Preface
RB, a well known tumor suppressor that functions in the control of cell cycle progression and proliferation has recently been shown to have additional functions in the maintenance of genomic stability such that inactivation of RB family proteins promotes chromosome instability (CIN) and aneuploidy. Several studies have provided potential explanations for these phenomena following RB loss and suggest this new function of RB may contribute to it's role in tumor suppression.
Introduction
The RB tumor suppressor is well known for its ability to repress transcription and to prevent cell proliferation by arresting cells either in G1, at the G1/S transition, or in S phase of the cell cycle. The functional inactivation of RB compromises the ability of cells to respond to signals that normally suppress cell proliferation and results in the mis-expression of genes that drive cell division. Lesions leading to the functional inactivation of RB are thought to occur in most cancer cells, creating a cellular environment that is permissive for inappropriate cell proliferation.
Whole chromosome aneuploidy is another common feature of cancer cells. The frequent gains and losses of whole chromosomes is termed chromosome instability (CIN)1-3. CIN, by definition, results in the generation of aneuploid cells4-9 and has important implications in cancer. For example, it has been demonstrated that ‘shuffling’ of genomic content by CIN can facilitate loss of heterozygosity (LOH) of tumor suppressors and increased copy number of oncogenes. Furthermore, the genomic diversity generated by CIN promotes the development of cancer cells that are resistant to therapeutics and are more prone to tumor relapse 1, 10-18. Consequently, CIN correlates with poor patient prognosis 1, 3, 18. The changes that result in aneuploidy are poorly understood. It is likely that there is not one cause, but multiple contributory factors. Identifying the events that are responsible for the mitotic defects underying CIN is an important goal.
Recent work has demonstrated that the inactivation of RB and RB-related proteins leads to defects in mitotic progression and increases the likelihood of chromosome mis-segregation (reviewed in 19). Given the frequency of RB-pathway defects in cancer cells, these studies raise the tantalizing idea that the mutational events that promote tumor cell proliferation may also contribute to much of the aneuploidy seen in tumor cells.
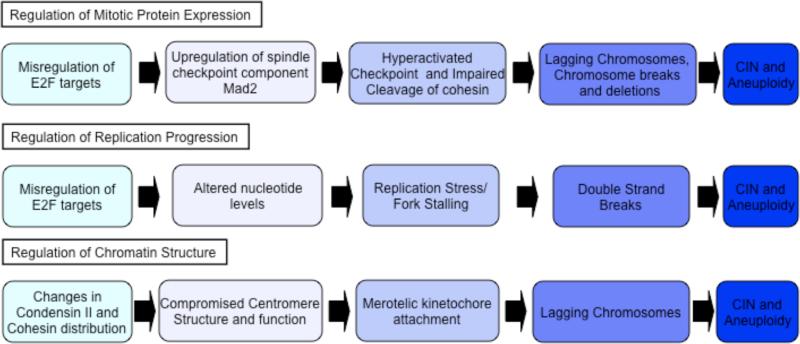
The mechanistic connection between RB family proteins and aneuploidy is not completely understood. Part of the puzzle arises from the fact that RB is regulated by cyclin-dependent kinase (CDK) phosphorylation and is generally thought to be converted to a functionally inactive state as cells progress towards S-phase, long before cells enter mitosis. If the conventional view of RB regulation/action is correct, then any link between RB and mitotic fidelity must be indirect. RB is a multifunctional protein and its inactivation has been shown to impact several different processes. There are multiple ways by which RB might plausibly influence mitotic progression and here we summarise three different explanations that have been proposed in a series of recent publications (Figure 1). We note that although the models are different, they are not mutually exclusive. Potentially, the different types of changes summarized below may have synergistic effects, with the cumulative effect promoting genomic instability in tumor cells.
Figure 1. Mechanisms of CIN.
Recent papers from several groups have proposed three different mechanisms to explain how the corruption of the RB pathway can promote chromosome segregation errors, chromsome instability (CIN) and aneuploidy.
Altered expression of genes with functions in mitosis
The best-characterized function of RB is its role in the regulation of the E2F transcription factor. Numerous genes are expressed at elevated levels in an E2F-dependent manner following the inactivation of RB. These include several genes with known roles in mitosis7, 20-22 and genes that have been linked to CIN 18, 23, 24. One of the most notable of these E2F targets is the spindle assembly checkpoint (SAC) protein MAD2. MAD2 overexpression is sufficient to induce CIN both in vivo and in vitro 18. Not only are MAD2 levels increased following the inactivation of RB1 in certain contexts, but recent experiments have also demonstrated that reducing the levels of MAD2 in a mouse tumor model delays tumor onset, reduces tumor burden, and results in tumors with a more stable karyotype9. Together these studies provide strong evidence that deregulated expression of MAD2 promotes CIN and that this, at least in some contexts, drives tumorigenesis. Interestingly MAD2 expression is upregulated by the inactivation of both the p53 and RB pathways. Recent experiments also indicate that loss of RB alone is not sufficient to de-regulate MAD2 expression as its regulation also depends on RB-related proteins. Thus, elevated expression of MAD2 may reflect the combined effects of pertubations in several tumor suppressor pathways.
