Abstract
The Drosophila bonus (bon) gene encodes a homolog of the vertebrate TIF1 transcriptional cofactors. bon is required for male viability, molting, and numerous events in metamorphosis including leg elongation, bristle development, and pigmentation. Most of these processes are associated with genes that have been implicated in the ecdysone pathway, a nuclear hormone receptor pathway required throughout Drosophila development. Bon is associated with sites on the polytene chromosomes and can interact with numerous Drosophila nuclear receptor proteins. Bon binds via an LxxLL motif to the AF-2 activation domain present in the ligand binding domain of βFTZ-F1 and behaves as a transcriptional inhibitor in vivo.
Introduction
Members of the nuclear receptor superfamily play important roles in cell differentiation, development, and homeostasis by binding to DNA response elements and positively or negatively regulating gene expression (Kastner et al. 1995 and Mangelsdorf et al. 1995). To perform this function, nuclear receptors require interactions with coactivators and corepressors. Although mechanisms of nuclear receptor coregulator action on the basis of cell culture assays, biochemical interactions, and structural studies are somewhat understood (McKenna et al., 1999), much remains to be learned about their in vivo significance. The existence of nuclear receptors and their cofactors in Drosophila allows these in vivo studies.
Transformation of larval tissues to adult structures during Drosophila metamorphosis is regulated by pulses of a steroid hormone, ecdysone (Riddiford, 1993). As shown in Figure 1A, an ecdysone pulse at the end of larval development drives the onset of prepupal development, pupariation, and eversion of larval imaginal discs to produce adult appendages (Robertson 1936 and Fristrom and Fristrom 1993). Twelve hours after pupariation, a second and smaller ecdysone pulse initiates the prepupa-to-pupa transition. This results in head eversion, leg and wing imaginal disc elongation, salivary gland cell death, and imaginal histoblasts proliferation to form the abdomen (Robertson 1936, Sliter and Gilbert 1992, Fristrom and Fristrom 1993 and Jiang et al. 1997). Responses to ecdysone are mediated by the ecdysone receptor (EcR) and its binding partner, ultraspiracle (USP), the Drosophila retinoid X receptor (RXR) homolog (Koelle et al. 1991, Yao et al. 1992 and Yao et al. 1993). Ecdysone binding to this nuclear receptor complex initiates a genetic cascade by activating transcription of a small set of “early” genes. These “early” genes encode transcription factors, including E74A, BR-C, and E75A, that regulate a larger set of “late” genes (Burtis et al. 1990, Segraves and Hogness 1990, DiBello et al. 1991, Urness and Thummel 1995 and Crossgrove et al. 1996). Transcription of “early-late” genes, such as DHR3, is initiated by the EcR/USP complex and is further upregulated by ecdysone-induced proteins (Koelle et al. 1992, Carney et al. 1997 and White et al. 1997). A second ecdysone pulse late in the prepupae reactivates the EcR/USP complex, which again upregulates E74A, E75A, BR-C, along with E93. Many of the proteins involved in this regulatory pathway encode members of the nuclear receptor superfamily.
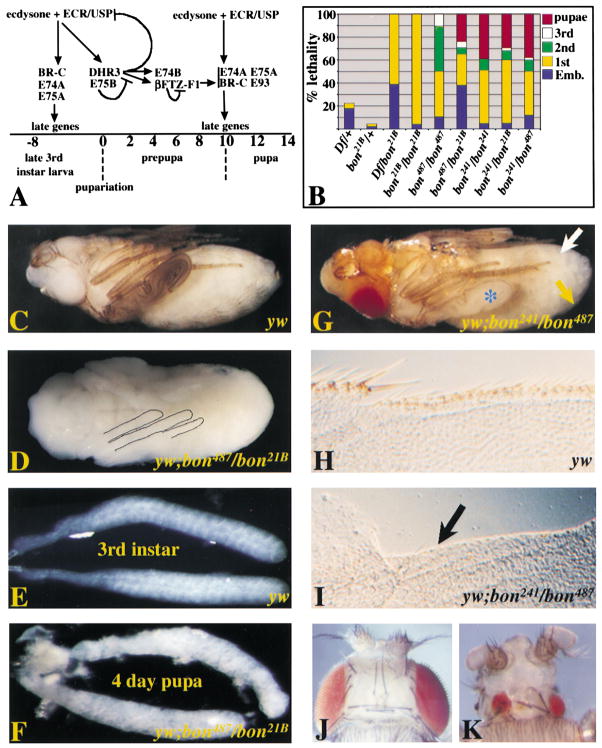
Figure 1. bon Plays a Role in Several Developmental Events and Is Required for Organismal Viability.
(A) Transcriptional responses to the larval and the prepupalecdysone pulses. Time points are shown hours prior to and after pupariation.(B) Animals with the listed genotypes were analyzed for their phase of lethality. The percentage of animals that died at each developmental stage is shown graphically (n = 200).(C and D) Animals dissected from the pupal case at ~4 days post-pupariation (pharate adult). (C) y w pharate adult compared to a similarly aged (D) y w; bon487/bon21B pupa initiate but fail to complete development of legs (outlined), wings, head, eyes, and cuticle.(E and F) Salivary glands of y w third instar larva and y w; bon487/bon21B pupa ( days post pupariation).(G) y w; bon241/bon487 pharate adult dissected from its pupal cases 4 days post-pupariation. Defects are seen in cuticle morphology, pigmentation of the wing (blue asterisk), and loss of tergite (yellow arrow) and sternal (white arrow) bristles (compare with Figure 1C).(H and I) y w and y w; bon241/bon487 wings. (I) y w; bon241/bon487 wings display a loss of bristles along the anterior wing margin (black arrow).(J and K) Adult heads of y w; FRT82B w+ clR3 (J) and y w eyeless-FLP glass-lacZ; FRT82B w+ clR3/FRT82B bon21B (K) flies. Note the severe loss of photoreceptors and cuticle.
We have identified and characterized bon, which encodes the Drosophila homolog of the mammalian transcription intermediary factors: TIF1α (Le Douarin et al., 1995a), TIF1β (also called KAP-1 [Friedman et al., 1996] or KRIP-1 [Kim et al., 1996]) (Le Douarin et al., 1996), and TIF1γ (Venturini et al., 1999). TIF1α, initially identified in a yeast genetic screen for proteins increasing the transactivation potential of RXRs (Le Douarin et al., 1995a), was found to interact via an LxxLL motif with several members of the nuclear receptor superfamily in a ligand- and activation function 2 (AF-2) integrity-dependent manner (Le Douarin et al., 1996; vomBaur et al., 1996). However, the biological relevance of these interactions has not yet been established. Moreover, once tethered to DNA through fusion to a heterologous DNA binding domain, TIF1α silences transcription, suggesting that it could play a dual role in transcription, being involved in both activation and repression (Le Douarin et al. 1996 and Nielsen et al. 1999).
In contrast to TIF1α, TIF1β and TIF1γ do not appear to have nuclear receptor binding activity. They possess an intrinsic transcriptional repression activity (Nielsen et al. 1999 and Venturini et al. 1999). TIF1β has been defined as a transcriptional co-repressor for the Krüppel Associated Box (KRAB) domains (Friedman et al. 1996, Kim et al. 1996 and Moosmann et al. 1996), which may function through association with (and/or formation of) heterochromatin (Nielsen et al., 1999). Interestingly, TIF1β is essential for early postimplantation mouse development (Cammas et al., 2000), implying that during early embryogenesis, a member of the TIF1 family is required. Hence, there is little information available at present about the in vivo roles of TIF1s and their relevant partners.
Here, we take advantage of the presence of a single TIF1 homolog in Drosophila to study the in vivo molecular interactions with its partners. Bon is a nuclear protein that is required for cell viability or proliferation. Bon plays a critical role in the development of several organs and tissues during embryogenesis and metamorphosis through regulation of genes involved in the ecdysone response. This regulation of gene expression seems to be based on its ability to interact with several nuclear receptors. In particular, we show that Bon interacts biochemically and genetically with βFTZ-F1 to negatively regulate its activity.
Results
Bon Is Required at Multiple Stages of Development
bon was isolated in a screen for mutations affecting embryonic peripheral nervous system (PNS) development (Salzberg et al., 1997). Three independent P-element alleles, bonS024108 (bon241), bonS024912 (bon249), and bonS048706 (bon487), which mapped to 92E8–14, failed to complement each other. Precise excisions of the P-elements reverts the phenotypes associated with their insertions demonstrating that the P-elements are responsible for the phenotypes. An additional bon allele, bon21B, was generated by imprecise excision of bon241. This allele fails to complement all bon alleles and a deficiency, Df(3R)HB79, which removes chromosomal region 92E.
To establish the strength of each allele, we performed complementation tests and defined the lethal phase associated with each allelic combination (Figure 1B). Df(3R)HB79/bon21B animals exhibit the earliest stage of lethality, while homozygous bon241/bon241 animals display the least severe phenotype, with 34% of the expected animals surviving to pharate adults. In all genetic combinations, some first instar larvae survive up to a week and fail to molt into second instar larvae. No male third instar larvae, pharate adults or adults survived in any bon genetic background, indicating that loss of bon has a more deleterious effect on males than females. Based on the complementation data, we ordered the bon alleles as follows: Df(3R)HB79>bon21B>bon487>bon241 = bon249.
