Summary
Mammalian microRNAs (miRNAs) pair to 3'UTRs of mRNAs to direct their posttranscriptional repression. Important for target recognition are ~7-nt sites that match the seed region of the miRNA. However, these seed matches are not always sufficient for repression, indicating that other characteristics help specify targeting. By combining computational and experimental approaches, we uncovered five general features of site context that boost site efficacy: AU-rich nucleotide composition near the site, proximity to sites for co-expressed miRNAs (which leads to cooperative action), proximity to residues pairing to miRNA nucleotides 13–16, and positioning within the 3'UTR at least 15 nt from the stop codon and away from the center of long UTRs. A model combining these context determinants quantitatively predicts site performance both for exogenously added miRNAs and for endogenous miRNA-message interactions. Because it predicts site efficacy without recourse to evolutionary conservation, the model also identifies effective nonconserved sites and siRNA off-targets.
Introduction
MicroRNAs are ~22-nt endogenous RNAs that pair to the messages of protein-coding genes to direct the posttranscriptional repression of these mRNAs (Bartel, 2004). A central goal for understanding their functions has been to understand how they recognize their target messages. Conserved Watson-Crick pairing to the 5' region of the miRNA, which includes the miRNA seed, enables prediction of targets above the background of false-positive predictions, indicating the importance of this region for miRNA target recognition (Lewis et al., 2003; Brennecke et al., 2005; Krek et al., 2005; Lewis et al., 2005). More than a third of human genes appear to have been under selective pressure to maintain their pairing to miRNA seeds (Lewis et al., 2005), and many messages that either decrease upon miRNA ectopic expression or increase upon miRNA knock-down have matches to the miRNA seed (Krutzfeldt et al., 2005; Lim et al., 2005; Giraldez et al., 2006; Rodriguez et al., 2007).
Messages downregulated after introducing a miRNA are most associated with four types of sites, which are in agreement with those anticipated from preferential conservation of sites in orthologous UTRs (Farh et al., in preparation). These include one 6mer, two 7mers, and one 8mer (Figure 1A). The 6mer is the perfect 6-nt match to the miRNA seed (miRNA nucleotides 2–7) (Lewis et al., 2005). The best 7mer site, referred to here as the 7mer-m8 site, contains the seed match augmented by a match to miRNA nucleotide 8 (Lewis et al., 2003; Brennecke et al., 2005; Krek et al., 2005; Lewis et al., 2005). Also effective is another 7mer, the 7mer-A1 site, which contains the seed match augmented by an A at target position 1 (Lewis et al., 2005). The 8mer site comprises the seed match flanked by both the match at position 8 and the A at position 1 (Lewis et al., 2005).
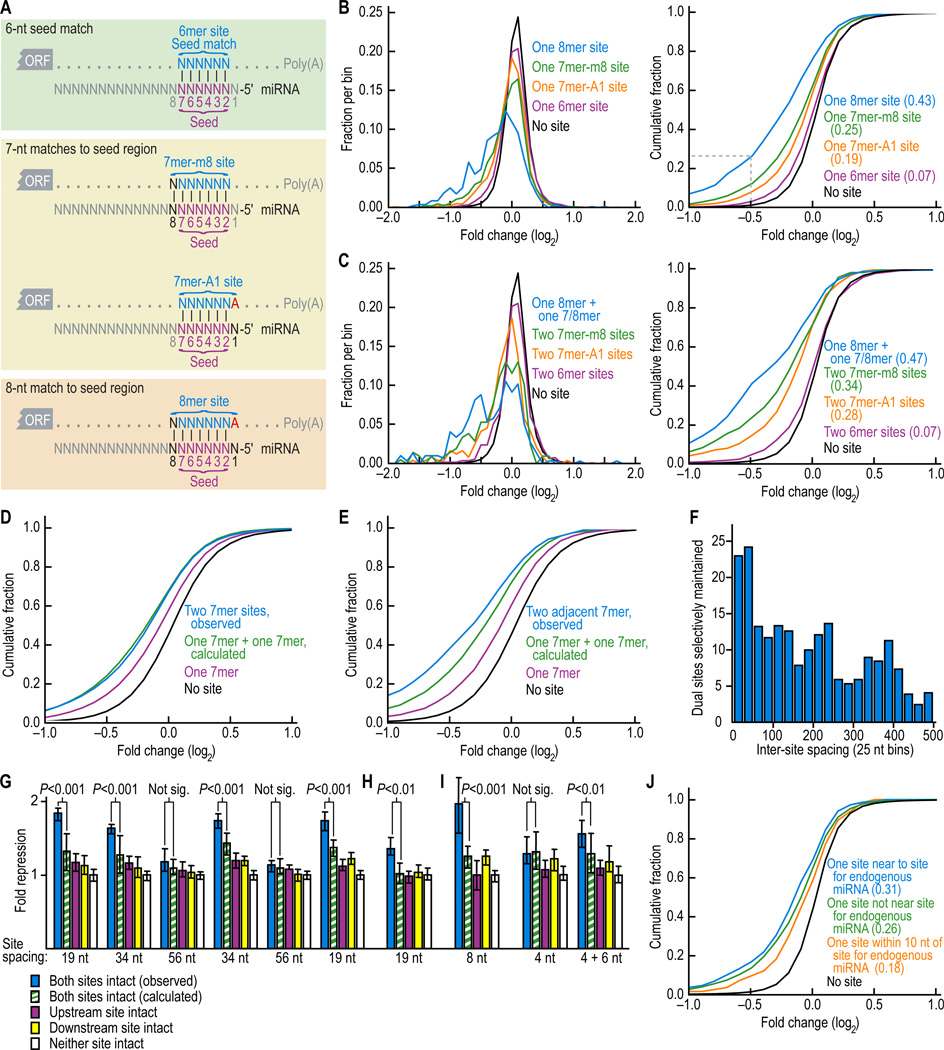
Figure 1. Downregulation of messages with 6-8mer sites.
