Abstract
It has been postulated that HIV-1 envelope properties, such as shorter and less glycosylated V1-V2 loops commonly observed among non-subtype B early – transmitted viruses, promote utilization of the gut homing integrin α4β7. This property potentially confers an advantage to some HIV-1 variants early after acquisition. We found that replication competent recombinant viruses incorporating HIV-1 subtype A compact and less glycosylated early versus chronic phase V1-V2 loops demonstrated no significant difference in binding to α4β7 high CD8+ T cells or replication in α4β7 high CD4+ T cells. Integrin α4β7 usage does not select for shorter less glycosylated envelopes during transmission.
Keywords: gut homing integrin α4β7, mucosal transmission, HIV-1 envelope, selection at transmission, gut associated lymphoid tissue
INTRODUCION
High level replication in gut associated lymphoid tissue (GALT) likely plays an important role in establishing a systemic infection early after HIV-1 acquisition.1, 2 Infection of CD4+ T cells expressing the gut homing integrin α4β7 potentially facilitates HIV-1 migration from mucosal sites to GALT.3, 4 Enhanced integrin α4β7 reactivity has been linked to specific envelope characteristics, such as smaller V1-V2 loops and transmission associated predicted N-linked glycosylation sites (PNGS).5 These envelope genotypes are commonly observed in subtype A, C, and D but not subtype B early – transmitted viruses.6-12 Enrichment of viruses with these envelope signatures suggests that specific viruses are preferentially favored for acquisition, and α4β7 integrin utilization potentially confers fitness for transmission.13
Studies suggesting that compact and less glycosylated envelope V1-V2 loops enhance α4β7 utilization have been primarily conducted with HIV-1 envelope surface unit monomer, gp120, and not native envelope trimers on virus particles.5 One recent study showed that replication of a small number (n = 6) of subtype C transmitted / founder (T/F) and unrelated chronic infection (n = 4) strains were not inhibited by blocking the α4β7 integrin suggesting that the infecting viruses do not use the α4β7 integrin more efficiently.14 Because the T/F and chronic isolates were obtained from different subjects, they did not examine α4β7 utilization differences among closely related viruses with and without the transmission associated genotypes, such as compact and less glycosylated V1-V2 loops. In this study, we directly assessed the influence of transmission associated envelope V1-V2 loop signatures on α4β7 utilization.
METHODS
Subjects and viruses
Demographics of the heterosexually infected subjects with subtype A HIV-1 and the envelope sequences examined in this study have been detailed previously.6, 15 We evaluated the most commonly amplified V1-V2 loop from both time-points and another atypical chronic sequence in two subjects (QA203 and QB424). The V1-V2 loops were placed in a Q23-17 subtype A HIV-1 envelope backbone as previously described.6, 15 The chimeric envelopes were incorporated into a plasmid containing Q23-17 HIV sequences from the primer binding site (PBS) to the 3’ long terminal repeat (LTR), pCMV-Q23-17-PBS?LTR, using yeast gap-repair homologous recombination as previously described.16-19 Replication competent viruses were generated by co-transfecting HEK293T cells with a plasmid containing the subject V1-V2 envelope within pCMV-Q23-17-PBS→LTR and another plasmid with Q23-17 sequences from 5’ LTR to early portion of gag, pCMV-Q23-17-LTR→Gag4.16 The 293T transfection supernatants were passaged on activated CD4+ T cells for a maximum of 7 days to generate high titer peripheral blood mononuclear cell (PBMC) derived viruses. Virus stocks were titered on TZM-bl cells as previously described.6, 20
Binding and replication assessment
Primary CD4+ and CD8+ T cells were isolated from HIV-1 negative blood donor’s buffy coats using antibody conjugated magnetic beads (Miltenyi Biotech) according to manufacturer’s instructions. Both CD8+ and CD4+ T cells were cultured with 2% phytohaemagglutinin (PHA), 20 ug/ml recombinant human IL-2 (r-IL-2) with or without 10 nM retinoic acid (RA) for 6 days. Approximately 1 × 105 infectious particles (IP) were incubated independently with 1 × 106 CD8+ T cells and 1 × 106 CD4+ T cells at 4˚ C for 1 hour in binding buffer (10mM HEPES, 150mM NaCl (HBS Buffer) buffer with100μM CaCl2 and 1mM MnCl2). In some cases, cells were pre-incubated with the specified antibodies at 37˚C for 30 minutes prior to virus exposure. The CD4+ and CD8+ T cells were washed with RPMI 3 times to remove unbound virus. RNA was isolated from the CD8+ T cell incubations using the QIAAMP Viral RNA kit (QIAGEN). HIV-1 copies were quantified using quantitative RT-PCR using previously described methods.21, 22 The CD4+ T cell cultures were incubated at 37˚C 5% C02. Supernatants were collected 3 days post infection and not at later times to probe early replication differences, which were more likely to be affected by α4β7 usage. Supernatant IP concentrations were estimated by infecting 1 × 104 TZM-bl cells with 8 serial two-fold dilutions, and cells were examined for beta-galactosidase production using Galacto-Light Plus System (Applied Biosystems) after 2 days. A linear interpolated curve of the relative light units (RLUs) versus supernatant dilution was used to estimate RLU/ul.
