Summary
Enzymatic heme catabolism by heme oxygenases is conserved from bacteria to humans and proceeds through a common mechanism leading to the formation of iron, carbon monoxide, and biliverdin. The first members of a novel class of heme oxygenases were recently identified in Staphylococcus aureus (IsdG and IsdI) and were termed the IsdG-family of heme oxygenases. Enzymes of the IsdG-family form tertiary structures distinct from those of the canonical heme oxygenase family, suggesting that IsdG-family members degrade heme via a unique reaction mechanism. Herein we report that the IsdG-family of heme oxygenases degrade heme to the oxo-bilirubin chromophore staphylobilin. We also present the crystal structure of heme-bound IsdI in which heme ruffling and constrained binding of oxygen is consistent with cleavage of the porphyrin ring at the β– or γ–meso carbons. Combined, these data establish that the IsdG-family of heme oxygenases degrade heme to a novel chromophore distinct from biliverdin.
Keywords: Staphylococcus aureus, heme oxygenase
Introduction
Most pathogenic bacteria have an absolute requirement for iron, a nutrient that is scarce within the human host (Bullen, 1999). The majority of vertebrate iron is found in the form of heme, which is bound by intracellular proteins, the most abundant of which is hemoglobin (Bullen, 1999). Heme-iron acquisition systems have been characterized in a number of pathogens; one such system is the iron-regulated surface determinant (Isd) system of S. aureus (Mazmanian et al., 2003). The Isd system is dedicated to heme-iron acquisition from host hemoglobin and includes nine proteins that bind hemoglobin, remove the heme cofactor, and traffic heme into the bacterium (Skaar and Schneewind, 2004). Once in the cytoplasm two paralagous heme oxygenases, IsdG and IsdI, degrade heme to release nutrient iron (Skaar et al., 2004). IsdG and IsdI represent the founding members of the IsdG-family of heme oxygenases. Orthologues have been identified in other staphylococci and several pathogenic species of Bacillus and Listeria, as well as Mycobacterium tuberculosis and the plant symbiont Bradyrhizobium japonicum (Chim et al., 2009; Puri and O'Brian, 2006; Skaar et al., 2004, 2006).
IsdG and IsdI share 64% sequence identity at the amino acid level (Reniere and Skaar, 2008). Moreover, the porphyrin-bound crystal structures are superimposable with a root mean square deviation of less than 1 Å, highlighting their significant structural similarity (Lee et al., 2008). However, both enzymes are required for staphylococcal growth on heme as a sole iron source, indicating that IsdG and IsdI are not functionally redundant (Reniere and Skaar, 2008). In support of this, S. aureus strains inactivated for either isdG or isdI are attenuated for virulence in a murine model of systemic infection (Reniere and Skaar, 2008). Insight into the requirement for two separate heme oxygenases came from studies into the regulation of these enzymes which revealed that IsdG and IsdI are differentially regulated depending upon iron and heme availability (Reniere and Skaar, 2008). In low iron conditions, transcription of isdG and isdI is increased due to the release of repression mediated by the canonical iron-dependent repressor Fur. A second layer of regulation exists post-transcriptionally as the stability of IsdG is affected by changes in exogenous heme concentrations. It has been modeled that this differential regulation provides a means for the bacteria to fine-tune the expression of heme oxygenase activity in the cell in order to rapidly adjust to changes in nutrient levels (Reniere and Skaar, 2008).
Heme oxygenases (HOs) are ubiquitous enzymes in nature and are required for many diverse processes, such as iron metabolism, oxidative stress response, and inflammation (Chung et al., 2009; Stocker et al., 1987; Tenhunen et al., 1969). Although HOs were identified in eukaryotes approximately forty years ago, it is only recently that bacterial heme oxygenases have been identified and studied. The first bacterial heme oxygenase described was HmuO from Corynebacterium diphtheriae, which was identified based on its homology to human heme oxygenase 1 (HO-1) (Schmitt, 1997). Since that initial report, numerous additional HO-1-like heme oxygenases have been identified in bacteria (Ratliff et al., 2001; Wilks and Schmitt, 1998; Zhu et al., 2000). Despite sharing little sequence identity, members of the HO-1 family exhibit significant structural similarity (Schuller et al., 2001). The HO-1 enzymes are monomeric α-helical proteins, each with a single active site and GXXXG catalytic motif (Schuller et al., 1999). Heme catabolism by the HO-1 family of enzymes has been studied extensively in both bacteria and vertebrates and the mechanism of heme degradation is believed to proceed similarly across most heme oxygenases, resulting in the production of α-biliverdin (Unno et al., 2004). One exception is pa-HO from Pseudomonas aeruginosa which differs in its regiospecificity, producing a mixture of biliverdin isomers (Friedman et al., 2004; Ratliff et al., 2001). In mammals biliverdin is further reduced to bilirubin by biliverdin reductase; however, the ultimate fate of biliverdin in bacteria is unknown as homologues of biliverdin reductase have not been discovered in bacteria.
The IsdG-family of heme oxygenases is structurally distinct from the HO-1 family. IsdG-family members form homodimeric β-barrel structures with two separate active sites, each containing three residues (Asn, Trp, His) that are required for activity (Wu et al., 2005). Initial reports demonstrated IsdG-catalyzed cleavage of the macrocyclic ring with concomitant release of free iron, although the precise mechanism of heme degradation by this class of heme oxygenases has not yet been delineated (Skaar et al., 2004). Recently it has been suggested that this family of enzymes degrade heme via a novel mechanism (Reniere et al., 2007). Porphyrin-bound structures of IsdG-family enzymes have revealed significant ruffling of the porphyrin ring in the active site. In fact, the porphyrin ring is distorted approximately 2 Å out of plane, far more than has been observed in any other heme-binding protein (Lee et al., 2008). Moreover, in vitro heme degradation catalyzed by recombinant IsdG results in the formation of a yellow reaction product distinct from the blue-green biliverdin chromophore (Lee et al., 2008; Reniere et al., 2007). Taken together, these structural and biochemical observations suggest that members of the IsdG-family of heme oxygenases degrade heme by a unique mechanism, resulting in the formation of a product that is distinct from biliverdin. Herein we present the crystal structure of heme-bound IsdI and demonstrate that the IsdG-family of enzymes degrade heme to the novel oxo-bilirubin chromophore staphylobilin.