(A) Canonical miRNA complementary sites.
(B) Effectiveness of single canonical sites. Changes in abundance of mRNAs following miRNA transfection were monitored with microarrays. Distributions of changes (0.1 unit bins) for messages containing the indicated single sites in their UTRs are shown (left), together with the cumulative distributions (right). The dashed line in the cumulative distributions indicates that 27% of mRNAs with UTRs containing a single 8mer were down-regulated at least 29% (2−0.5 = 0.71). Results of eleven experiments, each performed in duplicate and each transfecting a duplex for a different miRNA (Table S2), were consolidated. Results shown were an amalgam of the data from all 11 miRNAs; the relative strengths of the different sites were consistent when examining each transfection individually. For the cumulative plots, the minimal fraction of downregulated genes in that distribution is reported (parentheses), based on comparison with the no site distribution. Repression from UTRs containing an 8mer site was significantly more than that from UTRs with a 7mer-m1 site (P < 10−20, 1-sided K-S test); similar comparisons between UTRs containing a 7mer-m8 site versus a 7mer-A1 site, a 7mer-A1 versus a 6mer, and, a 6mer versus no site were also significant (P < 10−6, P < 10−20 and P < 10−31, respectively).
(C) Increased effectiveness of dual sites. Changes in mRNA abundance following miRNA transfection, represented as in B, except mRNAs with 3'UTRs containing the indicated pairs of sites were monitored. Repression from UTRs containing both an 8mer and either a 7mer or 8mer site was significantly more than that from UTRs with two 7mer-m8 sites (P < 10−3, 1-sided K-S test); similar comparisons between UTRs containing two 7mer-m8 sites versus two 7mer-A1 sites, two 7mer-A1 versus two 6mer, and, two 6mer versus no site were also significant (P = 0.034, P < 10−11 and P < 10−6 respectively).
(D) Independence of most dual sites. Cumulative distributions of changes in mRNA levels following miRNA transfection for messages containing the indicated combinations of miRNA binding sites. Simulated values for 3'UTRs containing two 7mer sites (green) were calculated by combining the effect of two single 7mers; actual values for 3'UTRs containing two 7mers are in blue and those with length-matched UTRs containing single 7mers are in purple; otherwise as shown for Figure 1B.
(E) Synergism between closely spaced sites. Cumulative distributions of changes in mRNA levels as for (D), except the plot for two observed sites (blue) only considered 3'UTRs containing two closely spaced 7mers (within 100 nt of each other). Repression from UTRs containing two adjacent sites was significantly increased compared to simulated UTRs containing one plus one site (P = 0.040, 1000 resampling iterations), whereas repression from UTRs containing two sites irrespective of distance did not significantly differ from simulated UTRs containing one plus one site (P = 0.81).