Bon Is Required for Metamorphosis
To pinpoint the phenotypes associated with bon mutations, we analyzed morphological defects associated with different allelic combinations. Df(3R)HB79/bon21B mutant embryos fully develop. However, many are unable to hatch from their egg case, and both Df(3R)HB79/bon21B embryos and first instar larvae exhibit disrupted fluid-filled trachea (data not shown). The embryonic/first instar lethality probably corresponds to the zygotic null or a severe loss of function phenotype as there is very little maternal protein remaining in mature Df(3R)HB79/bon21B embryos (see Immunohistochemistry).
Less severe loss of Bon function results in pupal defects. The majority of bon487/bon21B mutant pupae display an almost complete lack of pigmentation (Figures 1C and 1D). They initiate but fail to complete development of legs, wings, head, eyes, and cuticle. Salivary glands, which normally undergo apoptosis at 12 hr postpupariation, are present in 4 days post-pupariation bon487/bon21B pupae (Figure 1F) and are similar in size to third instar larval glands. In bon241/bon487 animals (Figure 1G), we observe defects in cuticle and bristle development. The abdominal cuticle of these flies appears immature and the tergite and sternal bristles are severely reduced or absent. Bristles of the anterior wing margin of bon241/bon487 pharate adults are almost entirely lacking (Figures 1H and 1I). Finally, there is a dramatic reduction in pigmentation in the mutant wing cuticle (compare Figure 1G to Figure 1C). This data indicates that bon is required for numerous developmental processes, including control of larval molting, cuticle deposition and pigmentation, bristle development, and elimination of salivary glands by cell death.
To determine the effect of complete loss of bon on the development of adult tissues, we used the FLP/FRT system to create mutant clones in the developing eye imaginal disc with the eyeless enhancer driving FLP(Newsome et al., 2000). Flies heterozygous for FRT bon21B and a cell lethal gene marked with w+ (FRT w+ cl3R) expressing FLP in the eye disc generated clones of bon21B/bon21B. Loss of bon in the eye causes a loss of all mutant photoreceptors (compare Figures 1J and 1K). A small patch of red photoreceptors remains because of a limited number of bon21B/cl3R cells. This indicates that bon is required for cell viability or proliferation of photoreceptors. In addition, much of the head cuticle is missing, indicating that most or all cells of the eye disc that produce cuticle are also lacking. In addition, no adult mutant bon clones were observed in FLP/FRT experiments using heat shock-FLP; FRT82 bon21B animals, even though numerous wild-type twin spots were observed (data not shown). Hence, early and complete loss of bon is lethal to cells or disrupts proliferation.
Bon Encodes the Drosophila Homolog of Vertebrate TIF1
To clone bon, genomic DNA flanking bon241 was isolated and used to identify cDNAs. The bon cDNA (AF210315) permitted isolation of genomic phages and determination of the structure of the locus (Figure 2A). Flanking sequences from bon241, bon249, and bon487 (Salzberg et al., 1997) were used to map the P-elements. Sequencing of bon21B revealed a deletion of most of exon 1 and the 5′ end of intron 1.
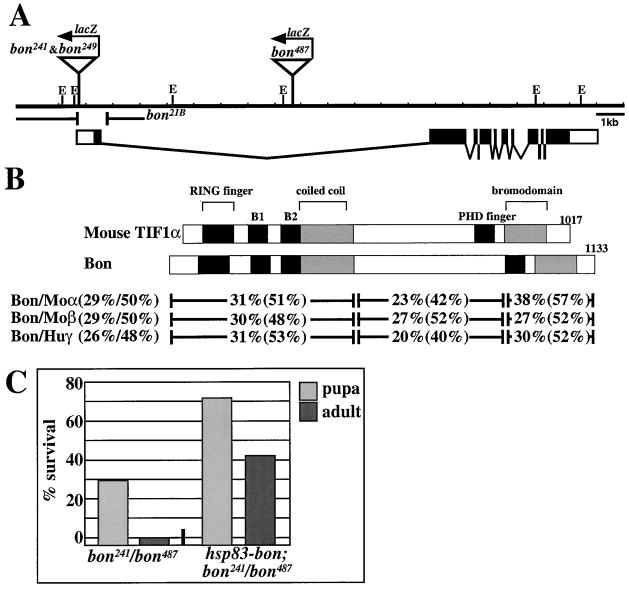
Figure 2. bon Encodes the Drosophila Homolog of Vertebrate TIF1.
(A) Approximately 20 kb of genomic DNA covering the bon locus (E = EcoRI restriction sites) is shown graphically. bon241 and bon249 P-elements map to the 5′ end of exon 1 and bon487 P-element maps to the middle of intron 1. bon21B removes all but 12 bp of exon 1 and the first 324 bp of intron 1.(B) Domain structures and homology between Bon and mouse TIF1α. Schematics of both proteins show conserved domains.(C) Rescue of bon mutations. A bon cDNA under the control of a Hsp83 ubiquitously expressing promoter rescues the bon241/bon487 phenotypes and lethality as observed in the increased percentage of both mutant pupae and adults (n = 400).
Database searches revealed that bon encodes the only Drosophila homolog of mammalian TIF1s. Bon exhibits 29% identity with mouse TIF1α (Le Douarin et al., 1995a) and mouse TIF1β (Friedman et al. 1996, Kim et al. 1996 and Le Douarin et al. 1996), and 26% identity with human TIF1γ (Venturini et al., 1999). The overall identity between Bon and TIF1s is similar to the identity observed between the TIF1 members (Venturini et al., 1999). A higher degree of identity is seen in the N- and C-terminal regions spanning the conserved domains (Figure 2B). At the N terminus, a C3HC4 zinc-finger motif or RING finger is followed by two cysteine-rich zinc binding regions (B-boxes) and a coiled coil domain forming a tripartite motif designated RBCC (Le Douarin et al. 1995a and Saurin et al. 1996). At the C terminus, a bromodomain (Jeanmougin et al., 1997) is preceded by a C4HC3 zinc-finger motif or PHD finger (Aasland et al., 1995).
Three lines of evidence suggest that the cDNA and phenotypes correspond to bon. First, three independently generated P-element alleles fail to complement each other and map to the 5′ UTR or first intron. Second, the severity of the mutant phenotypes directly correlates with the degree of loss of Bon (see Figure 3). Third, the P-elements are revertible to wild-type. Finally, expression of bon under the control of the ubiquitously expressing hsp83 promoter (Xiao and Lis, 1989) causes a partial rescue of bon241/bon487 animals (Figure 2C).
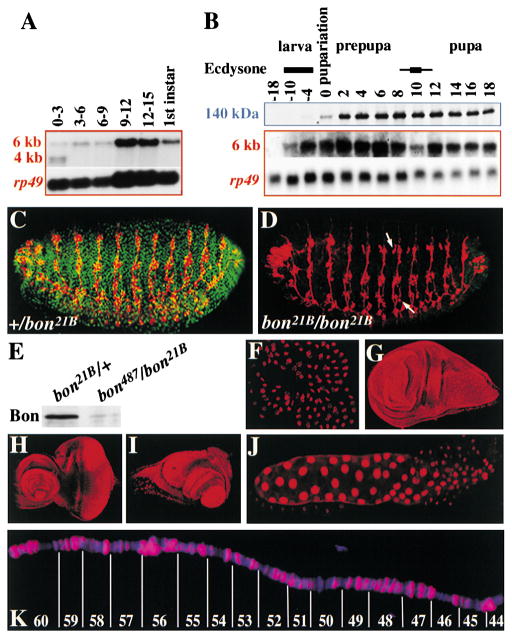
Figure 3. Bon Is a Nuclear Protein that Is Expressed throughout Development.
(A) Developmental Northern analysis of bon expression hours after egg lay.(B) Developmental Northern and Western analysis of bon mRNA and protein hours before and after pupariation. rp49 serves as a loading control.(C and D) Stage 17 embryos stained with anti-Bon antibody (green) and the neuronal specific MAB 22C10 (red). White arrows point to ectopic neurons in the dorsal and ventral clusters.(E) Western analysis of bon21B/+ (control) and bon21B/bon487 4 hr prepupae.(F–K) Immunohistochemistry using anti-Bon antibody (red) and DAPI (blue). Bon is found in fat body (F), wing disc (G), an eye/antennal disc (H), leg disc (I), and salivary gland (J) from a prepupa. Bon localization on polytene chromosome 2R (K) of a late third instar larva salivary gland.
Bon Is a Widely Expressed Nuclear Protein
Northern analysis demonstrates that bon produces one predominant 6 kb transcript and two 4 kb transcripts, which encode a protein of ~140 kDa. The two 4 kb transcripts are only present in 0–3 hr embryos (Figure 3A) and adult females (data not shown). It is therefore possible that the 4kb mRNAs are maternal components. bon is expressed throughout embryogenesis and in first instars (Figure 3A). Its levels increase in 9–12 hr embryos and are low during the second instar stage (data not shown). bon is upregulated in late third instar larvae (Figure 3B). The upregulation of bon during midembryogenesis and prior to pupariation correlates well with known high titer pulses of ecdysone (Riddiford, 1993).
To demonstrate the specificity of the Bon antiserum we performed immunohistochemical staining of late stage control and bon21B/bon21B embryos. Mutant embryos, which can easily be identified by the presence of extra neurons, contain little protein (Figure 3D). Similarly, Western analysis of bon487/bon21B prepupae (4 hr old) shows a significant decrease in protein levels compared to the bon21B/+ control (Figure 3E).
