Foundation Trust, Marlborough House Secure Unit, Milton Keynes, UK.
Center for Biomarker Research and Personalized Medicine, Virginia Commonwealth University, Richmond, Virginia, USA.
Instituto Nacional de Saude Dr Ricardo Jorge, Lisbon, Portugal.
BioFIG—Center for Biodiversity, Functional and Integrative Genomics, Campus da FCUL, Campo Grande, Lisbon, Portugal.
Instituto Gulbenkian de Cîencia, Lisbon, Portugal.
Battelle Center for Mathematical Medicine, Nationwide Children’s Hospital, Columbus, Ohio, USA.
The University of Queensland, Diamantina Institute, Brisbane, Queensland, Australia.
Howard Hughes Medical Institute, Children’s Hospital Boston, Boston, Massachusetts, USA.
Division of Genetics, Children’s Hospital Boston, Boston, Massachusetts, USA.
Department of Neurology, Harvard Medical School Center for Life Sciences, Boston, Massachusetts, USA.
Department of Pediatrics, Harvard Medical School Center for Life Sciences, Boston, Massachusetts, USA.
Columbia University College of Physicians and Surgeons, New York, New York, USA.
Faculty of Health and Medical Science, University of Copenhagen, Copenhagen, Denmark.
Institute of Medical Biometry, University of Bonn, Bonn, Germany.
Department of Biostatistics, University of Washington, Seattle, Washington, USA.
Department of Medicine, University of Washington, Seattle, Washington, USA.
Centre for Affective Disorders, Institute of Psychiatry, King’s College London, London, UK.
Division of Genetics, Children’s Hospital Boston, Harvard Medical School, Boston, Massachusetts, USA.
Department of Mental Health, Johns Hopkins University, Baltimore, Maryland, USA.
Department of Psychiatry, Leiden University Medical Center, Leiden, The Netherlands.
A list of members appears in the Supplementary Note.
Department of Psychiatry, Special Treatment and Evaluation Program (STEP), Veterans Affairs San Diego Healthcare System, San Diego, California, USA.
Department of Human and Molecular Genetics, Virginia Commonwealth University, Richmond, Virginia, USA.
Department of Psychiatry, Virginia Commonwealth University, Richmond, Virginia, USA.
Deceased.
These authors contributed equally to this work.
URLs. PGC, https://pgc.unc.edu/; Genetic Cluster Computer, http://www.geneticcluster.org/; GCTA, http://www.complextraitgenomics.com/software/gcta/.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
Project conception: K.S.K., N.R.W. and J.W.S. Analysis: S.H.L. and N.R.W. Writing of the manuscript: N.R.W., S.H.L., K.S.K. and S.V.F. Quality control for PGC data: S. Ripke and B.M.N. Revisions to the manuscript: S.M.P., J.W.S., R.H.P., B.J.M., P.F.S., A.T., C.O., M.J.D., R.D.O. and J.B. Statistical advice: M.E.G. and J.S.W. Data access: D.P. PGC Workgroup Chairs: M.J.D. (analysis), S.V.F. (ADHD), M.J.D. and B.D. (co-chairs ASD), J.K. and P. Sklar (co-chairs bipolar disorder), P.F.S. (major depressive disorder), M.C.O. (schizophrenia) and J.W.S. and K.S.K. (co-chairs cross-disorder group). Collection, genotyping and analysis for PGC Working Groups. PGC ADHD Working Group: B.M.N., S.V.F., A.T., R.A., P.A., T. Banaschewski, M. Bayés, J.B., J.K.B., M.C., B.C., J.C., A.E.D., R.P.E., J.E., B.F., C.M.F., L. Kent, J.K., K.-P.L., S.K.L., J.M., J.J.M., S.E.M., J.M.S., A. Miranda, S.F.N., R.D.O., J.A.R.-Q., A. Reif, M. Ribasés, H.R., A. Rothenberger, J.A.S., R.S., S.L. Smalley, E.J.S.S.-B., H.-C.S., A.A.T. and N.W. PGC ASD Working Group: R.A., D.E.A., A.J.B., A.B., C.B., J.D. Buxbaum, A. Chakravarti, E.H.C., H.C., M.L.C., G.D., E.D., S.E., E.F., C.M.F., L. Gallagher, D.H.G., M. Gill, D.E.G., J.L.H., H.H., J.H., V.H., S.M.K., L. Klei, D.H. Ledbetter, C. Lord, J.K.L., E.M., S.M.M., C.L.M., W.M.M., A.P.M., D.M.-D.-L., E.M.M., M. Murtha, G.O., A.P., J.R.P., A.D.P., M.A.P.