Results
The structure of heme-bound IsdI
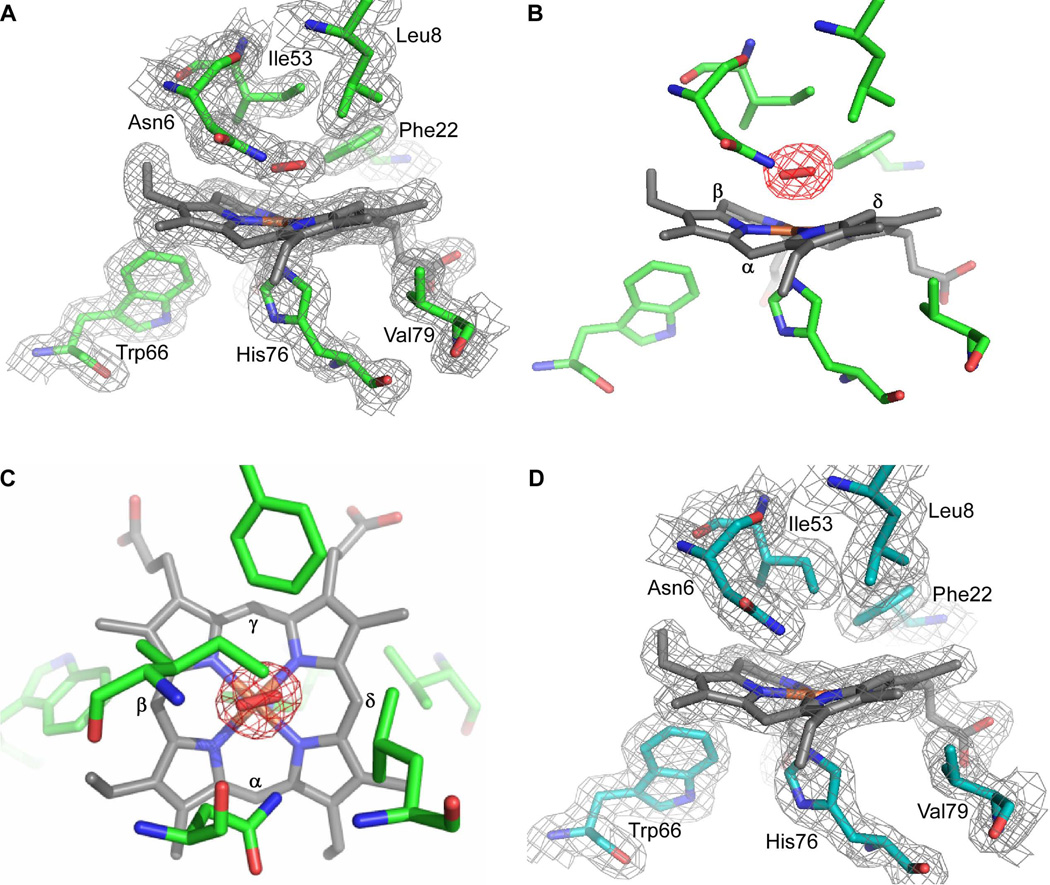
We have previously determined the crystal structures of a catalytically inactive variant of IsdG bound to heme (FePPIX) as well as native IsdI bound to the heme analogue cobalt protoporphyrin IX (CoPPIX) (Lee et al., 2008). However, studies into the mechanism of heme degradation have been hindered by the fact that the crystal structure of a catalytically active IsdG-family member bound to the substrate heme has not been reported. To gain insight into the mechanism of heme catabolism by IsdG-family enzymes, we crystallized catalytically active IsdI bound to heme by lowering the temperature to 4° C. The structure was solved to 1.5 Å resolution with one IsdI dimer in the asymmetric unit (Table 1). The fold of the IsdI-heme complex is similar to that observed in the IsdI-CoPPIX structure and the heme porphyrin ring is also highly ruffled. We observe elongated density for a sixth iron ligand on the distal side of heme, best modeled as a coordinating dioxygen species with an average Fe-O bond length of 2.05 Å (Fig. 1A–C). The presence of an oxygen species implies that heme-iron was photoreduced to Fe(II) in the X-ray beam during data collection. Further reduction of the oxygen species is suggested by the decreased Fe-O-O angle (108°). Notably, dithionite reduced crystals yield a structure without a density for a sixth ligand (Fig. 1D), likely due to the consumption of oxygen in the crystal by the reductant. No other significant differences are noted between the dithionite reduced and native structures.
Table 1.
Data collection and refinement statistics for IsdI-heme structures
| IsdI-heme | IsdI-heme (reduced) | |
|---|---|---|
| Data collectiona | ||
| Resolution range | 48.11 - 1.50 | 58.03 - 1.88 |
| Space group | P 212121 | P 212121 |
| Unit cell dimension (Å) | a=58.24 b=66.78 c=69.43 | a=58.03 b=67.93 c=70.67 |
| Unique reflections | 43991 (6351) | 25682 (3674) |
| Completeness (%) | 99.8 (100.0) | 99.8 (100.0) |
| Average I/σI | 13.1 (3.3) | 12.2 (4.1) |
| Redundancy | 6.8 (5.6) | 7.0 (6.9) |
| Rmerge | 0.077 (0.481) | 0.094 (0.487) |
| Refinement | ||
| Rwork(Rfree) | 0.181(0.214) | 0.201(0.242) |
| No. Atoms | 2256 | 2116 |
| Protein | 1884 | 1845 |
| Solvent | 282 | 185 |
| Heme | 86 | 86 |
| Dioxygen | 4 | - |
| Overall B-factor (Å2) | 21.7 | 29.6 |
| Protein | 19.7 | 28.8 |
| Solvent | 36.6 | 39.2 |
| Heme | 16.1 | 26.3 |
| Dioxygen | 14.4 | - |
| r.m.s.deviation | ||
| Bond length | 0.009 | 0.014 |
| Bond angles | 1.118 | 1.348 |
Values for the highest resolution shell are shown in parenthesis
Fig. 1.
Crystal structure of the active site of IsdI-heme. The structure of IsdI (green) bound to heme (gray) reveals density on the distal side modeled as a dioxygen species (red) liganded to the heme-iron. A. 2Fo - Fc map contoured at 1σ. B. A Fo - Fc omit map for oxygen is contoured at 3σ. The heme α-, β-, and δ-meso carbons are labeled. The γ-meso carbon is buried in the protein. C. The same figure as in b, but looking down perpendicular to the heme from the distal side. The heme α-, β-, γ-, and δ-meso carbons are labeled. D. Crystals of IsdI (cyan) bound to heme (gray) were reduced for 10 minutes in 50 mM (excess) dithionite. The 2Fo - Fc map (light gray) is contoured at 1σ. Heme and selected amino acids are depicted in sticks and are labeled. Nitrogen, oxygen and iron atoms are colored blue, red, and orange, respectively.
In contrast to the HO-1 family of enzymes, the oxygen species in the IsdI heme-binding pocket is aligned with the imidazole plane of the coordinating histidine, which lies along the β,δ-axis of the porphyrin ring (Fig. 1C). The orientation of the dioxygen is restricted by Phe22 and Asn6, which is also an H-bond donor to the oxygen species. In the HO-1 family, regiospecificity is also achieved by steric limitations of the dioxygen ligand such that only the α-meso carbon is accessible for hydroxylation (Unno et al., 2004). In the two IsdI heme-binding pockets of the asymmetric unit, the oxygen species are in opposite orientations such that the β- or δ-meso carbon is aligned for hydroxylation. The heme is bound predominantly in one orientation, suggesting that reaction at both meso-carbons is possible.
Due to the alignment of the oxygen species with respect to the heme ring in the active site, we hypothesized that the product of heme degradation by IsdG-family enzymes exhibits unique regiospecificity compared to the canonical HO-1 like enzymes. We therefore sought to determine the structures of the products of IsdG- and IsdI-mediated heme degradation and add insight into the mechanism by which these enzymes degrade heme.