Statistical Analysis
Values in the presence of RA and no antibody were compared to other cell culture conditions using the Wilcoxon rank sum test matched per donor cells. Early and chronic infection median values were compared among all subjects using the matched pairs Wilcoxon rank sum test. In 2 subjects (QA203 and QA424) where 2 chronic infection V1-V2 loops were examined a median of the 2 chronic infection values was used in the comparisons. All analysis was done with GraphPad Prism 5. All p-values were based on a two-sided test.
RESULTS
The α4β7 cell surface mean fluorescence intensity (MFI) increased in the presence as compared to the absence of RA in cells from 13 of the 17 blood donor’s buffy coats (Fig. 1A – B). We only used donor cells with FACS confirmed RA induced increase in α4β7 expression for further experiments. Although cells expressed α4β7 under both conditions, α4β7 MFI was significantly higher in the presence as compared to the absence of RA in CD4+ (mean fold change 2.5, range 1.3 – 4.5) and CD8+ T cells (mean fold change 2.1, range 1.5 – 3.2) (p = 0.0002 for both comparisons). We examined α4β7 binding in CD8+ as opposed to in CD4+ T cells with pre-blocked CD4 receptor because both cell types expressed relatively similar levels of α4β7. In addition, this strategy eliminated the possibility of low level virus binding to the unblocked CD4 receptor.
Figure 1.
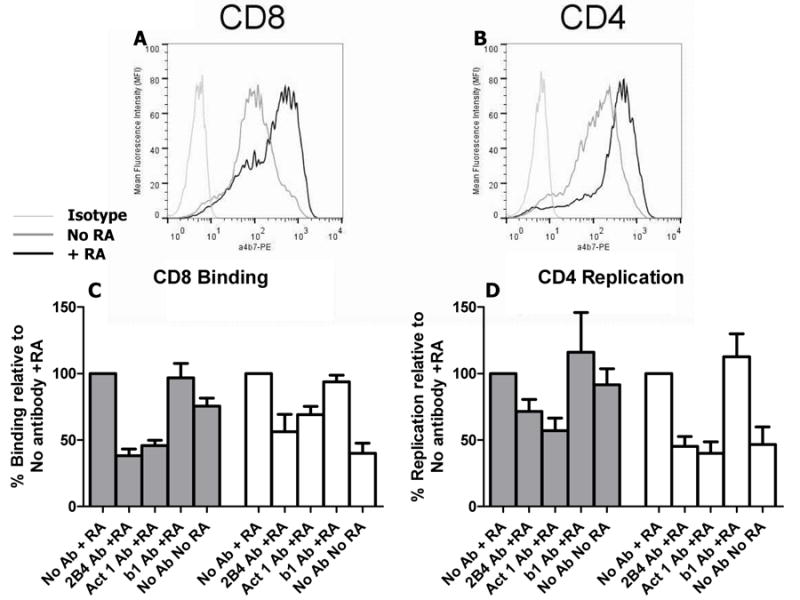
Virus binding and replication in cells expressing the α4β7 integrin. A and B, Flow cytometric analysis of α4β7 cell surface expression on CD8+ T cells (A) and CD4+ T cells (B) cultured with and without retinoic acid (RA). The α4β7 integrin was probed with phyocoerythrin (PE) conjugated anti-mouse integrin β7 antibody (clone FIB27) (BioLegend). An unrelated IgG1 antibody served as the isotype control. These are representative examples from multiple independent cell isolations. C and D, Bal envelope HIV-1 (gray bars) and SF162 (white bars) attachment to CD8+ T cells (C) and replication in CD4+ T cells (D) in the presence or absence of retinoic acid and antibodies specific for α4β7 (Act 1 (10ug) and 2B4 (2 ug) (R&D Systems)), and β1 (10 ug) (P4G11(Millipore)). Bars show mean values with standard error generated from 6 independent experiments with cells from 6 different donors. In each experiment, percent binding and replication is calculated relative to the cells cultured in the presence of RA and without any antibody (set at 100%).