(F) Selective maintenance of dual sites spaced at different intervals. Human 3'UTRs with exactly two 7mer sites to the same miRNA were binned based on inter-site distance (counting the number of nucleotides between the 3' nt of the first site and the 5' nt of the second site). The number of conserved dual sites exceeding the background (as estimated from the average of control cohorts) was plotted after performing for each bin site-conservation analysis analogous to that in Lewis et al (2005), using the miRNA families conserved broadly among vertebrates (Table S1).
(G) MicroRNA-mediated repression of luciferase reporter genes fused to 3'UTR fragments containing two miR-124 target sites with different spacing intervals. After normalizing to the transfection control, luciferase activity from HeLa cells cotransfected with each reporter construct and its cognate miRNA (miR-124) was normalized to that from cotransfection of each reporter with its non-cognate miRNA (miR-1). Plotted are the normalized values, with error bars representing the third largest and third smallest values among 12 replicates. P values (Wilcoxon rank-sum test) indicate whether repression from a reporter containing both sites (blue) was significantly greater than expected from multiplicative effects (green). For modeling independent activity of sites, repression expected from a reporter with two sites was the product of repression observed from otherwise identical reporters containing single intact sites (purple and yellow; pairing off the repression values in the order that they were generated). Reporters of the rightmost quintet were identical to those of the leftmost quintet, except the point substitutions disrupting target sites differed.
(H) Repression observed for the reporter constructs as in the leftmost quintet of (G) but modified such that both miR-124 sites were substituted with miR-1 sites. miR-196 served as the noncognate miRNA.
(I) Repression of reporter constructs containing 3'UTR fragments with naturally closely spaced miR-1 and miR-133 sites, compared to that of mutant derivatives of these fragments. A mixture of miR-1 and miR-133 was co-transfected as the cognate miRNA, and miR-196 served as the noncognate control.
(J) Cooperativity between sites to transfected and endogenous miRNAs in HeLa. Endogenous sites considered were those for let-7 RNA, miR-16, miR-21, miR-23, miR-24, miR-27, and miR-30 (Tom Tuschl, personal communication). 7mer-m8 sites at a cooperative distance (>7 nt and <40 nt) from an endogenous miRNA 7-8mer site were significantly more downregulated than sites that were either too close to an endogenous miRNA (≤7 nt, including overlapping sites; P = 0.0054, 1-sided K-S test), or not close to an endogenous site (≥40 nt, or no endogenous site, P = 0.036, 1-sided K-S test).
Reporter assays indicate that pairing to the seed is not only important for recognition but that in some cases 7-8mer sites appear sufficient (Doench and Sharp, 2004; Brennecke et al., 2005; Lai et al., 2005). However, seed sites do not always confer repression, and when repression occurs, the degree of repression is highly variable in different UTR contexts. In the animal, messages preferentially expressed in the same tissue as a highly expressed miRNA have 3'UTRs that are ~50% depleted in 7mer sites, presumably because these messages have important roles in that tissue, and during the course of evolution they have avoided acquiring sites to coexpressed miRNAs that would compromise their function (Farh et al., 2005). However, these UTRs are not completely devoid of 7mer sites, suggesting that even among miRNA sites in vivo, similar variability in site efficacy also exists, such that identical 7mer sites may or may not be consequential depending on the UTR context in which they arise. Thus, having a canonical 7-8mer site is clearly important but often not sufficient for detectable downregulation; unknown context determinants outside of seed sites must also be playing important roles for target recognition.
Without knowing these context determinants, experimentalists face a difficult challenge. Where should they begin when seeking to understand the molecular mechanisms of phenotypes arising from miRNA knockdown or ectopic expression studies? One approach is to focus only on the conserved sites, but so many messages are under selective pressure to maintain pairing to miRNAs—more than 200 messages, on average, for each highly conserved mammalian miRNA family—that this approach only serves to narrow the field, while potentially excluding some of the more rapidly evolving and species-specific interactions (Farh et al., 2005; Krek et al., 2005; Krutzfeldt et al., 2005; Lewis et al., 2005; Giraldez et al., 2006).