Immunohistochemical staining of numerous tissues show that Bon is a nuclear protein expressed in most and possibly all cells during embryogenesis (Figure 3C), in fat body (Figure 3F), imaginal discs (Figures 3G–3I), salivary glands (Figure 3J), brain, gut, Malpighian tubules, and trachea (data not shown). Bon is a chromatin-associated protein that localizes to ~10%–15% of the polytene chromosome bands (Figure 3K). This pattern is highly reproducible.
Ecdysone-Induced Genes Are Misregulated in bon Mutants
To determine whether the defects seen in bon mutants are due to disruptions in the ecdysone-regulated pathway, we examined the expression of several ecdysone-regulated genes in y w and bon241/bon241 larvae, prepupae, and pupae (Figure 4). In bon241/bon241 animals, levels of βFTZ-F1, EcR-A, EcR-B, E74A, E74B, and BR-C are reduced. It appears that each gene is upregulated in response to the ecdysone pulse, but is unable to maintain expression in the bon mutants. However, DHR3 transcripts are prematurely expressed and the overall level of expression is elevated in bon241/bon241 animals when compared to y w control animals. In addition, the EcR-A transcript levels appear slightly reduced in bon241/bon241 animals, while the EcR-B transcript levels are severely reduced when compared to controls. Similar observations were made for all of the above genes in bon21B/bon487 animals, except that DHR3 transcript levels are also reduced (data not shown). Based on these effects on gene expression, defects in larval molting and metamorphosis, and the temporal expression pattern of Bon, we propose that Bon plays an important role in the regulation of genes in the ecdysone response pathway.
Figure 4. Ecdysone-Induced Genes Are Misregulated in bon.
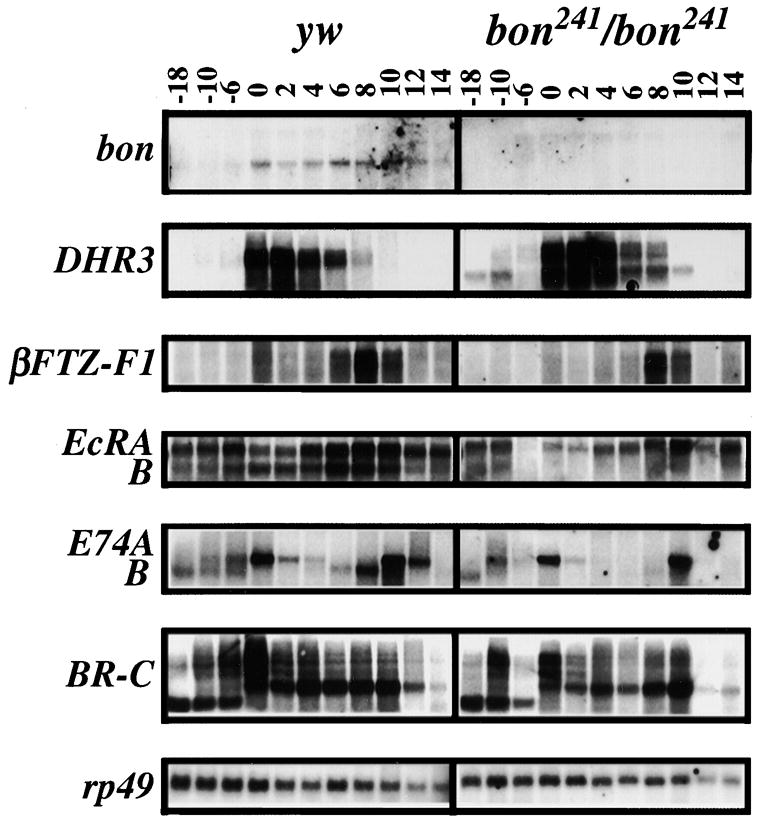
Total RNA was isolated from staged y w control and bon241/bon241 mutant animals and analyzed by Northern. Numbers on the top indicate hours pre and post pupariation at 25°C. In the bon241/bon241 background bon transcripts are severely reduced, although some transcripts of higher molecular weight, due to the P-element insertion, are detected. In comparison to y w, DHR3 transcripts are upregulated in the bon241/bon241 animals. βFTZ-F1, EcRA, EcRB, E74A, E74B, and BR-C transcript levels are reduced in the bon241/bon241 as compared to y w control background. Levels of rp49 transcripts serve as a control.
Bon Interacts with Several Nuclear Receptors
To better characterize the function of Bon, we sought to identify interacting proteins. A Drosophila (4–18 hr) embryonic cDNA library was screened using Bon as bait (see Experimental Procedures). Isolated cDNAs were classified as positive when retested in another version of the two-hybrid system using the DNA binding domain of the estrogen receptor fused to Bon (DBD-Bon) and an ERE-URA3 reporter gene (Le Douarin et al., 1995b). One positive clone encoded the 488 C-terminal residues of βFTZ-F1 (amino acids 315–802; Lavorgna et al., 1993). Coexpression of DBD-Bon with AAD–βFTZ-F1(315–802) transactivated the URA3 reporter (Figure 5A). Hence, Bon is able to interact with βFTZ-F1(315–802) in yeast cells.
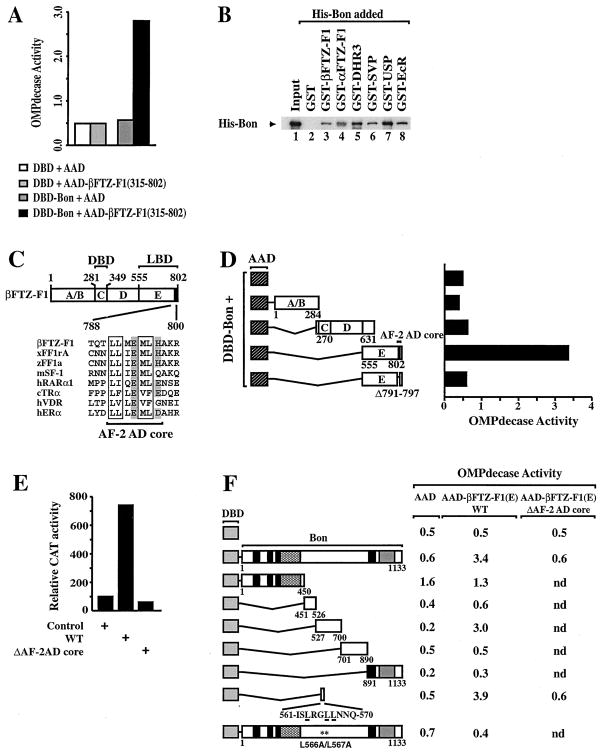
Figure 5. Bon Interacts with Several Nuclear Receptors and Selectively Interacts with the AF-2 AD Core Motif of βFTZ-F1 through a Conserved LxxLL Motif.
(A) Bon interacts with βFTZ-F1 (amino acids 315–802) in yeast. Plasmids expressing the unfused ERα DBD or DBD-Bon were introduced into PL3 together with the unfused VP16 AAD or AAD–βFTZ-F1 (315–802). Transformants were grown in liquid medium containing uracil. Extracts were prepared and assayed for OMP decase activity (nmol substrate/min/mg protein).(B) Bon interacts directly with full-length βFTZ-F1 and other Drosophila nuclear receptors in vitro. His-tagged Bon was incubated with control GST (lane 2) or GST fusions containing the indicated nuclear receptors (lanes 3–5) bound to glutathione-S-Sepharose beads. Bound Bon was detected by Western. Lane 1 shows one-fifth of the amount of input His-Bon.(C) Schematic representation of the structure of βFTZ-F1 with conserved regions of the receptor denoted A–E. Amino acid boundaries (numbers) were defined by sequence alignment with other nuclear receptors (Wurtz et al., 1996). A comparative alignment of the FTZ-F1 AF-2 AD core motif with other AF-2 AD core sequences is shown with the highly conserved pairs of hydrophobic residues boxed and charged residues shaded.(D) Bon interacts with the LBD of βFTZ-F1 in an AF-2-dependent manner. The indicated regions of βFTZ-F1 were fused to the VP16 AAD (hatched box) and assayed for activation with DBD-Bon in PL3. OMP decase activities are expressed as in Figure 5A. The values (±20%) are the average of at least three independent transformants. Expression of fusion proteins was confirmed by Western blotting using the antibody 2GV4 against VP16 (data not shown).(E) Characterization of the AF-2 activation domain of βFTZ-F1. S2 cells were transfected with a reporter construct containing a GAL4 binding site (17M-ERE-tk-CAT) and pCMV-β-gal, with or without vectors containing unfused GAL4 (control) or GAL4 fused to βFTZ-F1 (555–802) wild-type (WT) or to βFTZ-F1 (555–802)Δ791–797 lacking the AF-2 AD core motif (ΔAF-2AD core). CAT activities are expressed relative to that of the unfused GAL4 vector (taken as 100%). Values (±10%) represent the averages of three independent transfections after normalization to β-gal activities.(F) The LxxLL motif of Bon is both necessary and sufficient for interaction with the LBD/AF-2 of βFTZ-F1. The indicated mutants of Bon were fused to the ERαDBD and assayed for interaction with the “unfused” AAD or AAD fusions containing the entire E region of βFTZ-F1 (amino acids 555–802; AAD–βFTZ-F1 [E] WT) or a mutated E derivative in which the core of the AF-2 AD was deleted (Δ791 to 797; AAD–βFTZ-F1 [E] ΔAF-2AD core). OMPdecase activities are given in nmol substrate/min/mg protein. The values (±20%) are the average of at least three independent transformants. Expression of all the DBD and AAD fusion proteins indicated was confirmed by Western blotting using the antibodies F3 against the F region of ERα and 2GV4 against VP16, respectively (data not shown).