We first examined Bal envelope virus and HIV-1SF162, which have been shown to have α4β7 integrin reactivity.3, 4 Binding and replication in cells incubated with different antibodies specific for α4β7 (2B4 and Act 1) and a control antibody specific for β1 (P4G11) or in the absence of RA was compared to CD8+ and CD4+ T cells grown in the presence of RA and no antibody (set as 100%). Bal and SF162 RNA copies recovered from virus exposed CD8+ T cells was significantly lower in the presence of 2B4 (p = 0.03), Act 1 (p = 0.03), and in the absence of RA (p = 0.03) but was not significantly different in the presence of P4G11 (p > 0.05) (Fig. 1C). SF162 replication in CD4+ T cells was also significantly lower in the presence of Act 1 (p = 0.008), 2B4 (p = 0.008), and in the absence of RA (p = 0.02) but was not significantly different with P4G11 (p = 0.6) (Fig. 1D). Bal replication was significantly lower when CD4+ T cells were pre-exposed to Act 1 (p = 0.03), but not significantly different in the presence of 2B4 (p = 0.06), absence of RA (p = 0.4), or with P4G11 (p = 1.0) (Fig. 1D). Thus, virus binds to CD8+ T cells potentially because of baseline α4β7 expression, and this attachment increases with RA induced higher gut homing receptor levels. Similar to published reports, replication was not reduced among all virus isolates in the presence of α4β7 blocking antibodies or lower gut homing receptor levels.14, 23
A previous study showed that a HIV-1 subtype A envelope gp120 monomer incorporating V1-V2 loops from early in infection (QA203M1 also previously referred to as QA203D(M1)) showed around 20 fold greater binding to the α4β7 integrin compared to a gp120 with V1-V2 segments from the chronic phase of disease (QA203M41 also previously referred to as QA203B(M41)).5, 6 We compared binding and replication between the QA203D(M1) and QA203B(M41) V1-V2 loop envelope viruses as opposed to gp120 monomers. Because the producer cells determine the types of glycosylation present on a virus envelope, we derived all viruses with subject specific envelope V1-V2 loops from physiologically relevant PBMC cultures.5 QA203D(M1) and QA203B(M41) V1-V2 loops encode 61 and 73 amino acids with 5 and 8 PNGS respectively (Fig. 2A). Similar to the gp120 data, we found that QA203B(M41) virus attachment was lower compared to QA203D(M1) virus (set at 100%) (p = 0.06, n = 5 replicates) (Fig. 2B).5 The QA203B(M41) virus demonstrated significantly lower replication in CD4+ T cells expressing high levels of α4β7 compared to QA203D(M1) (set as 100%) (p = 0.008, n = 8 replicates) (Fig. 2C).
Figure 2.