We set out to discover the context features that help specify miRNA targeting, with the idea that such insights into target recognition would increase the ability to predict functional sites, both those that are conserved and those that are not. We found five independent features that influenced targeting, each of which had both experimental and computational support. Combining these determinants, we constructed a model of miRNA regulation capable of quantitatively predicting the performance of miRNA sites based solely on sequence, and we confirmed experimentally the predictive power of this model for both exogenously added miRNAs and for endogenous miRNA-message interactions. Because our approach accurately distinguishes effective from non-effective sites without regard to site conservation, it also provides the basis for identifying siRNA off-targets.
Results and Discussion
Closely Spaced Sites Often Act Synergistically
When examining mRNA microarray data from 11 miRNA transfection experiments, a large majority (75%) of the downregulated messages detected on microarrays have canonical 7-8mer sites in their 3'UTRs (Farh et al., in preparation). However, only a minority of the messages possessing single sites in their 3'UTRs displayed detectable destabilization on the array (19%, 25%, and 43% for the 7mer-A1, 7mer-m8, and 8mer, respectively, Figure 1B), which suggested that analyzing the differences between those messages that were detectably downregulated and those that were not could help identify context features important for target recognition.
As anticipated from previous studies (Doench and Sharp, 2004; Brennecke et al., 2005; Lai et al., 2005), multiple sites were associated with greater mRNA destabilization in our transfection experiments (Figures 1B and 1C). When considering all genes, the repression observed for those with two-sites was almost exactly that expected if the two sites had contributed independently to repression; that is, the repression for a gene with two sites matched the result anticipated by multiplying the repression from two single sites (Figure 1D). This multiplicative effect, a hallmark of independent and noncooperative action, was observed previously in a heterologous reporter assay designed to model miRNA repression (Doench and Sharp, 2004).
We observed one notable exception to the overall tendency of apparently independent action: when the two sites were close together, the repression tended to be greater than that expected from the independent contributions of two single sites (Figure 1E). Examining the conservation of sites in orthologous UTRs of human, mouse, rat and dog, we found that when plotting the number of co-conserved sites for authentic miRNAs, after subtracting the average number of co-conserved sites for control cohorts, the greatest enrichment was for closely spaced dual sites (Figure 1F). Although most of the co-conserved sites were at longer intervals (because longer intervals greatly outnumber shorter intervals), the observed enrichment compared to any other specific intervals indicated a detectable biological preference for short intervals. Such cases with short inter-site spacing appear to be the more effective ones and thus would be easiest to identify genetically. In agreement with this idea, the C. elegans lin-4:lin-14 and lsy-6:cog-1 interactions and the Drosophila miR-4:Bearded interactions all involve dual conserved 8mer sites with short inter-site spacing (Lee et al., 1993; Wightman et al., 1993; Johnston and Hobert, 2003; Lai et al., 2005); see targetscan.org for minor refinements to C. elegans site annotations).
We investigated this phenomenon further using reporter assays, examining UTR fragments containing two sites and asking whether the observed repression from these two sites was greater than that expected from the sites individually (calculating expected repression as the product of the repression values when measured for each of the two sites in isolation). The two UTR fragments for which observed repression deviated most from that expected corresponded to the two shortest inter-site distances examined (Figure S1). For example, two proximal miR-124 sites individually mediated only subtle down-regulation, whereas both sites together mediated more robust down-regulation, which was significantly cooperative (Figure 1G, left-most). Increasing spacing from 19 to 56 intervening nt, using either of two different insertion sequences, fully abrogated cooperativity, whereas increasing it to only 34 nt did not (Figure 1G). Cooperativity was maintained when both sites were changed to miR-1 sites (Figure 1H). Importantly, repression from UTRs containing either an intact upstream or downstream site alone (purple or yellow bars, respectively) was not significantly altered by the different inter-site sequences employed.
The opportunities for cooperative miRNA function would dramatically increase if sites to two different miRNAs could act cooperatively. To test this possibility, we tested reporters based on UTR fragments containing closely spaced sites for miR-1 and miR-133, two miRNAs co-expressed in muscle cells (Figure 1I). The one with 8-nt inter-site spacing exhibited cooperative repression in response to a mixture of miR-1 and miR-133. The one with 4-nt spacing did not, which we attributed to spacing that was too close, in that cooperativity was achieved by extending the spacing by another 6 nt. To test more generally whether sites to different miRNAs act cooperatively when closely spaced, we examined whether sites to our transfected miRNAs were more effective if they were near to sites for miRNAs endogenously expressed in the cells. Repression was strongest when spacing between the endogenous and the transfected miRNA sites was 8 to ~40 nt (Figure 1J).