MAD2 regulates mitotic progression by inhibiting the anaphase promoting complex/cyclosome (APC/C)–cell division cycle 20 (CDC20) complex (an E3 ubiquitin ligase) and promoting the activity of the SAC. The SAC prevents the premature protease-dependent cleavage of cyclin B and cohesin, which is required to maintain sister chromatid cohesion. Ultimately, the cleavage of cohesin initiates anaphase progression and allows chromosome segregation. Precisely why elevated levels of MAD2 cause errors in chromosome segregation is not certain. Shvartman et al. have suggested that elevated MAD2 may delay cohesin cleavage, resulting in the improper segregation of sister chromatids and leading to DNA damage 9. This hypothesis has yet to be directly tested and it is also possible that MAD2-induced aneuploidy involves activities of MAD2 that extend beyond the SAC and/or mitosis. In cells in which MAD2 expression is deregulated, increased MAD2 is seen throughout the cell cycle. In addition, recent studies have uncovered a SAC-independent role for MAD2 in the regulation of kinetochore microtubule stability and merotelic attachments that potentially may be very important in the generation of CIN (Compton and colleagues. unpublished data).
Although MAD2 is clearly important, it is unlikely to be the sole E2F target that links RB-pathway defects to the generation of aneuploidy. Segregation errors have been described following RB depletion under conditions where MAD2 expression is not detectably increased 25, and even in the experiments where reducing MAD2 has been shown to reduce chromosome segregation errors, the effect is partial. Other E2F-targets also have well defined roles in mitosis (Table 1, Figure 2) and promote chromosome segregation errors when overexpressed (including centromere protein A (CENPA) and HEC1 (also known as NDC80)) 4, 26, 27. Indeed, several E2F target genes were listed among the most highly misregulated genes in an expression profile signature of CIN tumors 22, 28. It is unclear whether this association occurs because of a specific link between E2F-deregulation and CIN, or whether the gene expression changes simply reflect changes in rates of in cell proliferation. In most cases, the functional consequences of E2F-driven changes in the expression of mitotic genes are not known, and some of these changes may even promote proper segregation rather than suppress it. In addition, in some cell types the inactivation of pRB undermines the normal coupling of replication and mitotic progression29-31. The resulting endoreduplication produces polyploid cells, which, if competent to proceed through subsequent mitosis, can lead to CIN irrespective of additional changes in gene expression32. Given that no two expression profiling experiments show precisely the same lists of deregulated E2F targets in different tumor cells, it is easy to imagine that the importance of individual E2F targets might vary in different tumor cells, depending on the combinations of mutations that are present and the extent to which different genes are deregulated. Nevertheless, these studies indicate that the altered expression of mitotic proteins is one way by which the inactivation of RB family proteins influences progression through mitosis and reduces the fidelity of chromosome segregation.
Table 1.
This table provides examples of the types of mitotic proteins that have been shown to be upregulated when pRB proteins are inactivated and E2F is deregulated. (for details see 7, 21, 22). Deregulated expression of several of these genes has been reported to be a component of an expression profile signature seen in CIN tumors28 (indicated with *), and some of these proteins have been shown to physically associate with pRB 34, 51 (indicated with #). While there are many potential changes in the levels and/or activity of mitotic proteins, we note that the relative importance of the individual changes is not known, and it is unclear how many of these events lead to specific mitotic defects.
| Gene | Attributed function | Mitotic Localization | Influence on Chromosome segregation |
|---|---|---|---|
| AurA/Stk6* | kinase; regulates localization and activity of numerous mitotic proteins | enriched at spindle poles | Promotes correction of erroneous kinet-MT attachments (merotely) |
| Brca1# | Promotes spindle bipolarity | centrosomes | Depletion causes multipolar spindles and segregation errors |
| Bub1 | kinase; mitotic checkpoint protein; regulates recruitment and activity of numerous kinetochore proteins | Kinetochore | Depletion compromises the SAC and promotes segregation errors |
| Cdc20*# | regulates APC/C activity and mitotic exit | mitotic spindles, centrosomes and kinetochores | depletion delays anaphase progression/mitotic exit |
| Cyclins A/B; Cdk1*#/Cdk2# | Regulate mitotic entry, NEB, spindle assembly and SAC | Cytoplasmic, enrichment at spindle poles, spindle midbody | Regulates mitotic timing to allow proper chromosome attachment/alignment prior to anaphase onset |
| Hec1 | Required for stable kinetochore-microtubule attachment | Outer kinetochore | stabilizes kinetochore -MT attachments in a phosphorylation-dependent manner |
| HSET/KIFC1 | minus-end directed kinesin; promotes spindle bipolarity, mitotic spindle pole focusing | binds microtubules, enriched at spindle poles | promotes clustering of supernumary centrosomes, may promote merotely when extra centrosomes are present |
| KID/KIF22 | Plus-end directed microtubule motor | On chromatin | Promotes chromosome oscillations and metaphase alignment |
| Mad2* | mitotic checkpoint protein; prevents mitotic exit by inhibiting APC/Cdc20 activity | Outer Kinetochore | OE stabilizes kinet-MT attachments and promotes merotelic attachments |
| MCAK/KIF2C | Induces plus-end microtubule deploymerization | Kinetochore | Promotes correction of erroneous kinet-MT attachments merotely) |