Purification of the IsdG- and IsdI-catalyzed heme degradation products
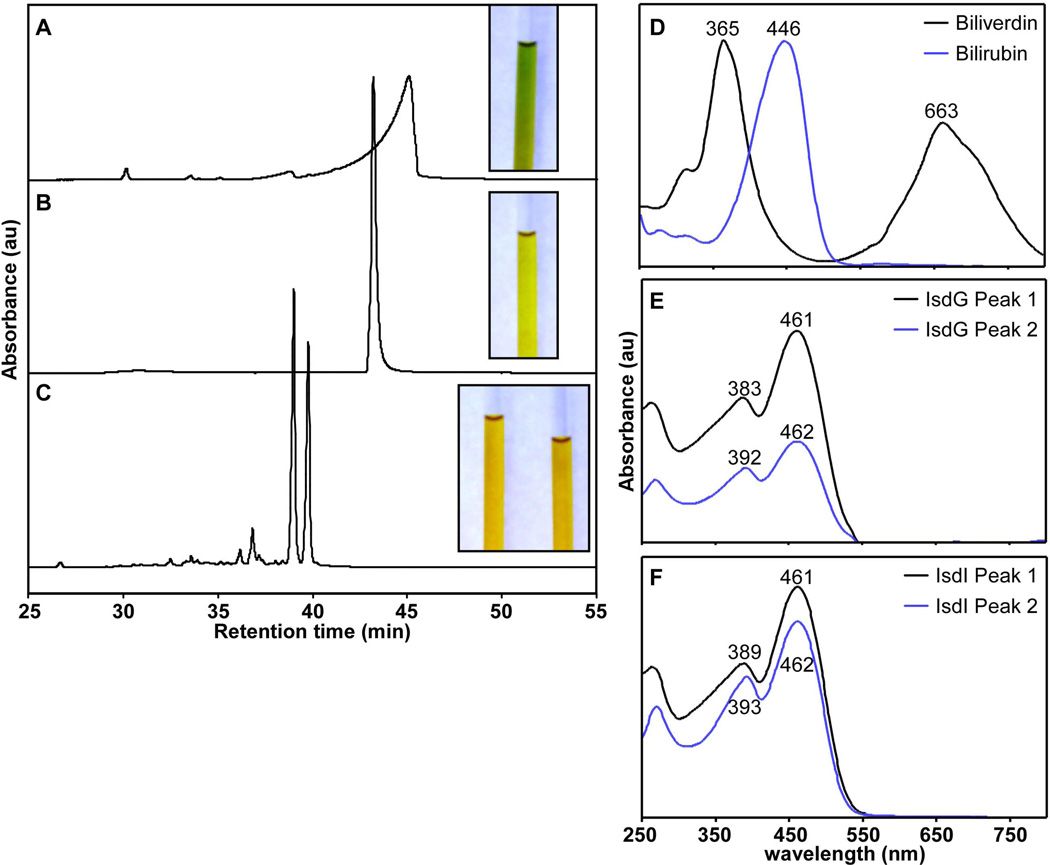
As previous attempts to crystallize the product-bound enzymes were unsuccessful, we sought to isolate the product of IsdG-mediated heme degradation for structural studies. In vitro heme degradation reactions were performed with purified recombinant IsdG and IsdI using ascorbate as the electron donor and catalase to prevent coupled oxidation (see Experimental Procedures). Purification of the products of IsdG- and IsdI-catalyzed heme degradation results in a red-orange solid that becomes yellow when dissolved in dimethyl sulfoxide (DMSO) or water/acetonitrile (1:1). This is in stark contrast to the blue-green color of the canonical heme degradation product biliverdin (Fig. 2A–C, insets). HPLC-separation of the IsdG and IsdI reaction products results in two distinct peaks which elute earlier than both biliverdin and bilirubin, indicating a higher degree of polarity (Fig. 2A–C). Product peaks from both IsdG- and IsdI-mediated degradation exhibit similar optical spectra with one maximum at ~390 nm and one at ~460 nm. The peak maxima of biliverdin are 365 and 663 nm and the peak maximum of bilirubin is 446 nm (Fig. 2D–F). Together, these data suggest that the IsdG-family of oxygenases catabolize heme to a unique chromophore that is more polar than biliverdin and bilirubin.
Fig. 2.
Purification and optical spectra of heme degradation products. HPLC tracings of heme degradation products separated on a linear acetonitrile gradient. Insets are images of the heme degradation products dissolved in DMSO. The contrast of the photos was adjusted to present the clearest images. A. Biliverdin purification monitored at 405 nm. B. Bilirubin purification monitored at 405 nm. C. The product of IsdI-catalyzed heme degradation monitored at 465 nm. The two prominent peaks were collected separately and both product images are shown (inset). IsdG- and IsdI-catalyzed heme degradation products have overlapping spectra so only that of IsdI is shown for simplicity. D–F. Optical spectra of each fraction as measured by the photodiode array detector upon elution.
IsdG and IsdI degrade heme to staphylobilin
In order to determine the molecular composition of the products of IsdG- and IsdI-mediated heme degradation, the HPLC-purified samples were subjected to high resolution electrospray ionization mass spectrometry (HRESIMS). Each of the eluting product peaks gave a mass of 598.25 Da (m/z 599.2605 [M + H], Δ = 3.0 – 4.2 ppm), corresponding to a molecular formula of C33H34N4O7 (Fig. S1). As biliverdin and bilirubin have molecular masses of 582.6 and 584.7 Da, respectively, these data conclusively demonstrate that heme degradation catalyzed by IsdG and IsdI does not produce biliverdin or bilirubin.
This hypothesis is supported by analysis of 1H NMR spectra of the purified fractions acquired in d6-DMSO. Inspection of the downfield region indicates that both eluting fractions possess two pyrrol amide (δ 10.4 – 10.5) and two pyrrol amine (δ 11.0 – 11.3) resonances; however notable chemical shift differences in this region were evident between the two fractions (Fig. S2). Furthermore, both products of IsdG- and IsdI-mediated heme degradation have distinct 1H NMR spectra in comparison to reported spectra of biliverdin and bilirubin (Fig. S4–S7) (Kaplan, 1981). The most significant dissimilarity is again in the NH region in which the pyrrol amines of the IsdG and IsdI products are shifted as much as 0.8 ppm in comparison to reported hemes (Fig. S2). Chemical shift values in this range are also observed for 10-oxo-mesobilirubin which was chemically synthesized by Chen et al. in order to investigate possible oxidative pathways of bilirubin elimination in vivo (Chen, 1999). Due to the addition of a carbonyl group at the γ-meso carbon of bilirubin, 10-oxo-mesobilirubin is more stable than the parent molecule and the pyrrole NHs are considerably more deshielded than the corresponding NHs of bilirubin (Chen, 1999). These similarities further suggest that the products of IsdG-mediated heme degradation may be oxidized forms of bilirubin.
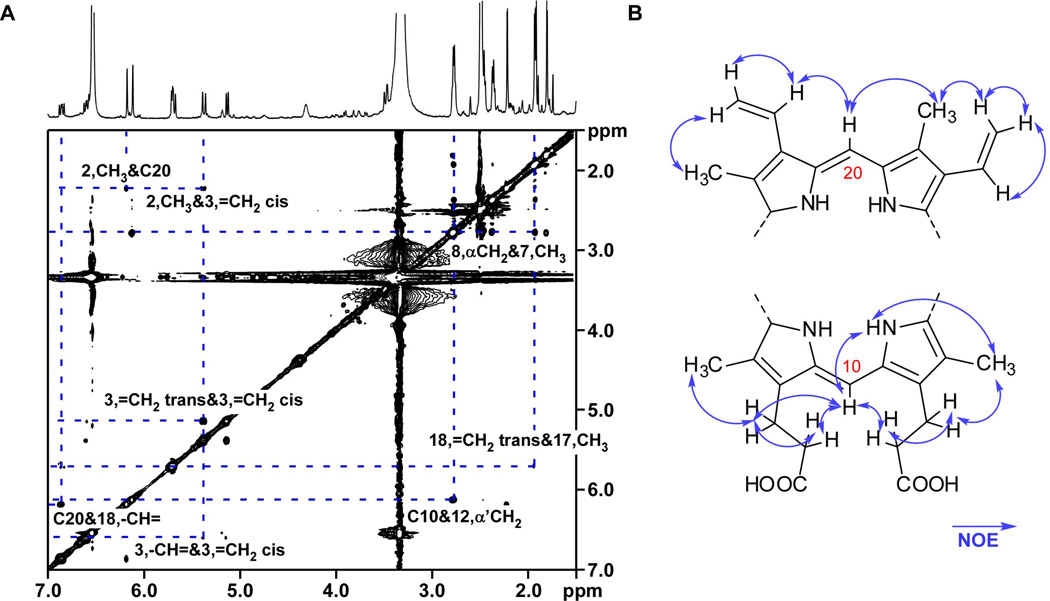
The 1H NMR spectra also establish several regions of biliverdin and bilirubin that are conserved in the IsdG-family products. All side-chain resonances present comparable chemical shifts and coupling values to canonical hemes (Tables 2 and 3), indicating that the vinyl side chains and the α- and β-CH2 positions of the propionate group chains are intact and that the methyl substituents are unmodified. Side-chain functional groups are readily positioned in their corresponding pyrrol rings using two-dimensional NMR data (Fig. S3–S7). The observed nuclear Overhauser effect (NOE) interactions between C20-CH and C3-CH3 and also C18-CH= indicate that the α-bridge is intact, and correlations between C10-CH and α,α’-CH2, β,β’-CH2 indicate that the γ-bridge is intact (Fig. 3A). These spectral data allow us to assemble two partial structures, one hemisphere containing the intact α-bridge and the other encompassing the intact γ-bridge (Fig. 3B). Therefore, although the heme degradation products of the IsdG-family share structural similarities with bilirubin, these products are not cleaved at the α-meso carbon.