We conclude that one of the important context determinants that influences efficacy of sites is their proximity to sites for coexpressed miRNAs. A recent report, published while our paper was in review, reached similar conclusions (Saetrom et al., 2007). Cooperative miRNA function implies a mechanism whereby repression can become more sensitive to small changes in miRNA levels. Moreover, cooperativity of sites for co-expressed miRNAs greatly enhances the regulatory effect and utility of combinatorial miRNA expression.
Additional Watson-Crick Pairing at Nucleotides 12–17 Enhances miRNA Targeting
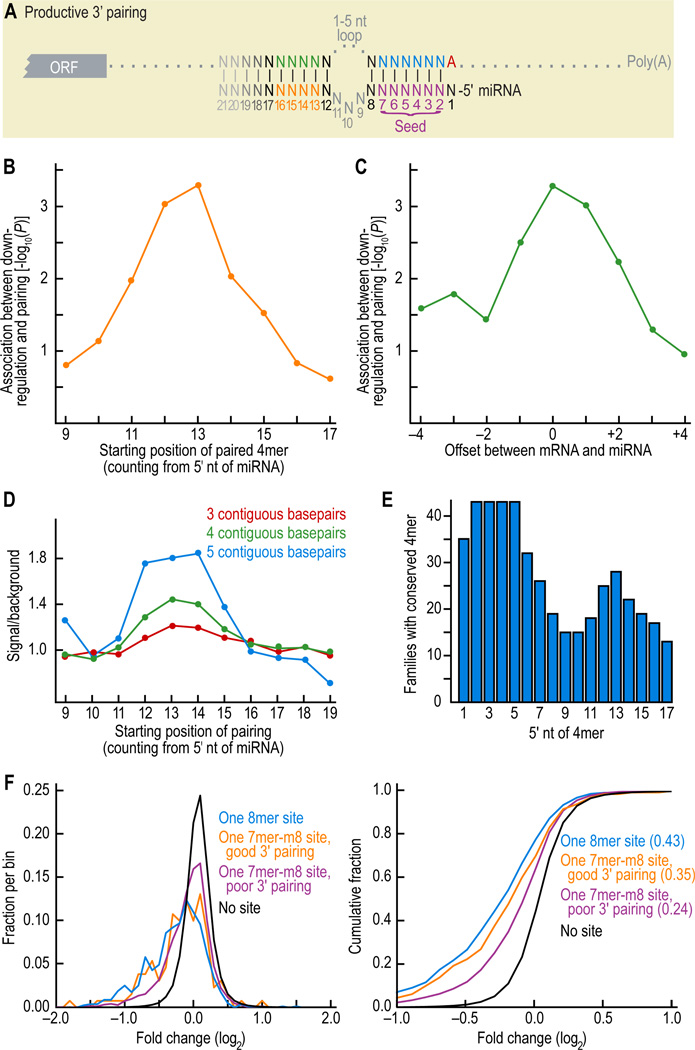
Pairing to the 3' portion of the miRNA is often thought to enhance repression of canonical 7- or 8mer sites. However, when we used previously developed energy-based rubrics for predicting and scoring 3' supplementary pairing (Lewis et al., 2003; John et al., 2004; Krek et al., 2005), those sites with better scores were no more effective; indeed, increased supplementary pairing, as measured by these rubrics, appeared to disfavor efficacy (Figure S2A). We also examined the results of a different approach that predicted sites with extensive Watson-Crick and G:U pairing along the length of the miRNA and tolerated imperfect seed matches (Miranda et al., 2006). However, after excluding those predictions with canonical seed sites, the remainder performed no better than messages without sites (Figure S2B). We therefore searched for a rubric that accomplished its intended purpose of identifying productive 3' pairing, evaluating site efficacy while systematically varying position, continuity, and identity (± G:U pairs) of pairing. Overall, consequential pairing preferentially involved Watson-Crick pairing to miRNA nucleotides 12–17, most especially nucleotides 13–16 (Figure 2A). Watson-Crick pairing to four contiguous nucleotides was most associated with down-regulation if it started at position 13 (Figure 2B). Regarding the position of the UTR segment that paired to the miRNA 3' region, the most optimal arrangement placed it directly opposite the miRNA strand, although several neighboring registers were also effective (Figure 2C).
