Abstract
Dyslexia is a common pediatric disorder that affects 5-17% of schoolchildren in the United States. It is marked by unexpected difficulties in fluent reading despite adequate intelligence, opportunity, and instruction. Classically, neuropsychologists have studied dyslexia using a variety of neurocognitive batteries to gain insight into the specific deficits and impairments in affected children. Since dyslexia is a complex genetic trait with high heritability, analyses conditioned on performance on these neurocognitive batteries have been used to try to identify associated genes. This has led to some successes in identifying contributing genes, although much of the heritability remains unexplained. Additionally, the lack of relevant human brain tissue for analysis and the challenges of modeling a uniquely human trait in animals are barriers to advancing our knowledge of the underlying pathophysiology. In vivo imaging technologies, however, present new opportunities to examine dyslexia and reading skills in a clearly relevant context in human subjects. Recent investigations have started to integrate these imaging data with genetic data in attempts to gain a more complete and complex understanding of reading processes. In addition to bridging the gap from genetic risk variant to a discernible neuroimaging phenotype and ultimately to the clinical impairments in reading performance, the use of neuroimaging phenotypes will reveal novel risk genes and variants. In this article, we briefly discuss the genetic and imaging investigations and take an in-depth look at the recent imaging-genetics investigations of dyslexia.
Keywords: Dyslexia, neuroimaging, genetics, imaging-genetics
1. Introduction
1.1 Background of Dyslexia
Language based learning disabilities are the most common learning disabilities in schoolchildren in the United States [1]. Dyslexia, also known as reading disability, comprises a majority of these language based learning disabilities. Prevalence estimates vary depending on diagnostic criteria, with estimates ranging from 5-17% in western countries including the United States and the United Kingdom [2]. Nonetheless, dyslexia is common in pediatric populations across the globe and remains a lifelong impairment. These unexplained difficulties in reading can negatively impact a child's academic performance, reduce self-perception of cognitive abilities, and yield various undesirable socioeconomic consequences [2-3]. Neuropsychologists have investigated the specific reading and language processes that underlie dyslexia. Phonological processing is widely viewed as the core deficit in dyslexia, although deficits in reading comprehension, orthography, auditory stimuli integration, and semantic processing are also often observed [2, 4-9].
Interventions are available to remediate these reading deficits and help in the development of proficient reading and academic skills. Interventions are most successful when they are applied at younger ages, making early diagnosis a priority for optimal outcomes [9-10]. Currently, to make a diagnosis, trained neuropsychologists and educational professionals perform an exhaustive series of expensive neurocognitive assessments on each child. This, however, requires that the child has started to develop reading and language skills, which can delay early detection. Additionally, the expenses of diagnostic neurocognitive testing and need for trained professionals are significant roadblocks to the delivery of effective treatment to affected children. The challenges of early detection of a behavioral disorder make testing for genetic factors, detectable biomarkers, and/or neuroimaging signatures attractive alternatives.
1.2. Genetic Etiologies of Dyslexia
Family studies have long shown that dyslexia and overall reading abilities have significant genetic components, with heritability estimated at 54-84% [11-12]. Over the past two decades, genetic studies have examined which loci and specific genes contribute to dyslexia and reading skills. As with most complex and neurobehavioral traits, these investigations have produced both successes and failures. Genetic linkage studies have identified nine dyslexia genetic loci termed DYX1-DYX9 spanning 1-20 million bases on 8 different chromosomes, each with varying degrees of evidence supporting their role (Table 1). Within these loci, several dyslexia risk genes have been identified including DCDC2, KIAA0319, TTRAP, and THEM2 on chromosome 6 [13-15], DYX1C1 and CYP19A1 on chromosome 15 [16-18], C2orf3 and MRPL19 on chromosome 2 [19-20], ROBO1 on chromosome 3 [21-22], and KIAA0319L on chromosome 1 [23]. Outside of these DYX loci, other genes are also associated with dyslexia and performance on reading tasks, including FOXP2 and CNTNAP2 on chromosome 7 as well as ATP2C2 and CMIP on chromosome 16 [24-29]. Of these, the most replicated and well-studied are DCDC2 and KIAA0319 on chromosome 6, DYX1C1 on chromosome 15, and FOXP2 and CNTNAP2 on chromosome 7. In-depth reviews discussing the genetics of dyslexia and related language disorders have been published elsewhere [30-34].
Table 1.
Nine dyslexia (DYX) loci identified by genetic analyses
| Locus | Location | Candidate Genes | Locus Replicated? | Imaging-Genetics? |
|---|---|---|---|---|
| DYX1 | 15q21.3 | DYX1C1, CYP19A1 | Yes | Yes |
| DYX2 | 6p22 | DCDC2, KIAA0319, TTRAP, THEM2 | Yes | Yes |
| DYX3 | 2p16-p15 | C2orf3/MRPL19 | Yes | Yes |
| DYX4 | 6q13-16.2 | N/A | Yes | No |
| DYX5 | 3p12-q13 | ROBO1 | Yes | No |
| DYX6 | 18p11.2 | N/A | Yes | No |
| DYX7 | 11p15.5 | DRD4 | No | No |
| DYX8 | 1p36-p34 | KIAA0319L | Yes | No |
| DYX9 | Xq27.3 | N/A | No | No |
To define phenotypes in genetic studies of dyslexia and reading processes, investigators have primarily used neurobehavioral phenotypes, similar to those used to diagnose children in schools and clinics. There are advantages to using a solely behavioral approach in defining phenotypes. First, neurobehavioral reading measures match the clinical presentation of cases in schools and the tests used to diagnose affected individuals. Thus, genetic variants that show association with these phenotypes are likely to be relevant to the clinical neurobehavioral outcome of interest. Second, the neurobehavioral batteries used to define dyslexia are normalized, validated measures that have been used in research and clinically for decades. This allows for reliability and accuracy in ascertaining reading abilities and allows for results to be compared across disciplines and investigations. Third, researchers have developed these neurobehavioral batteries to examine several components of reading and language, such as phonological awareness and semantic processing, to ascertain the effects of various factors on different disorder subtypes and sub-processes. These specific batteries can examine the effects of genetic factors on specific reading and language processes.
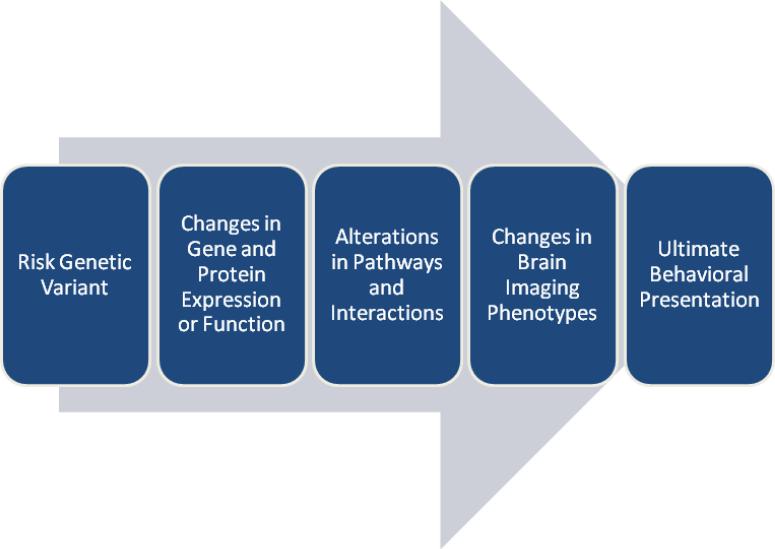
However, when looking specifically at genetic and biological investigations of dyslexia, the use of neurobehavioral reading tests can have disadvantages. There are numerous intermediary factors that separate genes from the downstream reading processes. Genetic factors can lead to changes in gene expression, protein expression, protein folding, and protein signaling, along with many other functional changes. Additionally, recent studies have demonstrated the importance of non-coding elements including miRNAs and other non-coding RNAs which can influence gene and protein expression as well protein structure itself. These changes in gene and protein function have direct effects on protein-protein, protein-DNA, protein-RNA, and other protein-species interactions. These changed interactions then have a larger effect on cellular and tissue functionality. It is then changes in these functionalities that can give rise to reading impairments and the ultimate observed neurobehavioral phenotype (Figure 1). Using only neurobehavioral measures to characterize dyslexia and reading processes can overlook the mechanistic implications of risk genes and variants, and ultimately the ascertainment of the molecular mechanisms, pathways, and gene networks.
Figure 1.
The path from risk genetic variants to the ultimate clinical behavioral phenotype of impaired reading performance. Risk genetic variants lead to changes in the expression and/or functionality of genes and proteins. These changes in expression and function alter the performance and functionality of pathways and species that interact with the altered gene and protein. These fundamental changes in pathways and interactions alter brain imaging phenotypes in a gross level which then lead to the observed impairments in reading performance.
With these limitations in mind, researchers have begun to look to other means to bridge the gap between gene, mechanism, and clinical presentation. One common method is to use animal models to functionally interrogate genes identified by human studies. These include knockdown, knockout, and knock-in genetic models in systems including Drosophila, mouse, rat, C. elegans, among numerous others. These systems allow for the in vivo examination of gene and protein function to characterize molecular and functional mechanisms underlying disease. Unfortunately, modeling human-specific genetic elements and human-specific phenotypes in animal models can prove difficult, especially for non-essential traits such as reading and language that have not evolved in many species and do not influence overall reproductive fitness. Although much can be and has been learned about specific gene function by modeling these human-specific non-essential phenotypes in animal models, there remains a need for a means to examine these processes in vivo that is directly related to reading and language abilities.
1.3. Use of Imaging in Dyslexia
One strategy researchers have begun to explore is in vivo neuroimaging techniques to gain structural, connectivity, and functional insights into the reading and language centers of the brain (Table 2). These studies have detected neuroimaging differences between subjects with dyslexia and non-impaired controls. Subjects with dyslexia have structural differences compared to non-impaired controls, both in grey matter density and in white matter microstructure connectivity and integrity [35-47]. Functional magnetic resonance imaging (fMRI) studies show that subjects with dyslexia display different patterns of brain activation during reading tasks compared to non-impaired controls [48-56]. Generally, impaired subjects show increased bilateral, symmetric brain activation patterns as opposed to the left-stereotyped activation patterns in non-impaired individuals [57]. Detailed reviews of dyslexia imaging studies are published elsewhere [58].
Table 2.
Imaging techniques used in imaging-genetics studies of dyslexia
| Technique | What it measures | Genes | References |
|---|---|---|---|
| Structural Magnetic Resonance Imaging (MRI) |
Volume and thickness of cerebral regions, including grey and white matter volumes |
DCDC2
KIAA0319 FOXP2 CNTNAP2 DYX1C1 C2orf3/MRPL19 |
Meda et al.81 Jamadar et al.85 Darki et al.86 Belton et al.113 Watkins et al.114 Vargha-Khadem et al.115 Tan et al.131 Scerri et al.20 |
| Functional magnetic resonance imaging (fMRI) | Blood-oxygen-level-dependent (BOLD) signal indicating neural activity patternings during reading-related tasks performed in scanner |
DCDC2
KIAA0319 FOXP2 CNTNAP2 |
Cope et al.87 Pinel et al.89 Liegeois et al.116 Wilcke et al.25 Scott-Zealand et al.132 Whalley et al.133 |
| Resting State functional magnetic resonance imaging | Blood-oxygen-level-dependent (BOLD) signal depicting blood flow indicating possible regional interactions and connections while not performing tasks |
KIAA0319
DCDC2 |
Jamadar et al.88 |
| Fractional Anisotropy (FA) |
Diffusion Tensor Imaging (DTI) by MRI to infer white-matter connectivity among various regions of the brain |
CNTNAP2 | Tan et al.131 |
| Positron Emission Tomography (PET) |
Measures 3 dimensional activity throughout the brain by detection of an injected radiotracer (e.g. glucose, neurotransmitter) | FOXP2 | Vargha-Khadem et al.115 |
Although studies have shown various neuroimaging differences between subjects with and without dyslexia, imaging techniques are currently not sensitive or reliable enough to be used as a diagnostic tool in the clinic. However, in research, the prospect of integrating these neuroimaging data with genetic and neurobehavioral data to gain a mechanistic model of gene, brain function and structure, and ultimate behavioral phenotype is attractive, although difficult and complex. Determining whether these neuroimaging differences cause reading deficits or are a result of having dyslexia can be difficult. The use of various data modalities including human genetic, neuroimaging, and animal model studies can help untangle these complex relationships. The remainder of this review aims to discuss the investigations that have attempted to integrate neuroimaging data with genetic and neurobehavioral data (termed imaging-genetics), followed by comments on the promise and future of imaging-genetics in examining dyslexia. To date, imaging-genetic studies of dyslexia have focused on the following genes: (1) DCDC2, (2) KIAA0319, (3) FOXP2, (4) CNTNAP2, and (5) DYX1C1/C2orf3/MRPL19 (Table 3).