Table 2.
1H and 13C data of 5-oxo-δ-bilirubin
| Positiona |
1H, m (J in Hz) |
13C (ppm) |
COSY | HMBC | NOESY |
|---|---|---|---|---|---|
| 13, CH3 | 1.80, s | 7.9 | - | 126.7, 144.3, 172.0 | 2.79 |
| 7, CH3 | 1.92, s | 9.1 | - | 125.3, 131.7 | 2.76, 11.30 |
| 17, CH3 | 1.93, s | 9.1 | - | 140.3, 171.6 | 5.71 |
| 2, CH3 | 2.22, s | 10.8 | - | - | 5.37, 6.18 |
| 8, α CH2 | 2.36, t (7.1) | 34.4 | 2.76 | 126.7, 173.4 | 2.76, 6.12 |
| 12, α' CH2 | 2.46, t (7.5) | 33.5 | 2.79 | 144.3, 173.1 | 2.79, 6.12 |
| 8, β CH2 | 2.76, d (7.0) | 18.5 | 2.36 | - | 1.92, 2.36 |
| 12, β' CH2 | 2.79, d (7.0) | 18.5 | 2.46 | 144.3, 173.1 | 1.80, 2.46 |
| 3, =CH2 cis | 5.13, dd (1.6, 11.7) | 115.2 | 6.60 | 125.1 | 5.37, 6.60 |
| 3, =CH2trans | 5.37, dd (1.7, 18.0) | 115.2 | 6.60 | 125.1 | 2.22, 5.13 |
| 18, =CH2 trans | 5.68, dd (1.4, 13.5) | 122.7 | 6.86 | 140.3 | 1.93 |
| 18, =CH2 cis | 5.71, dd (1.5, 7.4) | 122.7 | 6.86 | 140.3 | 6.86 |
| 5 | - | 174.7 | - | - | - |
| 10 | 6.12, s | 95.9 | - | 126.7, 144.3 | 2.36, 2.46, 2.79, 11.30 |
| 20 | 6.18, s | 96.8 | 121.4, 140.3 | 2.22, 6.86 | |
| 3, -CH= | 6.60, dd (11.7, 18.0) | 128.2 | 5.13, 5.37 | 121.4 | 5.13 |
| 18, -CH= | 6.86, dd (11.6, 17.6) | 126.5 | 5.68, 5.71 | 125.8 | 5.71, 6.18 |
| 23 NH | 10.40, s | - | - | 126.7, 132.5, 144.3 | 11.04, 11.30 |
| 24 NH | 10.48, s | - | - | 125.8, 131.7, 140.3 | 11.04, 11.30 |
| 21 NH | 11.04, s | - | - | 125.1, 127.6, 131.6 | 10.40, 10.48 |
| 22 NH | 11.30, s | - | - | 120.4, 125.3 | 1.92, 6.12, 10.48, 10.40 |
| 8, COOH | 12.09, bs | 173.4 | - | - | - |
| 12, COOH | 12.09, bs | 173.1 | - | - | - |
Position based on numbering scheme in Fig. 4.
Table 3.
1H and 13C data of 15-oxo-β-bilirubin
| Positiona |
1H, m (J in Hz) |
13C (ppm) |
COSY | HMBC | NOESY |
|---|---|---|---|---|---|
| 7, CH3 | 1.81, s | 7.8 | - | 126.6, 144.3, 172.0 | 2.79 |
| 13, CH3 | 1.99, s | 9.5 | - | 124.8, 127.5, 131.3 | 2.79 |
| 17, CH3 | 2.05, s | 11.0 | - | 122.9, 132.5 | - |
| 2, CH3 | 2.18, s | 8.8 | - | 124.2, 133.6, 142.1 | - |
| 12, α CH2 | 2.37, m (1.8) | 34.3 | 2.79 | 18.4, 173.3 | 2.79 |
| 8, α' CH2 | 2.46, d (7.6) | 33.4 | 2.79 | 18.6, 172.9 | - |
| 8, β CH2 12, β' CH2 | 2.79, q (7.2, 12.0) | 18.4, 18.6 | 2.37, 2.46 | 33.4, 126.6, 144.3, 173.3 | 1.81, 1.99, 2.37 |
| 18, =CH2 cis | 5.35, dd (1.5, 11.6) | 115.8 | 6.87 | 124.8 | - |
| 3, =CH2 cis | 5.38, d (2.3) | 118.6 | 6.60 | 124.2 | 6.24 |
| 18, =CH2 trans | 5.41, dd (1.6, 17.6) | 115.8 | 6.87 | - | 6.24, 6.87 |
| 10 | 6.12, s | 95.8 | - | 126.6, 144.3 | 2.79 |
| 15 | - | 174.9 | - | - | - |
| 20 | 6.24, s | 97.1 | - | 124.2, 142.1 | 2.18, 5.38, 6.87 |
| 3, =CH2 trans | 6.25, dd (2.4, 17.5) | 118.6 | 6.60 | 124.2 | 2.18, 5.38 |
| 3, -CH= | 6.60, dd (11.6, 17.6) | 126.2 | 5.38 | - | - |
| 18, -CH= | 6.87, dd (11.6, 17.8) | 128.3 | 5.35, 5.41 | - | 5.41, 6.24 |
| 21 NH | 10.47, s | - | - | 124.2, 132.4, 142.1 | 11.05, 11.28 |
| 22 NH | 10.47, s | - | - | 124.2, 132.4, 142.1 | 11.05, 11.28 |
| 23 NH | 11.05, s | - | - | 126.6 | 10.47, 11.28 |
| 24 NH | 11.28, s | - | - | 122.9, 124.8 | 10.47, 11.05 |
| 8, COOH | 12.09, bs | 172.9 | - | - | - |
| 12, COOH | 12.09, bs | 173.3 | - | - | - |
Position based on numbering scheme in Fig. 4.
Fig. 3.
NOESY correlations of IsdG product peak 1. A. NOESY spectrum (in d6-DMSO) of the first eluting product focused on key regions that demonstrate correlations between the α- and γ-bridge CHs and neighboring protons. The second eluting product from the IsdG-catalyzed reaction and the IsdI-catalyzed products have similar NOESY spectra, and are shown in Fig. S5–S7. B. Two fragments of a bilirubin-like molecule with NOE correlations (arrows). Carbon 10 and 20 are labeled according to standard bilirubin nomenclature in which C10 corresponds to the γ-meso carbon and C20 corresponds to the α-meso carbon of the porphyrin ring.