| Nek2* | regulates centrosome separation/bipolar spindle assembly; phosphorylates HEC1 to regulate kinetochore-microtubule attachments | centrosomes, kinetochores | destabilizes kinet-MT attachments through Hec1 phosphorylation |
| NuSAP* | Contributes to nucleation, stabilization and crosslinking of microtubules near chromosomes | spindle midbody | Promotes kinetochore-fiber formation |
| PLK1 | kinase; regulates activity of numerous mitotic proteins | Centrosomes, centromeres, central spindle | phosphorylates SA1/2 components of cohesin complex, facilitating it's removal from chromatin |
| Prc1* | Crosslinks anti-parallel spindle microtubules | central spindle; spindle midbody | promotes cytokinesis |
| Rad21# | Component of the cohesin complex, regulates sister chromatid cohesion | initially all along chromosomes, enriched at centromeres following prometaphase | Maintains sister chromatid cohesion to allow for proper biorientation and segregation |
| SMC2/4 | Component of Condensin I and II complexes, promotes chromatin condensation | Along chromosomes; condensin II is enriched at centromeres | Maintains chromatid condensation and promotes sister resolution, biorientation and proper segregation |
| Stathmin/OP18* | Regulates mitotis entry and spindle assembly; promotes microtubule dissasembly | binds MT dimers | Promotes microtubule dynamics required for spindle assembly |
also upregulated as a part of CIN expression profile
has been reported to be in a complex with pRB
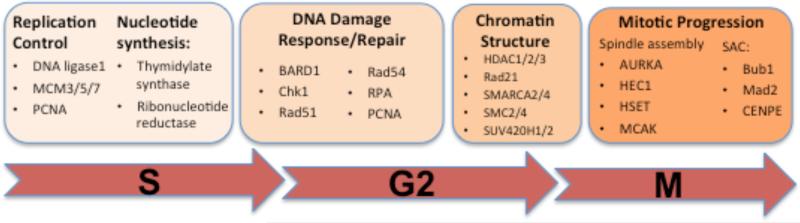
Figure 2. pRB-mediated effects on progression of S through M phase.
The RB/E2F pathway has well characterized roles in the regulation of quiescence, senescence and S phase entry. RB also regulates the expression of genes that act at later stages of the cell cycle and has been shown to physically interact with proteins that function during these stages 7, 20-22, 34, 35, 40-42. As a result, the functional inactivation of pRB does not simply affect G1 regulation, but influences cell cycle events that occur later in the cell cycle. Changes that occur at any of these steps can be detrimental to genomic stability.
Influence on replication progression
Recent work from Bestor and colleagues has highlighted a different type of cellular stress that occurs when RB-family members are inactivated and E2F is deregulated 33. In this study, the authors demonstrated that the expression of human papillomavirus (HPV) E7, a viral oncoprotein that targets RB, p107 and p130, alters S phase progression and promotes the stalling of replication forks. Stalling is especially prevalent at repetitive regions of the genome, including fragile sites, and is associated with increased levels of markers of DNA damage and with a high frequency of LOH 33. The list of E2F-regulated genes that are deregulated following E7 expression includes several genes that are necessary for nucleotide synthesis. Bestor et al. proposed that the altered replication dynamics are largely a consequence of suboptimal nucleotide pools. In support of this, they show that adding nucleosides to the cell culture media gives a strikingly strong suppression of the replication defects in E7-expressing cells 33.
This work adds to a series of studies that have linked RB with the control of DNA replication. Through its effects on E2F, the loss of RB alters the expression of numerous proteins that are needed for S-phase and RB/E2F proteins have also been shown to physically interact with replication factors 21, 34, 35,31 (Figure 2). Changes in the levels and/or activity of replication proteins in may influence the process of DNA replication in RB-deficient cells in multiple ways. RB loss has been shown to alter the spatial organization of DNA synthesis within the nucleus 36 and to affect the slowing of replication fork progression in the presence of genotoxic stress 37. The normal slowing of replication forks is thought to suppress the formation of double strand breaks (DSBs)38. Although there is an unexplained contradiction in the observations that the disruption of the RB pathway promotes replication fork stalling 33 and at the same time prevents slowing of replication fork progression in response to replication stress 37, such studies highlight the fact that the loss of RB disrupts the normal process of genome replication.
Analysis of replication fork progression in HPV E7-expressing cells shows that stalling is especially prevalent at repetitive regions of the genome, including fragile sites, and results in DSBs 33. In normal cells, DSBs would be repaired prior to entry into mitosis. However, the E2F pathway also regulates expression of numerous components of the DNA damage response and repair pathways 35 and various studies have shown that DNA damage checkpoints are compromised when the RB pathway is corrupted and cells can enter mitosis with unrepaired breaks 39-42 (Figure 1). The presence of DSBs during mitosis has several potential consequences. DSBs can activate the SAC 43 and sustained activation may influence mitotic fidelity in a similar manner to that proposed for MAD2 overexpression. Additionally, unresolved damage, particularly damage at centromeres or telomeres, may directly compromise chromosome segregation during mitosis and result in the generation of aneuploid daughter cells.
These results raise the possibility that the mitotic abnormalities seen in RB-deficient cells are a consequence of changes that occurred much earlier in the cell cycle. The idea that RB is necessary for normal replication, and that its loss causes replication stalling and DSBs, is consistent with multiple studies showing that loss of RB results in increased DNA damage, and that elevated DNA damage in turn results in increased genomic instability 23, 39, 44-47. Indeed, Bestor et al. suggest that much of the DNA damage, CIN and genome instability seen when RB is inactivated is an indirect consequence of replication fork stalling resulting from deregulated E2F.