Figure 2. Characteristics of beneficial 3' pairing.
(A) Pairing scheme, highlighting the core region of productive 3' pairing between the miRNA (orange) and 3'UTR (green).
(B) Preferential position of supplementary pairing within the miRNA 3' region. Starting with 7mer-m8 sites for representative vertebrate miRNAs of Table S1, a 4mer window was slid across the 3' end of the miRNA, searching for its Watson-Crick match in the opposing region of the message. A 3' pairing score was assigned to each miRNA 4mer, crediting one point for each contiguous pair within the 4mer, a half point for extending the contiguous pairing 1 nt upstream and a half point for extending it 1 nt downstream. The position of the miRNA 4mer and its complement in the message were allowed to be offset, but a ½ point penalty was assessed for each nucleotide of offset beyond ± 2 nt, and pairing to message positions already paired to the miRNA seed region (1–8) was disallowed. When the 4mer had alternative regions of contiguous pairing, it was assigned the highest of the alternative scores. For each position, the Spearman correlation between the score and down-regulation on the array was determined and its P value plotted.
(C) Preferred offsets between paired regions of miRNA and mRNA. Analysis of (B) was repeated using a 4mer fixed at miRNA nucleotides 13–16, and the correlation between score and down-regulation was evaluated for alternative pairing offsets. A positive offset shifts the miRNA 3' pairing segment to the right, relative to the message.
(D) Preferential positions of conserved pairing. Human 3'UTRs were scanned for sites with at least a 6mer seed match to miRNAs of Table S1. For each contiguous 4mer beginning at nucleotide 9 of the miRNA, we searched for the complementary Watson-Crick 4mer directly opposite in the message, allowing for a ± 2-nt offset and excluding overlap into nucleotides 1–8. Those cases in which the seed and its supplemental 4mer were co-conserved were considered conserved instances of 3' pairing. This was compared to control chimeric miRNA sequences with the same 5' seed sequence but with the 3' end of a different miRNA, and signal/background was calculated. The analysis was repeated for 3mers and 5mers.
(E) 4mers conserved among paralogous human miRNAs. For each position the number of human miRNA families that have a perfectly conserved 4mer is indicated; families with only a single human paralog were excluded.
(F) Effectiveness of 7mer-m8 sites compensated with either good or poor 3' pairing. Distributions (left) and cumulative distributions (right) of changes in abundance of mRNAs following miRNA transfection were monitored with microarrays, and are displayed as in Figure 1B, including for reference the results of canonical 8mer sites. Sites with good pairing were those with scores ≥ 4 and sites with poor pairing were those with scores ≤ 2. Pairing score was determined as in (B) but using a fixed miRNA 4mer corresponding to nucleotides 13–16 and crediting contiguous pairing elsewhere with ½ point per pair. Sites with good pairing were significantly more effective than sites with poor pairing (P = 0.007, 1-sided K-S test).
Turning to evolutionary conservation, Watson-Crick pairing to four contiguous miRNA nucleotides was substantially more conserved when this pairing started at miRNA positions 12, 13, or 14 (Figure 2D), and those same nucleotides at the core of effective 3' pairing were the best conserved miRNA nucleotides outside of the seed region (Figure 2E). Thus, site conservation results were in accord with the experimental results from the array, as would be expected if supplemental pairing, as identified by our rubric, influences protein output in the animal.
The similarities between 3' pairing and seed pairing were striking. Analogous to seed pairing, 3' pairing was relatively insensitive to predicted thermodynamic stability and instead quite sensitive to geometry, preferring contiguous Watson-Crick pairs uninterrupted by wobbles, bulges, or other mismatches. Also like seed pairing, 3' pairing was sensitive to position, with pairing at the 3' core (positions 13–16) being more important for efficacy than pairing to other positions.
Using the guidelines derived from analyses of site efficacy and conservation, we developed a simple scheme for scoring 3' pairing based on rewarding pairing throughout the 3' end of the miRNA, but with particular emphasis on the 13–16 nucleotide region. Comparing the efficacy of high-scoring sites with that of low-scoring sites revealed that 3' pairing was an effective determinant, most particularly for 7mer-m8 sites (Figure 2F), although the magnitude was less than the difference between a 7mer site and an 8mer site.