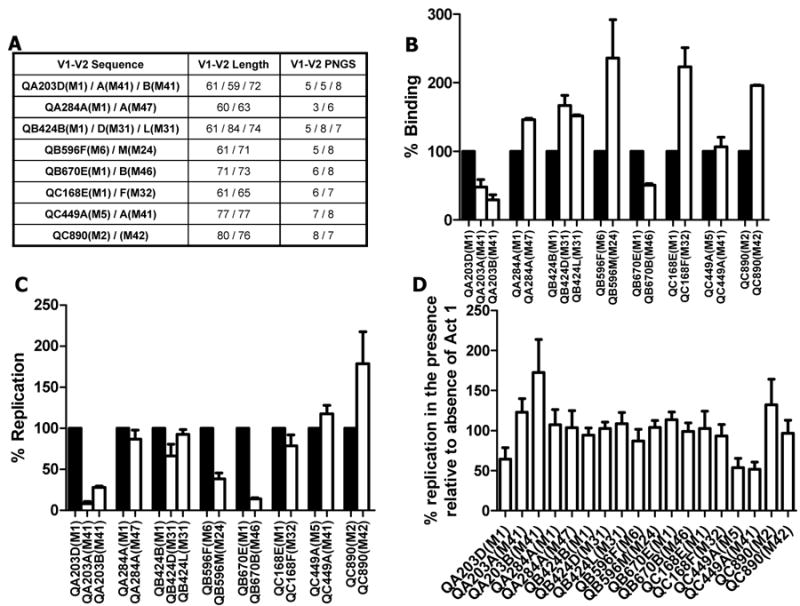
Influence of longitudinal V1-V2 loop changes on α4β7 utilization. A, Table lists the V1-V2 loop ID, length and number of predicted N-linked glycosylation sites (PNGS). In each V1-V2 loop name, the first part lists the subject ID. The duration (in months) from estimated acquisition date to the time of sample collection from which the V1-V2 loop was obtained is indicated in the brackets of each V1-V2 loop name. The envelope sequences have been described in detail previously.6, 15 B and C, Attachment to CD8+ T cells (B) and replication in CD4+ T cells (C) among early (black bars) and chronic (white bars) infection V1-V2 envelope loop chimeras. In each experiment, percent binding and replication is calculated relative to the envelope chimera with the early infection V1-V2 loop (set at 100%). D, Percent replication in the presence relative to the absence of 10 ug of Act 1. Bars show mean values with standard error generated from a minimum of 3 and 6 independent binding and replication experiments respectively. In each independent experiment, cells were obtained from different donors.
To assess the generalizability of this finding, we examined the influence of other longitudinally isolated V1-V2 loops on α4β7 utilization (Fig. 2A). All envelopes possessed the tripeptide V2 motif associated with α4β7 binding.3 The chronic as compared to the early infection V1-V2 loops contained significantly more PNGS (p = 0.03) and were longer (p = 0.09) although length differences did not reach statistical significance. In only 2 of the 8 subjects (QA203 and QB670), higher amounts of RNA was recovered from the viruses with early relative to the chronic infection V1-V2 (Fig. 2B). In aggregate, early as compared to chronic infection viruses showed no significant differences in binding to α4β7 high CD8+ T cells (p = 0.2). In all subjects, except QC449 and QC890, viruses with early relative to chronic infection V1-V2 replicated more efficiently in α4β7 high CD4+ T cells (Fig. 2C). These chronic versus early differences were significant (p < 0.05) for some isolates (QA203A(M41), QA203B(M41), QB424D(M31), QB596M(M24), and QB670B(M46)). In aggregate, however, there was no significant replication difference among the viruses with longitudinally collected V1-V2 (p = 0.3). Because majority of early compared to chronic infection viruses demonstrated increased replication in α4β7 high CD4+ T cells, we examined if they were more sensitive to α4β7 blocker, Act 1. Early versus chronic virus V1V2 replication was not inhibited to a greater extent in the presence of Act 1 (p = 0.15). Thus, Act 1 did not inhibit replication among most isolates, and a number of V1V2 chimeric viruses actually demonstrated enhanced replication, similar to a previous published report.14 Infectious virus in α4β7 high CD4+ T cell culture day 3 supernatant did not have significant correlation with number of RNA recovered from α4β7 high CD8+ T cells (ρ = 0.04, p = 0.8, Spearman rank correlation).