Influence of RB-loss on chromosome structure
A third connection between RB and chromosome segregation has been provided by studies describing chromatin changes in cells in which the RB-family members are inactivated. Such changes include general defects in chromatin condensation, as well as more specific changes in the architecture of the telomeric and centromeric regions 5, 8, 48-51.
The ability of RB to recruit regulatory complexes to chromatin is one of the molecular features that enable it to regulate transcription. RB-family members are thought to localise primarily to promoter regions. The reason why RB loss affects higher-order chromosome structures is uncertain, but may involve both the loss of RB-recruited complexes as well as changes in the expression of RB-regulated genes. A wide variety of chromatin regulators have been shown to be recruited to chromatin by RB, including several proteins with important roles in heterochromatin formation 49, 52-56. In addition, the RB family of proteins have also been shown to physically interact with components of the condensin II complex and to influence the localization of both condensin and cohesin complexes to chromatin 8, 51, with the changes being most evident at the centromere.
Changes in centromere structure may be particularly relevant to the generation of chromosome segregation errors. The mitotic defects resulting from the specific depletion of RB from non-transformed cells include merotely, an improper attachment where a single kinetochore associates with microtubules from both spindle poles. Changes in centromeric structure, including changes caused by a reduction in the centromeric composition of condensin and cohesin complexes, similar to the defects seen in RB-depleted cells, are known to promote mal-attachment of the chromosome to the mitotic spindle, and thereby promote segregation errors 57, 58.
Although deformation of centromeric structure is associated with defects during mitosis, the loading of condensin and cohesin complexes onto chromatin begins much earlier in the cell cycle and changes in the stability of their chromatin-association on DNA are tightly coordinated with cell cycle progression. Condensin II and cohesin complexes are loaded onto chromatin as early as telophase, with cohesin additionally loaded onto newly synthesized chromatin concurrent with replication. However, chromatin association alone is not sufficient for function and both complexes are additionally regulated by modifications. For example, acetylation of the structural maintenance of chromosomes 3 (SMC3) subunit of cohesin converts what was merely chromatin association to a functional cohesion between replicated sister chromosomes 59. Whether the altered patterns of DNA replication seen in RB-depleted cells changes either the loading or stability of cohesin or condensin complexes is not yet known.
Characterization of mutant alleles of mouse Rb1 that is competent for regulation of E2F-dependent transcription but defective for interaction with chromatin regulators including the condensin II protein CAPD3, suggests that the ability of RB to recruit these complexes to chromatin contributes to its tumor suppressor activity, at least when combined with mutation of p53 5. Taken together, these studies suggest that inactivation of RB proteins causes changes in chromosome structure, that these defects promote segregation errors, and that this property of RB is relevant to tumor suppression.
A CINful path
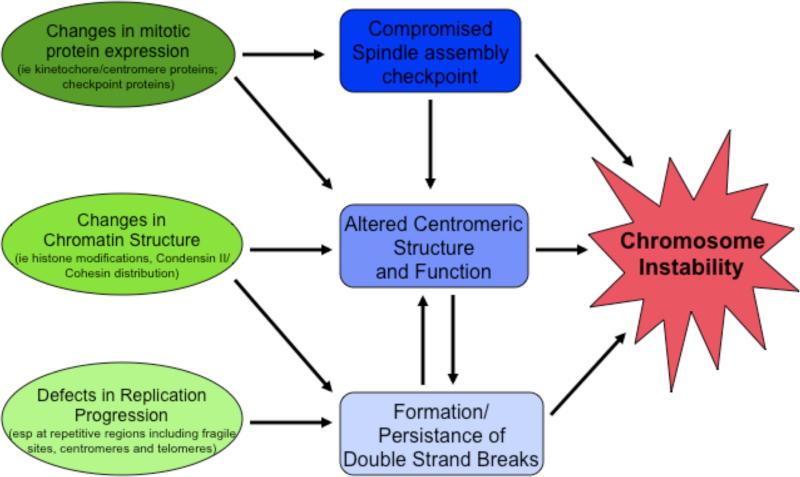
Given the multifaceted roles of RB in the control of gene expression and chromatin structure, it would be remarkable if there were a single feature of RB-deficient cells that would be sufficient to explain all of the mitotic defects and the propensity for CIN. A more likely scenario is that these properties represent the accumulated effect of multiple changes. As described above, the inactivation of RB leads to the mis-expression of proteins with important mitotic functions, the stalling of replication forks, elevated DNA damage, and altered chromosome structure. Each phenotype can be studied in isolation, but all are present when the RB-family is inactivated (Figure 2), and we note that there are several ways in which these different types of changes are interconnected and may synergise with one another (Figure 3).
Figure 3. The CINful path.
Previous models to explain CIN and aneuploidy when pRB is inactivated have proposed that specific cellular changes (green), such as changes in the expression of mitotic proteins, changes in chromatin structure, or defects in replication fork progression ultimately cause defects during mitosis (blue). This figure illustrates the fact that these different aspects of the pRB-loss of function phenotype do not exist in isolation, but are highly interconnected and are likely to influence one another. We suggest that changes in chromosome segregation and genome stability seen when pRB-proteins are inactivated represent the combined effect of mutiple defects.