We suspect that very extensive 3' pairing might be more effective than that observed in Figure 2F, but that very extensive pairing was too rare to be reliably evaluated in our array analysis. Extensive 3' pairing also appears to be utilized relatively rarely during biological targeting in mammals. For example, the numbers of canonical sites (6-8mers) with extensive 3' pairing (comprising ≥ 5 contiguous, well-positioned pairs) that were conserved above background averaged two per miRNA family. Nonetheless, we anticipate our guidelines for detecting consequential 3' pairing will help identify unusual but important cases in which extensive 3' pairing is crucial for mammalian target repression. For example, 3' pairing can help compensate for imperfect seed pairing (Doench and Sharp, 2004; Brennecke et al., 2005), and a few sites with extensive 3' compensatory pairing, including the let-7 sites in C. elegans lin-41, and the miR-196 site in mammalian HoxB8 are known to function in mammalian cells (Reinhart et al., 2000; Lewis et al., 2003; Yekta et al., 2004). In all of these cases, the sites would have exceptionally high scores using our rubric (≥10 contiguous Watson-Crick base pairs).
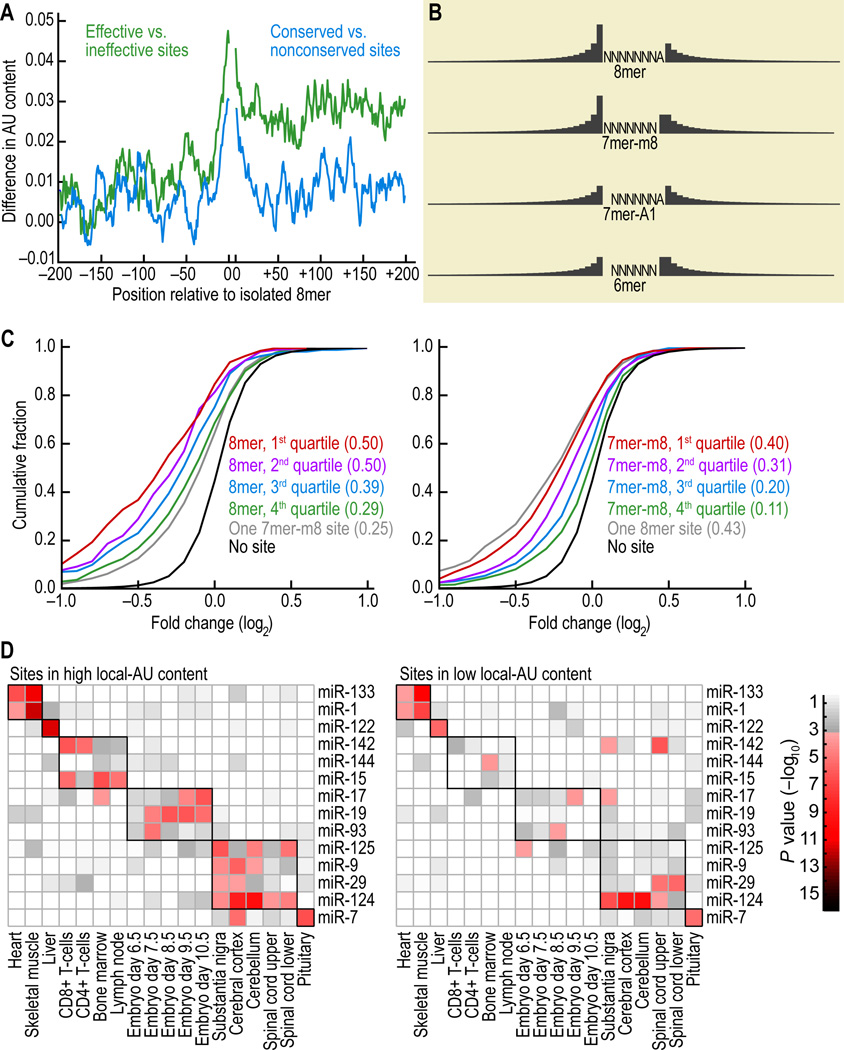
Effective Sites Preferentially Reside Within a Locally AU-rich Context
When sites were divided into functional and nonfunctional sites based on their performance on the microarray, we found that the nucleotides immediately flanking the functional sites were highly enriched for A and U content relative to the nonfunctional sites (Figure 3A, green plot). This phenomenon of high local AU content was important in the immediate vicinity of the site and then fell off quickly. A comparison of the local nucleotide composition adjacent to conserved and nonconserved miRNA sites concurred with the array data; the nucleotides immediately flanking the conserved sites were highly enriched for A and U content relative to those flanking the nonconserved sites (Figure 3A, blue plot).