-V., J. Piven, F.P., K. Rehnström, K. Roeder, G.R., S.J.S., S.L. Santangelo, G.D.S., S.W.S., M. State, J.S. Sutcliffe, P. Szatmari, A.M.V., V.J.V., C.A.W., T.H.W., E.M.W., A.J.W., T.W.Y., B.D. and M.J.D. PGC BPD Working Group: S.M.P., D.A., H.A., O.A.A., A.A., L.B., J.A.B., J.D. Barchas, T.B.B., N.B., M. Bauer, F.B., S.E.B., W.B., D.H.R.B., C.S.B., M. Boehnke, G.B., R. Breuer, W.E.B., W.F.B., S. Caesar, K. Chambert, S. Cichon, D.A.C., A. Corvin, W.H.C., D.W.C., R.D., F. Degenhardt, S. Djurovic, F. Dudbridge, H.J.E., B.E., A.E.F., I.N.F., M. Flickinger, T.F., J.F., C.F., L.F., E.S.G., M. Gill, K.G.-S., E.K.G., T.A.G., D.G., W.G., H.G., M.L.H., M. Hautzinger, S. Herms, M. Hipolito, P.A.H., C.M.H., S.J., E.G.J., I.J., L.J., R. Kandaswamy, J.L.K., G.K.K., D.L.K., P.K., M. Landén, N.L., M. Lathrop, J. Lawrence, W.B.L., M. Leboyer, P.H.L., J. Li, P.L., D.-Y.L., C. Liu, F.W.L., S.L., P.B.M., W.M., N.G.M., M. Mattheisen, K.M., M. Mattingsdal, K.A.M., P.M., M.G.M., A. McIntosh, R.M., A.W.M., F.J.M., A. McQuillin, S.M., I.M., F.M., G.W.M., J.L.M., G.M., D.W.M., V. Moskvina, P.M., T.W.M., W.J.M., B.M.-M., R.M.M., C.M.N., I.N., V.N., M.M.N., J.I.N., E.A.N., C.O., U.O., M.J.O., B.S.P., J.B.P., P.P., E.M.Q., S. Raychaudhuri, A. Reif, J.P.R., M. Rietschel, D. Ruderfer, M. Schalling, A.F.S., W.A.S., N.J.S., T.G.S., J. Schumacher, M. Schwarz, E.S., L.J.S., P.D.S., E.N.S., D.S.C., M. Steffens, J.S. Strauss, J. Strohmaier, S.S., R.C.T., F.T., J.T., J.B.V., S.J.W., T.F.W., S.H.W., W.X., A.H.Y., P.P.Z., P.Z., S. Zöllner, J.R.K., P. Sklar, M.J.D., M.C.O. and N.C. PGC MDD Working Group: M.R.B., T. Bettecken, E.B.B., D.H.R.B., D.I.B., G.B., R. Breuer, S. Cichon, W.H.C., I.W.C., D. Czamara, E.J.D.G., F. Degenhardt, A.E.F., J.F., S.D.G., M. Gross, S.P.H., A.C.H., A.K.H., S. Herms, I.B.H., F.H., W.J.H., S. Hoefels, J.-J.H., M.I., I.J., L.J., J.-Y. T., J.A.K., M.A.K., A.K., W.B.L., D.F.L., C.M.L., D.-Y.L., S.L., D.J.M., P.A.F.M., W.M.,. N.G.M., M. Mattheisen, P.J.M., P.M., A. McIntosh, A.W.M., C.M.M., L.M., G.W.M., P.M., B.M.-M., W.A.N., M.M.N., D.R.N., B.W.P., M.L.P., J.B.P., M. Rietschel, W.A.S., T.G.S., J. Shi, S.I.S., S.L. Slager, J.H.S., M. Steffens, F.T., J.T., M.U., E.J.C.G.v.d.O., G.V.G., M.M.W., G.W., F.G.Z., P.F.S. and N.R.W. PGC SCZ Working Group: S. Ripke, B.M.N., S.M.P., B.J.M., I.A., F.A., O.A.A., M.H.A., N.B., D.W.B., D.H.R.B., R. Bruggeman, N.G.B., W.F.B., W.C., R.M.C., K. Choudhury, S. Cichon, C.R.C., P.C., A. Corvin, D. Curtis, S. Datta, S. Djurovic, G.J.D., J.D., F. Dudbridge, A.F., R.F., N.B.F., M. Friedl, P.V.G., L. Georgieva, I.G., M. Gill, H.G., L.D.H., M.L.H., T.F.H., A.M.H., P.A.H., C.M.H., A.I., A.K.K, R.S.K., M.C.K., E.K., Y.K., G.K.K., B.K., L. Krabbendam, R. Krasucki, J. Lawrence, P.H.L., T.L., D.F.L., J.A.L., D.-Y.L., D.H. Linszen, P.K.E.M., W.M., A.K.M., M. Mattheisen, M. Mattingsdal, S.M., S.A.M., A. McIntosh, A. McQuillin, H.M., I.M., V. Milanova, D.W.M., V. Moskvina, I.M.-G., M.M.N., C.O., A.O., L.O., R.A.O., M.J.O., C.N.P., M.T.P., B.S.P., J. Pimm, D.P., V.P., D.J.Q., H.B.R., M. Rietschel, L.R., D. Ruderfer, D. Rujescu, A.R.S., T.G.S., J. Shi, J.M.S., D.S.C., T.S.S., S.T., J.V.O., P.M.V., T.W., S. Zammit, P. Sklar, M.J.D., M.C.O., N.C., P.F.S. and K.S.K. PGC Cross-Disorder Group Working Group: S.H.L., S. Ripke, B.M.N., S.M.P., R.H.P., A.T., A.F., M.C.N., J.I.N., B.W.P., M. Rietschel, T.G.S., N.C., S.L. Santangelo, P.F.S., J.W.S., K.S.K. and N.R.W. PGC Analysis Working Group: S.H.L., S. Ripke, B.M.N., S.M.P., V.A., E.M.B., P.H.L., S.E.M., M.C.N., D.P., M.J.D. and N.R.W.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Kendler KS, Eaves LJ. Psychiatric Genetics (Review of Psychiatry) American Psychiatric Association; Arlington, VA: 2005. [Google Scholar]
- 2.Tsuang M, Faraone S. The Genetics of Mood Disorders. Johns Hopkins University Press; Baltimore, MD: 1990. [Google Scholar]
- 3.Smoller JW, Finn CT. Family, twin, and adoption studies of bipolar disorder. Am J Med Genet C Semin Med Genet. 2003;123C:48–58. doi: 10.1002/ajmg.c.20013. [DOI] [PubMed] [Google Scholar]