Table 3.
Candidate genes examined in imaging-genetic studies of dyslexia
| Gene | Location | Function | Imaging Techniques Performed | References |
|---|---|---|---|---|
| DCDC2 | 6p22 | Neuronal Migration Cilia |
MRI fMRI Resting State |
Meda et al.81 Jamadar et al.85 Darki et al.86 Cope et al.87 Jamadar et al.88 |
| KIAA0319 | 6p22 | Neuronal Migration | MRI, fMRI Resting State |
Jamadar et al.85 Darki et al.86 Jamadar et al.88 Pinel et al.89 |
| FOXP2 | 7q31 | Transcriptional Regulation Neurogenesis |
MRI fMRI PET |
Belton et al.113 Watkins et al.114 Vargha-Khadem et al.115 Pinel et al.89 Liegeois et al.116 Wilcke et al.25 |
| CNTNAP2 | 7q35 | Cell Adhesion Voltage Gated Channels |
MRI fMRI FA |
Tan et al.131 Scott-Zealand et al.132 Whalley et al.133 |
| DYX1C1 | 15q21.3 | Neuronal Migration Cilia |
MRI | Jamadar et al.85 Darki et al.86 |
| C2orf3IMRPL19 | 2p11-q11.2 | Unknown | MRI | Scerri et al.20 |
2. Imaging-Genetics Studies of Dyslexia
2.1. The DYX2 Locus: DCDC2 and KIAA0319
The DYX2 locus is located on chromosome 6p22 in which two dyslexia risk genes, DCDC2 and KIAA0319, are encoded (Table 1). DCDC2 and KIAA0319 are the two most replicated dyslexia risk genes, having been associated with dyslexia and overall reading skills in numerous studies [13-15, 29, 59-73]. Animal models of the effects of mutations in these genes have been examined in mice, rats, and C. elegans. KIAA0319 and DCDC2 knockout mice have shown several behavioral impairments including spatial memory and response to acoustic stimuli, but again underscore the difficulties in modeling a uniquely human trait in non-human systems [74]. In utero RNAi knockdowns of DCDC2 and KIAA0319 in embryonic rats exhibited aberrant neuronal migration, suggesting that dyslexia may result in dysfunction in neuronal migration during fetal development [13, 75-77]. In utero RNAi Kiaa0319 knockdowns in rats have also shown the influence of this gene on dendrite morphology, migratory defects resulting in heterotopias in white matter, and changes in corpus collosum area along with deficits in responses to acoustic stimuli and impaired spatial learning [76, 78-79]. Preliminary work in C. elegans showed that DCDC2 affects ciliated neurons, suggesting specific neuron types that may be affected by risk variants [80]. Therefore, the functional implications on neuronal migration and other neural phenotypes suggest that DCDC2 and KIAA0319 have distinct neuroimaging impacts. These neuroimaging implications of DCDC2 and KIAA0319 may mediate their effects on reading performance. To this end, recent investigations have examined DCDC2 and KIAA0319 using various neuroimaging techniques.
Imaging-genetic studies have started to examine the contributions of DCDC2 to neuroimaging phenotypes using structural and functional MRI data. Our group found a relationship between grey matter volume in subjects with dyslexia and a microdeletion located within intron 2 of DCDC2 [81]. This microdeletion contains a functional variant, termed READ1, which is a highly polymorphic complex tandem repeat with over 30 observed alleles [13, 73, 82]. The READ1 element has been associated with dyslexia in numerous independent cohorts [13, 73, 83-84]. The READ1 element has the capability to modulate DCDC2 expression in vitro and specifically binds the transcription factor ETV6 [73, 82]. Meda et al. showed that this variant has downstream effects upon grey matter volume in dyslexic subjects, possibly mediated by its effect on DCDC2 and/or other gene expression [81]. Further, Jamadar et al. demonstrated an association between SNPs within DCDC2 and grey matter volumes in the superior prefrontal, temporal, and occipital networks in subjects with schizophrenia, defined using parallel independent component analysis (ICA) [85]. Darki et al. recently reported association of DCDC2 SNPs with white matter volume in the left temporo-parietal region [86]. The authors then connected white matter volume to reading skills in the same subjects, suggesting that DCDC2 influences white matter volume which then influences reading performance. This investigation attempts to connect the dots from genetic variants, to neuroimaging phenotype, to the ultimate neurobehavioral clinical presentation. These studies indicate that grey and white matter volumes are influenced by DCDC2, possibly by changes in gene expression that influence neuronal migration, and then exert an effect upon reading performance. Regionally, volumetric differences in the temporo-parietal, superior prefrontal, temporal, and occipital areas appear to be influenced by the DYX2 locus.
READ1 in DCDC2 was further characterized in Cope et al. 2012 using fMRI during four reading-related tasks [87]. Alleles 4 and 8 of READ1 were associated with brain activation patterns in the left anterior inferior parietal lobe and right lateral occipital temporal gyrus during semantic processing tasks. These associations were hypothesized to be protective in nature as their direction of effect was similar to brain activation patterns of non-impaired individuals. The READ1 element appears to have mediatory effects on gene expression and neuroimaging phenotypes that then influence reading performance. Using resting state fMRI data, Jamadar et al. also showed association of DCDC2 variants with a Broca-medial parietal network in both schizophrenic and non-impaired cohorts [88]. In addition to grey/white matter volume, DCDC2 appears to alter brain activation patterns and regional connections in various anatomic language and reading centers.
Similar to DCDC2, KIAA0319 associated with differences in grey matter volumes in the Darki et al. and Jamadar et al. studies, once again tying the DYX2 locus to volumetric differences in white/grey matter volume [85-86]. Examining grey matter volumes using voxel-based morphometry, Jamadar et al. found superior and inferior cerebellar networks were related to KIAA0319 [85]. Darki et al. also reported association of KIAA0319 with white matter volume in the left temporo-parietal region [86]. These structural studies depict a connection from KIAA0319 to white/grey matter volumes, which then appear to influence reading skills.
Using fMRI data, Pinel et al. observed an association between a variant in THEM2 and lower asymmetric activation of the posterior superior temporal sulcus during reading and phonology tasks [89]. The observed lower asymmetry of brain activation patterns by genetic marker is similar to the observed asymmetry in affected individuals. Although the associated marker in the Pinel et al. study was located within THEM2, it appears that this marker captures variation from the KIAA0319 risk haplotype. This risk haplotype has been associated with dyslexia numerous times and encompasses the 5’ portion of KIAA0319, along with neighboring genes TTRAP and THEM2 [64, 66, 70, 72, 77, 90]. This KIAA0319 risk-haplotype has been associated with lower KIAA0319 gene expression [91]. In fact, the DCDC2 READ1 element and KIAA0319 risk haplotype have been shown to interact genetically in a synergistic manner [66, 73, 91]. However, whether these genomic loci interact physically remains to be determined, although the possibility of an interaction between a regulatory element and a promoter region is intriguing. Using resting state fMRI in schizophrenia and non-impaired groups, Jamadar et al. found a left Broca-superior/inferior parietal network was related to two KIAA0319 SNPs in non-impaired individuals. In both schizophrenic and non-impaired individuals, a left Wernicke-fronto-occipital network was related to two KIAA0319 SNPs [88]. A bilateral Wernicke-fronto-parietal network was related to one KIAA0319 SNP only in non-impaired individuals. These differences between schizophrenia and non-impaired groups indicate that non-impaired individuals may utilize different parts of the brain during reading tasks than those with a distinct neurobehavioral or neuropsychiatric disorder. Nonetheless, KIAA0319 contributes to a variety of imaging phenotypes particularly in terms of grey/white matter volumes, brain circuitry, and brain activation patterns during reading-related tasks.
2.2. FOXP2
FOXP2 and CNTNAP2 both reside on chromosome 7 and were implicated in other disorders before being examined in dyslexia. FOXP2 was first implicated in a family segregating a severe form of dyspraxia of speech, designated the KE family [26, 93-96]. Following this discovery, FOXP2 has become the most widely studied gene related to language, including a myriad of approaches from animal models such as mice and finches to in vitro molecular work to examine its regulatory capabilities [97-102]. In fact, many have examined the human specific nature of FOXP2 expression and function and correlated this with the evolution of language in humans [103-107]. FOXP2 is a regulator of neurogenesis during embryonic development [108]. Mouse models have been generated carrying mutant Foxp2. These animals exhibit abnormal ultrasonic vocalizations, thought to be a phenotype to model human speech development, as well as other disorders including developmental delay, motor function, and brain development [108-111]. Expression of human, but not mouse, FOXP2 enhanced progenitor and neuron development [111-112]. However, how the expression and function of FOXP2 influences the brain and language skills mechanistically still remains to be elucidated.
The first imaging investigations of FOXP2 were of structural differences among affected and unaffected members of the KE family. Although speech impairments and speech dyspraxia are distinct disorders from dyslexia, the neuroimaging implication for a Mendelian disorder of a single gene (in this case FOXP2) can make a significant contribution to the pathobiology of the FOXP2 gene and where/how this gene functions. As this family is affected with a single gene disorder, the use of related individuals can aid to remove other genetic and environmental factors that may have influenced neuroimaging measures. Multiple imaging studies of this family have found structural abnormalities, particularly in grey matter density [113-115]. These include decreased grey matter density bilaterally in the caudate nucleus, cerebellum, and inferior frontal, and increased grey matter density bilaterally in the planum temporal gyrus [113-115]. These changes in neuroimaging measures, particularly in grey matter density, appear to be a consequence of the FOXP2 mutation and consequently, could influence language in these affected family members.
Investigators have also performed fMRI studies of KE family members [116]. Non-impaired family members who did not have the causal FOXP2 variant exhibited normal left dominant patterning of brain activation during reading tasks. Affected individuals with the mutant FOXP2 exhibited different brain activation patterns with increased posterior and bilateral brain activation. These affected family members also exhibited reduced activation of Broca's area, as well as other cortical language related regions and the putamen. A PET functional imaging study showed underactivation of the left supplemental motor area (SMA), left subjacent cingulate cortex, and left preSMA/cingulate cortex and overactivation of the left caudate nucleus and left ventral prefrontal region in affected family members [115]. These studies of this one family demonstrate the utility of and amount of insight gained from examining rare variants with large effect. Beyond the KE family, new studies have examined the role of FOXP2 in reading. Recent studies by Wilcke et al. in subjects with dyslexia and Pinel et al. in non-impaired individuals found differences among variants in FOXP2 in regional brain activation during reading tasks including temporo-parietal, inferior frontal, and precentral brain regions [25, 89]. Together, these studies of rare and more common variants within FOXP2 show the important role it plays in overall language and reading skills throughout the brain.
2.3. CNTNAP2
FOXP2 is a transcription factor and modulates DNA transcription at numerous loci throughout the genome, underscoring the importance of transcriptional regulation in dyslexia and other neurobehavioral disorders. Interestingly, one of its gene targets is the cell adhesion gene CNTNAP2. CNTNAP2 has been linked and associated with numerous neurobehavioral and neuropsychiatric disorders including Tourette's syndrome, schizophrenia, speech delay, and autism spectrum disorders [117-124]. Recently, the associations of CNTNAP2 were expanded to included language impairment and dyslexia [24, 29, 125]. CNTNAP2 is expressed in the frontal and temporal lobes of developing brain and in frontal cortex of adults [126]. These areas are vital to reading and language development. CNTNAP2 encodes the Caspr2 protein, which functions in voltage gated channels at Nodes of Ranvier and is important in cellular migration [127-129]. This function suggests that variants in CNTNAP2 may cause abnormalities in the propagation of action potentials throughout neural circuits and integration of written language stimuli. It has also been hypothesized that CNTNAP2 helps to mediate interactions between neurons and glia [130]. Connecting CNTNAP2 molecular neural functions to the observed language associations by imaging-genetics would help explain how changes in CNTNAP2 may cause impairments in reading and language.
As CNTNAP2 encodes a protein that functions in cellular migration, Tan et al. used structural MRI to examine grey and white matter volume implications of a previously associated variant, rs7794745 [131]. Most samples in imaging-genetic studies are small in size; however, this study examined MRI measures in over 300 children. This larger sample size, in addition to increasing statistical power, also allowed the investigators to compare subjects homozygous for the risk allele with subjects homozygous for the nonrisk allele. There were differences in gray and white matter volumes in regions of interest between these two groups. Specifically, they observed reduced grey matter in fusiform, posterior cerebellar hemispheres, posterior occipital cortices, cerebellar vermis, left superior cerebellum, and right frontal pole. There were reductions in white matter volume in posterior thalamic radiation (thalamus to posterior parietal/occipital cortex), right caudal inferior fronto-occipital fasciculus (frontal/occipital lobe, fusiform), and right rostral cingulum. To expand on their findings with grey/white matter, Tan et al. used DTI to examine the fractional anisotropy (FA) implications of the rs7794745 variant [131]. There were sex-specific reductions in FA, specifically in the right rostral inferior fronto-occipital fasciculus in males and the right anterior thalamic radiation/putaminal head in females. These sex-specific differences in white matter tracts may contribute to gender differences of dyslexia and language disorders.