DISCUSSION
In this study, we examined the influence of longitudinally collected envelope V1-V2 loops on binding to α4β7 high CD8+ T cells and replication in α4β7 high CD4+ T cells. The majority of early infection V1-V2 loops were smaller with a significantly lower number of PNGS compared to the chronic phase variants. The early as compared to chronic V1-V2 loop viruses did not consistently demonstrate higher binding or replication among the cells with high levels of the gut homing receptor. In aggregate, our results suggest that attachment to the integrin or replication in α4β7 expressing CD4+ T cells does not influence the observed enrichment of subtype A viruses with compact and less glycosylated V1-V2 loop envelopes early after HIV-1 acquisition.
Reactivity to the α4β7 integrin has been primarily examined among different gp120s and not infectious viruses. Functional trimers on replication competent virus are likely structurally different than envelope surface unit monomers. Interestingly, both a chimeric gp120 and recombinant functional envelope on infectious virus with an early as compared to a chronic QA203 V1-V2 demonstrated greater binding to α4β7 expressing cells.5 This suggests that α4β7 binding may not be immensely different between gp120s and envelope trimers although this needs to be confirmed in more isolates. One recent study has examined α4β7 utilization among subtype C T/F and unrelated chronic phase viruses.14 In contrast to this study, we assessed intra-subject subtype A envelope V1-V2 loop influence on α4β7 usage. Combined results from both studies suggest that gut homing receptor utilization does not favor the selection of variants with specific envelope genotypes, such as compact and less glycosylated V1-V2 loops commonly observed during non-subtype B HIV-1 transmission.6-12
The biological mechanism for the observed selection of specific viruses remains unclear. Because infection efficiency in α4β7 high CD4+ T cells is likely not a transmission phenotype, prevention strategies aimed at blocking HIV-1 envelope - α4β7 interaction may not stop acquisition. Furthermore, recently described V2 directed antibodies shown as a correlate of protection in an HIV-1 vaccine trial likely do not prevent infection by blocking α4β7 binding.24-26
Acknowledgments
We thank all the subjects who have contributed samples for these studies. We thank the NIH AIDS Research and Reference Reagent Program for TZM-bl cells and Act 1 antibody.
Source of Funding: This study was supported by NIH grant AI1077473 (MS).
Footnotes
Conflict of Interest: No author has a commercial or other association that might pose a conflict of interest.
References
- 1.Mehandru S, Poles MA, Tenner-Racz K, et al. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J Exp Med. 2004;200:761–770. doi: 10.1084/jem.20041196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haase AT. Perils at mucosal front lines for HIV and SIV and their hosts. Nat Rev Immunol. 2005;5:783–792. doi: 10.1038/nri1706. [DOI] [PubMed] [Google Scholar]
- 3.Arthos J, Cicala C, Martinelli E, et al. HIV-1 envelope protein binds to and signals through integrin alpha4beta7, the gut mucosal homing receptor for peripheral T cells. Nat Immunol. 2008;9:301–309. doi: 10.1038/ni1566. [DOI] [PubMed] [Google Scholar]
- 4.Cicala C, Martinelli E, McNally JP, et al. The integrin alpha4beta7 forms a complex with cell-surface CD4 and defines a T-cell subset that is highly susceptible to infection by HIV-1. Proc Natl Acad Sci U S A. 2009;106:20877–20882. doi: 10.1073/pnas.0911796106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nawaz F, Cicala C, Van Ryk D, et al. The genotype of early-transmitting HIV gp120s promotes alpha (4) beta(7)-reactivity, revealing alpha (4) beta(7) +/CD4+ T cells as key targets in mucosal transmission. PLoS Pathog. 2011;7:e1001301. doi: 10.1371/journal.ppat.1001301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sagar M, Wu X, Lee S, et al. HIV-1 V1-V2 envelope loop sequences expand and add glycosylation sites over the course of infection and these modifications affect antibody neutralization sensitivity. J Virol. 2006;80:9586–9598. doi: 10.1128/JVI.00141-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derdeyn CA, Decker JM, Bibollet-Ruche F, et al. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science. 2004;303:2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- 8.Chohan B, Lang D, Sagar M, et al. Selection for human immunodeficiency virus type 1 envelope glycosylation variants with shorter V1-V2 loop sequences occurs during transmission of certain genetic subtypes and may impact viral RNA levels. J Virol. 