It is likely that some of the mitotic defects associated with RB loss originate in S-phase. Defects in replication fork progression may leave unreplicated regions of the genome and promote DSBs that, if not resolved, interfere with mitotic progression and chromosome segregation 60. Replication fork stalling is especially prevalent at repetitive regions of the genome. Given that centromeric and telomeric regions are composed of highly repetitive sequences, these regions may be common locations for stalled replication forks when RB is inactivated. In addition, changes in replication dynamics may also alter the distribution (or activity) of cohesin and condensin II components. Since these complexes serve an essential role at the mitotic centromere, this region might be particularly vulnerable to altered levels. However, cohesin and condensin complexes have also been shown to serve important roles in replication fork progression, as well as prevention and efficient repair of DSBs61-63, any of which could contribute to genomic stability. These overlapping and interconnected roles of replication and chromatin structure are one example of how various defects resulting from RB misregulation may cooperate to promote CIN.
An additional way in which the various defects may interact is in the promotion of merotely. Merotelic attachment is thought to be a prominent cause of CIN in cell lines and chromosome segregation errors in tumors 64-66. Chromosome association with microtubules of the mitotic spindle is a complex process and is regulated at multiple levels. This complexity has made it difficult to discern the mechanism(s) behind such erroneous attachments60. RB-depleted cells show evidence of merotelic attachments and at least two distinct changes may cooperate to promote this. Potentially, the altered centromere structure in RB-deficient cells may be more prone to formation of merotelic attachments. In addition, the overexpression of kinetochore components (for example, HEC1 and MAD2) may stabilize erroneous attachments, therby preventing correction. Finally, E2F-dependent upregulation of MAD2 expression has been shown to delay progression through mitosis 23, a change that may enhance defects in sister chromatid cohesion in RB-depleted cells 8, 67, further promoting erroneous attachments. Together, these changes may make RB-deficient cells prone to the formation of merotelic attachments, and/or slower to correct such problems. In addition to promoting whole chromosome instability, merotelic attachments have also recently been shown to lead to structural chromosome abnormalities as a result of breaks during cytokinesis68.
These connections illustrate how defects in DNA replication might cause and be compounded by changes in chromosome structure, and how these changes, together with persistant DSBs, may exacerbate problems resulting from the mis-expression of mitotic proteins. The idea that all these changes are interconnected raises the key question of whether correcting any one of these defects would be sufficient to prevent aneuploidy and CIN. Using viral oncoproteins to inactivate RB, published work has shown that reducing MAD2 levels is sufficient to partially suppress aneuploidy and CIN 9, and that nucleoside supplementation is able to reduce the frequency of DSBs 33. Although it is an important point, it is not yet known whether manipulations that correct chromosome defects in RB-deficient cells impact rates of aneuploidy and CIN. Clearly further studies are needed to compare each of these approaches and to determine which strategy can be most easily exploited to suppress CIN.
Regulation of mitotic fidelity and tumor suppression by RB
There is compelling evidence that the inactivation of RB causes mitotic defects and promotes aneuploidy but, for several reasons, it is currently unknown how often these defects contribute directly to the pathogenesis of human tumors.
First, although mitotic defects, CIN, and genomic changes occur when RB is inactivated these changes are often more obvious when multiple family members are targeted, and this seems to be particularly true in mouse studies. In some cancer cells pRB is inactivated by deletion/mutation of the RB1 gene, but in others pRB is functionally inactivated through the expression of viral oncoproteins or through the increased activity of cdk's that not only inactivate pRB but also affect the pocket protein family members p107 and p130. While different types of lesions in the pRB pathway can promote aneuploidy 19, it is not clear that that the underlying mechanism will be the same in each case. It is also not yet known whether the mitotic phenotypes summarised here will be most relevant in human tumors with RB1 mutations, or in cells with deregulated CDKs that act on all three RB family proteins, or whether they will primarily occur in tumor cells that express viral oncoproteins such as HPV E7 that directly target all three RB-family members.
Second, for CIN to promote tumorigenesis, tumor cells need to be able to tolerate genomic change. The ability of cells to tolerate aneuploidy depends heavily on the presence of additional mutations, such as p53 inactivation, and this suggests that the genetic background of a tumor may be very important. It is noteworthy that several studies that have examined the effects of targeting RB have used cells that lack functional p53 5, 9 and or in which all three pocket protein family members are functionally inactivated. Interestingly, one form of cancer that typically fails to mutate or lose the p53 gene is retinoblastoma. A recent study of retinoblastoma tumors showed that the genome of these tumors is relatively stable, even though these cells exibit chromosomal changes and mitotic defects, such as high incidence of lagging chromosomes, that are associated with aneuploidy in other cell types 69.
Third, understanding how studies in cell lines and tumor models relate to human cancer is complicated by the fact that currently there is no simple way to assess the frequency of chromosome mis-segregation in human tumors. A commonly used substitute for measurement of CIN is the degree of aneuploidy. However, aneuploidy is not an accurate read out of CIN: the segregation errors that are characteristic of CIN may, or may not, result in aneuploidy, depending on tolerance or survival of the altered genome; moreover, aneuploid cells can also be generated by a one-time defect in mitosis, without the high rate of segregation errors characteristic of CN. Tumors can be highly aneuploid but genomically stable, or exhibit low levels of aneuploidy but be highly CIN. This makes it difficult to say which tumor samples exhibit CIN and which do not.