In addition to CNTNAP2's effects on brain structure and circuitry, other studies have examined the effects of CNTNAP2 risk variants on brain activation patterns. Scott-Van Zeeland et al. 2010 examined frontal lobar connectivity and the CNTNAP2 variant rs2710102 using fMRI [132]. There was reduced activity in medial prefrontal cortex, along with more widespread and bilateral connectivity in frontal cortex and anterior temporal pole in risk subjects. Subjects with the non-risk variant had a stereotypical left-lateralized network, including the left inferior frontal gyrus, insula, anterior temporal pole, superior temporal gyrus and angular gyrus, usually seen in non-impaired subjects. These associations were replicated in two additional small samples, including the right middle frontal gyrus and left interacalcarine cortex. These replication cohorts also displayed the same network of activation patterns. Whalley et al. examined the rs7794745 and rs2710102 risk variants of CNTNAP2 and fMRI brain activation patterns [133]. They found that healthy individuals with the risk allele had a significant increase in activation of right inferior frontal gyrus [analogous to Broca's area] and right lateral temporal cortex, similar to increased right (i.e. bilateral) brain activation in risk allele groups in other studies. These studies implicated CNTNAP2 as altering brain activation patterns in a similar fashion to those seen in subjects with dyslexia. With its associations with multiple disorders, however, it remains to be seen whether these changes in brain activation specifically contribute to dyslexia or contribute to core deficits observed in multiple neurobehavioral disorders.
2.4. Other Genes: DYX1C1 and MRPL19/C2orf3
There have been limited imaging-genetics studies of other dyslexia loci and genes, namely DYX1C1 on chromosome 15 and the DYX3 locus on chromosome 2. DYX1C1 on chromosome 15q21 was first identified in a family with dyslexia co-segregating with a translocation disrupting the DYX1C1 gene [16]. Efforts at replicating the association of this gene have been mixed, but positive replication has been achieved in several cohorts [17, 134-138]. Most in vivo work with DYX1C1 has occurred using in utero RNAi knockdown models in rats, similar to those completed with KIAA0319 and DCDC2 [71, 139-143]. Behaviorally, these animals display impairments in analogous domains including working memory, auditory processing, and spatial reasoning [144]. Anatomically, there are neuronal migratory and laminar disruptions, and a subset of the rats display hippocampal heterotopias [145-146]. Pathway analyses have bolstered these neurological implications by linking DYX1C1 with other major genes involved in neuronal migration and brain development pathways, along with cytoskeletal proteins similar to DCDC2 and KIAA0319 [147-148]. DYX1C1 has also been linked to ciliated neurons and cilary function in migration [148-149]. Investigators have found associations of DYX1C1 with grey and white matter volumes in a superior cerebral network and temporo-parietal regions, respectively [85-86]. The role of DYX1C1 in neuronal migration may have an impact on the development of grey and white matter.
The DYX3 locus on chromosome 2 is less studied than the other genes presented in this review, but is centered on MRPL19 and C2orf3 with mixed results in replication analyses [19, 150-153]. SNPs in these genes have shown association with reading and language performance in multiple cohorts, including a study by Scerri et al [20]. Scerri et al also found an association of variants in DYX3 and white matter structure in the posterior of the corpus callosum and cingulum, regions that connect large portions of the cortex in the parietal, occipital, and temporal lobes [20]. Although not yet as widely studied, these studies offer a promising start into understanding the implications of DYX1C1 and the DYX3 locus on brain imaging phenotypes.
3. Where Imaging-Genetics of Dyslexia Currently Stands
3.1 By Neuroimaging Method
Neuroimaging technology can provide a wide array of data, including those on structural volume, grey/white matter volume, resting state, brain activation during specific reading-related tasks, connectivity, DTI, and FA. They may be inherently interrelated, but each reveals a nuanced, unique aspect of brain functionality. Therefore, a gene's association with these various imaging measures can reveal nuanced, specific as well as universal, widespread roles of these genes in brain development and function.
So far, there have been a variety of associations between the specific genes presented in this review and neuroimaging measures. All studied genes in this review (DCDC2, KIAA0319, FOXP2, and CNTNAP2, DYX1C1, and C2orf3/MRPL19) associated with grey and white matter volumes. These studies suggest that the genes that impart risk to dyslexia fundamentally influence and alter brain development. Changes in grey and white matter volume as well as overall structure may influence the proper propagation and/or integration of language stimuli throughout critical brain regions of interest, yielding the behavioral deficits in phonology, semantics, spelling, and overall reading comprehension. These genetic associations with grey and white matter volume differences lend credence to the current neuronal migration hypothesis underlying dyslexia. Animal models of DCDC2, KIAA0319, and DYX1C1 all exhibit abnormalities in the migration of neurons from the ventricular zone into the cortex. These abnormalities in neuronal migration in animal models may mirror the differences in grey and white matter volumes observed in imaging-genetics studies in humans. These changes in volumes may bring about a yet unidentified effect, perhaps alterations in brain activation patterns and/or efficient integration and propagation of action potentials that influence reading skills.
The four more widely studied genes (DCDC2, KIAA0319, FOXP2, and CNTNAP2) presented in this review have shown association with fMRI brain activation patterns during reading-related tasks. These changes in brain activation patterns suggest that risk variants contribute to the ability of specific regions of the brain to fluently process and respond to written language stimuli. Resting state fMRI identified networks connecting neural language centers where DCDC2 and/or KIAA0319 risk variants may also contribute to deficits in reading and language fluency. Changes in both brain activation patterns and networks may be a consequence of aberrant neuronal migration that results from altered gene expression as modeled in experimental animals. Failure of neurons and supporting cells to migrate to proper locations may alter the brains ability to efficiently integrate language stimuli and propagate the appropriate neural response in specific critical brain regions for fluent language. Other imaging technology including PET and FA has been less widely studied. However, FOXP2 variants have been shown to associate with differences in activation during PET, which supplements and bolsters functional studies using fMRI. CNTNAP2 showed associations with FA, again showing that CNTNAP2 influences brain circuitry important in reading and language. Imaging-genetic studies have shown that risk genes identified using neurobehavioral phenotypes have implications for brain structure, grey/white matter, brain connectivity and circuitry, and brain activation during reading-related tasks that influence reading and language performance and deficits.
3.2 By Brain Region
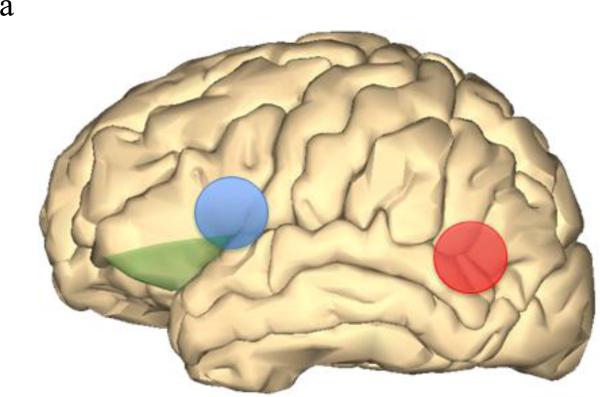
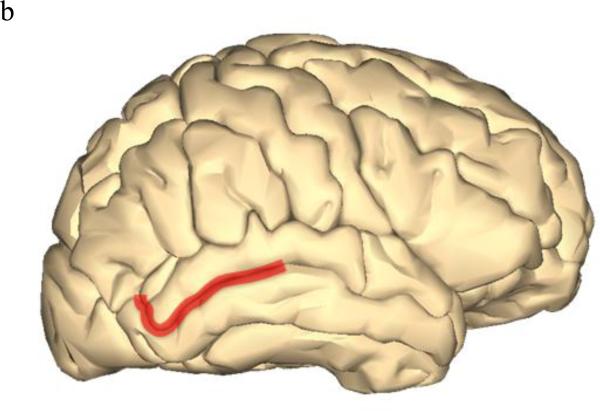
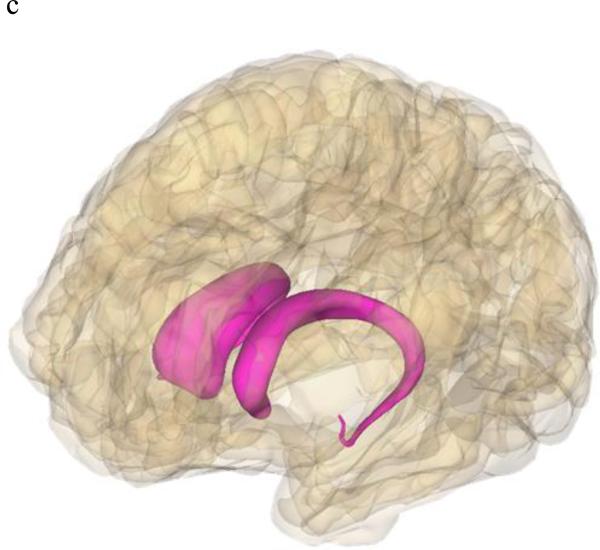
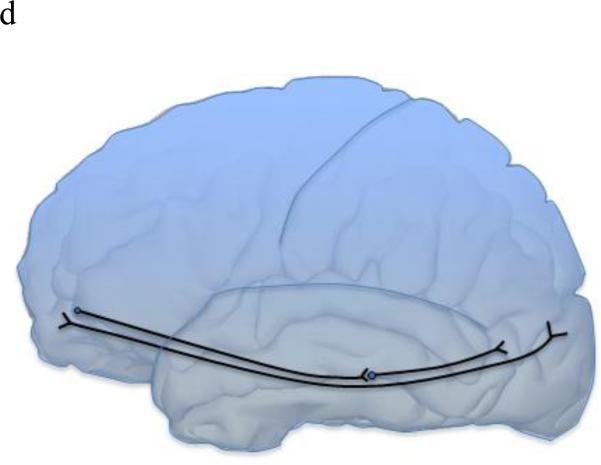
As imaging-genetic studies of dyslexia increase in number, how and where dyslexia risk genes function will become more apparent. In addition to brain gene expression data recently available, imaging data can aid in identifying where risk genes normally function, where gene dysfunction can lead to disorders, and possible signatures of the disorder state. There also may be gene-specific, location-specific dysfunctions that are related to dyslexia and other higher-order cognitive disorders. By integrating behavioral, genetic, and imaging data, we can identify brain regions with gene-specific effects (i.e. only a small subset of risk genes function in this region) or with universal effects (i.e. all/many risk gene function in this region). For example, DCDC2, KIAA0319, FOXP2, and CNTNAP2 have all shown associations in Broca's area and the inferior frontal gyrus (Figure 2a). Broca's area and inferior frontal gyrus are main language centers in the brain, so it is logical for many dyslexia risk genes to be associated with various imaging measures in these areas. Similarly, increased right brain activation during reading-related tasks was observed in risk allele variants of KIAA0319, FOXP2, and CNTNAP2, as opposed to the typical left-side dominant brain activation seen in non-impaired and non-risk groups. However, with KIAA0319, this loss of asymmetry was seen only in the posterior superior temporal sulcus (Figure 2b). More widespread, bilateral brain activation patterns were seen in FOXP2 and CNTNAP2 risk variants, which may explain why FOXP2 and CNTNAP2 are associated with a litany of neurological disorders and KIAA0319 is somewhat specific to reading, language, and cognition. FOXP2 was the only gene linked with the caudate nucleus and showed associations with grey matter and PET scan overactivation (Figure 2c). DCDC2, KIAA0319, and DYX1C1 (white matter) along with FOXP2 (fMRI activation) associated with the temporo-parietal regions of the brain (Figure 2a). The associations with Broca's area/inferior frontal gyrus as well as the temporal-parietal regions suggests that connections between these areas, possibly including the inferior fronto-occipito fasiculus which showed a relationship with CNTNAP2, will be important areas for imaging-genetics research in the future (Figure 2d). However, lack of association does not necessarily mean that these genes do not function or relate to these regions or neuroimaging data of interest. Imaging-genetic studies of dyslexia are still limited in number and many hypotheses remain to be tested with sufficient statistical and biological power.
Figure 2.
Brain models showing brain locations that displayed associations with dyslexia risk genes. Images were generated using BodyParts3D/Anatomography service by DBCLS, Japan.
Figure 2a Left brain depicting Broca's area (blue), inferior frontal gyrus (green), and temporo-parietal area (red) which all were associated with multiple dyslexia risk genes.
Figure 2b: KIAA0319 showed specific loss of asymmetric activation of the right posterior superior temporal sulcus.