2005;79:6528–6531. doi: 10.1128/JVI.79.10.6528-6531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sagar M, Laeyendecker O, Lee S, et al. Selection of HIV variants with signature genotypic characteristics during heterosexual transmission. J Infect Dis. 2009;199:580–589. doi: 10.1086/596557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frost SD, Liu Y, Pond SL, et al. Characterization of human immunodeficiency virus type 1 (HIV-1) envelope variation and neutralizing antibody responses during transmission of HIV-1 subtype B. J Virol. 2005;79:6523–6527. doi: 10.1128/JVI.79.10.6523-6527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gnanakaran S, Bhattacharya T, Daniels M, et al. Recurrent signature patterns in HIV-1 B clade envelope glycoproteins associated with either early or chronic infections. PLoS Pathog. 2011;7:e1002209. doi: 10.1371/journal.ppat.1002209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li M, Salazar-Gonzalez JF, Derdeyn CA, et al. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J Virol. 2006;80:11776–11790. doi: 10.1128/JVI.01730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sagar M. HIV-1 transmission biology: selection and characteristics of infecting viruses. J Infect Dis. 2010;202(Suppl 2):S289–296. doi: 10.1086/655656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parrish N, Wilen C, Banks L, et al. Transmitted/founder and chronic subtype C HIV-1 use CD4 and CCR5 receptors with equal efficiency and are not inhibited by blocking the integrin a4b7. PLoS Pathog. 2012:8. doi: 10.1371/journal.ppat.1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sagar M, Kirkegaard E, Lavreys L, et al. Diversity in HIV-1 envelope V1-V3 sequences early in infection reflects sequence diversity throughout the HIV-1 genome but does not predict the extent of sequence diversity during chronic infection. AIDS Res Hum Retroviruses. 2006;22:430–437. doi: 10.1089/aid.2006.22.430. [DOI] [PubMed] [Google Scholar]
- 16.Chatziandreou N, Arauz Belen A, Freitas I, et al. Sensitivity changes over the course of infection increases the likelihood of resistance against fusion but not CCR5 receptor blockers. AIDS Research and Human Retroviruses. 2012 doi: 10.1089/aid.2011.0319. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Etemad B, Fellows A, Kwambana B, et al. Human immunodeficiency virus type 1 V1-to-V5 envelope variants from the chronic phase of infection use CCR5 and fuse more efficiently than those from early after infection. J Virol. 2009;83:9694–9708. doi: 10.1128/JVI.00925-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dudley DM, Gao Y, Nelson KN, et al. A novel yeast-based recombination method to clone and propagate diverse HIV-1 isolates. Biotechniques. 2009;46:458–467. doi: 10.2144/000113119. [DOI] [PubMed] [Google Scholar]
- 19.Marozsan AJ, Arts EJ. Development of a yeast-based recombination cloning/system for the analysis of gene products from diverse human immunodeficiency virus type 1 isolates. J Virol Methods. 2003;111:111–120. doi: 10.1016/s0166-0934(03)00166-6. [DOI] [PubMed] [Google Scholar]
- 20.Wei X, Decker JM, Liu H, et al. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother. 2002;46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rousseau CM, Nduati RW, Richardson BA, et al. Association of levels of HIV-1-infected breast milk cells and risk of mother-to-child transmission. J Infect Dis. 2004;190:1880–1888. doi: 10.1086/425076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sagar M, Lavreys L, Baeten JM, et al. Infection with multiple human immunodeficiency virus type 1 variants is associated with faster disease progression. J Virol. 2003;77:12921–12926. doi: 10.1128/JVI.77.23.12921-12926.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pauls E, Ballana E, Moncunill G, et al. Evaluation of the anti-HIV activity of natalizumab, an antibody against integrin alpha4. AIDS. 2009;23:266–268. doi: 10.1097/qad.0b013e328320a7f8. [DOI] [PubMed] [Google Scholar]
- 24.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 25.Haynes BF, Gilbert PB, McElrath MJ, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med. 2012;366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rolland M, Edlefsen PT, Larsen BB, et al. Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature. 2012 doi: 10.1038/nature11519. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]