To test whether Bon interacted with βFTZ-F1 as well as other Drosophila nuclear receptors in vitro, we performed binding assays using purified recombinant proteins. Glutathione-S transferase (GST)-fused βFTZ-F1, αFTZ-F1 (amino acids 154–1029), Seven-up (SVP), DHR3, USP, and EcR were immobilized on glutathione-Sepharose and incubated with purified N-terminally His-tagged Bon (His-Bon). His-Bon bound to GST-βFTZ-F1, GST-αFTZ-F1, GST-DHR3, GST-SVP, GST-USP, and GST-EcR (Figure 5B, lanes 3–8), but not to GST alone (lane 2). Thus, Bon can bind directly to many members of the nuclear receptor family in vitro.
Bon Interacts with the LBD/AF-2 Domain of βFTZ-F1 through a Conserved LxxLL Motif
To define the domain(s) of βFTZ-F1 responsible for Bon interaction, a deletion analysis of βFTZ-F1 was performed using the yeast two-hybrid system. Various segments of βFTZ-F1 (Figures 5C and 5D) were fused to the VP16 AAD and assayed for DBD-Bon interaction. No increase in reporter activity was observed with the N-terminal A/B region or with a fusion protein containing residues 270–631, which include the DNA binding domain and the hinge region of the receptor (Figure 5D). In contrast, a 7-fold activation was detected in the presence of AAD–βFTZ-F1(555–802), indicating that the E region encompassing the putative ligand binding domain (LBD) is sufficient for interaction with Bon.
Sequence analysis of the E region of βFTZ-F1 revealed a conserved transcriptional activation domain 2 core motif (AF-2 AD core) between residues 791 and 797 (Figure 5C). To investigate its activity, we cotransfected an expression vector encoding the E region of βFTZ-F1 fused to the yeast GAL4 DNA binding domain into Drosophila Schneider (S2) cells together with a GAL4 reporter plasmid. We observed an increase in reporter gene activity, whereas no transactivation was detected with a GAL4 βFTZ-F1 construct lacking the AF-2 AD core (Figure 5E). Deletion of the AF-2 AD core also abolished Bon interaction with the βFTZ-F1 (E) in yeast (Figure 5D). Thus, the LBD of βFTZ-F1 contains an AF-2 activation domain, whose integrity is required for Bon interaction.
To determine which domain of Bon interacts with βFTZ-F1, a series of DBD-Bon deletion constructs were generated and assayed for interaction with the E region of βFTZ-F1. No significant increase in reporter activity was observed when fusion proteins of the RBCC motif (1–450) and the PHD/bromodomain (891–1133) of Bon were coexpressed with AAD–βFTZ-F1 (E) (Figure 5F). In contrast, a 15-fold enhancement was observed in the presence of DBD-Bon (527–700) domain. Analysis of this region revealed a predicted α-helical segment extending from residues 561 to 570. This domain contains an LxxLL consensus sequence, originally identified in the nuclear receptor-interacting domain of TIF1α (Le Douarin et al., 1996) and subsequently found in many other AF-2 mediators (Heery et al., 1997). In the presence of AAD–βFTZ-F1 (E) wild-type, but not AAD–βFTZ-F1(E)ΔAF-2 AD-core, residues 561 to 570 of Bon fused to the ERα DBD activated the reporter gene ~8-fold above the level of unfused AAD (Figure 5F). Thus, Bon contains an LxxLL motif that is sufficient to interact with the LBD of βFTZ-F1 in an AF-2-integrity-dependent manner. To investigate whether Bon actually binds βFTZ-F1 through this LxxLL motif, we generated mutations in Bon that eliminate the conserved leucine residues at positions 566 and 567. The replacement of these leucines by alanine residues abolishes the interaction with the LBD of βFTZ-F1 in yeast (Figure 5F; see DBD-BonL566A/L5467A). Hence, Bon interacts with the AF-2 of βFTZ-F1 through an LxxLL motif.
Loss of Bon Suppresses Phenotypes Associated with Loss of βFTZ-F1 Function
βFTZ-F1 plays an important role in the stage-specific response to the prepupal ecdysone pulse by positively regulating the expression of E74A, E75B, BR-C, EDG84A, and E93, and negatively regulating its own expression (Figure 1A) (Lavorgna et al. 1993, Broadus et al. 1999 and Murata et al. 1996). Mutant βFTZ-F1 animals display variable defects in early pupal events such as adult head eversion, leg elongation, and salivary gland cell death. Similar phenotypes are observed in bon mutant pupae.
The phenotypes associated with βFTZ-F1ex17/Df(3L)CatDh104 mutants have been categorized into three lethal pupal classes (Figures 6B–6D)(Broadus et al., 1999): 38% die as pharate adults with short malformed legs (Figure 6B); 45% undergo head eversion, but arrest early in pupal development (Figure 6C); and 17% fail to undergo head eversion, but continue developing into cryptocephalic pharate adults (Figure 6D). All βFTZ-F1 mutants have deformed legs.
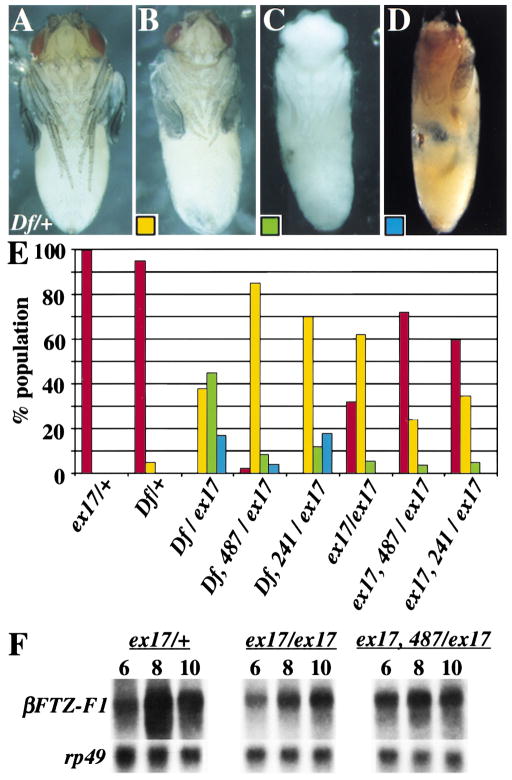
Figure 6. Loss of Bon Suppresses Phenotypes Associated with βFTZ-F1.
(A) Df(3L)CatDH104/+ (Df/+) control animal prior to eclosion.(B–D) Df(3L)CatDH104/βFTZ-F1ex17 mutant animals four days after pupariation: (B) Pharate adult stage with malformed legs.(C) Pupa that has completed head eversion, but arrested in development.(D) Cryptocephalic pharate adult with malformed legs.(E) Shows the percentage of animals of listed genotypes that are adults (red), pharate adults (yellow), arrested pupae (green), or cryptocephalic adults (blue) (n = 400). ex17 = βFTZ-F1ex17, Df = Df(3L)CatDH104, 487 = bon487, 241 = bon241.(F) Northern blot analysis of βFTZ-F1ex17/+, βFTZ-F1ex17/βFTZ-F1ex17, and βFTZ-F1ex17, bon487/βFTZ-F1ex17 animals at 6, 8, and 10 hr after pupariation.
Because Bon is able to interact with βFTZ-F1 in vitro, we attempted to establish whether Bon interacts with βFTZ-F1 in vivo. Flies were generated with either bon241 or bon487 in the Df(3L)CatDh104/βFTZ-F1ex17 or βFTZ-F1ex17/βFTZ-F1ex17 mutant backgrounds and assessed for their effect on βFTZ-F1 phenotypes. Loss of one copy of bon is able to suppress the phenotypes associated with loss of βFTZ-F1(Figure 6E). In the Df(3L)CatDh104/βFTZ-F1ex17 background, partial loss of Bon rescued the majority of mutant animals to pharate adult stages: 87% for bon487 and 70% for bon241. In the βFTZ-F1ex17 homozygotes, partial loss of Bon dramatically increased the number of adult escapers: 72% for bon487 and 60% for bon241, compared to 31% in a wild-type background. In addition, one mutant copy of bon also strongly suppressed the leg phenotypes associated with loss of βFTZ-F1. In summary, these data indicate that partial loss of Bon suppresses the phenotypes associated with a partial loss of βFTZ-F1.
βFTZ-F1ex17 has been shown to be a hypomorphic allele that is the result of a deletion of a positive regulatory element. Northern analysis has demonstrated that βFTZ-F1ex17/βFTZ-F1ex17 animals exhibit low levels of βFTZ-F1 transcripts (Broadus et al., 1999). We therefore hypothesized that bon suppression of the βFTZ-F1ex17 phenotypes may be the result of βFTZ-F1 upregulation. To test this hypothesis, we performed Northern analysis on βFTZ-F1ex17/+, βFTZ-F1ex17/βFTZ-F1ex17, and βFTZ-F1ex17bon487/βFTZ-F1ex17 staged prepupae and estimated the levels of βFTZ-F1 expression. One mutant copy of bon487 resulted in a 1.8- and a 1.9-fold upregulation of βFTZ-F1 in βFTZ-F1ex17 mutants (Figure 6F, n = 2). These results suggest that suppression by bon is at least partially due to the upregulation of βFTZ-F1 and that Bon seems to play a direct role in repressing βFTZ-F1 expression. These data appear in contrast to, but are not inconsistent with the general loss of Bon function that affects the transcription of most nuclear receptors negatively. Because βFTZ-F1 is a downstream effector in the ecdysone pathway, the specificity of the interaction between βFTZ-F1 and Bon is probably masked in a severe loss of function bon animal (see Discussion).