Figure 2c: FOXP2 showed associations with grey matter and activation in the caudate nucleus.
Figure 2d: CNTNAP2 showed association with the inferior fronto-occipito fasciculus (IFOF), which connects two main language areas: the inferior frontal gyrus and temporo-parietal area.
3.3 Current Limitations of Imaging Genetics
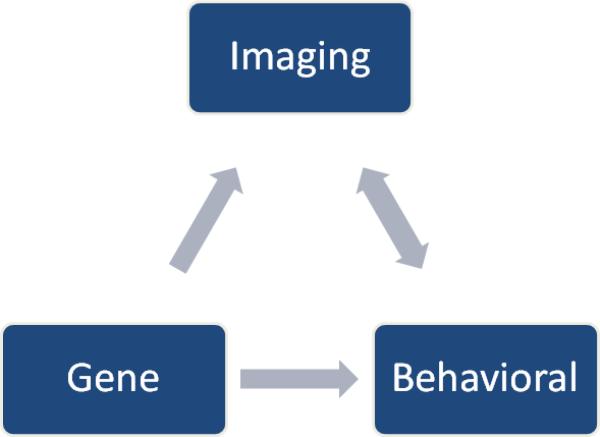
As with every emerging field and technology, there are various limitations of the initial imaging-genetics studies of dyslexia. (1) The first limitation in most imaging-genetics studies is the relatively small sample size. Small sample size in genetics studies can be especially worrisome due to population stratification and lower minor allele frequencies, which can also lower statistical power. The largest imaging study reported to date is that of Tan et al., which included approximately 300 subjects [131].. Although imaging-genetic studies can be daunting in terms of expense, time, and the amount of data to interpret, larger studies following the models of the Alzheimer's Disease Neuroimaging Initiative (ADNI) (http://www.loni.ucla.edu/ADNI/) and Pediatric Imaging Neurocognition Genetics study (PING) (http://ping.chd.ucsd.edu/) are necessary to expand the promise of imaging-genetic studies. New study designs including longitudinal imaging-genetics investigations on both impaired and non-impaired individuals could provide a means to temporally dissect the genetic contributions to brain and neurocognitive development. (2) Another difficulty is that subjects are from a relatively healthy pediatric population. Recruitment of these individuals can be difficult since there is little to no incentive to participate. (3) Interpreting what imaging data and associations with genetic and behavioral factors actually mean in a biological context is challenging. Although there are numerous hypotheses regarding the actual meaning of increased brain activation in fMRI, brain circuitry revealed through resting state fMRI and FA studies, and connectivity measures through DTI, it is still difficult to make definitive conclusions about the biological and behavioral implications of these data. Discerning whether these neuroimaging phenotypes are causal of or resulting from case status is challenging due to the inherent complexity and plasticity of the brain. Further neurophysiological and molecular interrogation using cell-based and organism models as well as longitudinal imaging-genetic studies can help in determining causal and temporal relationships. Adding to the inherent difficulty of these studies are the nuanced nature of neuroimaging, and the currently underdeveloped methods and standards for reporting findings to other scientists and the lay public. (4) All of the presented studies started with a small subset of genetic variants and then examined the implications of these specific genetic factors on brain imaging measures. Selection bias along with ignoring most of the genome could lead to misleading and incomplete results and hypotheses. (5) With the exception of Darki et al., most of the presented imaging-genetic studies of dyslexia examine the effects of genetic variants on behavior and brain imaging data independently, instead of fully integrating them together (Figure 3). Such an analytical strategy does not take advantage of the vast amount of data collected on single subjects. Integration of these data could be powerful in unraveling the mystery and for developing risk models of neurobehavioral disorders such as dyslexia.
Figure 3.
A schematic of the relationship between gene, behavior, and imaging, in which neuroimaging measures are a mediatory step between risk genetic variants and the ultimate neurobehavioral phenotype.
4. The Future of Imaging-Genetics in Dyslexia
Currently, we have an incomplete view of the biological etiologies underlying dyslexia. Neuropsychological, genetic, molecular, and imaging studies have made much progress into identifying impairments, candidate genes/signatures, and possible pathways that may contribute to the deficits observed in dyslexia and related disorders. However, how these varying levels of phenotype interact and relate to each other to lead to the ultimate phenotype remains elusive. Additionally, currently, only neurobehavioral batteries are used as a diagnostic tool for dyslexia. Imaging and genetic tools are currently unable to specifically and sensitively identify children with and without dyslexia. Future studies are needed to identify and to develop risk models to accurately use imaging and genetic data as a diagnostic or screening tool. Currently, imaging-genetics studies are used to gain a more mechanistic understanding into the pathophysiology of dyslexia and reading-related processes.
The studies presented in this review represent some of the first attempts to integrate genetic, molecular, imaging, and neurobehavioral data to characterize dyslexia. These studies attempt to connect the dots from genes, to molecular and cellular function, to changes at the neurological and brain level, and finally to the ultimate neurobehavioral clinical presentation of impaired reading (Figure 1, Figure 3). Imaging-genetics provides an ethical means to gain mechanistic insight into the pathophysiology in human subjects in vivo. Instead of relying upon animal models that may approximate the behavioral and biological deficits seen in dyslexia, human imaging-genetics allow for the direct examination of human risk genetic variants with imaging data directly related to reading and language processes. This could represent the beginnings of a logical progression of risk factors to neuroimaging signatures to neurobehavioral presentation that may allow for earlier detection and remediation of affected individuals. Early detection in dyslexia is vital for optimal remediation and academic outcomes.
The advent of cheaper, more advanced imaging and genetic technologies has made the integration of human brain imaging and genetics to investigate dyslexia possible. Imaging-genetics remains an immature field, but the studies presented in this review have begun the promising use of imaging and genetics to gain in vivo insights. So far, imaging-genetic studies of dyslexia have relied on the positive results from neurobehavioral genetic studies to identify candidate genes and variants, which are then used to analyze independent imaging data. Although producing interesting results, hypothesis driven studies informed by past neurobehavioral studies limit the extent to which pathophysiology can be explored. These studies can only reveal possible implications of these very specific genetic variants on brain imaging phenotypes. They cannot reveal other genetic contributors to the overall neurobehavioral reading deficits nor the imaging phenotype changes associated with dyslexia. Hypothesis-free genetic studies, similar to those seen commonly in human genetics such as genome-wide association studies (GWAS) and sequencing studies, could reveal new genes and pathways that contribute to alterations in brain imaging phenotypes, and how they contribute to the ultimate neurobehavioral phenotypes. In addition, new imaging technologies, such as Nuclear Magnetic Resonance (NRM) and Single Photon Emission Computed Tomography (SPECT), can detect metabolites and neurotransmitters in vivo. Recent genetic and neurobehavioral analyses have begun to implicate neurotransmitter signaling in language disorders [154-155]. Therefore, directly examining these signaling molecules and other metabolites in the brain could lead to further characterization of mechanisms underlying reading and language impairments.
A major hurdle underlying imaging-genetics as a field is the analyses of such large and diverse datasets. Typical GWAS datasets contain over 1 million markers and can be increased drastically with the imputation of non-genotyped SNPs. Sequencing data contain even larger amounts of data. Imaging data can comprise thousands of data points depending on which level analysis is performed (voxel-based, regions of interest, various circuits and networks). Examining each of these data points in a hypothesis-free manner would result in an extraordinary number of tests, which would require an exorbitant correction for multiple testing. Integrating these together in addition to neurobehavioral phenotypes can result in millions of hypotheses and an inordinate amount of computation costs and data to analyze. Data reduction methods, such as parallel ICA performed in the Jamadar studies, as well as new analytical methods to collapse and/or integrate a variety of data types into relevant risk models would facilitate larger, hypothesis-free analyses of neurobehavioral disorders.
In addition to accounting for the shear amount of data accumulated in imaging-genetics studies, specific consequences of rare and common genetic variation can complicate findings. Common variants typically have a relatively small, yet significant individual effect on phenotypes of interest, and identify genes and proteins fundamental to these processes. Many of these common variants reside in non-coding regions including miRNAs, long non-coding RNAs, and enhancer/silencer elements. Variation in these elements can have a substantial impact upon gene expression and function, as hypothesized with READ1 in DCDC2, which can lead to changes in neuroimaging and behavioral phenotypes. Small effect sizes require either a large number of subjects or a strong association to detect the relationship between phenotype and genetic marker. The need for large number of subjects could prove difficult in imaging studies due to costs and time investment, making large collaborative projects an attractive choice in the future. Recent studies have determined the importance of rare variants in common disease, particularly in neurobehavioral disorders such as autism spectrum disorders, intellectual disability, and bipolar disorder. These rare variants can be SNPs, usually in coding regions, or copy number variants, including deletions and duplications (e.g. 16p11.2 deletions and duplications). Although these rare variants appear to have a large effect size, the fact that they are rare can make it difficult to ascertain subjects or to make valid statistical conclusions due to sample size. Ensuring adequate size of comparison groups and screening for rare variation with the dwindling cost of DNA sequencing can allow for the use of both common and rare variants in examining neurobehavioral disorders using imaging, genetic, and behavioral data.
5. Conclusion
This review aimed to highlight the relatively new imaging-genetic studies of dyslexia and the promise this rising field has to unlock new insights behind the mechanisms and pathophysiology underlying this widely prevalent disorder. Although the field is immature, much has been learned regarding the possible downstream effects of dyslexia risk genes on brain structure, function, and circuitry. Current hypotheses into the underlying biology of dyslexia include: (1) risk genes leading to abnormalities to white/grey matter volumes and (2) risk genes disrupting brain activation patterns during reading-related tasks. These hypotheses appear to be rooted in aberrant neuronal migration, although other phenotypes including electrophysiological paradigms need to be explored. The future of imaging-genetics looks promising as more cohorts collecting a wide range of neurobehavioral, genetic, and imaging data start to become more widely available as has been seen in the ADNI and PING studies. Larger samples and new analytical strategies should reveal new mechanisms (genetic and imaging) that contribute to reading deficits in dyslexia. These insights will aid in the earlier detection of children with dyslexia and aid their overall academic and remediation potential.
Highlights.
Dyslexia is a complex, polygenic disorder affecting 5-17% of US schoolchildren.
Imaging-genetics aims to integrate neuroimaging, cognitive, and genetic data.
Imaging-genetics can identify novel risk genes and gene function in the brain.
The use of imaging-genetics will reveal new mechanisms of dyslexia.
Acknowledgements
The authors wish to acknowledge funding by the National Institutes of Health (Grant ref: R01 NS043530 awarded to JRG, and F31 DC012270 awarded to JDE) and Manton Family Foundation. We also thank Dr. Natalie R. Powers for thoughtful discussion and for editing the text.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare competing financial interests: Yale University has applied for a patent covering the complex tandem repeat and deletion in READ1 (inventor: J.R.G.), and sublicensed it to JS Genetics, Inc. J.R.G. is a founder and equity holder of JS Genetics, Inc.
References
- 1.National Institute for Child Health and Development Learning Disabilities: Overview. Website of the National Institute of Child Health and Development. 2012 http://www.nichd.nih.gov/health/topics/learning/Pages/default.aspx#f1.
- 2.Peterson RL, Pennington BF. Developmental dyslexia. Lancet. 2012;379:1997–2007. doi: 10.1016/S0140-6736(12)60198-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.National Center for Education Statistics Educational Finance Statistics Center, Special Education spending: what we are spending in special education in the U.S.? 2001.