- 4.Ronald A, Simonoff E, Kuntsi J, Asherson P, Plomin R. Evidence for overlapping genetic influences on autistic and ADHD behaviours in a community twin sample. J Child Psychol Psychiatry. 2008;49:535–542. doi: 10.1111/j.1469-7610.2007.01857.x. [DOI] [PubMed] [Google Scholar]
- 5.Rommelse NN, Franke B, Geurts HM, Hartman CA, Buitelaar JK. Shared heritability of attention-deficit/hyperactivity disorder and autism spectrum disorder. Eur Child Adolesc Psychiatry. 2010;19:281–295. doi: 10.1007/s00787-010-0092-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lichtenstein P, Carlstrom E, Rastam M, Gillberg C, Anckarsater H. The genetics of autism spectrum disorders and related neuropsychiatric disorders in childhood. Am J Psychiatry. 2010;167:1357–1363. doi: 10.1176/appi.ajp.2010.10020223. [DOI] [PubMed] [Google Scholar]
- 7.Rapoport J, Chavez A, Greenstein D, Addington A, Gogtay N. Autism spectrum disorders and childhood-onset schizophrenia: clinical and biological contributions to a relation revisited. J Am Acad Child Adolesc Psychiatry. 2009;48:10–18. doi: 10.1097/CHI.0b013e31818b1c63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.King BH, Lord C. Is schizophrenia on the autism spectrum? Brain Res. 2011;1380:34–41. doi: 10.1016/j.brainres.2010.11.031. [DOI] [PubMed] [Google Scholar]
- 9.Sullivan PF, et al. Family history of schizophrenia and bipolar disorder as risk factors for autism. Arch Gen Psychiatry. 2012;69:1099–1103. doi: 10.1001/archgenpsychiatry.2012.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crespi B, Stead P, Elliot M. Comparative genomics of autism and schizophrenia. Proc Natl Acad Sci USA. 2010;107:1736–1741. doi: 10.1073/pnas.0906080106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mortensen PB, Pedersen MG, Pedersen CB. Psychiatric family history and schizophrenia risk in Denmark: which mental disorders are relevant? Psychol Med. 2010;40:201–210. doi: 10.1017/S0033291709990419. [DOI] [PubMed] [Google Scholar]
- 12.Faraone SV, Biederman J, Wozniak J. Examining the comorbidity between attention deficit hyperactivity disorder and bipolar disorder: a meta-analysis of family-genetic studies. Am J Psychiatry. 2012;169:1256–1266. doi: 10.1176/appi.ajp.2012.12010087. [DOI] [PubMed] [Google Scholar]
- 13.Cole J, Ball HA, Martin NC, Scourfield J, McGuffin P. Genetic overlap between measures of hyperactivity/inattention and mood in children and adolescents. J Am Acad Child Adolesc Psychiatry. 2009;48:1094–1101. doi: 10.1097/CHI.0b013e3181b7666e. [DOI] [PubMed] [Google Scholar]
- 14.Craddock N, O’Donovan MC, Owen MJ. Genes for schizophrenia and bipolar disorder? Implications for psychiatric nosology. Schizophr Bull. 2006;32:9–16. doi: 10.1093/schbul/sbj033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green EK, et al. The bipolar disorder risk allele at CACNA1C also confers risk of recurrent major depression and of schizophrenia. Mol Psychiatry. 2010;15:1016–1022. doi: 10.1038/mp.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams NM, et al. Genome-wide analysis of copy number variants in attention deficit/hyperactivity disorder confirms the role of rare variants and implicates duplications at 15q13.3. Am J Psychiatry. 2012;169:195–204. doi: 10.1176/appi.ajp.2011.11060822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363:166–176. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- 18.Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee SH, Yang J, Goddard ME, Visscher PM, Wray NR. Estimation of pleiotropy between complex diseases using SNP-derived genomic relationships and restricted maximum likelihood. Bioinformatics. 2012;28:2540–2542. doi: 10.1093/bioinformatics/bts474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Psychiatric GWAS Consortium Bipolar Disorder Working Group. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near. ODZ4 Nat Genet. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2013;18:497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anney R, et al. Individual common variants exert weak effects on the risk for autism spectrum disorderspi. Hum Mol Genet. 