Bon Negatively Regulates Activation by βFTZ-F1 AF-2
To determine whether Bon is able to repress transcription, we fused the coding sequence of Bon to the yeast GAL4 DNA binding domain. The resulting fusion protein was tested for its ability to repress transcription activated by ER(C)-VP16, a chimeric activator containing the DBD of ERα fused to VP16. GAL4-Bon and ER(C)-VP16 were transiently transfected into S2 cells with a reporter containing a GAL4 binding site (17M) and an estrogen response element (ERE) in front of a thymidine kinase (tk) promoter-CAT fusion (17M-ERE-tk-CAT). As shown in Figure 7A, GAL4-Bon efficiently repressed transcription in a dose-dependent manner. In contrast, coexpression of Bon without the GAL4 DNA binding domain caused a reproducible increase in CAT activity, indicating that repression by Bon is entirely dependent on DNA binding.
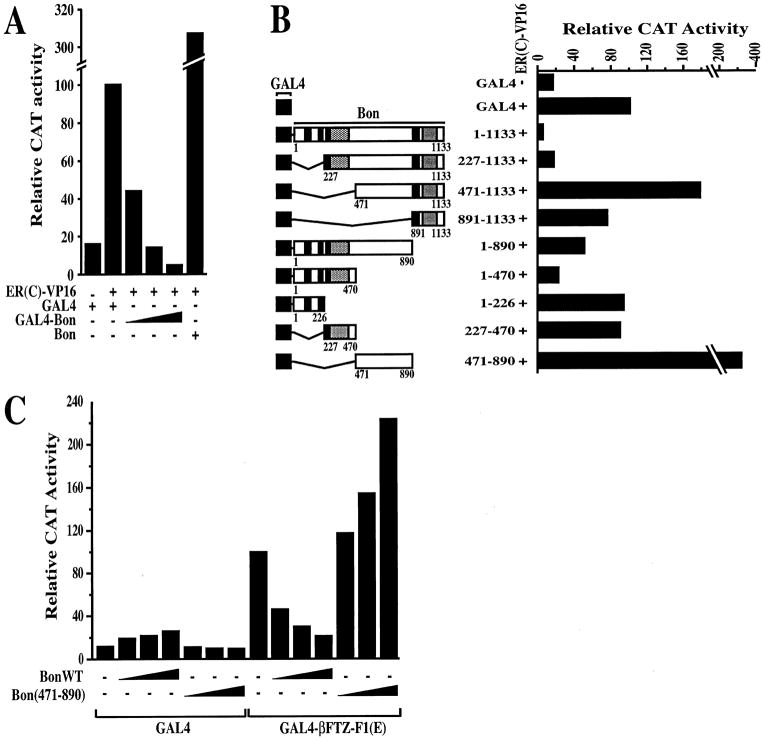
Figure 7. Bon Has an Autonomous Repression Function and Negatively Regulates Activation by βFTZ-F1 AF-2.
(A) Bon represses activated transcription in a dose-dependent manner when targeted to the promoter. The 17M-ERE-tk-CAT (2 μg) reporter and 2 μg of pCMV–β–gal plasmid (as internal control) were cotransfected into S2 cells with the indicated pPAC5C-based vectors expressing the activator ER(C)-VP16 (2 μg) and the unfused GAL4 DBD (GAL4: 2 μg) or Bon fused to the GAL4 DBD (GAL4-Bon: 0.02, 0.2, or 2 μg) or Bon tagged with the FLAG epitope (Bon: 2 μg). CAT activities are expressed relative to controls of ER(C)-VP16 and the unfused GAL4 (taken as 100%). Values (±10%) represent the averages of three independent duplicated transfections after normalization to β-galactosidase activities.(B) The RBCC motif of Bon is required for repression. Cotransfection assays were conducted as described in (A) with GAL4-Bon expression vector. Expression of the fusion proteins was confirmed by Western using the antibody 2GV3 against the GAL4 DBD (data not shown).(C) Effect of Bon overexpression on transactivation by βFTZ-F1 AF-2. S2 cells were transfected with the 17M-ERE-tk-CAT reporter plasmid (3 μg), 2 μg of pCMV–β–gal plasmid, and 6 μg of GAL4 expression vectors encoding GAL4 or GAL4–βFTZ-F1 (E), together with 1, 3, and 6 μg of either FLAG-tagged Bon wild-type or FLAG-tagged Bon (471–890). CAT activities are expressed relative to those of GAL4–βFTZ-F1 (E) (taken as 100%). Values (±0%) represent the averages of three independent duplicated transfections after normalization to β–gal activities.
To map the domain of Bon responsible for transcriptional repression, a set of N- and C-terminally truncated derivatives were assayed for their ability to repress VP16-activated transcription in S2 cells. In the absence of the RBCC motif, the GAL4-Bon fusion protein (Figure 7B, GAL4-Bon [471–1133]) failed to repress transcription, indicating that the N-terminal region of Bon is required for repression. However, this region was not sufficient for full repression (compare GAL4-Bon with GAL4-Bon [1–470]). Consistent with this, a C-terminal truncation, GAL4-Bon (1–890), was a less potent repressor indicating that the C-terminal residues of the protein including the PHD finger and the bromodomain also contribute to the repression potential of Bon. However, this domain on its own exhibited little repression (GAL4 [891–1133]). A 3- to 4-fold increase in CAT activity was observed with the central region between the coiled-coil and the PHD finger (GAL4-Bon [471–890]), suggesting that Bon may also contain a “masked” activation domain. Note, however, that no significant activation was observed with GAL4-Bon (471–890) tested in the absence of ER(C)-VP16 (data not shown). Taken together, these results indicate that most of the repression activity of Bon resides within the N-terminal RBCC domain.
To investigate functional consequences of the Bon-βFTZ-F1 interaction, the transcriptional activity of βFTZ-F1 AF-2 was assayed alone or in combination with overexpressed Bon in transiently transfected cells. Bon and the GAL4-βFTZ-F1(E) derivative were cotransfected into S2 cells together with the GAL4-responsive reporter, 17M-ERE-tk-CAT. GAL4-βFTZ-F1(E) exerted a trans-stimulation activity that was repressed by the addition of Bon (Figure 7C). In control experiments, coexpression of Bon did not suppress either basal or VP16-stimulated transcription in the presence of GAL4 alone (Figure 7C, lanes 2–4) or ER(C)-VP16 (Figure 7A, lane 6), respectively. We next examined whether N- and C-terminally truncated Bon (471–890), which contains the βFTZ-F1-interacting domain, but has no repressive activity (Figure 7B), was able to serve as a dominant-negative inhibitor of endogenous Bon function. Expression of this portion of Bon increased the transcriptional activity of GAL4-βFTZ-F1(E) in a dose-dependent manner (Figure 7C). No major effect on reporter gene expression in the absence of GAL4-βFTZ-F1(E) was observed. Taken together, these results provide support for the hypothesis that Bon plays a role in downregulating βFTZ-F1-dependent transcription.
Discussion
We have identified and characterized a Drosophila homolog of mammalian TIF1α (Le Douarin et al., 1995a), TIF1β (Friedman et al. 1996, Kim et al. 1996 and Le Douarin et al. 1996), and TIF1γ (Venturini et al., 1999). BLAST analysis of the annotated Drosophila genomic sequence (Myers et al., 2000) did not reveal any other closely related sequences, suggesting that Bon is the sole member of the TIF1 family in Drosophila. In addition, we found no TIF1 ortholog in Caenorhabditiselegans and S. cerevisiae. Thus, Bon may be a prototype of the TIF1 family.
A Role for Bon in Chromatin-Mediated Transcriptional Repression
Bon and TIF1s contain an N-terminal RBCC (RING finger/B boxes/coiled coil) motif. In the absence of the RBCC motif, the GAL4-Bon protein, unlike the full-length protein, fails to repress transcription. The TIF1β RBCC domain has been shown to be necessary for the oligomerization of TIF1β and KRAB binding (Peng et al., 2000). Because Bon is able to homodimerize (J.A.O. and R.L., unpublished data), this domain may be involved in formation of protein complexes.
The PHD finger and bromodomain are characteristic features of nuclear proteins known to be associated with chromatin and/or to function at the chromatin level (Aasland et al. 1995, Jeanmougin et al. 1997 and Winston and Allis 1999). For instance, the chromosomal proteins Trithorax and Polycomb-like contain multiple PHD fingers (Aasland et al., 1995), while the histone acetyltransferases CBP and GCN5 as well as the chromatin-remodeling factor SWI2/SNF2 are also bromodomain containing proteins (Jeanmougin et al., 1997). It has recently been demonstrated that bromodomains bind to acetyl-lysine and specifically interact with the amino-terminal tails of histones H3 and H4, suggesting a chromatin-targeting function for this highly evolutionarily conserved domain (Dhalluin et al., 1999). Because Bon is localized to hundreds of chromatin bands on Drosophila polytene chromosomes, it is probably involved in chromatin-mediated regulation of transcription of numerous genes.