Rather than aneuploidy, a more accurate representation of CIN may be a measure of numerical heterogeneity within a population. Analysis of a set of tumor lines for which numerical heterogeneity has been analyzed70 shows no simple correlation with RB1 status. This may not be surprising since analysis of this same set of tumor cell lines also fails to show a correlation with p53 status, even though mutation of p53 has been shown in numerous model systems to influence genome stability. The lack of a correlation between CIN and the mutational status of either p53 or pRB is perhaps understandable given that most established tumors have lesions in both the p53 and pRB pathways. While a propensity for chromosome segregation errors may be directly linked to pRB inactivation, the degree of CIN or aneuploidy seen in a developed tumor presumably reflects a more complex balance of factors, including the tolerance of genomic change and the selection for or against specific genotypes, and is unlikely to be attributed to a single gene.
Finally, it is clear that there is not a single answer that applies to all situations. CIN and aneuploidy are not required for tumorigenesis but appear to become important in contexts where increased genomic variation is advantageous, such as during tumor evolution and relapse. Although RB pathway lesions are found in most tumor cells, these changes are an initiating event in some tumors but a late event in others. A potential explanation is that RB pathway lesions have multiple consequences and that different aspects of the mutant phenotype are advantageous in specific cell types or at different stages of tumor development. In cells in which the p53 pathway is active, high levels of CIN may be detrimental, perhaps even lethal, in early stages of tumorigenesis. Lesions in the RB pathway may have very different consequences when additional mutations have occurred that allow proliferation of aneuploid cells. In situations where RB-inactivation is an initiating event, the loss of G1/S regulation by RB may simply outweigh the mitotic defects. Alternatively, these cells may be intrinsically more tolerant to genomic change, or may suppress the mitotic defects.
The challenge for future studies is threefold: to understand more completely why inactivation of RB proteins causes CIN/aneuploidy, to find ways to prevent these mitotic defects, and to apply this information to specific tumor contexts where suppression of CIN/aneuploidy is beneficial. Such a comprehensive analysis of how different types of lesions in the RB pathway influence genome stability may help delineate a more individualized approach to suppressing CIN and tumor progression.
Acknowledgements
We thank D. Compton and L. Kabeche for sharing their unpublished data, N. Ganem for critical reading and suggestions on the manuscript, and the reviewers for their helpful comments. This work was supported by an American Cancer Society Fellowship to A.L.M., the Massachusetts General Hospital Cancer Center Saltonstall Foundation Scholarship to N.J.D., and funding from AstraZeneca and NIH grants GM81607 and CA64402 to N.J.D.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
References
- 1.Baker DJ, Jin F, Jeganathan KB, van Deursen JM. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell. 2009;16:475–86. doi: 10.1016/j.ccr.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sotillo R, et al. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Amato A, Schillaci T, Lentini L, Di Leonardo A. CENPA overexpression promotes genome instability in pRb-depleted human cells. Mol Cancer. 2009;8:119. doi: 10.1186/1476-4598-8-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coschi CH, et al. Mitotic chromosome condensation mediated by the retinoblastoma protein is tumor-suppressive. Genes Dev. 2010;24:1351–63. doi: 10.1101/gad.1917610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dimaras H, et al. Loss of RB1 induces non-proliferative retinoma: increasing genomic instability correlates with progression to retinoblastoma. Hum Mol Genet. 2008;17:1363–72. doi: 10.1093/hmg/ddn024. [DOI] [PubMed] [Google Scholar]
- 7.Iovino F, Lentini L, Amato A, Di Leonardo A. RB acute loss induces centrosome amplification and aneuploidy in murine primary fibroblasts. Mol Cancer. 2006;5:38. doi: 10.1186/1476-4598-5-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manning AL, Longworth MS, Dyson NJ. Loss of pRB causes centromere dysfunction and chromosomal instability. Genes Dev. 2010;24:1364–76. doi: 10.1101/gad.1917310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schvartzman JM, Duijf PH, Sotillo R, Coker C, Benezra R. Mad2 Is a Critical Mediator of the Chromosome Instability Observed upon Rb and p53 Pathway Inhibition. Cancer Cell. 2011;19:701–14. doi: 10.1016/j.ccr.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi CM, et al. Chromosomal instability is a risk factor for poor prognosis of adenocarcinoma of the lung: Fluorescence in situ hybridization analysis of paraffin-embedded tissue from Korean patients. Lung Cancer. 2009;64:66–70. doi: 10.1016/j.lungcan.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 11.Gao C, et al. Chromosome instability, chromosome transcriptome, and clonal evolution of tumor cell populations. Proc Natl Acad Sci U S A. 2007;104:8995–9000. doi: 10.1073/pnas.0700631104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heilig CE, et al. Chromosomal instability correlates with poor outcome in patients with myelodysplastic syndromes irrespectively of the cytogenetic risk group. J Cell Mol Med. 2010;14:895–902. doi: 10.1111/j.1582-4934.2009.00905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuukasjarvi T, et al. Genetic heterogeneity and clonal evolution underlying development of asynchronous metastasis in human breast cancer. Cancer Res. 1997;57:1597–604. [PubMed] [Google Scholar]