- 4.Liberman IY, Shankweiler D. Liberman IY, editor. Phonology and the problems of learning to read and write. Remedial and Special Education. 1985;6(6):8–17. In Topical Issue. [Google Scholar]
- 5.Wagner RK, Torgeson JK. The nature of phonological awareness and its causal role in the acquisition of reading skills. Psychological Bulletin. 1987;101:192–212. [Google Scholar]
- 6.Shaywitz SE. Dyslexia. N Engl J Med. 1998;338(5):307–12. doi: 10.1056/NEJM199801293380507. [DOI] [PubMed] [Google Scholar]
- 7.Hornickel J, Kraus N. Unstable representation of sound: a biological marker of dyslexia. J Neurosci. 2013;33(8):3500–4. doi: 10.1523/JNEUROSCI.4205-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pennington BF. From single to multiple deficit models of developmental disorders. Cognition. 2006;101(2):385–413. doi: 10.1016/j.cognition.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 9.Foorman BR, Torgesen J. Critical elements of classroom and small-group instruction promote reading success in all children. Learning Disabilities Research and Practice. 2001;16(4):203–12. [Google Scholar]
- 10.Lovett MW, Steinbach KA, Frijters JC. Remediating the core deficits of developmental reading disability: a double-deficit perspective. J Learn Disabil. 2000;33(4):334–58. doi: 10.1177/002221940003300406. [DOI] [PubMed] [Google Scholar]
- 11.Astrom RL, Wadsworth SJ, DeFries JC. Etiology of the stability of reading difficulties: the longitudinal study of reading disabilities. Twin Res Hum Genet. 2007;10(3):434–9. doi: 10.1375/twin.10.3.434. [DOI] [PubMed] [Google Scholar]
- 12.DeFries JC, Fulker DW, LaBuda MC. Evidence for a genetic aetiology in reading disability of twins. Behav Genet. 1987;17(3):263–77. doi: 10.1038/329537a0. [DOI] [PubMed] [Google Scholar]
- 13.Meng H, Smith SD, Hager K, Held M, Liu J, Olson RK, Pennington BF, DeFries JC, Gelernter J, O'Reilly-Pol T, Somlo S, Skudlarski P, Shaywitz SE, Shaywitz BA, Marchione K, Wang Y, Paramasivam M, LoTurco JJ, Page GP, Gruen JR. DCDC2 is associated with reading disability and modulates neuronal development in the brain. Proc Natl Acad Sci U S A. 2005;102:17053–17058. doi: 10.1073/pnas.0508591102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Francks C, Paracchini S, Smith SD, Richardson AJ, Scerri TS, Cardon LR, Marlow AJ, MacPhie IL, Walter J, Pennington BF, Fisher SE, Olson RK, DeFries JC, Stein JF, Monaco AP. A 77-kilobase region of chromosome 6p22.2 is associated with dyslexia in families from the United Kingdom and from the United States. Am J Hum Genet. 2004;75:1046–1058. doi: 10.1086/426404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deffenbacher KE, Jenyon JB, Hoover DM, Olson RK, Pennington BF, DeFries JC, Smith SD. Refinement of the 6p21.3 quantitative trait locus influencing dyslexia: linkage and association analysis. Hum Genet. 2004;115(2):128–38. doi: 10.1007/s00439-004-1126-6. [DOI] [PubMed] [Google Scholar]
- 16.Nopola-Hemmi J, Taipale M, Haltia T, Lehesjoki AE, Voutilainen A, Kere J. Two translocations of chromosome 15q associated with dyslexia. J Med Genet. 2000;37(10):771–5. doi: 10.1136/jmg.37.10.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wigg KG, Couto JM, Feng Y, Anderson B, Cate-Carter TD, Macciardi F, Tannock R, Lovett MW, Humphries TW, Barr CL. Support for EKN1 as the susceptibility locus for dyslexia on 15q21. Mol Psychiatry. 2004;9(12):1111–21. doi: 10.1038/sj.mp.4001543. [DOI] [PubMed] [Google Scholar]
- 18.Anthoni H, Sucheston LE, Lewis BA, Tapia-Páez I, Fan X, Zucchelli M, Taipale M, Stein CM, Hokkanen ME, Castrén E, Pennington BF, Smith SD, Olson RK, Tomblin JB, Schulte-Körne G, Nöthen M, Schumacher J, Müller-Myhsok B, Hoffmann P, Gilger JW, Hynd GW, Nopola-Hemmi J, Leppanen PH, Lyytinen H, Schoumans J, Nordenskjöld M, Spencer J, Stanic D, Boon WC, Simpson E, Mäkelä S, Gustafsson JA, Peyrard-Janvid M, Iyengar S, Kere J. The aromatase gene CYP19A1: several genetic and functional lines of evidence supporting a role in reading, speech, and language. Behav Genet. 2012;42(4):509–27. doi: 10.1007/s10519-012-9532-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anthoni H, Zucchelli M, Matsson H, Müller-Myhsok B, Fransson I, Schumacher J, Massinen S, Onkamo P, Warnke A, Griesemann H, Hoffmann P, Nopola-Hemmi J, Lyytinen H, Schulte-Körne G, Kere J, Nöthen MM, Peyrard-Janvid M. A locus on 2p12 containing the co-regulated MRPL19 and C2orf3 genes is associated to dyslexia. Hum Mol Genet. 2007;16(6):667–77. doi: 10.1093/hmg/ddm009. [DOI] [PubMed] [Google Scholar]
- 20.Scerri TS, Darki F, Newbury DF, Whitehouse AJ, Peyrard-Janvid M, Matsson H, Ang QW, Pennell CE, Ring S, Stein J, Morris AP, Monaco AP, Kere J, Talcott JB, Kingberg T, Paracchini S. The dyslexia candidate locus on 2p12 is associated with general cognitive ability and white matter structure. PLoS One. 2012;7(11):e50312. doi: 10.1371/journal.pone.0050321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hannula-Jouppi K, Kaminen-Ahola N, Taipale M, Eklund R, Nopola-Hemmi J, Kääriäinen H, Kere J. The axon guidance receptor gene ROBO1 is a candidate gene for developmental dyslexia. PLoS Genet. 2005;1(4):e50. doi: 10.1371/journal.pgen.0010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bates TC, Luciano M, Medland SE, Montgomery GW, Wright MJ, Martin NG. Genetic variance in a component of the language acquisition device: ROBO1 polymorphisms associated with phonological buffer deficits. Behav Genet. 2011;41(1):50–7. doi: 10.1007/s10519-010-9402-9. [DOI] [PubMed] [Google Scholar]
- 23.Couto JM, Gomez L, Wigg K, Cate-Carter T, Archibald J, Anderson B, Tannock R, Kerr EN, Lovett MW, Humphries T, Barr CL. The KIAA0319-like (KIAA0319L) gene on chromosome 1p34 as a candidate for reading disabilities. J Neurogenet. 2008;22(4):295–313. doi: 10.1080/01677060802354328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peter B, Raskind WH, Matsushita M, Lisowski M, Vu T, Berninger VW, Wijsman EM, Brkanac Z. Replication of CNTNAP2 association with nonword repetition and support for FOXP2 association with timed reading and motor activities in a dyslexia family sample. J Neurodev Disord. 2011;3(1):39–49. doi: 10.1007/s11689-010-9065-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilcke A, Ligges C, Burkhardt J, Alexander M, Wolf C, Quente E, Ahnert P, Hoffmann P, Becker A, Muller-Myhsok B, Cichon S, Boltze J, Kirsten H. Imaging genetics of FOXP2 in dyslexia. Eur J Hum Genet. 2011;20(2):224–9. doi: 10.1038/ejhg.2011.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacDermot KD, Bonora E, Sykes N, Coupe AM, Lai CS, Vernes SC, Vargha- Khadem F, McKenzie F, Smith RL, Monaco AP, Fisher SE. Identification of FOXP2 truncation as a novel cause of developmental speech and language deficits. Am J Hum Genet. 2005;76(6):1074–80. doi: 10.1086/430841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaminen N, Hannula-Jouppi K, Kestilä M, Lahermo P, Muller K, Kaaranen M, Myllyluoma B, Voutilainen A, Lyytinen H, Nopola-Hemmi J, Kere J. A genome scan for developmental dyslexia confirms linkage to chromosome 2p11 and suggests a new locus on 7q32. J Med Genet. 2003;40(5):340–5. doi: 10.1136/jmg.40.5.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scerri TS, Morris AP, Buckingham LL, Newbury DF, Miller LL, Monaco AP, Bishop DV, Paracchini S. DCDC2, KIAA0319 and CMIP are associated with reading- related traits. Biol Psychiatry. 2011;70:237–245. doi: 10.1016/j.biopsych.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newbury DF, Paracchini S, Scerri TS, Winchester L, Addis L, Richardson AJ, Walter J, Stein JF, Talcott JB, Monaco AP. Investigation of dyslexia and SLI risk variants in reading- and language-impaired subjects. Behav Genet. 2011;41(1):90–104. doi: 10.1007/s10519-010-9424-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith SD. Genes, language development, and language disorders. Ment Retard Dev Disabil Res Rev. 2007;13(1):96–105. doi: 10.1002/mrdd.20135. [DOI] [PubMed] [Google Scholar]
- 31.Graham SA, Fisher SE. Decoding the genetics of speech and language. Curr Opin Neurobiol. 2013;23(1):43–51. doi: 10.1016/j.conb.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 32.Pennington BF, Bishop DV. Relations among speech, language, and reading disorders. Annu Rev Psychol. 2009;60:283–306. doi: 10.1146/annurev.psych.60.110707.163548. [DOI] [PubMed] [Google Scholar]
- 33.Gibson CJ, Gruen JR. The human lexinome: genes of language and reading. J Commun Disord. 2008;41(5):409–20. doi: 10.1016/j.jcomdis.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newbury DF, Monaco AP. Genetic advances in the study of speech and language disorders. Neuron. 2010;68(2):309–20. doi: 10.1016/j.neuron.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eliez S, Rumsey JM, Giedd JN, Schmitt JE, Patwardhan AJ, Reiss AL. Morphological alteration of temporal lobe gray matter in dyslexia: an MRI study. J Child Psychol Psychiatry. 2000;41(5):637–44. doi: 10.1111/1469-7610.00650. [DOI] [PubMed] [Google Scholar]
- 36.Klingberg T, Hedehus M, Temple E, Salz T, Gabrieli JD, Moseley ME, Poldrack RA. Microstructure of temporo-parietal white matter as a bassi for reading ability: evidence from diffusion tensor magnetic resonance imaging. Neuron. 2000;25(2):493–500. doi: 10.1016/s0896-6273(00)80911-3. [DOI] [PubMed] [Google Scholar]
- 37.Brown WE, Eliez S, Menon V, Rumsey JM, White CD, Reiss AL. Preliminary evidence of widespread morphological variations of the brain in dyslexia. Neurology. 2001;56:781–783. doi: 10.1212/wnl.56.6.781. [DOI] [PubMed] [Google Scholar]
- 38.Paulesu E, Démonet JF, Fazio F, McCrory E, Chanoine V, Brunswick N, Cappa SF, Cossu G, Habib M, Frith CD, Frith U. Dyslexia: cultural diversity and biological unity. Science. 2001;291(5511):2165–7. doi: 10.1126/science.1057179. [DOI] [PubMed] [Google Scholar]
- 39.Brambati SM, Termine C, Ruffino M, Stella G, Fazio F, Cappa SF, Perani D. Regional reductions of gray matter volume in familial dyslexia. Neurology. 2004;63(4):742–5. doi: 10.1212/01.wnl.0000134673.95020.ee. [DOI] [PubMed] [Google Scholar]
- 40.Silani G, Frith U, Demonet JF, Fazio F, Perani D, Price C, Frith CD, Paulesu E. Brain abnormalities underlying altered activation in dyslexia: a voxel based morphometry study. Brain. 2005;128(Pt 10):2453–61. doi: 10.1093/brain/awh579. [DOI] [PubMed] [Google Scholar]
- 41.Vinckenbosch E, Robichon F, Eliez S. Gray matter alteration in dyslexia: converging evidence from volumetric and voxel-by-voxel MRI analyses. Neuropsychologia. 2005;43(4):324–31. doi: 10.1016/j.neuropsychologia.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 42.Deutsch GK, Dougherty RF, Bammer R, Siok WT, Gabrieli JD, Wandell B. Children's reading performance is correlated with white matter structure measured by diffusion tensor imaging. Cortex. 2005;41(3):354–63. doi: 10.1016/s0010-9452(08)70272-7. [DOI] [PubMed] [Google Scholar]
- 43.Beaulieu C, Plewes C, Paulson LA, Roy D, Snook L, Concha L, Phillips L. Imaging brain connectivity in children with diverse reading ability. Neuroimage. 2005;25(4):1226–71. doi: 10.1016/j.neuroimage.2004.12.053. (2005) [DOI] [PubMed] [Google Scholar]