Bon can repress both basal and activated transcription when recruited to the promoter region of a target gene, similar to TIF1α, -β, and -γ (Figure 7A). For TIF1α and TIF1β, a link between silencing and histone modification has been established (Nielsen et al., 1999), and TIF1β is part of a large multiprotein complex that possesses histone deacetylase activity (Underhill et al., 2000). Moreover, TIF1β was also reported to colocalize and interact directly with members of the heterochromatin protein 1 (HP1) family (Nielsen et al., 1999). Similar to TIF1β, TIF1α can bind the HP1 proteins in vitro (Le Douarin et al. 1996 and Nielsen et al. 1999). However, TIF1α-mediated repression in transfected cells does not require the integrity of the HP1 interaction domain (Nielsen et al., 1999), nor is there any significant subnuclear colocalization of HP1α and TIF1α (Remboutsika et al., 1999). No interactions were observed between Bon and HP1 in a yeast two-hybrid assay, nor did we find evidence for genetic interactions (data not shown). However, in a yeast two-hybrid screen, Bon interacted with members of the Polycomb group (C. S., P. C., and R. L., unpublished data), suggesting that Bon may also be part of heterochromatin-like complexes and/or may require some of the members of the Polycomb group genes to repress transcription. This would imply that Bon has a dual role, similar to some members of the Polycomb group family: transcriptional repression and heterochromatin formation (Jacobs and van Lohuizen, 1999). Both of these roles may be required in transcriptional repression.
Bon Directly Interacts with Nuclear Receptors and Inhibits Their Activity
Upon ecdysone binding, the EcR/USP complex upregulates the expression of a group of transcription factors, many of which are nuclear receptors (Koelle et al. 1991, Yao et al. 1992 and Yao et al. 1993). During this ecdysone regulatory cascade, both induction and repression of transcription are required to regulate the timing and the response to the ecdysone signal. Bon is able to interact with many members of the nuclear receptor family, suggesting it may have a role in multiple steps during metamorphosis and affect expression of many ecdysone regulated genes. For example, DHR3, a key component of the ecdysone response, is required for patterning and integrity of the adult cuticle (Lam et al., 1997), and DHR3 mutant clones exhibit a loss of pigmentation, cuticle defects, and missing bristles, similar to a partial loss of Bon (Figures 1D and 1G). In addition, mutations in βFTZ-F1(Broadus et al., 1999), E74B(Fletcher and Thummel, 1995), and BR-C(Kiss et al., 1988) exhibit malformed legs, which are a result of failure in the ecdysone response pathway. Again, we observe very similar defects in bon mutants (Figure 1D). Salivary glands in βFTZ-F1(Broadus et al., 1999), BR-C(Ristifo and White, 1992), and bon mutant pupae also fail to undergo apoptosis. The ability of bon mutations to cause phenotypes that resemble defects associated with mutations with multiple members of the pathway suggests that Bon is interacting with several members of the pathway at several stages, in agreement with the biochemical observations.
The interaction of Bon with nuclear receptors is similar to TIF1α (Le Douarin et al., 1996) but unlike TIF1β and TIF1γ (Le Douarin et al. 1997 and Venturini et al. 1999). This interaction requires the integrity of the nuclear receptor AF-2 activation domain and is mediated by the Bon/TIF1α LxxLL motif. These observations suggest that Drosophila nuclear receptors and Bon have co-evolved to maintain their interaction. It is therefore likely that the biological role of this interaction has been conserved in mammals.
Our data provide genetic evidence for the biological relevance of the interaction between Bon and the nuclear receptor βFTZ-F1. We find that reduction in the level of Bon, but not the complete loss of Bon, which affects the entire pathway, suppresses the phenotypes associated with a regulatory loss of function mutation of βFTZ-F1(Broadus et al., 1999). This suppression is likely to be the result of an increase in the transcription of βFTZ-F1, suggesting that Bon plays a role in the repression of βFTZ-F1. Because βFTZ-F1 represses its own transcription (Woodard et al., 1994), it is likely that a protein complex containing βFTZ-F1 and Bon is required for this repression. Removal of a copy of Bon may therefore lead to an up-regulation of βFTZ-F1 transcription. Although these data appear to contrast the loss of βFTZ-F1 transcription in bon241/bon241 mutants (Figure 4), they are not inconsistent. In the bon241/bon241 mutant background, loss of two copies of bon severely affects the entire ecdysone pathway. This is clearly not the case when one copy of bon is mutated. Therefore, removal of one copy of bon in the βFTZ-F1 mutant background allows for the detection of protein:protein interactions between βFTZ-F1 and Bon. Thus the phenotypic suppression (Figure 6) and the S2 cell transcription data (Figure 6D) are in agreement with Bon functioning as a negative regulator of βFTZ-F1-dependent transcription. It is therefore tempting to speculate by analogy that TIF1α may also interact with and inhibit transactivation by nuclear receptors in mammals. We favor a model in which Bon (or TIF1α), once recruited to particular regions of chromatin containing acetylated histones via their bromodomain, interacts via their LxxLL motif with the AF-2 domain of DNA-bound nuclear receptors. This complex then represses transcription from cognate target genes, possibly via an effect on chromatin structure.
Experimental Procedures
Fly strains, cloning of bonus, plasmid constructs and yeast two hybrid screen can be accessed at http://www.molecule.org/cgi/content/full/7/4/753/DC1.
Animal Staging and Phenotypic Analysis
For phenotypic and Northern analyses the Tb+ marker was used to distinguish mutant animals. Third instar larvae, prepupae, and pupae were staged according to Andres and Thummel (1994). First instar larvae were staged by collecting embryos for 20 hr followed by aging for 24 hr. For phenotypic analysis, whole pupae were dissected out of the pupal case and photographed. Wings from pharate adults were dissected in water, allowed to inflate, mounted, and photographed.
Clonal Analysis
y w; FRT82B bon21B/TM6B, Tb flies were crossed to y w ey-FLP; FRT82B cl3R3/TM6B, Tb to generate eye discs lacking Bon. y w ey-FLP; FRT82B bon21B/FRT82B cl3R3 and y w ey-FLP; FRT82B/TM6B, Tb (control) animals were dissected out of their pupal cases as pharate adults and photographed. To induce bon mutant clones, we subjected hs-FLP; FRT82B bon21B/FRT82B w+ and hs-FLP; FRT82B bon21B/FRT82B Sb animals to a 38°C heat shock for 2 hr.
Antibodies
Monoclonal antibodies (mAbs) used: anti-VP16 mAb, 2GV4 (Le Douarin et al., 1995a); anti-ERα F mAb; F3, raised against the F region of human ERα (Le Douarin et al., 1995a); anti-GAL4 (1–147); 2GV3 (Le Douarin et al., 1996); ANTI-FLAG® M2 mAb (Sigma-Aldrich, St. Louis, MO); and Mab 22C10 (Zipursky et al., 1985). Polyclonal antibodies used: Alexa Fluor® 488 goat anti-guinea pig IgG (H+L) conjugate (Molecular Probes, Eugene, OR); Cy3 goat anti-mouse IgG (H+L) conjugate (Jackson ImmunoResearch Laboratories, West Grove, PA); Cy3 goat anti-guinea pig IgG (H+L) conjugate (Jackson ImmunoResearch Laboratories); and biotinylated goat anti-guinea pig IgG (H+L) conjugate (Jackson ImmunoResearch Laboratories).
A bon DNA fragment corresponding to amino acids 506–866 was cloned into the pET-28a (+) expression vector (Novagen, Madison, WI). Guinea pig immune serum (Cocalico Biologicals, Inc., Reamstown, PA) was used at 1:10,000 for Western analysis and 1:5000 for immunohistochemistry.
Southern, Northern, and Western Analyses
Total RNA was isolated from third instar larvae, prepupae, and pupae using TRI-Reagent (Molecular Research Center, Inc., Cincinnati, OH). Southern and Northern analyses were performed according to standard protocols. For Northern analysis the following probes were used: full-length bon, open reading frame of DHR3(Koelle et al., 1992), full-length EcR B1(Talbot et al., 1993), common region of E74A and B(Burtis et al. 1990 and Thummel et al. 1990), common region of BR-C(DiBello et al., 1991), 1.6 kb EcoRI fragment from βFTZ-F1(Lavorgna et al., 1993), 1.6 kb AccI fragment from E93(Baehrecke and Thummel, 1995), and full length rp49(O’Connell and Rosbash, 1984).
Combined densitometry reading, as equalized by rp49 expression, of βFTZ-F1 expression for 6, 8, and 10 hr time points for Figure 6F: experiment 1, βFTZ-F1ex17/βFTZ-F1ex17(190 counts) and βFTZ-F1ex17bon487/βFTZ-F1ex17 (340 counts); experiment 2, βFTZ-F1ex17/βFTZ-F1ex17 (140 counts) and βFTZ-F1ex17bon487/βFTZ-F1ex17(266 counts).
For Western analysis, 20 μg of proteins isolated from third instar larvae, prepupae, and pupae were resolved on SDS-PAGE and transferred to a nitrocellulose membrane for immunoblotting. Antibody binding to the membrane was detected by enhanced chemiluminescence (Amersham Pharmacia Biotech Inc., Piscataway, NJ).