2012;21:4781–4792. doi: 10.1093/hmg/dds301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cross-Disorder Group of the Psychiatric GWAS Consortium. Genome-wide analysis identifies loci with shared effects on five major psychiatric disorders. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neale BM, et al. Meta-analysis of genome-wide association studies of attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry. 2010;49:884–897. doi: 10.1016/j.jaac.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stergiakouli E, et al. Investigating the contribution of common genetic variants to the risk and pathogenesis of ADHD. Am J Psychiatry. 2012;169:186–194. doi: 10.1176/appi.ajp.2011.11040551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lionel AC, et al. Rare copy number variation discovery and cross-disorder comparisons identify risk genes for ADHD. Sci Transl Med. 2011;3:95ra75. doi: 10.1126/scitranslmed.3002464. [DOI] [PubMed] [Google Scholar]
- 29.Hinney A, et al. Genome-wide association study in German patients with attention deficit/hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:888–897. doi: 10.1002/ajmg.b.31246. [DOI] [PubMed] [Google Scholar]
- 30.Ribasés M, et al. Exploration of 19 serotoninergic candidate genes in adults and children with attention-deficit/hyperactivity disorder identifies association for 5HT2A, DDC and MAOB. Mol Psychiatry. 2009;14:71–85. doi: 10.1038/sj.mp.4002100. [DOI] [PubMed] [Google Scholar]
- 31.Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sinauer Associates; Sunderland, MA: 1998. [Google Scholar]
- 32.Purcell SM, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee SH, Goddard ME, Wray NR, Visscher PM. A better coefficient of determination for genetic profile analysis. Genet Epidemiol. 2012;36:214–224. doi: 10.1002/gepi.21614. [DOI] [PubMed] [Google Scholar]
- 34.Raychaudhuri S, et al. Accurately assessing the risk of schizophrenia conferred by rare copy-number variation affecting genes with brain function. PLoS Genet. 2010;6:e1001097. doi: 10.1371/journal.pgen.1001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee SH, et al. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet. 2012;44:247–250. doi: 10.1038/ng.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lubke GH, et al. Estimating the genetic variance of major depressive disorder due to all single nucleotide polymorphisms. Biol Psychiatry. 2012;72:707–709. doi: 10.1016/j.biopsych.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klei L, et al. Common genetic variants, acting additively, are a major source of risk of autism. Mol Autism. 2012;3:9. doi: 10.1186/2040-2392-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Browning SR, Browning BL. Population structure can inflate SNP-based heritability estimates. Am J Hum Genet. 2011;89:191–193. doi: 10.1016/j.ajhg.2011.05.025. author reply 193–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang J, et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet. 2011;43:519–525. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee SH, et al. Estimation and partitioning of polygenic variation captured by common SNPs for Alzheimer’s disease, multiple sclerosis and endometriosis. Hum Mol Genet. 2013;22:832–841. doi: 10.1093/hmg/dds491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Constantino JN, Todd RD. Intergenerational transmission of subthreshold autistic traits in the general population. Biol Psychiatry. 2005;57:655–660. doi: 10.1016/j.biopsych.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 42.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loftus EV, Jr, et al. Increased risks of developing anxiety and depression in young patients with Crohn’s disease. Am J Gastroenterol. 2011;106:1670–1677. doi: 10.1038/ajg.2011.142. [DOI] [PubMed] [Google Scholar]