- 44.Niogi SN, McCandliss BD. Left lateralized white matter microstructure accounts for individual differences in reading ability and disability. Neuropsychologia. 2006;44(11):2178–88. doi: 10.1016/j.neuropsychologia.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 45.Hoeft F, Meyler A, Hernandez A, Juel C, Taylor-Hill H, Martindale JL, McMillon G, Kolchugina G, Black JM, Faizi A, Deutsch GK, Siok WT, Reiss AL, Whitfield-Gabrieli S, Gabrieli JD. Functional and morphometric brain dissociation between dyslexia and reading ability. Proc Natl Acad Sci U S A. 2007;104(10):4234–9. doi: 10.1073/pnas.0609399104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gebauer D, Enzinger C, Kronbichler M, Schurz M, Reishofer G, Koschutnig K, Kargl R, Purgstaller C, Fazekas F, Fink A. Distinct patterns of brain function inchildren with isolated spelling impairment: new insights. Neuropsuchologia. 2012;50(7):1353–61. doi: 10.1016/j.neuropsychologia.2012.02.020. [DOI] [PubMed] [Google Scholar]
- 47.Vandermosten M, Boets B, Poelmans H, Sunaert S, Wouters J, Ghesquiere P. A tractography study in dyslexia: neuroanatomic correlates of orthographic, phonological and speech processing. Brain. 2012;135(Pt 3):935–48. doi: 10.1093/brain/awr363. [DOI] [PubMed] [Google Scholar]
- 48.Rumsey JM, Andreason P, Zametkin AJ, Aquino T, King AC, Hamburger SD, Pikus A, Rapoport JL, Cohen RM. Failure to activate the left temporoparietal cortex in dyslexia. An oxygen 15 positron emission tomographic study. Arch Neurol. 1992;49(5):527–34. doi: 10.1001/archneur.1992.00530290115020. [DOI] [PubMed] [Google Scholar]
- 49.Paulesu E, Frith U, Snowling M, Gallagher A, Morton J, Frackowiak RS, Frith CD. Is developmental dyslexia a disconnection syndrome? Evidence from PET scanning. Brain. 1996;119(Pt 1):143–57. doi: 10.1093/brain/119.1.143. [DOI] [PubMed] [Google Scholar]
- 50.Rumsey JM, Nace K, Donohue B, Wise D, Maisog JM, Andreason P. A positron emission tomographic study of impaired word recognition and phonological processing in dyslexic men. Arch Neurol. 1997;54(5):562–73. doi: 10.1001/archneur.1997.00550170042013. [DOI] [PubMed] [Google Scholar]
- 51.Shaywitz SE, Shaywitz BA, Pugh KR, Fulbright RK, Constable RT, Mencl WE, Shankweiler DP, Liberman AM, Skudlarski P, Fletcher JM, Katz L, Marchione KE, Lacadie C, Gatenby C, Gore JC. Functional disruption in the organization of the brain for reading in dyslexia. Proc Natl Acad Sci U S A. 1998;95(5):2636–41. doi: 10.1073/pnas.95.5.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Horwitz B, Rumsey JM, Donohue BC. Functional connectivity of the angular gyrus in normal reading and dyslexia. Proc Natl Acad Sci USA. 1998;95:8939–8944. doi: 10.1073/pnas.95.15.8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brunswick N, McCrory E, Price CJ, Frith CD, Frith U. Explicit and implicit processing of words and pseudowords by adult developmental dyslexics: A search for Wernicke's Wortschatz? Brain. 1999;122(Pt 10):1901–17. doi: 10.1093/brain/122.10.1901. [DOI] [PubMed] [Google Scholar]
- 54.Steinbrink C, Groth K, Lachmann T, Riecker A. Neural correlates of temporal auditory processing developmental dyslexia during German vowel length discrimination: an fMRI study. Brain Lang. 2011;121(1):1–11. doi: 10.1016/j.bandl.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 55.Raschle NM, Zuk J, Gaab N. Functional characteristics of developmental dyslexia in left-hemispheric posterior brain regions predate reading onset. Proc Natl Acad Sci U S A. 2012;109(6):2156–61. doi: 10.1073/pnas.1107721109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Welcome SE, Joanisse MF. Individual differences in skilled adult readers reveal dissociable patterns of neural activity associated with component processes of reading. Brian Lang. 2012;120(3):360–71. doi: 10.1016/j.bandl.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 57.Maisog JM, Einbinder ER, Flowers DL, Turkeltaub PE, Eden GF. A meta-analysis of functional neuroimaging of dyslexia. Ann N Y Acad Sci. 2008;1145:237–59. doi: 10.1196/annals.1416.024. [DOI] [PubMed] [Google Scholar]
- 58.Shaywitz SE, Shaywitz BA. Paying attention to reading: the neurpbiology of reading and dyslexia. Dev Psychopathol. 2008;20(4):1329–49. doi: 10.1017/S0954579408000631. [DOI] [PubMed] [Google Scholar]
- 59.Cardon LR, Smith SD, Fulker DW, Kimberling WJ, Pennington BF, DeFries JC. Quantitative trait locus for reading disability on chromosome 6. Science. 1994;266(5183):276–9. doi: 10.1126/science.7939663. [DOI] [PubMed] [Google Scholar]
- 60.Gayán J, Smith SD, Cherny SS, Cardon LR, Fulker DW, Brower AM, Olson RK, Pennington BF, DeFries JC. Quantitative-trait locus for specific language and reading deficits on chromosome 6p. Am J Hum Genet. 1999;64(1):157–64. doi: 10.1086/302191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaplan DE, Gayán J, Ahn J, Won TW, Pauls D, Olson RK, DeFries JC, Wood F, Pennington BF, Page GP, Smith SD, Gruen JR. Evidence for linkage and association with reading disability on 6p21.3-22. Am J Hum Genet. 2002;70(5):1287–98. doi: 10.1086/340449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Deffenbacher KE, Kenyon JB, Hoover DM, Olson RK, Pennington BF, DeFries JC, Smith SD. refinement of the 6p21.3 quantitative trait locus influencing dyslexia: linkage and association analyses. Hum Genet. 2004;115(2):128–38. doi: 10.1007/s00439-004-1126-6. [DOI] [PubMed] [Google Scholar]
- 63.Meng H, Smith SD, Hager K, Held M, Liu J, Olson RK, Pennington BF, DeFries JC, Gelernter J, O'Reilly-Pol T, Somlo S, Skudlarski P, Shaywitz SE, Shaywitz BA, Marchione K, Wang Y, Paramasivam M, LoTurco JJ, Page GP, Gruen JR. DCDC2 is associated with reading disability and modulates neuronal development in the brain. Proc Natl Acad Sci U S A. 2005;102(47):17053–8. doi: 10.1073/pnas.0508591102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cope N, Harold D, Hill G, Moskvina V, Stevenson J, Holmans P, Owen MJ, O'Donovan MC, Williams J. Strong evidence that KIAA0319 on chromosome 6p is a susceptibility gene for developmental dyslexia. Am J Hum Genet. 2005;76(4):581–91. doi: 10.1086/429131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schumacher J, Anthoni H, Dahdouh F, König IR, Hillmer AM, Kluck N, Manthey M, Plume E, Warnke A, Remschmidt H, Hülsmann J, Cichon S, Lindgren CM, Propping P, Zucchelli M, Ziegler A, Peyrard-Janvid M, Schulte-Körne G, Nöthen MM, Kere J. Strong genetic evidence of DCDC2 as a susceptibility gene for dyslexia. Am J Hum Genet. 2006;78(1):52–62. doi: 10.1086/498992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harold D, Paracchini S, Scerri T, Dennis M, Cope N, Hill G, Moskvina V, Walter J, Richardson AJ, Owen MJ, Stein JF, Green ED, O'Donovan MC, Williams J, Monaco AP. Further evidence that the KIAA0319 gene confers susceptibility to developmental dyslexia. Mol Psychiatry. 2006;11(12):1085–91. doi: 10.1038/sj.mp.4001904. [DOI] [PubMed] [Google Scholar]
- 67.Couto JM, Livne-Bar I, Huang K, Xu Z, Cate-Carter T, Feng Y, Wigg K, Humphries T, Tannock R, Kerr EN, Lovett MW, Bremner R, Barr CL. Association of reading disabilities with regions marked by acetylated H3 histones in KIAA0319. Am J Med Genet B Neuropsychatr Genet. 2010;153B(2):447–62. doi: 10.1002/ajmg.b.30999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lind PA, Luciano M, Wright MJ, Montgomery GW, Martin NG, Bates TC. Dyslexia and DCDC2: normal variation in reading and spelling is associated with DCDC2 polymorphisms in an Australian population sample. Eur J Hum Genet. 2010;18(6):668–73. doi: 10.1038/ejhg.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhong R, Yang B, Tang H, Zou L, Song R, Zhu LQ, Miao X. Meta-analysis of the association between DCDC2 polymorphisms and risk of dyslexia. Mol Neurobiol. 2013;47(1):435–42. doi: 10.1007/s12035-012-8381-7. [DOI] [PubMed] [Google Scholar]
- 70.Luciano M, Lind PA, Duffy DL, Castles A, Wright MJ, Montgomery GW, Martin NG, Bates TC. A haplotype spanning KIAA0319 and TTRAP is associated with normal variation in reading and spelling ability. Biol Psychiatry. 2007;62:811–817. doi: 10.1016/j.biopsych.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 71.Zou L, Chen W, Shao S, Sun Z, Zhong R, Shi J, Miao X, Song R. Genetic variant in KIAA0319, but not in DYX1C1, is associated with risk of dyslexia: an integrated meta-analysis. Am J Med Genet B Neuropsychiatr Genet. 2012;159B(8):970–6. doi: 10.1002/ajmg.b.32102. [DOI] [PubMed] [Google Scholar]
- 72.Elbert A, Lovett MW, Cate-Carter T, Pitch A, Kerr EN, Barr CL. Genetic variation in the KIAA0319 5’ region as a possible contributor to dyslexia. Behav Genet. 2011;41(1):77–89. doi: 10.1007/s10519-010-9434-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Powers NR, Eicher JD, Butter F, Kong Y, Miller LL, Ring SM, Mann M, Gruen JR. Alleles of a polymorphic ETV6 binding site in DCDC2 confer risk of reading and language impairment. Am J Hum Genet. 2013 doi: 10.1016/j.ajhg.2013.05.008. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gabel LA, Marin I, LoTurco JJ, Che A, Murphy C, Manglani M, Kass S. Mutation of the dyslexia-associated gene Dcdc2 impairs LTM and visuo-spaatial performance in mice. Genes Brain Behav. 2011;10(8):868–75. doi: 10.1111/j.1601-183X.2011.00727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Burbridge TJ, Wang Y, Volz AJ, Peschansky VJ, Lisann L, Galaburda AM, LoTurco JJ, Rosen GD. Postnatal analysis of the effect of embryonic knockdown and overexpression of candidate dyslexia susceptibility gene homolog Dcdc2 in the rat. Neuroscience. 2008;152(3):723–33. doi: 10.1016/j.neuroscience.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Szalkowski CE, Fiondella CG, Galaburda AM, Rosen GD, Loturco JJ, Fitch RH. Neocortical disruption and behavioral impairments in rats following in utero RNAi of candidate dyslexia risk gene Kiaa0319. Int J Dev Neurosci. 2012;30(4):293–302. doi: 10.1016/j.ijdevneu.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Paracchini S, Thomas A, Castro S, Lai C, Paramasivam M, Wang Y, Keating BJ, Taylor JM, Hacking DF, Scerri T, Francks C, Richardson AJ, Wade-Martins R, Stein JF, Knight JC, Copp AJ, LoTurco J, Monaco AP. The chromosome 6p22 haplotype associated with dyslexia reduces the expression of KIAA0319, a novel gene involved in neuronal migration. Hum Mol Genet. 2006;15:1659–1666. doi: 10.1093/hmg/ddl089. [DOI] [PubMed] [Google Scholar]
- 78.Peschansky VJ, Burbridge TJ, Volz AJ, Fiondella C, Wissner-Gross Z, Galaburda AM, LoTurco JJ, Rosen GD. The effect of variation in expression of the candidate dyslexia susceptibility gene homolog Kiaa0319 on neuronal migration and dendritic morphology in the rat. Cereb Cortex. 2010;20(4):884–97. doi: 10.1093/cercor/bhp154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Szalkowski CE, Fiondella CF, Truong DT, Rosen GD, LoTurco JJ, Fitch RH. The effects of Kiaa0319 knockdown on cortical and subcortical anatomy in male rats. Int J Dev Neurosci. 2013;31(2):116–22. doi: 10.1016/j.ijdevneu.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Massinen S, Hokkanen ME, Matsson H, Tammimies K, Tapia-Páez I, Dahlström-Heuser V, Kuja-Panula J, Burghoorn J, Jeppsson KE, Swoboda P, Peyrard-Janvid M, Toftgård R, Castrén E, Kere J. Increased expression of the dyslexia candidate gene DCDC2 affects length and signaling of primary cilia in neurons. PLoS One. 2011;6(6):e20580. doi: 10.1371/journal.pone.0020580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meda SA, Gelernter J, Gruen JR, Calhoun VD, Meng H, Cope NA, Pearlson GD. Polymorphism of DCDC2 reveals differences in cortical morphology of healthy individuals-a preliminary voxel based morphometry study. Brian Imaging Behav. 2008;2(1):21–26. doi: 10.1007/s11682-007-9012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Meng H, Powers NR, Tang L, Cope NA, Zhang PX, Fuleihan R, Gibson C, Page GP, Gruen JR. A dyslexia-associated variant in DCDC2 changes gene expression. Behav Genet. 2011;41:58–66. doi: 10.1007/s10519-010-9408-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marino C, Meng H, Mascheretti S, Rusconi M, Cope N, Giorda R, Molteni M, Gruen JR. DCDC2 genetic variants and susceptibility to developmental dyslexia. Psychiatr Genet. 2012;22:25–30. doi: 10.1097/YPG.0b013e32834acdb2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wilcke A, Weissfuss J, Kirsten H, Wolfram G, Boltze J, Ahnert P. The role of gene DCDC2 in German dyslexics. Ann Dyslexia. 2009;59:1–11. doi: 10.1007/s11881-008-0020-7. [DOI] [PubMed] [Google Scholar]