Immunohistochemistry and Confocal Microscopy
Staged embryos were collected, dechorionated, fixed in 4% formaldehyde/1X PBS, and devitellinized in methanol. Pupal tissues were isolated by dissection and fixed in 4% formaldehyde/1X PBS. Salivary gland polytene chromosomes were fixed and squashed according to a standard protocol (Andrew and Scott, 1994). Embryo and pupal tissue images were captured using a Bio-Rad MRC 600 laser scanning confocal microscope. Polytene chromosome images were captured using a Zeiss Axiovert microscope and Hamamatsu digital camera.
In Vitro Binding Assays
GST and GST fusion proteins were expressed in E. coli and purified on gluthathione-Sepharose (Amersham Pharmacia Biotech Inc.). 6 His-tagged Bon was expressed in recombinant Baculovirus-infected Sf9 insect cells and purified on Ni2+-chelating columns (Amersham Pharmacia Biotech Inc.). Purified proteins were quantified by Coomassie staining after SDS-PAGE separation and by Bradford protein assay. 200 ng of purified His-Bon was incubated with 1 μg of unfused GST or equimolar amounts of bacterially expressed GST fusion proteins loaded on glutathione-S-sepharose beads for 1 hr at 20°C in a final volume of 200 μl binding buffer (BB: 50 mMTris-HCl [pH 7.5], 100 mMKCl, 0.3 mM DTT, 1 mM PMSF, 10 mM MgCl2, 5% glycerol, 0.5% NP40, and protease inhibitor mixture). The beads were washed three times with 200 μl of BB buffer, resuspended in a SDS-containing buffer, boiled for 10 min, and proteins were analyzed by SDS-PAGE. The ECL detection system was used for immunodetection (Amersham Pharmacia Biotech Inc.).
Transactivation Assays
Yeast transformants were grown for ~16 hr to a cell titer of 2.5 × 107 in selective minimal medium. β-galactosidase assays on individual L40 transformants were carried out as previously described (vomBaur et al., 1996). PL3 cell-free extracts were assayed for OMP decase activity according to Le Douarin (1995b). S2 cells were grown at 25°C in Schneider medium (GIBCO) supplemented with 10% heat-inactivated fetal calf serum. Cells were transfected with 20 μg DNA as described by Krasnow et al. (1989). Cells recovered from each of the duplicate plates were split and half was used for Western blotting and half lysed for CAT activity measurements. CAT assays were performed with the CAT-ELISA kit (Roche Molecular Biochemicals, Indianapolis, IN).
Acknowledgments
We thank C. Thummel, A. Baniahmad, M. Koelle, R.M. Evans, S.M. Hollenberg, and the Bloomington Stock Center for reagents and flies. We thank S. Goode, M. Tsai, S. Tsai, G. Richards, and members of the Bellen and Chambon labs for comments on the manuscript. We thank L. Chetkovich, Y. He, M. Cerviño, T. Lerouge, S. Vicaire, D. Stephan, A. Staub, and F. Ruffenach for technical assistance. We also thank J. M. Garnier for construction of the Drosophila embryo cDNA VP16 fusion library. This work was supported by the Centre National de la Recherche Scientifique, the Institut National de la Santé et de la Recherche Médicale, l’Hôpital Universitaire de Strasbourg, the association pour la Recherchesur le Cancer (ARC), the Collège de France, the Fondation pour la Recherche Médicale, and Bristol-Myers-Squibb. J. A. O. was supported by the Ministerio de Educación y Ciencia de España and a Marie-Curie research training grant from the European Commission and C. S. by the Fundacionpara el Futuro de Colombia and the ARC. R. B. was supported by a NSF training grant of the Program in Developmental Biology.
References
- Aasland R, Gibson TJ, Stewart AF. The PHD finger implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci. 1995;20:56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- Andres AJ, Thummel CS. Methods for quantitative analysis of transcription in larvae and prepupae. Methods Cell Biol. 1994;44:565–573. doi: 10.1016/s0091-679x(08)60932-2. [DOI] [PubMed] [Google Scholar]
- Andrew DJ, Scott MP. Immunological Method for Mapping Protein Distributions on Polytene Chromosomes. Vol. 44. Academic Press, Inc; San Diego, CA: 1994. [DOI] [PubMed] [Google Scholar]
- Baehrecke EH, Thummel CS. The Drosophila E93 gene from the 93F early puff displays stage- and tissue-specific regulation by 20-hydroxyecdysone. Dev Biol. 1995;171:85–97. doi: 10.1006/dbio.1995.1262. [DOI] [PubMed] [Google Scholar]
- Broadus J, McCabe JR, Endrizzi B, Thummel CS, Woodard CT. The Drosophila beta FTZ-F1 orphan nuclear receptor provides competence for stage-specific responses to the steroid hormone ecdysone. Mol Cell. 1999;3:143–149. doi: 10.1016/s1097-2765(00)80305-6. [DOI] [PubMed] [Google Scholar]
- Burtis KC, Thummel CS, Jones CW, Karim FD, Hogness DS. The Drosophila 74EF early puff contains E74, a complex ecdysone-inducible gene that encodes two ets-related proteins. Cell. 1990;61:85–99. doi: 10.1016/0092-8674(90)90217-3. [DOI] [PubMed] [Google Scholar]
- Cammas F, Mark M, Dolle P, Dierich A, Chambon P, Losson R. Mice lacking the transcriptional corepressor TIF1b are defective in early postimplantation development. Development. 2000;127:2955–2963. doi: 10.1242/dev.127.13.2955. [DOI] [PubMed] [Google Scholar]
- Carney GE, Wade AA, Sapra R, Goldstein ES, Bender M. DHR3, an ecdysone-inducible early-late gene encoding a Drosophila nuclear receptor, is required for embryogenesis. Proc Natl Acad Sci USA. 1997;94:12024–12029. doi: 10.1073/pnas.94.22.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossgrove K, Bayer CA, Fristrom JW, Guild GM. The Drosophila Broad-Complex early gene directly regulates late gene transcription during the ecdysone-induced puffing cascade. Dev Biol. 1996;180:745–758. doi: 10.1006/dbio.1996.0343. [DOI] [PubMed] [Google Scholar]
- Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- DiBello PR, Withers DA, Bayer CA, Fristrom JW, Guild GM. The Drosophila Broad-Complex encodes a family of related proteins containing zinc fingers. Genetics. 1991;129:385–397. doi: 10.1093/genetics/129.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JC, Thummel CS. The Drosophila E74 gene is required for the proper stage- and tissue-specific transcription of ecdysone-regulated genes at the onset of metamorphosis. Development. 1995;121:1411–1421. doi: 10.1242/dev.121.5.1411. [DOI] [PubMed] [Google Scholar]
- Friedman JR, Fredericks WJ, Jensen DE, Speicher DW, Huang XP, Neilson EG, Rauscher FJ., 3rd KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- Fristrom D, Fristrom JW. The metamorphic development of the adult epidermis. Vol. 2. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1993. [Google Scholar]
- Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- Jacobs JJ, van Lohuizen M. Cellular memory of transcriptional states by Polycomb-group proteins. Semin Cell Dev Biol. 1999;10:227–235. doi: 10.1006/scdb.1999.0304. [DOI] [PubMed] [Google Scholar]
- Jeanmougin F, Wurtz JM, Le Douarin B, Chambon P, Losson R. The bromodomain revisited. Trends Biochem Sci. 1997;22:151–153. doi: 10.1016/s0968-0004(97)01042-6. [DOI] [PubMed] [Google Scholar]
- Jiang C, Baehrecke EH, Thummel CS. Steroid regulated programmed cell death during Drosophila metamorphosis. Development. 1997;124:4673–4683. doi: 10.1242/dev.124.22.4673. [DOI] [PubMed] [Google Scholar]
- Kastner P, Mark M, Chambon P. Nonsteroid nuclear receptors what are genetic studies telling us about their role in real life? Cell. 1995;83:859–869. doi: 10.1016/0092-8674(95)90202-3. [DOI] [PubMed] [Google Scholar]
- Kim SS, Chen YM, O’Leary E, Witzgall R, Vidal M, Bonventre JV. A novel member of the RING finger family, KRIP-1, associates with the KRAB-A transcriptional repressor domain of zinc finger proteins. Proc Natl Acad Sci USA. 1996;93:15299–15304. doi: 10.1073/pnas.93.26.15299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss I, Beaton AH, Tardiff J, Fristrom D, Fristrom JW. Interactions and developmental effects of mutations in the Broad-Complex of Drosophila melanogaster. Genetics. 1988;118:247–259. doi: 10.1093/genetics/118.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelle MR, Segraves WA, Hogness DS. DHR3a Drosophila steroid receptor homolog. Proc Natl Acad Sci USA. 1992;89:6167–6171. doi: 10.1073/pnas.89.13.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelle MR, Talbot WS, Segraves WA, Bender MT, Cherbas P, Hogness DS. The Drosophila EcR gene encodes an ecdysone receptor, a new member of the steroid receptor superfamily. Cell. 1991;67:59–77. doi: 10.1016/0092-8674(91)90572-g. [DOI] [PubMed] [Google Scholar]
- Krasnow MA, Saffman EE, Kornfeld K, Hogness DS. Transcriptional activation and repression by Ultrabithorax proteins in cultured Drosophila cells. Cell. 1989;57:1031–1043. doi: 10.1016/0092-8674(89)90341-3. [DOI] [PubMed] [Google Scholar]