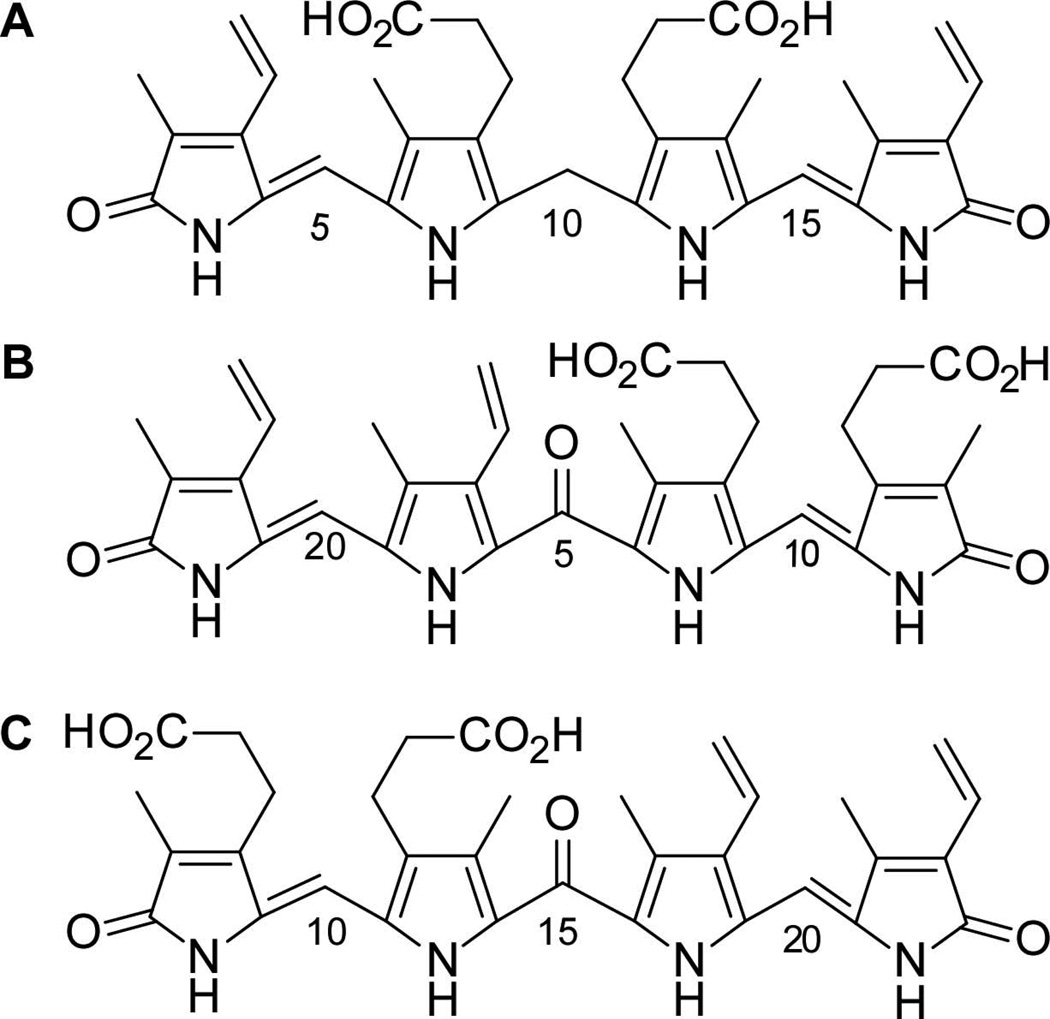
From the molecular formula obtained by HRESIMS and the NMR data described above, we have been able to propose structures of the products of IsdG- and IsdI-mediated heme degradation. In accordance with the 10-oxo-mesobilirubin nomenclature (Chen, 1999), we have named these unprecedented molecules 5-oxo-δ-bilirubin (elution peak 1) and 15-oxo-β-bilirubin (elution peak 2) (Fig. 4). Collectively, we refer to these molecules as staphylobilins. The β- and δ-meso carbon cleavage is in agreement with the alignment of molecular oxygen in two orientations along the β,δ-heme axis in the crystal structure of heme-bound IsdI (Fig. 1). Alternatively, the heme molecule may bind in either orientation about the α,γ-axis in the active site, resulting in either the β- or δ-isomer of oxo-bilirubin, as reported previously for the CoPPIX and inactive IsdG variant (Lee et al., 2008). Similarly, in human HO-1, heme is orientationally disordered about the α,γ-axis, although in that case α-cleavage occurs with either heme seating (Schuller et al., 1999).
Fig. 4.
Linear representations of heme degradation products. A. Bilirubin. B. 5-oxo-δ-bilirubin, the first eluting product. C. 15-oxo-β-bilirubin, the second product to elute upon HPLC purification. The bilirubin carbon numbering scheme was used for the IsdG and IsdI products for simplicity.
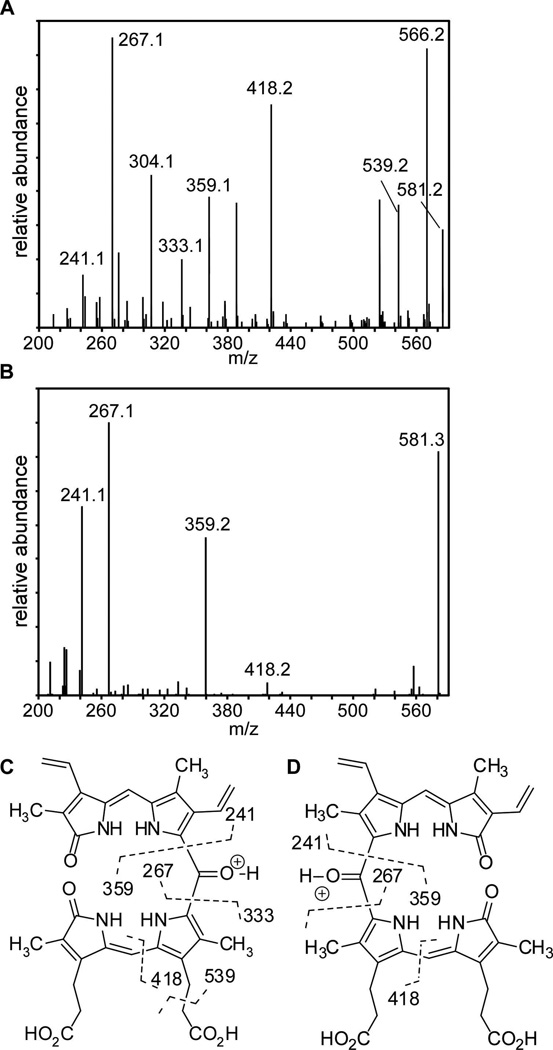
The position of the remaining oxygen atom in the proposed structures of β- and δ-oxo-bilirubins was confirmed by tandem ESI-MS analyses (Fig. 5). Fragmentation generated by collision-induced dissociation results in cleavage on either side of the proposed dipyrryl ketone and is diagnostic for the presence of the 5- and 15-oxo-bridges in these structures. Fragmentation at these positions results in the formation of ion pairs consisting of an oxonium ion (m/z 359, 267 [M+]) and pyrrolium ion (m/z 241, 333 [M’+H]). Pyrrolium ions M’ correspond to ions resulting from intramolecular proton transfer reactions, possibly facilitated by propionate groups which are proximal in the ridge-tile conformation (Chen, 1999). Beta-fragmentation of the β-propionic acid side chains is also evident (m/z 539.2), as has been noted in the mass spectral analysis of analogous mesobiliverdin (Cole et al., 1968). Lastly, we observe cleavage at the C-10 methine bridge (m/z 418.2), similar to that observed in mesobiliverdin (Cole et al., 1968). The 5- and 15-oxo-bridges in these structures were further confirmed by observation of a fifth carbonyl resonance in 5-oxo-bilirubin at 175.1 ppm via cryogenic 13C-NMR (Fig. S4C), consistent with the reported shift of the oxo-bridge in 10-oxo-bilirubin at 176 ppm (Chen, 1999).
Fig. 5.
Tandem LC- HRESIMS. A. ESI-MS/MS spectrum of fragment ions selecting for 599.3 m/z for 5-oxo-δ-bilirubin. B. ESI-MS/MS spectrum of fragment ions selecting for 599.3 m/z for 15-oxo-β-bilirubin. C,D. Fragmentation scheme corresponding to the masses and molecular formulas obtained in A and B respectively. ESI-MS/MS spectra for IsdI-catalyzed products are similar.