- 44.Kohane IS, et al. The co-morbidity burden of children and young adults with autism spectrum disorders. PLoS ONE. 2012;7:e33224. doi: 10.1371/journal.pone.0033224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benach JL, Li E, McGovern MM. A microbial association with autism. mBio. 2012;3:e00019–12. doi: 10.1128/mBio.00019-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wray NR, Lee SH, Kendler KS. Impact of diagnostic misclassification on estimation of genetic correlations using genome-wide genotypes. Eur J Hum Genet. 2012;20:668–674. doi: 10.1038/ejhg.2011.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bromet EJ, et al. Diagnostic shifts during the decade following first admission for psychosis. Am J Psychiatry. 2011;168:1186–1194. doi: 10.1176/appi.ajp.2011.11010048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laursen TM, Agerbo E, Pedersen CB. Bipolar disorder, schizoaffective disorder, and schizophrenia overlap: a new comorbidity index. J Clin Psychiatry. 2009;70:1432–1438. doi: 10.4088/JCP.08m04807. [DOI] [PubMed] [Google Scholar]
- 49.Tsuang MT, Woolson RF, Winokur G, Crowe RR. Stability of psychiatric diagnosis. Schizophrenia and affective disorders followed up over a 30- to 40-year period. Arch Gen Psychiatry. 1981;38:535–539. doi: 10.1001/archpsyc.1980.01780300047005. [DOI] [PubMed] [Google Scholar]
- 50.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wray NR, et al. Genome-wide association study of major depressive disorder: new results, meta-analysis, and lessons learned. Mol Psychiatry. 2012;17:36–48. doi: 10.1038/mp.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Falconer D, Mackay T. Introduction to Quantitative Genetics. 4. Longman Scientific & Technical; Harlow, UK: 1996. [Google Scholar]
- 53.Van Snellenberg JX, de Candia T. Meta-analytic evidence for familial coaggregation of schizophrenia and bipolar disorder. Arch Gen Psychiatry. 2009;66:748–755. doi: 10.1001/archgenpsychiatry.2009.64. [DOI] [PubMed] [Google Scholar]
- 54.Lichtenstein P, et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet. 2009;373:234–239. doi: 10.1016/S0140-6736(09)60072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McGuffin P, et al. The heritability of bipolar affective disorder and the genetic relationship to unipolar depression. Arch Gen Psychiatry. 2003;60:497–502. doi: 10.1001/archpsyc.60.5.497. [DOI] [PubMed] [Google Scholar]
- 56.Moreno-De-Luca D, et al. Deletion 17q12 is a recurrent copy number variant that confers high risk of autism and schizophrenia. Am J Hum Genet. 2010;87:618–630. doi: 10.1016/j.ajhg.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 58.Nijmeijer JS, et al. Identifying loci for the overlap between attention-deficit/ hyperactivity disorder and autism spectrum disorder using a genome-wide QTL linkage approach. J Am Acad Child Adolesc Psychiatry. 2010;49:675–685. doi: 10.1016/j.jaac.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mulligan A, et al. Autism symptoms in attention-deficit/hyperactivity disorder: a familial trait which correlates with conduct, oppositional defiant, language and motor disorders. J Autism Dev Disord. 2009;39:197–209. doi: 10.1007/s10803-008-0621-3. [DOI] [PubMed] [Google Scholar]
- 60.Ripke SA, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. doi: 10.1038/ng.2742. in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sullivan PF, Daly MJ, O’Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet. 2012;13:537–551. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reich T, James JW, Morris CA. The use of multiple thresholds in determining the mode of transmission of semi-continuous traits. Ann Hum Genet. 1972;36:163–184. doi: 10.1111/j.1469-1809.1972.tb00767.x. [DOI] [PubMed] [Google Scholar]
- 63.Cochran WG. The combination of estimates from different experiments. Biometrics. 1954;10:101–129. [Google Scholar]
- 64.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Br Med J. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for Genome-wide Complex Trait Analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