- Lam GT, Jiang C, Thummel CS. Coordination of larval and prepupal gene expression by the DHR3 orphan receptor during Drosophila metamorphosis. Development. 1997;124:1757–1769. doi: 10.1242/dev.124.9.1757. [DOI] [PubMed] [Google Scholar]
- Lavorgna G, Karim FD, Thummel CS, Wu C. Potential role for a FTZ-F1 steroid receptor superfamily member in the control of Drosophila metamorphosis. Proc Natl Acad Sci USA. 1993;90:3004–3008. doi: 10.1073/pnas.90.7.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin B, Nielsen AL, Garnier JM, Ichinose H, Jeanmougin F, Losson R, Chambon P. A possible involvement of TIF1 alpha and TIF1 beta in the epigenetic control of transcription by nuclear receptors. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- Le Douarin B, Nielsen AL, You J, Chambon P, Losson R. TIF1 alphaa chromatin-specific mediator for the ligand-dependent activation function AF-2 of nuclear receptors? Biochem Soc Trans. 1997;25:605–612. doi: 10.1042/bst0250605. [DOI] [PubMed] [Google Scholar]
- Le Douarin B, Pierrat B, vomBaur E, Chambon P, Losson R. A new version of the two-hybrid assay for detection of protein-protein interactions. Nucleic Acids Res. 1995;23:876–878. doi: 10.1093/nar/23.5.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin B, Zechel C, Garnier JM, Lutz Y, Tora L, Pierrat P, Heery D, Gronemeyer H, Chambon P, Losson R. The N-terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. EMBO J. 1995;14:2020–2033. doi: 10.1002/j.1460-2075.1995.tb07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. The nuclear receptor superfamily. The second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna NJ, Lanz RB, O’Malley BW. Nuclear receptor coregulators. Cellular and molecular biology. Endocr Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- Moosmann P, Georgiev O, Le Douarin B, Bourquin JP, Schaffner W. Transcriptional repression by RING finger protein TIF1 beta that interacts with the KRAB repressor domain of KOX1. Nucleic Acids Res. 1996;24:4859–4867. doi: 10.1093/nar/24.24.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T, Kageyama Y, Hirose S, Ueda H. Regulation of the EDG84A gene by FTZ-F1 during metamorphosis in Drosophila melanogaster. Mol Cell Biol. 1996;16:6509–6515. doi: 10.1128/mcb.16.11.6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, Flanigan MJ, Kravitz SA, Mobarry CM, Reinert KH, Remington KA, et al. A whole-genome assembly of Drosophila. Science. 2000;287:2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- Newsome TP, Asling B, Dickson BJ. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development. 2000;127:851–860. doi: 10.1242/dev.127.4.851. [DOI] [PubMed] [Google Scholar]
- Nielsen AL, Ortiz JA, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, Chambon P, Losson R. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 1999;18:6385–6395. doi: 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell PO, Rosbash M. Sequence, structure, and codon preference of the Drosophila ribosomal protein 49 gene. Nucleic Acids Res. 1984;12:5495–5513. doi: 10.1093/nar/12.13.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Begg GE, Schultz DC, Friedman JR, Jensen DE, Speicher DW, Rauscher FJ., 3rd Reconstitution of the KRAB-KAP-1 repressor complexa model system for defining the molecular anatomy of RING-B box-coiled-coil domain-mediated protein-protein interactions. J Mol Biol. 2000;295:1139–1162. doi: 10.1006/jmbi.1999.3402. [DOI] [PubMed] [Google Scholar]
- Remboutsika E, Lutz Y, Gansmuller A, Vonesch JL, Losson R, Chambon P. The putative nuclear receptor mediator TIF1alpha is tightly associated with euchromatin. J Cell Sci. 1999;112:1671–1683. doi: 10.1242/jcs.112.11.1671. [DOI] [PubMed] [Google Scholar]
- Riddiford LM. Hormones and Drosophila development. Vol. 2. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1993. [Google Scholar]
- Ristifo LL, White K. Mutations in a steroid hormone-regulated gene disrupt the metamorphosis of internal tissues in Drosophila. Salivary glands, muscle, and gut. Roux’s Arch Dev Biol. 1992;201:221–234. doi: 10.1007/BF00188753. [DOI] [PubMed] [Google Scholar]
- Robertson CW. The metamorphosis of Drosophila melanogaster, including and accurately timed account of the principal morphological changes. J Morphol. 1936;59:351–399. [Google Scholar]
- Salzberg A, Prokopenko SN, He Y, Tsai P, Pal M, Maroy P, Glover DM, Deak P, Bellen HJ. P-element insertion alleles of essential genes on the third chromosome of Drosophila melanogaster mutations affecting embryonic PNS development. Genetics. 1997;147:1723–1741. doi: 10.1093/genetics/147.4.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin AJ, Borden KL, Boddy MN, Freemont PS. Does this have a familiar RING? Trends Biochem Sci. 1996;21:208–214. [PubMed] [Google Scholar]
- Segraves WA, Hogness DS. The E75 ecdysone-inducible gene responsible for the 75B early puff in Drosophila encodes two new members of the steroid receptor superfamily. Genes Dev. 1990;4:204–219. doi: 10.1101/gad.4.2.204. [DOI] [PubMed] [Google Scholar]
- Sliter TJ, Gilbert LI. Developmental arrest and ecdysteroid deficiency resulting from mutations at the dre4 locus of Drosophila. Genetics. 1992;130:555–568. doi: 10.1093/genetics/130.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot WS, Swyryd EA, Hogness DS. Drosophila tissues with different metamorphic responses to ecdysone express different ecdysone receptor isoforms. Cell. 1993;73:1323–1337. doi: 10.1016/0092-8674(93)90359-x. [DOI] [PubMed] [Google Scholar]
- Thummel CS, Burtis KC, Hogness DS. Spatial and temporal patterns of E74 transcription during Drosophila development. Cell. 1990;61:101–111. doi: 10.1016/0092-8674(90)90218-4. [DOI] [PubMed] [Google Scholar]
- Underhill C, Qutob MS, Yee SP, Torchia J. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J Biol Chem. 2000;275:40463–40470. doi: 10.1074/jbc.M007864200. [DOI] [PubMed] [Google Scholar]
- Molecular analysis of a steroid-induced regulatory hierarchy the Drosophila E74A protein directly regulates L71–6 transcription. EMBO J. 1995;14:6239–6246. doi: 10.1002/j.1460-2075.1995.tb00314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturini L, You J, Stadler M, Galien R, Lallemand V, Koken MH, Mattei MG, Ganser A, Chambon P, Losson R, de The H. TIF1gamma, a novel member of the transcriptional intermediary factor 1 family. Oncogene. 1999;18:1209–1217. doi: 10.1038/sj.onc.1202655. [DOI] [PubMed] [Google Scholar]
- vom Baur E, Zechel C, Heery D, Heine MJ, Garnier JM, Vivat V, Le Douarin B, Gronemeyer H, Chambon P, Losson R. Differential ligand-dependent interactions between the AF-2 activating domain of nuclear receptors and the putative transcriptional intermediary factors mSUG1 and TIF1. EMBO J. 1996;15:110–124. [PMC free article] [PubMed] [Google Scholar]
- White KP, Hurban P, Watanabe T, Hogness DS. Coordination of Drosophila metamorphosis by two ecdysone-induced nuclear receptors. Science. 1997;276:114–117. doi: 10.1126/science.276.5309.114. [DOI] [PubMed] [Google Scholar]
- Winston F, Allis CD. The bromodomaina chromatin-targeting module? Nat Struct Biol. 1999;6:601–604. doi: 10.1038/10640. [DOI] [PubMed] [Google Scholar]
- Woodard CT, Baehrecke EH, Thummel CS. A molecular mechanism for the stage specificity of the Drosophila prepupal genetic response to ecdysone. Cell. 1994;79:607–615. doi: 10.1016/0092-8674(94)90546-0. [DOI] [PubMed] [Google Scholar]
- Wurtz JM, Bourguet W, Renaud JP, Vivat V, Chambon P, Moras D, Gronemeyer H. A canonical structure for the ligand-binding domain of nuclear receptors. Nat Struct Biol. 1996;3:87–94. doi: 10.1038/nsb0196-87. [DOI] [PubMed] [Google Scholar]
- Xiao H, Lis JT. Heat shock and developmental regulation of the Drosophila melanogaster hsp83 gene. Mol Cell Biol. 1989;9:1746–1753. doi: 10.1128/mcb.9.4.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao TP, Forman BM, Jiang Z, Cherbas L, Chen JD, McKeown M, Cherbas P, Evans RM. Functional ecdysone receptor is the product of EcR and Ultraspiracle genes. Nature. 1993;366:476–479. doi: 10.1038/366476a0. [DOI] [PubMed] [Google Scholar]
- Yao TP, Segraves WA, Oro AE, McKeown M, Evans RM. Drosophila ultraspiracle modulates ecdysone receptor function via heterodimer formation. Cell. 1992;71:63–72. doi: 10.1016/0092-8674(92)90266-f. [DOI] [PubMed] [Google Scholar]
- Zipursky SL, Venkatesh TR, Benzer S. From monoclonal antibody to gene for a neuron-specific glycoprotein in Drosophila. Proc Natl Acad Sci USA. 1985;82:1855–1859. doi: 10.1073/pnas.82.6.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]