Discussion
In this study we have presented the crystal structure of native IsdI bound to heme and described HRESIMS and NMR studies on the products of IsdG- and IsdI-catalyzed heme degradation. Combined, these data demonstrate that IsdG and IsdI degrade heme to release a mixture of β- and δ- isomers of oxo-bilirubin. A few cases of heme oxygenases that degrade heme to non-alpha biliverdins or modified biliverdins have been demonstrated (Paiva-Silva et al., 2006; Ratliff et al., 2001; Zhang et al., 2004). However, to the best of our knowledge, the oxo-bilirubin molecules described here have not been previously observed or characterized.
It has been reported that IsdG-mediated heme degradation results in the formation of biliverdin based on similar HPLC elution profiles between these molecules (Skaar et al., 2004). However, the prior study utilized standard methods for isolating biliverdin, which includes a chloroform extraction. We have found that staphylobilin is not very soluble in chloroform, and therefore, the previously published chromatograms are not representative of purified staphylobilin. Moreover, the previous chromatographic method used a methanol:water mobile phase and monitored absorbance at 380 nm. In the current study we have used an acetonitrile:water mobile phase and 465 nm, which better distinguishes the two products. We believe it is for these reasons that the product of IsdG-mediated heme degradation was previously misassigned as biliverdin.
The HRESIMS data provide conclusive evidence that heme degradation catalyzed by IsdG-family enzymes does not result in the canonical heme degradation product biliverdin or the eukaryotic metabolite bilirubin (Fig. S1). The downfield regions of the 1H NMR spectra demonstrate that the IsdG-family products share similarities with 10-oxo-mesobilirubin (Fig. S2) (Chen, 1999). Moreover, the HMBC and NOESY spectra demonstrate an intact correlation network between the α-meso carbon proton and the vinyl protons at C18 and methyl protons at C2. The γ-meso proton also displays correlations with the propionate side chain protons of rings B and C (Fig. 3 and S3). These data provide strong evidence that the α- and γ-meso carbons of the porphyrin ring are intact, accounting for all but 28 Daltons of the isolated heme degradation products. ESI-MS/MS data and 13C NMR spectra demonstrate that the remaining mass comprises a bridging carbonyl (C=O) between the α/γ positions in the two isomers (Fig. S4C and S5C).
The mechanism of the HO-1 family of enzymes has been studied extensively, although surprisingly many details remain unclear (Fig. S8) (Unno et al., 2007; Wilks, 2002). After the enzyme binds ferric-heme, one electron is donated by the reducing substrate, converting heme-iron to the ferrous state. Molecular oxygen then binds the reduced pentacoordinate heme to form a meta-stable oxy complex. From there, a one electron reduction and protonation yields a ferric-hydroperoxo complex. This complex then self-hydroxylates at the α-meso carbon, although it is unknown how the protonation activates the hydroperoxo intermediate to promote meso-hydroxylation. Ferric α-meso-hydroxyheme is then converted to biliverdin through multiple oxidoreductive steps and a verdoheme intermediate (Matsui et al., 2005; Schuller et al., 1999; Unno et al., 2007). The conversion of verdoheme to biliverdin is incompletely understood, although it is known to be the rate-limiting step of the reaction in vivo (Unno et al., 2007). However, in vitro the rate-limiting step is biliverdin release. In mammals this step is accelerated in vivo by biliverdin reductase, which removes biliverdin from the active site and reduces it to bilirubin. All characterized bacterial heme oxygenases catalyze single-turnover reactions in vitro, suggesting that another protein is required to remove the heme degradation product from bacterial heme oxygenases in vivo (Puri and O'Brian, 2006; Wegele et al., 2004; Wilks and Schmitt, 1998; Zhu et al., 2000).
Both IsdG-like and HO-1-like enzymes produce biliverdin derivatives suggesting that both reactions may proceed similarly up to formation of a hydroperoxo intermediate. However, in the case of IsdG-like enzymes the complex would self-hydroxylate at the β- or δ-meso carbon, rather than the α-meso carbon. Importantly, HO-1-like enzymes contain an ordered set of water molecules in the binding pocket that funnel protons to the catalytic site and stabilize the hydroperoxy intermediate (Friedman et al., 2003; Lad et al., 2003). This water network is absolutely required for catalysis in all HO-1 enzymes (Matsui et al., 2005; Schuller et al., 1999; Schuller et al., 2001; Sugishima et al., 2000); however, a similar water network is not present in IsdG-family enzymes (Lee et al., 2008). Therefore, it is currently unclear how intermediates are activated and stabilized during IsdG-mediated catalysis.
It is possible that the meso-hydroxyheme intermediate may then go through a verdoheme intermediate that is hydroxylated and then degraded to staphylobilin, or through formation of a β,δ-dihydroxyheme intermediate that is subsequently degraded to staphylobilin (Fig. S8). Addition of biliverdin to purified IsdG in the presence of ascorbate does not lead to formation of staphylobilin (data not shown), indicating that α-biliverdin is not an intermediate in the pathway. However, more experiments are necessary to investigate the mechanism of staphylobilin formation.
In the active site of IsdI there are several amino acids that are completely conserved among members of the IsdG-family of enzymes, including Asn6. We hypothesize that the conserved asparagine has two roles in heme catalysis. The asparagine amide is observed to form an H-bond to the bound oxygen species and potentially stabilizes intermediate(s) in the pathway. Asn6 along with Phe22 may also sterically direct the sites of hydroxylation, as they are only 3.5 Å from the α- and γ-meso carbons, respectively (Fig.1). Though the precise nature of the oxygen species observed in the crystal structure remains to be characterized, the steric constraints imposed by Asn6 and Phe22 are likely to be structurally conserved leaving only the β- and δ-meso carbons accessible for hydroxylation. Phe22 is conserved in all IsdG members with the exception of HmuD and HmuQ from B. japonicum, which encode for tryptophan at that site. Phe22 is one of the residues implicated in directing heme ruffling such that the β- or δ-meso carbons are tilted towards the oxygen species (Lee et al., 2008). Considering that the pathway of heme degradation has not been experimentally determined, identification of the reaction products of IsdG-family enzymes raises many mechanistic questions to be investigated in future studies.
Interestingly, we noted that 67.5 ± 1.9% of the IsdG-catalyzed product elutes in the first peak as the δ-isomer, whereas only 56.2 ± 1.6% of the IsdI-catalyzed product elutes as the δ-isomer. This may indicate that the heme-binding pocket of IsdG is more constricted than that of IsdI, and therefore preferentially allows for one heme seating over another. The ramifications for this regiospecificity in vivo and the fate and function of staphylobilin inside the cell are currently under investigation.
Bilirubin, the terminal product of heme degradation in mammals, is one of the most potent antioxidants in the serum (Stocker et al., 1987). The other products of vertebrate heme catabolism, biliverdin and carbon monoxide, also have beneficial anti-oxidative and anti-inflammatory properties (Kirkby and Adin, 2006). In contrast, the role of bacterial heme degradation products in vivo has yet to be uncovered. Considering the important roles of biliverdin and bilirubin within vertebrate cells, it is tempting to speculate that staphylobilins may have important functions within bacterial pathogens. This is particularly intriguing considering that IsdG and IsdI are differentially regulated in response to environmental changes experienced by the bacterium (Reniere and Skaar, 2008). It is possible that the different isomers of staphylobilin are required under different environmental conditions. Moreover, the significant difference in protein structure and mechanism between human HO-1 and the IsdG-family of enzymes, combined with the known requirement for IsdG-mediated heme degradation during bacterial pathogenesis (Reniere and Skaar, 2008; Skaar et al., 2006), suggests that inhibiting this class of bacterial heme oxygenases is an attractive therapeutic strategy.
Experimental Procedures
Protein purification
Recombinant IsdG and IsdI were purified from Escherichia coli BL21(DE3) as previously described (Skaar et al., 2004). Purity was assessed by SDS-PAGE and concentrations were determined using a BCA protein assay.