- 14.McClelland SE, Burrell RA, Swanton C. Chromosomal instability: a composite phenotype that influences sensitivity to chemotherapy. Cell Cycle. 2009;8:3262–6. doi: 10.4161/cc.8.20.9690. [DOI] [PubMed] [Google Scholar]
- 15.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–8. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 16.Rajagopalan H, Lengauer C. Aneuploidy and cancer. Nature. 2004;432:338–41. doi: 10.1038/nature03099. [DOI] [PubMed] [Google Scholar]
- 17.Swanton C, et al. Chromosomal instability determines taxane response. Proc Natl Acad Sci U S A. 2009;106:8671–6. doi: 10.1073/pnas.0811835106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sotillo R, Schvartzman JM, Socci ND, Benezra R. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature. 2010;464:436–40. doi: 10.1038/nature08803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manning AL, Dyson NJ. pRB, a tumor suppressor with a stabilizing presence. Trends Cell Biol. 2011;21:433–41. doi: 10.1016/j.tcb.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chakraborty S, et al. Identification of genes associated with tumorigenesis of retinoblastoma by microarray analysis. Genomics. 2007;90:344–53. doi: 10.1016/j.ygeno.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 21.Ishida S, et al. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Molecular and cellular biology. 2001;21:4684–99. doi: 10.1128/MCB.21.14.4684-4699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knudsen ES, Knudsen KE. Tailoring to RB: tumour suppressor status and therapeutic response. Nature reviews. Cancer. 2008;8:714–24. doi: 10.1038/nrc2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hernando E, et al. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 2004;430:797–802. doi: 10.1038/nature02820. [DOI] [PubMed] [Google Scholar]
- 24.Michel LS, et al. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–9. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- 25.Amato A, Lentini L, Schillaci T, Iovino F, Di Leonardo A. RNAi mediated acute depletion of retinoblastoma protein (pRb) promotes aneuploidy in human primary cells via micronuclei formation. BMC Cell Biol. 2009;10:79. doi: 10.1186/1471-2121-10-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diaz-Rodriguez E, Sotillo R, Schvartzman JM, Benezra R. Hec1 overexpression hyperactivates the mitotic checkpoint and induces tumor formation in vivo. Proc Natl Acad Sci U S A. 2008;105:16719–24. doi: 10.1073/pnas.0803504105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng L, Chen Y, Riley DJ, Chen PL, Lee WH. Retinoblastoma protein enhances the fidelity of chromosome segregation mediated by hsHec1p. Mol Cell Biol. 2000;20:3529–37. doi: 10.1128/mcb.20.10.3529-3537.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–8. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 29.Mayhew CN, et al. RB loss abrogates cell cycle control and genome integrity to promote liver tumorigenesis. Gastroenterology. 2007;133:976–84. doi: 10.1053/j.gastro.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 30.Niculescu AB, 3rd, et al. Effects of p21(Cip1/Waf1) at both the G1/S and the G2/M cell cycle transitions: pRb is a critical determinant in blocking DNA replication and in preventing endoreduplication. Molecular and cellular biology. 1998;18:629–43. doi: 10.1128/mcb.18.1.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srinivasan SV, Mayhew CN, Schwemberger S, Zagorski W, Knudsen ES. RB loss promotes aberrant ploidy by deregulating levels and activity of DNA replication factors. J Biol Chem. 2007;282:23867–77. doi: 10.1074/jbc.M700542200. [DOI] [PubMed] [Google Scholar]
- 32.Storchova Z, Kuffer C. The consequences of tetraploidy and aneuploidy. Journal of cell science. 2008;121:3859–66. doi: 10.1242/jcs.039537. [DOI] [PubMed] [Google Scholar]
- 33.Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, Bensimon A, Zamir G, Shewach DS, Kerem B. Nucleotide Deficiency Promotes Genomic Instability in Early Stages of Cancer Development. Cell. 2011;145:435–446. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morris EJ, Dyson NJ. Retinoblastoma protein partners. Adv Cancer Res. 2001;82:1–54. doi: 10.1016/s0065-230x(01)82001-7. [DOI] [PubMed] [Google Scholar]
- 35.Ren B, et al. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes & development. 2002;16:245–56. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barbie DA, et al. Nuclear reorganization of mammalian DNA synthesis prior to cell cycle exit. Mol Cell Biol. 2004;24:595–607. doi: 10.1128/MCB.24.2.595-607.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knudsen KE, et al. RB-dependent S-phase response to DNA damage. Mol Cell Biol. 2000;20:7751–63. doi: 10.1128/mcb.20.20.7751-7763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bosco EE, et al. RB signaling prevents replication-dependent DNA double-strand breaks following genotoxic insult. Nucleic Acids Res. 2004;32:25–34. doi: 10.1093/nar/gkg919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Harn T, et al. Loss of Rb proteins causes genomic instability in the absence of mitogenic signaling. Genes Dev. 2010;24:1377–88. doi: 10.1101/gad.580710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harrington EA, Bruce JL, Harlow E, Dyson N. pRB plays an essential role in cell cycle arrest induced by DNA damage. Proc Natl Acad Sci U S A. 1998;95:11945–50. doi: 10.1073/pnas.95.20.11945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cobrinik D. Pocket proteins and cell cycle control. Oncogene. 2005;24:2796–809. doi: 10.1038/sj.onc.1208619. [DOI] [PubMed] [Google Scholar]