- 85.Jamadar S, Powers NR, Meda SA, Gelernter J, Gruen JR, Pearlson GD. Genetic influences of cortical gray matter in language-related regions in healthy controls and schizophrenia. Schizophr Res. 2011;129(2-3):141–8. doi: 10.1016/j.schres.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Darki F, Peyrard-Janvid M, Matsson H, Kere J, Klingberg T. Three dyslexia susceptibility genes, DYX1C1, DCDC2, and KIAA0319, affect temporo-parietal white matter structure. Biol Psychiatry. 2012;72(8):671–6. doi: 10.1016/j.biopsych.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 87.Cope N, Eicher JD, Meng H, Gibson CJ, Hager K, Lacadie C, Fulbright RK, Constable RT, Page GP, Gruen JR. Variants in the DYX2 locus are associated with altered brain activation in reading-related brain regions in subjects with reading disability. Neuroimage. 2012;63(1):148–56. doi: 10.1016/j.neuroimage.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jamadar S, Powers NR, Meda SA, Calhoun VD, Gelernter J, Gruen JR, Pearlson GD. Genetic influences of resting state fMRI activity in language-related brain regions in healthy controls and schizophrenia patients: a pilot study. Brain Imaging Behav. 2013;7(1):15–27. doi: 10.1007/s11682-012-9168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pinel P, Fauchereau F, Moreno A, Barbot A, Lathrop M, Zelenika D, LeBihan D, Poline JB, Bourgeron T, Dehaene S. Genetics variants of FOXP2 and KIAA0319/TTRAP/THEM2 locus are associated with altered brain activation in distinct language-related regions. J Neurosci. 2012;32(3):817–25. doi: 10.1523/JNEUROSCI.5996-10.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Paracchini S, Steer CD, Buckingham LL, Morris AP, Ring S, Scerri T, Stein J, Pembrey ME, Ragoussis J, Golding J, Monaco AP. Association of the KIAA0319 dyslexia susceptibility gene with reading skills in the general population. Am J Psychiatry. 2008;165:1576–1584. doi: 10.1176/appi.ajp.2008.07121872. [DOI] [PubMed] [Google Scholar]
- 91.Dennis MY, Paracchini S, Scerri TS, Prokunina-Olsson L, Knight JC, Wade-Martins R, Coggill P, Beck S, Green ED, Monaco AP. A common variant associated with dyslexia reduces expression of the KIAA0319 gene. PLoS Genet. 2009;5:e1000436. doi: 10.1371/journal.pgen.1000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ludwig KU, Schumacher J, Schulte-Korne G, Konig IR, Warnke A, Plume E, Anthoni H, Peyrard-Janvid M, Meng H, Ziegler A, Remschmidt H, Kere J, Gruen JR, Muller-Myhsok B, Nothen MM, Hoffmann P. Investigation of the DCDC2 intron 2 deletion/compound short tandem repeat polymorphism in a large German dyslexia sample. Psychiatr Genet. 2008;18:310–312. doi: 10.1097/YPG.0b013e3283063a78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fisher SE, Vargha-Khadem F, Watkins KE, Monaco AP, Pembrey ME. Localisation of a gene implicated in a severe speech and language disorder. Nat Genet. 1998;18(2):168–70. doi: 10.1038/ng0298-168. [DOI] [PubMed] [Google Scholar]
- 94.Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead- domain gene is mutated in a severe speech and language disorder. Nature. 2001;413(6855):519–23. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- 95.Watkins KE, Dronkers NF, Vargha-Khadem F. Behavioural analysis of an inherited speech and language disorder: comparison with acquired aphasia. Brain. 2012;125(Pt 3):452–64. doi: 10.1093/brain/awf058. [DOI] [PubMed] [Google Scholar]
- 96.Vernes SC, Nicod J, Elahi FM, Coventry JA, Kenny N, Coupe AM, Bird LE, Davies KE, Fisher SE. Functional genetic analysis of mutations implicated in a human speech and language disorder. Hum Mol Genet. 2006;15(21):3154–67. doi: 10.1093/hmg/ddl392. [DOI] [PubMed] [Google Scholar]
- 97.French CA, Groszer M, Preece C, Coupe AM, Rajewsky K, Fisher SE. Generation of mice with a conditional Foxp2 null allele. Genesis. 2007;45(7):440–6. doi: 10.1002/dvg.20305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fujita E, Tanabe Y, Shiota A, Ueda M, Suwa K, Momoi MY, Momoi T. Ultrasonic vocalization impairment of Foxp2 (R552H) knockin mice related to speech-language disorder and abnormality of Purkinje cells. Prco Natl Sci U S A. 2008;105(8):3117–22. doi: 10.1073/pnas.0712298105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Groszer M, Keays DA, Deacon RM, de Bono JP, Prasad-Mulcare S, Gaub S, Baum MG, French CA, Nicod J, Coventry JA, Enard W, Fray M, Brown SD, Nolan PM, Pääbo S, Channon KM, Costa RM, Eilers J, Ehret G, Rawlins JN, Fisher SE. Impaired synaptic plasticity and motor learning in mice with a point mutation implicated in human speech deficits. Curr Biol. 2008;18(5):354–62. doi: 10.1016/j.cub.2008.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vernes SC, Spiteri E, Nicod J, Groszer M, Taylor JM, Davies KE, Geschwind DH, Fisher SE. high-throughput analysis of promoter occupancy reveals direct neural targets of FOXP2, a gene mutated in speech and language disorders. Am J Hum Genet. 2007;81(6):1232–50. doi: 10.1086/522238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hilliard AT, Miller JE, Fraley ER, Horvath S, White SA. Molecular microcircuitry underlies functional specification in a basal ganglia circuit dedicated to vocal learning. Neuron. 2012;73(3):537–52. doi: 10.1016/j.neuron.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schulz SB, Haesler S, Scharff C, Rochefort C. Knockdown of FoxP2 alters spine density in Area X of the zebra finch. Genes Brain Behav. 2010;9(7):732–40. doi: 10.1111/j.1601-183X.2010.00607.x. [DOI] [PubMed] [Google Scholar]
- 103.Enard W, Przeworski M, Fisher SE, Lai CS, Wiebe V, Kitano T, Monaco AP, Pääbo S. Molecular evolution of FOXP2, a gene involved in speech and language. Nature. 2002;418(6900):869–72. doi: 10.1038/nature01025. [DOI] [PubMed] [Google Scholar]
- 104.Enard W. FOXP2 and the role of cortico-basal ganglia circuits in speech and language evolution. Curr Opin Neurobiol. 2011;21(3):415–24. doi: 10.1016/j.conb.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 105.Krause J, Lalueza-Fox C, Orlando L, Enard W, Green RE, Burbano HA, Hublin JJ, Hänni C, Fortea J, de la Rasilla M, Bertranpetit J, Rosas A, Pääbo S. The derived FOXP2 variant of modern humans was shared with Neandertals. Curr Biol. 2007;17(21):1908–12. doi: 10.1016/j.cub.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 106.Ptak SE, Enard W, Wiebe V, Hellmann I, Krause J, Lachmann M, Pääbo S. Linkage disequilibrium extends across putative selected sites in FOXP2. Mol Biol Evol. 2009;26(10):2181–4. doi: 10.1093/molbev/msp143. [DOI] [PubMed] [Google Scholar]
- 107.Coop G, Bullaughey K, Luca F, Przeworski M. The timing of selection at the human FOXP2 gene. Mol Biol Evol. 2008;25(7):1257–9. doi: 10.1093/molbev/msn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tsui D, Vessey JP, Tomita H, Kaplan DR, Miller FD. FoxP2 regulated neurogenesis during embryonic cortical development. J Neurosci. 2013;33(1):244–58. doi: 10.1523/JNEUROSCI.1665-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shu W, Cho JY, Jiang Y, Zhang M, Weisz D, Elder GA, Schmeidler J, De Gasperi R, Sosa MA, Rabidou D, Santucci AC, Perl D, Morrisey E, Buxbaum JD. Altered ultrasonic vocalization in mice with a disruption in the Foxp2 gene. Proc Natl Acad Sci U S A. 2005;102(27):9643–8. doi: 10.1073/pnas.0503739102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gaub S, Groszer M, Fisher SE, Ehret G. The structure of innate vocalizations in Foxp2-deficient mouse pups. Genes Brain Behav. 2010;9(4):390–401. doi: 10.1111/j.1601-183X.2010.00570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Enard W, Gehre S, Hammerschmidt K, Hölter SM, Blass T, Somel M, Brückner MK, Schreiweis C, Winter C, Sohr R, Becker L, Wiebe V, Nickel B, Giger T, Müller U, Groszer M, Adler T, Aguilar A, Bolle I, Calzada-Wack J, Dalke C, Ehrhardt N, Favor J, Fuchs H, Gailus-Durner V, Hans W, Hölzlwimmer G, Javaheri A, Kalaydjiev S, Kallnik M, Kling E, Kunder S, Mossbrugger I, Naton B, Racz I, Rathkolb B, Rozman J, Schrewe A, Busch DH, Graw J, Ivandic B, Klingenspor M, Klopstock T, Ollert M, Quintanilla-Martinez L, Schulz H, Wolf E, Wurst W, Zimmer A, Fisher SE, Morgenstern R, Arendt T, de Angelis MH, Fischer J, Schwarz J, Pääbo S. A humanized version of Foxp2 affects cortico- basal ganglia circuits in mice. Cell. 2009;137(5):961–71. doi: 10.1016/j.cell.2009.03.041. [DOI] [PubMed] [Google Scholar]
- 112.Reimers-Kipping S, Hevers W, Pääbo S, Enard W. Humanized Foxp2 specifically affects cortico-based ganglia circuits. Neuroscience. 2011;175:75–84. doi: 10.1016/j.neuroscience.2010.11.042. [DOI] [PubMed] [Google Scholar]
- 113.Belton E, Salmond CH, Watkins KE, Vargha-Khadem F, Gadian DG. Bilateral brain abnormalities associated with dominantly inherited verbal and orofacial dyspraxia. Hum Brain Mapp. 2003;18(3):194–200. doi: 10.1002/hbm.10093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Watkins KE, Vargha-Khadem F, Ashburner J, Passingham RE, Connelly A, Friston KJ, Frackowiak RS, Mishkin M, Gadian DG. MRI analysis of an inherited speech and language disorder: structural brain abnormalities. Brain. 2002;125(pt 3):465–78. doi: 10.1093/brain/awf057. [DOI] [PubMed] [Google Scholar]
- 115.Vargha-Khadem F, Watkins KE, Price CJ, Ashburner J, Alcock KJ, Connelly A, Frackowiak RS, Friston KJ, Pembrey ME, Mishkin M, Gadian DG, Passingham RE. Neural basis of an inherited speech and language disorder. Proc Natl Acad Sci U S A. 1998;95(21):12695–700. doi: 10.1073/pnas.95.21.12695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liegeois F, Baldeweg T, Connelly A, Gadian DG, Mishkin M, Vargha-Khadem F. Language fMRI abnormalities associated with FOXP2 gene mutation. Nat Neurosci. 2003;6(11):1230–7. doi: 10.1038/nn1138. [DOI] [PubMed] [Google Scholar]
- 117.Vernes SC, Newbury DF, Abrahams BS, Winchester L, Nicod J, Groszer M, Alarcón M, Oliver PL, Davies KE, Geschwind DH, Monaco AP, Fisher SE. A functional genetic link between distinct developmental language disorders. N Engl J Med. 2008;359(22):2337–45. doi: 10.1056/NEJMoa0802828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vernes SC, Oliver PL, Spiteri E, Lockstone HE, Puliyadi R, Taylor JM, Ho J, Mombereau C, Brewer A, Lowy E, Nicod J, Groszer M, Baban D, Sahgal N, Cazier JB, Ragoussis J, Davies KE, Geschwind DH, Fisher SE. Foxp2 regulates gene networks implicated in neurite outgrowth in the developing brain. PLoS Genet. 2011;7(7):e1002145. doi: 10.1371/journal.pgen.1002145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Verkerk AJ, Mathews CA, Joosse M, Eussen BH, Heutink P, Oostra BA, Tourette Syndrome Association International Consortium for Genetics CNTNAP2 is disrupted ina family with Gilles de la Tourette syndrome and obsessive compulsive disorder. Genomics. 2003;82(1):1–9. doi: 10.1016/s0888-7543(03)00097-1. [DOI] [PubMed] [Google Scholar]