Crystallography
Crystals of recombinant IsdI-heme were prepared by sitting drop vapour diffusion at 4° C. Protein (15 mg/ml in 20 mM Tris-HCl, 0.2 M NaCl) was mixed 1:1 with a reservoir of 25% polyethylene glycol 3350, 0.2 M MgCl2, and 0.1 M Bis-Tris buffer pH 5.5. Prior to data collection crystals were looped from a cryoprotectant of 10% ethylene glycol in reservoir solution on ice and immersed in liquid nitrogen. Reduced crystals were obtained by soaking the ferric crystals in 50 mM sodium dithionite in cryoprotectant solution for 10 minutes before immersion in liquid nitrogen. Diffraction data sets were collected under a cryostream at 100 K on beamline 08ID-1 at the Canadian Light Source (Saskatoon, SK). The programs in the CCP4 suite (Potterton et al., 2003) were used to solve and refine the structures. Data were processed and scaled with MOSFLM (Leslie, 1992) and SCALA (Evans, 2006), respectively. Initial phases were obtained with the program MolRep (Vagin and Teplyakov, 1997), using a search model of the protein moiety from the structure of IsdI in complex with cobalt protoporphyrin IX (CoPPIX, PDB ID: 2ZDP). Model building was carried out using COOT visualization software (Lohkamp, 2005) and the structure was refined with the program Refmac5 (Murshudov et al., 1997). Final structure validation was done with the program PROCHECK (Laskowski et al., 1993) and figures were prepared with the program PyMOL (DeLano, 2002).
Heme degradation and product purification
Heme degradation reactions were performed in tris-buffered saline (50 mM tris, pH 7.5, 150 mM NaCl) at room temperature protected from light. IsdG or IsdI (50 µM) were incubated with equimolar hemin (Sigma) and catalase (from bovine liver, Sigma) at a 0.5:1 molar ratio of catalase:hemoprotein. We also performed the reactions in the presence of superoxide dismutase (20 units/mL) to test the role of superoxides in the heme degradation mechanism. Products and product distributions in reactions performed in the presence of catalase and/or superoxide dismutase were unchanged. Ascorbic acid was added to a final concentration of 1 mM and spectral changes between 300–800 nm were monitored. Following completion of the reaction the mixtures were filtered through Centricon Plus spin filters (MWCO 5 kDa, Millipore) by centrifugation at 3,220 × g to remove the buffer, salts, and non-degraded heme. The heme degradation product(s) were eluted from the proteins by addition of water:acetonitrile (1:1), 0.1% trifluoroacetic acid (TFA) followed by centrifugation. The eluate was lyophilized and resuspended in a minimum volume of water:acetonitrile (1:1), 0.1% TFA, and further purified by reverse-phase chromatography on a Varian ProStar HPLC using a Microsorb-MV C-18 column. Biliverdin (MP Biomedicals) and bilirubin (Sigma) were dissolved in 0.1 M NaOH and diluted in water:acetonitrile (1:1), 0.1% TFA for HPLC analysis.
HPLC analysis was performed using 95% water / 5% acetonitrile with 0.1% TFA as the mobile phase with a flow rate of 1 mL/min. After a 10 min equilibration period, a linear 40 min acetonitrile gradient (5%–80%) was employed and the final concentration was maintained for an additional 20 min. The eluant was monitored using a photodiode array detector from 200–900 nm and two peaks (λmax: 465nm) were individually collected and lyophilized for further analysis. The ratio of isomers was calculated by integration of the peaks and averaged over ten injections (p < 0.0001).
LC-MS/MS Analysis
Each lyophilized product was dissolved in acetonitrile/water (50:50) with 0.25% formic acid at a concentration of 80 ng/µL prior to injection. The LC-MS/MS analysis was performed on a Waters Acquity UPLC system (Waters, Milford, MA) integrated with a Thermo Finnigan LTQ linear ion trap mass spectrometer (Thermo Fisher Scientific, Waltham, MA) using a Thermo Hypersil Gold C18 column (1.9 µm, 2.1 × 150 mm). Prior to MS analysis, LC separation was performed using water:acetonitrile (95:5) with 0.25% formic acid as the mobile phase. After a 10 µl sample injection, a 20 minute linear acetonitrile gradient (5%–100%) was performed at a flow rate of 300 µl/min. The following optimized parameters were used for the detection of analyte: N2 sheath gas 36 psi; N2 auxiliary gas 20 psi; capillary temperature 300° C; source voltage 3.8 kV; source current 100 MA; skimmer offset 0.00 V; capillary offset −44.00 V; tube lens offset −103.30 V; activation time 30 ms (MS), 50 ms (MS2); isolation width 1 m/z (MS), 2 m/z (MS2). Data acquisition and quantitative spectral analyses were conducted using the Thermo-Finnigan Xcaliber software, version 2.0 Sur1.
NMR
See Supplementary Material for NMR methods.
Acknowledgements
We thank Melissa D. Carter from the Wright Laboratory for technical assistance with mass spectral data collection. This research was supported by the Searle Scholars Program, United States Public Health Service Grants AI69233 and AI073843 from the National Institute of Allergy and Infectious Diseases, and a Canadian Institute of Health Research Grant MOP-49597. Portions of this research were carried out at the Canadian Light Source, which is supported by NSERC, NRC, CIHR, and the University of Saskatchewan. Eric P. Skaar, Ph.D. holds an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Welcome Fund. Michelle L. Reniere was funded by NIH Training Grant in Mechanisms of Vascular Disease, 5 T32 HL07751. Drs. Bachmann, Skaar, Stec and Wright are members of the Vanderbilt Institute for Chemical Biology. Coordinates have been deposited in the Protein Data Bank under the accession numbers 3LGN and 3LGM for the IsdI-heme and IsdI-heme (reduced) structures, respectively.