- 42.Bourgo RJ, et al. RB restricts DNA damage-initiated tumorigenesis through an LXCXE-dependent mechanism of transcriptional control. Molecular cell. 2011;43:663–72. doi: 10.1016/j.molcel.2011.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mikhailov A, Cole RW, Rieder CL. DNA damage during mitosis in human cells delays the metaphase/anaphase transition via the spindle-assembly checkpoint. Curr Biol. 2002;12:1797–806. doi: 10.1016/s0960-9822(02)01226-5. [DOI] [PubMed] [Google Scholar]
- 44.Frame FM, Rogoff HA, Pickering MT, Cress WD, Kowalik TF. E2F1 induces MRN foci formation and a cell cycle checkpoint response in human fibroblasts. Oncogene. 2006;25:3258–66. doi: 10.1038/sj.onc.1209352. [DOI] [PubMed] [Google Scholar]
- 45.Pickering MT, Kowalik TF. Rb inactivation leads to E2F1-mediated DNA double-strand break accumulation. Oncogene. 2006;25:746–55. doi: 10.1038/sj.onc.1209103. [DOI] [PubMed] [Google Scholar]
- 46.Eguchi T, Takaki T, Itadani H, Kotani H. RB silencing compromises the DNA damage-induced G2/M checkpoint and causes deregulated expression of the ECT2 oncogene. Oncogene. 2007;26:509–20. doi: 10.1038/sj.onc.1209810. [DOI] [PubMed] [Google Scholar]
- 47.Jackson MW, et al. p130/p107/p105Rb-dependent transcriptional repression during DNA-damage-induced cell-cycle exit at G2. J Cell Sci. 2005;118:1821–32. doi: 10.1242/jcs.02307. [DOI] [PubMed] [Google Scholar]
- 48.Garcia-Cao M, Gonzalo S, Dean D, Blasco MA. A role for the Rb family of proteins in controlling telomere length. Nat Genet. 2002;32:415–9. doi: 10.1038/ng1011. [DOI] [PubMed] [Google Scholar]
- 49.Gonzalo S, et al. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat Cell Biol. 2005;7:420–8. doi: 10.1038/ncb1235. [DOI] [PubMed] [Google Scholar]
- 50.Isaac CE, et al. The retinoblastoma protein regulates pericentric heterochromatin. Mol Cell Biol. 2006;26:3659–71. doi: 10.1128/MCB.26.9.3659-3671.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Longworth MS, Herr A, Ji JY, Dyson NJ. RBF1 promotes chromatin condensation through a conserved interaction with the Condensin II protein dCAP-D3. Genes Dev. 2008;22:1011–24. doi: 10.1101/gad.1631508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brehm A, et al. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 53.Luo RX, Postigo AA, Dean DC. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–73. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 54.Magnaghi-Jaulin L, et al. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–5. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 55.Lai A, et al. RBP1 recruits both histone deacetylase-dependent and -independent repression activities to retinoblastoma family proteins. Mol Cell Biol. 1999;19:6632–41. doi: 10.1128/mcb.19.10.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McCabe MT, Davis JN, Day ML. Regulation of DNA methyltransferase 1 by the pRb/E2F1 pathway. Cancer Res. 2005;65:3624–32. doi: 10.1158/0008-5472.CAN-04-2158. [DOI] [PubMed] [Google Scholar]
- 57.Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43:525–58. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 58.Ng TM, Waples WG, Lavoie BD, Biggins S. Pericentromeric sister chromatid cohesion promotes kinetochore biorientation. Mol Biol Cell. 2009;20:3818–27. doi: 10.1091/mbc.E09-04-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Skibbens RV. Establishment of sister chromatid cohesion. Curr Biol. 2009;19:R1126–32. doi: 10.1016/j.cub.2009.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cimini D. Merotelic kinetochore orientation, aneuploidy, and cancer. Biochim Biophys Acta. 2008;1786:32–40. doi: 10.1016/j.bbcan.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 61.Terret ME, Sherwood R, Rahman S, Qin J, Jallepalli PV. Cohesin acetylation speeds the replication fork. Nature. 2009;462:231–4. doi: 10.1038/nature08550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mannini L, Menga S, Musio A. The expanding universe of cohesin functions: a new genome stability caretaker involved in human disease and cancer. Hum Mutat. 2010;31:623–30. doi: 10.1002/humu.21252. [DOI] [PubMed] [Google Scholar]
- 63.Lightfoot J, Testori S, Barroso C, Martinez-Perez E. Loading of Meiotic Cohesin by SCC-2 Is Required for Early Processing of DSBs and for the DNA Damage Checkpoint. Current biology : CB. 2011;21:1421–30. doi: 10.1016/j.cub.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 64.Cimini D, et al. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol. 2001;153:517–27. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–82. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol. 2008;180:665–72. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Daum JR, et al. Cohesion fatigue induces chromatid separation in cells delayed at metaphase. Current biology : CB. 2011;21:1018–24. doi: 10.1016/j.cub.2011.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Janssen A, van der Burg M, Szuhai K, Kops GJ, Medema RH. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science. 2011;333:1895–8. doi: 10.1126/science.1210214. [DOI] [PubMed] [Google Scholar]
- 69.Zhang J, et al. A Novel Retinoblastoma Therapy from Genomic and Epigenitic Analyses. Nature. 2012 doi: 10.1038/nature10733. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Roschke AV, et al. Karyotypic complexity of the NCI-60 drug-screening panel. Cancer research. 2003;63:8634–47. [PubMed] [Google Scholar]