- 120.Belloso JM, Bache I, Guitart M, Caballin MR, Halgren C, Kirchhoff M, Ropers HH, Tommerup N, Tümer Z. Disruption of the CNTNAP2 gene in a t(7;15) translocation family without symptoms of Gilees ed la Tourette syndrome. Eur J Hum Genet. 2007;15(6):711–3. doi: 10.1038/sj.ejhg.5201824. [DOI] [PubMed] [Google Scholar]
- 121.Friedman JI, Vrijenhoek T, Markx S, Janssen IM, van der Vliet WA, Faas BH, Knoers NV, Cahn W, Kahn RS, Edelmann L, Davis KL, Silverman JM, Brunner HG, van Kessel AG, Wijmenga C, Ophoff RA, Veltman JA. CNTNAP2 gene dosage variation is associated with schizophrenia and epilepsy. Mol Psychiatry. 2008;13(3):261–6. doi: 10.1038/sj.mp.4002049. [DOI] [PubMed] [Google Scholar]
- 122.Alarcón M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, Bomar JM, Sebat J, Wigler M, Martin CL, Ledbetter DH, Nelson SF, Cantor RM, Geschwind DH. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism- susceptibility gene. Am J Hum Genet. 2008;82(1):150–9. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Arking DE, Cutler DJ, Brune CW, Teslovich TM, West K, Ikeda M, Rea A, Guy M, Lin S, Cook EH, Chakravarti A. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet. 2008;82(1):160–4. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bakkaloglu B, O'Roak BJ, Louvi A, Gupta AR, Abelson JF, Morgan TM, Chawarska K, Klin A, Ercan-Sencicek AG, Stillman AA, Tanriover G, Abrahams BS, Duvall JA, Robbins EM, Geschwind DH, Biederer T, Gunel M, Lifton RP, State MW. Molecular cytogenetic analysis and resequencing of contactin associated protein-liked 2 in autism spectrum disorders. Am J Hum Genet. 2008;82(1):165–73. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Whitehouse AJ, Bishop DV, Ang QW, Pennell CE, Fisher SE. CNTNAP2 variants affect early language development in the general population. Genes Brain Behav. 2011;10(4):451–6. doi: 10.1111/j.1601-183X.2011.00684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Abrahams BS, Tentler D, Perederiy JV, Oldham MC, Coppola G, Geschwind DH. Genome-wide analyses of human perisylvian cerebral cortical patterning. Prco Natl Acad U S A. 2007;104(45):17849–54. doi: 10.1073/pnas.0706128104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Poliak S, Gollan L, Martinez R, Custer A, Einheber S, Salzer JL, Trimmer JS, Shrager P, Peles E. Caspr2, a new member of the neurexin superfamily, is localized at the juxtaparanodes of myelinated axons and associates with K+ channels. Neuron. 1999;24(4):1037–47. doi: 10.1016/s0896-6273(00)81049-1. [DOI] [PubMed] [Google Scholar]
- 128.Poliak S, Salomon D, Elhanany H, Sabanay H, Kiernan B, Pevny L, Stewart CL, Xu X, Chiu SY, Shrager P, Furley AJ, Peles E. Juxtaparanodal clustering of Shaker-like K+ channels in myelinated axons depends on Caspr2 and TAG-1. J Cell Biol. 2003;162(6):1149–60. doi: 10.1083/jcb.200305018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Peñagarikano O, Abrahams BS, Herman EI, Winden KD, Gdalyahu A, Dong H, Sonnenblick LI, Gruver R, Almajano J, Bragin A, Golshani P, Trachtenberg JT, Peles E, Geschwind DH. Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell. 2011;147(1):235–46. doi: 10.1016/j.cell.2011.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Corfas G, Velardez MO, Ko CP, Ratner N, Peles E. Mechanisms and roles of axon-Schwann cell interactions. J Neurosci. 2004;24(42):9250–60. doi: 10.1523/JNEUROSCI.3649-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tan GC, Doke TF, Ashburner J, Wood NW, Frackowiak RS. Normal variation in fronto-occipital circuitry and cerebellar structure with an autism-associated polymorphism of CNTNAP2. Neuroimage. 2010;53(3):1030–42. doi: 10.1016/j.neuroimage.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Scott-Van Zeeland AA, Abrahams BS, Alvarez-Retuerto AI, Sinnenblick LI, Rudie JD, Ghahremani D, Mumford JA, Poldrack RA, Dapretto M, Geschwind DH, Bookheimer SY. Altered functional connectivity in frontal lobe circuits is associated with variation in the autism risk gene CNTNAP2. Sci Transl Med. 2010;2(56):56ra80. doi: 10.1126/scitranslmed.3001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Whalley HC, O'Connell G, Sussmann JE, Peel A, Stanfield AC, Hayiou-Thomas ME, Johnstone EC, Lawrie SM, McIntosh AM, Hall J. Genetic variation in CNTNAP2 alters brain function during linguistic processing in healthy individuals. Am J Med Genet B Neuropsychiatr Genet. 2011;156B(8):941–8. doi: 10.1002/ajmg.b.31241. [DOI] [PubMed] [Google Scholar]
- 134.Marino C, Giorda R, Luisa Lorusso M, Vanzin L, Salandi N, Nobile M, Citterio A, Beri S, Crespi V, Battaglia M, Molteni M. A family-based association study does not support DYX1C1 on 15q21.3 as a candidate gene in developmental dyslexia. Eur J Hum Genet. 2005;13(4):491–9. doi: 10.1038/sj.ejhg.5201356. [DOI] [PubMed] [Google Scholar]
- 135.Meng H, Hager K, Held M, Page GP, Olson RK, Pennington BF, DeFries JC, Smith SD, Gruen JR. TDT-association analysis of EKN1 and dyslexia in a Colorado twin cohort. Hum Genet. 2005;118:87–90. doi: 10.1007/s00439-005-0017-9. [DOI] [PubMed] [Google Scholar]
- 136.Bellini G, Bravaccio C, Calamoneri F, Donatella Cocuzza M, Fiorillo P, Gagliano A, Mazzone D, del Giudice EM, Scuccimarra G, Militerni R, Pascotto A. No evidence for association between dyslexia and DYX1C1 functional variants in a group of children and adolescents from Southern Italy. J Mol Neurosci. 2005;27(3):311–4. doi: 10.1385/jmn:27:3:311. [DOI] [PubMed] [Google Scholar]
- 137.Marino C, Citterio A, Giorda R, Facoetti A, Menozzi G, Vanzin L, Lorusso ML, Nobile M, Molteni M. Association of short-term memory with a variant within DYX1C1 in developmental dyslexia. Genes Brain Behav. 2007;6(7):640–6. doi: 10.1111/j.1601-183X.2006.00291.x. [DOI] [PubMed] [Google Scholar]
- 138.Brkanac Z, Chapman NH, Matsushita MM, Chun L, Nielsen K, Cochrane E, Berninger VW, Wijsman EM, Raskind WH. Evaluation of candidate genes for DYX1 and DYX2 in families with dyslexia. Am J Med Genet B Neuropsychatr Genet. 2007;144B(4):556–60. doi: 10.1002/ajmg.b.30471. [DOI] [PubMed] [Google Scholar]
- 139.Wang Y, Paramasivam M, Thomas A, Bai J, Kaminen-Ahola N, Kere J, Voskuil J, Rosen GD, Galaburda AM, Loturco JJ. DYX1C1 functions in neuronal migration in developing neocortex. Neuroscience. 2006;143(2):515–22. doi: 10.1016/j.neuroscience.2006.08.022. [DOI] [PubMed] [Google Scholar]
- 140.Threlkeld SW, McClure MM, Bai J, Wang Y, LoTurco JJ, Rosen GD, Fitch RH. Developmental disruptions and behavioral impairments in rats following in utero RNAi of Dyx1c1. Brain Res Bull. 2007;71(5):508–14. doi: 10.1016/j.brainresbull.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Dahdouh F, Anthoni H, Tapia-Páez I, Peyrard-Janvid M, Schulte-Körne G, Warnke A, Remschmidt H, Ziegler A, Kere J, Müller-Myhsok B, Nöthen MM, Schumacher J, Zucchelli M. Further evidence for DYX1C1 as a susceptibility factor for dyslexia. Psychiatr Genet. 2009;19(2):59–63. doi: 10.1097/YPG.0b013e32832080e1. [DOI] [PubMed] [Google Scholar]
- 142.Bates TC, Lind PA, Luciano M, Montgomery GW, Martin NG, Wright MJ. Dyslexia and DYX1C1: deficits in reading and spelling associated with a missense mutation. Mol Psychiatry. 2010;15(12):1190–6. doi: 10.1038/mp.2009.120. [DOI] [PubMed] [Google Scholar]
- 143.Tran C, Gagnon F, Wigg KG, Feng Y, Gomez L, Cate-Carter TD, Kerr EN, Field LL, Kaplan BJ, Lovett MW, Barr CL. A family-based association analysis and meta-analysis of the reading disabilities candidate gene DYX1C1. Am J Med Genet B Neuropsychiatr Genet. 2013;162B(2):146–56. doi: 10.1002/ajmg.b.32123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Szalkowski CE, Hinman JR, Threlkeld SW, Wang Y, LePack A, Rosen GD, Chrobak JJ, LoTurco JJ, Fitch RH. Persistent spatial working memory deficits in rats following in utero RNAi of Dyx1c1. Genes Brain Behav. 2011;10(2):244–52. doi: 10.1111/j.1601-183X.2010.00662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rosen GD, Bai J, Wang Y, Fiondella CG, Threlkeld SW, LoTurco JJ, Galaburda AM. Disruption of neuronal migration by RNAi of Dyx1c1 results in neocortical and hippocampal malformations. Cereb Cortex. 2007;17(11):2562–72. doi: 10.1093/cercor/bhl162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Currier TA, Etchegaray MA, Haight JL, Galaburda AM, Rosen GD. The effects of embryonic knockdown of the candidate dyslexia susceptibility gene homologue Dyx1c1 on the distribution of GABAergic neurons in the cerebral cortex. Neuroscience. 2011;172:535–46. doi: 10.1016/j.neuroscience.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tammimies K, Tapia-Páez I, Rüegg J, Rosin G, Kere J, Å. Gustafsson J, Nalvarte I. The rs3743205 SNP is important for the regulation of the dyslexia candidate gene DYX1C1 by estrogen receptor-B and DNA methylation. Mol Endocrinol. 2012;26(4):619–29. doi: 10.1210/me.2011-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tammimies K, Vitezic M, Matsson H, Le Guyader S, Bürglin TR, Ohman T, Strömblad S, Daub CO, Nyman TA, Kere J, Tapia-Páez I. Molecular networks of DYX1C1 gene show connection to neuronal migration genes and cytoskeletal proteins. Biol Psychiatry. 2013;73(6):583–90. doi: 10.1016/j.biopsych.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 149.Hoh RA, Stowe TR, Turk E, Stearns T. Transcriptional program of ciliated epithelial cells reveals new cilium and centrosome components and links to human disease. PLoS One. 2013;7(12):e52166. doi: 10.1371/journal.pone.0052166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Fagerheim T, Raeymaekers P, Tønnessen FE, Pedersen M, Tranebjaerg L, Lubs HA. A new gene (DYX3) for dyslexia is located on chromosome 2. J Med Genet. 1999;36(9):664–9. [PMC free article] [PubMed] [Google Scholar]
- 151.Kaminen N, Hannula-Jouppi K, Kestilä M, Lahermo P, Muller K, Kaaranen M, Myllyluoma B, Voutilainen A, Lyytinen H, Nopola-Hemmi J, Kere J. A genome scan for developmental dyslexia confirms linkage to chromosome 2p11 and suggests a new locus on 7q32. J Med Genet. 2003;40(5):340–5. doi: 10.1136/jmg.40.5.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Peyrard-Janvid M, Anthoni H, Onkamo P, Lahermo P, Zucchelli M, Kaminen N, Hannula-Jouppi K, Nopola-Hemmi J, Voutilainen A, Lyytinen H, Kere J. Fine mapping of the 2p11 dyslexia locus and exclusion of TACr1 as a candidate gene. Hum Genet. 2004;114(5):510–6. doi: 10.1007/s00439-004-1103-0. [DOI] [PubMed] [Google Scholar]
- 153.Paracchini S, Ang QW, Stanley FJ, Monaco AP, Pennell CE, Whitehouse AJ. Analysis of dyslexia candidate genes in the Raine cohort representing the general Australian population. Genes Brain Behav. 2011;10(2):158–65. doi: 10.1111/j.1601-183X.2010.00651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Eicher JD, Powers NR, Cho K, Miller LL, Mueller KL, Ring SM, Tomblin JB, Gruen JR. Associations of prenatal nicotine exposure and the dopamine related genes ANKK1 and DRD2 to verbal language. PLoS One. 2013;8(5):e63762. doi: 10.1371/journal.pone.0063762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Beaver KM, Delisi M, Vaughn MG, Wright JP. Association between the A1 allele of the DRD2 gene and reduced verbal abilities in adolescence and early adulthood. J Neural Transm. 2010;117(7):827–30. doi: 10.1007/s00702-010-0421-8. [DOI] [PubMed] [Google Scholar]