References
- Bullen JJ, Griffiths E. Iron and Infection: Molecular, Physiological and Clinical Aspects. New York: John Wiley and Sons; 1999. [Google Scholar]
- Chen Q, Huggins MT, Lightner DA, Norona W, McDonagh AF. Synthesis of a 10-Oxo-Bilirubin: Effects of the Oxo Group on Conformation, Transhepatic Transport, and Glucuronidation. Journal of the American Chemical Society. 1999;121:9253–9264. [Google Scholar]
- Chim N, Iniguez A, Nguyen TQ, Goulding CW. Unusual Diheme Conformation of the Heme-Degrading Protein from Mycobacterium tuberculosis. J Mol Biol. 2009 doi: 10.1016/j.jmb.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SW, Hall SR, Perrella MA. Role of haem oxygenase-1 in microbial host defence. Cell Microbiol. 2009;11:199–207. doi: 10.1111/j.1462-5822.2008.01261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole WJ, Chapman DJ, Siegelman HW. The structure and properties of phycocyanobilin and related bilatrienes. Biochemistry. 1968;7:2929–2935. doi: 10.1021/bi00848a033. [DOI] [PubMed] [Google Scholar]
- DeLano WL, editor. The PyMOL Molecular Graphics System. 2002. [Google Scholar]
- Evans P. Scaling and assessment of data quality. Acta Crystallographica Section D. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- Friedman J, Lad L, Deshmukh R, Li H, Wilks A, Poulos TL. Crystal structures of the NO- and CO-bound heme oxygenase from Neisseriae meningitidis Implications for O2 activation. J Biol Chem. 2003;278:34654–34659. doi: 10.1074/jbc.M302985200. [DOI] [PubMed] [Google Scholar]
- Friedman J, Lad L, Li H, Wilks A, Poulos TL. Structural basis for novel delta-regioselective heme oxygenation in the opportunistic pathogen Pseudomonas aeruginosa. Biochemistry. 2004;43:5239–5245. doi: 10.1021/bi049687g. [DOI] [PubMed] [Google Scholar]
- Kaplan D, Navon G. Nuclear magnetic resonance studies of the conformation of bilirubin and its derivatives in solution. J.C.S. Perkin II. 1981;10:1374–1383. [Google Scholar]
- Kirkby KA, Adin CA. Products of heme oxygenase and their potential therapeutic applications. Am J Physiol Renal Physiol. 2006;290:F563–F571. doi: 10.1152/ajprenal.00220.2005. [DOI] [PubMed] [Google Scholar]
- Lad L, Wang J, Li H, Friedman J, Bhaskar B, Ortiz de Montellano PR, Poulos TL. Crystal structures of the ferric, ferrous, and ferrous-NO forms of the Asp140Ala mutant of human heme oxygenase-1: catalytic implications. J Mol Biol. 2003;330:527–538. doi: 10.1016/s0022-2836(03)00578-3. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- Lee WC, Reniere ML, Skaar EP, Murphy ME. Ruffling of metalloporphyrins bound to IsdG and IsdI, two heme-degrading enzymes in Staphylococcus aureus. J Biol Chem. 2008;283:30957–30963. doi: 10.1074/jbc.M709486200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. 1992;26 [Google Scholar]
- Lohkamp B, Emsley P, Cowtan K. Coot News. CCP4 Newsletter. 2005;42 [Google Scholar]
- Matsui T, Furukawa M, Unno M, Tomita T, Ikeda-Saito M. Roles of distal Asp in heme oxygenase from Corynebacterium diphtheriae HmuO: A water-driven oxygen activation mechanism. J Biol Chem. 2005;280:2981–2989. doi: 10.1074/jbc.M410263200. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Skaar EP, Gaspar AH, Humayun M, Gornicki P, Jelenska J, Joachmiak A, Missiakas DM, Schneewind O. Passage of heme-iron across the envelope of Staphylococcus aureus. Science. 2003;299:906–909. doi: 10.1126/science.1081147. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallographica Section D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Paiva-Silva GO, Cruz-Oliveira C, Nakayasu ES, Maya-Monteiro CM, Dunkov BC, Masuda H, Almeida IC, Oliveira PL. A heme-degradation pathway in a blood-sucking insect. Proc Natl Acad Sci U S A. 2006;103:8030–8035. doi: 10.1073/pnas.0602224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- Puri S, O'Brian MR. The hmuQ and hmuD genes from Bradyrhizobium japonicum encode heme-degrading enzymes. J Bacteriol. 2006;188:6476–6482. doi: 10.1128/JB.00737-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratliff M, Zhu W, Deshmukh R, Wilks A, Stojiljkovic I. Homologues of neisserial heme oxygenase in gram-negative bacteria: degradation of heme by the product of the pigA gene of Pseudomonas aeruginosa. J. Bacteriol. 2001;183:6394–6403. doi: 10.1128/JB.183.21.6394-6403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reniere ML, Torres VJ, Skaar EP. Intracellular metalloporphyrin metabolism in Staphylococcus aureus. Biometals. 2007;20:333–345. doi: 10.1007/s10534-006-9032-0. [DOI] [PubMed] [Google Scholar]
- Reniere ML, Skaar EP. Staphylococcus aureus haem oxygenases are differentially regulated by iron and haem. Mol Microbiol. 2008;69:1304–1315. doi: 10.1111/j.1365-2958.2008.06363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt MP. Utilization of host iron sources by Corynebacterium diphtheriae: identification of a gene whose product is homologous to eukaryotic heme oxygenases and is required for acquisition of iron from heme and hemoglobin. J. Bacteriol. 1997;179:838–845. doi: 10.1128/jb.179.3.838-845.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller DJ, Wilks A, Ortiz de Montellano PR, Poulos TL. Crystal structure of human heme oxygenase-1. Nat Struct Biol. 1999;6:860–867. doi: 10.1038/12319. [DOI] [PubMed] [Google Scholar]
- Schuller DJ, Zhu W, Stojiljkovic I, Wilks A, Poulos TL. Crystal structure of heme oxygenase from the gram-negative pathogen Neisseria meningitidis and a comparison with mammalian heme oxygenase-1. Biochemistry. 2001;40:11552–11558. doi: 10.1021/bi0110239. [DOI] [PubMed] [Google Scholar]
- Skaar EP, Gaspar AH, Schneewind O. IsdG and IsdI, heme-degrading enzymes in the cytoplasm of Staphylococcus aureus. J Biol Chem. 2004;279:436–443. doi: 10.1074/jbc.M307952200. [DOI] [PubMed] [Google Scholar]
- Skaar EP, Schneewind O. Iron-regulated surface determinants (Isd) of Staphylococcus aureus: stealing iron from heme. Microbes Infect. 2004;6:390–397. doi: 10.1016/j.micinf.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Skaar EP, Gaspar AH, Schneewind O. Bacillus anthracis IsdG, a heme-degrading monooxygenase. J Bacteriol. 2006;188:1071–1080. doi: 10.1128/JB.188.3.1071-1080.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker R, Yamamoto Y, McDonagh AF, Glazer AN, Ames BN. Bilirubin is an antioxidant of possible physiological importance. Science. 1987;235:1043–1046. doi: 10.1126/science.3029864. [DOI] [PubMed] [Google Scholar]
- Sugishima M, Omata Y, Kakuta Y, Sakamoto H, Noguchi M, Fukuyama K. Crystal structure of rat heme oxygenase-1 in complex with heme. FEBS Lett. 2000;471:61–66. doi: 10.1016/s0014-5793(00)01353-3. [DOI] [PubMed] [Google Scholar]
- Tenhunen R, Marver HS, Schmid R. Microsomal heme oxygenase. Characterization of the enzyme. J Biol Chem. 1969;244:6388–6394. [PubMed] [Google Scholar]
- Unno M, Matsui T, Chu GC, Couture M, Yoshida T, Rousseau DL, Olson JS, Ikeda-Saito M. Crystal structure of the dioxygen-bound heme oxygenase from Corynebacterium diphtheriae: Implications for heme oxygenase function. J Biol Chem. 2004 doi: 10.1074/jbc.M400491200. [DOI] [PubMed] [Google Scholar]
- Unno M, Matsui T, Ikeda-Saito M. Structure and catalytic mechanism of heme oxygenase. Nat Prod Rep. 2007;24:553–570. doi: 10.1039/b604180a. [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. Journal of Applied Crystallography. 1997;30:1022–1025. [Google Scholar]
- Wegele R, Tasler R, Zeng Y, Rivera M, Frankenberg-Dinkel N. The heme oxygenase(s)-phytochrome system of Pseudomonas aeruginosa. J Biol Chem. 2004;279:45791–45802. doi: 10.1074/jbc.M408303200. [DOI] [PubMed] [Google Scholar]
- Wilks A, Schmitt MP. Expression and characterization of a heme oxygenase (Hmu O) from Corynebacterium diphtheriae Iron acquisition requires oxidative cleavage of the heme macrocycle. J. Biol. Chem. 1998;273:837–841. doi: 10.1074/jbc.273.2.837. [DOI] [PubMed] [Google Scholar]
- Wilks A. Heme oxygenase: evolution, structure, and mechanism. Antioxid Redox Signal. 2002;4:603–614. doi: 10.1089/15230860260220102. [DOI] [PubMed] [Google Scholar]
- Wu R, Skaar EP, Zhang R, Joachimiak G, Gornicki P, Schneewind O, Joachimiak A. Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases. J Biol Chem. 2005;280:2840–2846. doi: 10.1074/jbc.M409526200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Sato M, Sasahara M, Migita CT, Yoshida T. Unique features of recombinant heme oxygenase of Drosophila melanogaster compared with those of other heme oxygenases studied. Eur J Biochem. 2004;271:1713–1724. doi: 10.1111/j.1432-1033.2004.04077.x. [DOI] [PubMed] [Google Scholar]
- Zhu W, Wilks A, Stojiljkovic I. Degradation of heme in gram-negative bacteria: the product of the hemO gene of Neisseriae is a heme oxygenase. J. Bacteriol. 2000;182:6783–6790. doi: 10.1128/jb.182.23.6783-6790.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]