Figure 3. Substantial influence of local AU content.
(A) Preferred nucleotide composition in the vicinity of effective and conserved 8mer sites. For the site-efficacy analysis (green), sites associated with the greatest down-regulation in the expression arrays (top third of sites when ranking for down-regulation) were compared with those associated with least down-regulation (bottom third of sites). At each position, counting from the site, the fractional difference in AU composition within a ±5-nt window is plotted. The analysis was repeated for conservation (blue), comparing nucleotide composition flanking conserved sites versus nonconserved sites for the 11 miRNA families (Table S1).
(B) Weighting of the AU composition for the different types of sites. For each position within 30 nt upstream and downstream of the site, the presence of an A or a U increased the score for the site by an amount proportional to the height of the bar for that nucleotide. When moving away from the site, the weight (bar height) decreased with the inverse of the distance from the site. For example, the weight of the nucleotides downstream of the 8mer were 1/2, 1/3, 1/4, 1/5 … that of the nucleotide upstream of the 8mer.
(C) Effectiveness of 8mer (left) and 7mer-m8 (right) sites with varying local AU content. Sites were separated into four quartiles according to rubrics of (B). For reference, the results of another canonical site is included (grey), otherwise as in Figure 1B. For both site types, the quartile with the highest local AU content was significantly more downregulated than the quartile with the lowest local AU content (P < 10−7 for 8mer sites, P < 10−29 for 7mer-m1 sites).
(D) Site-depletion analysis implying greater efficacy of sites emerging with high local AU composition. To evaluate the efficacy of sites in different local AU contexts, sites were partitioned at the median into two equal-sized groups based on their local AU content, and the depletion analysis was performed for each tissue-miRNA pair. P values indicate the extent of depletion for individual pairs. The boxes on the grid indicate those pairs in which the miRNA is expressed, which was where highly expressed messages were expected to be most depleted in sites to that miRNA.
We developed a rubric that considered the composition of residues 30 nt upstream and 30 nt downstream of the seed site, with weighting tailing off with the inverse of the distance from the seed site (Figure 3B). The 7mers scoring in the top quartile by our rubric appeared at least as effective as 8mers scoring in the bottom two quartiles, illustrating the substantial influence of local AU composition for site efficacy (Figures 3C).
As an additional test of whether sites within high local AU density were more effective in the animal, we performed site-depletion analysis. This analysis was based on the finding that messages preferentially expressed in the same tissue as the miRNA are depleted for sites matching that miRNA, because these highly expressed messages have been under selective pressure to avoid deleterious repression (Farh et al., 2005). Sites within high local AU density, as measured by our rubric, were significantly more depleted in messages preferentially co-expressed with the miRNA compared to sites within low AU density (Figure 3D, P < 10−6, 1-sided K-S test). Because site depletion, as with site conservation, is a function of protein downregulation in the animal, our results indicated that local AU content impacts not only mRNA destabilization but also protein expression.
High global AU composition within 3'UTRs also correlates with a higher density of conserved miRNA complementary sites, as well as higher 3'UTR conservation more broadly (Robins and Press, 2005). When considering nucleotide composition of the entire 3'UTR, we observed a correlation between global AU content and efficacy on the array. However, this correlation was not as strong as for local composition, and after accounting for local AU context, no significant residual correlation remained for global AU composition, indicating that global AU composition does not directly influence site efficacy.
Because the preference for A’s and U’s in the vicinity of the site appeared to involve no more than fortuitous Watson-Crick pairing, this local AU determinant resembled the preference for A across from nucleotide 1, thereby extending the apparent non-Watson-Crick component of site recognition far beyond position 1. Indeed, the preference for A at this position might be considered a component of the local AU effect, skewed to favor A over U. Local AU density also explained why previous methods designed to identify and score 3' pairing were counterproductive for finding more effective sites (Figure S2A). Each of these methods uses predicted folding free energy to score 3' pairing (Lewis et al., 2003; John et al., 2004; Krek et al., 2005). Seed sites present in regions of higher local GC content will tend to have predicted 3' pairing with better folding free energy due to fortuitous GC base-pairing, when in fact they are significantly less effective.
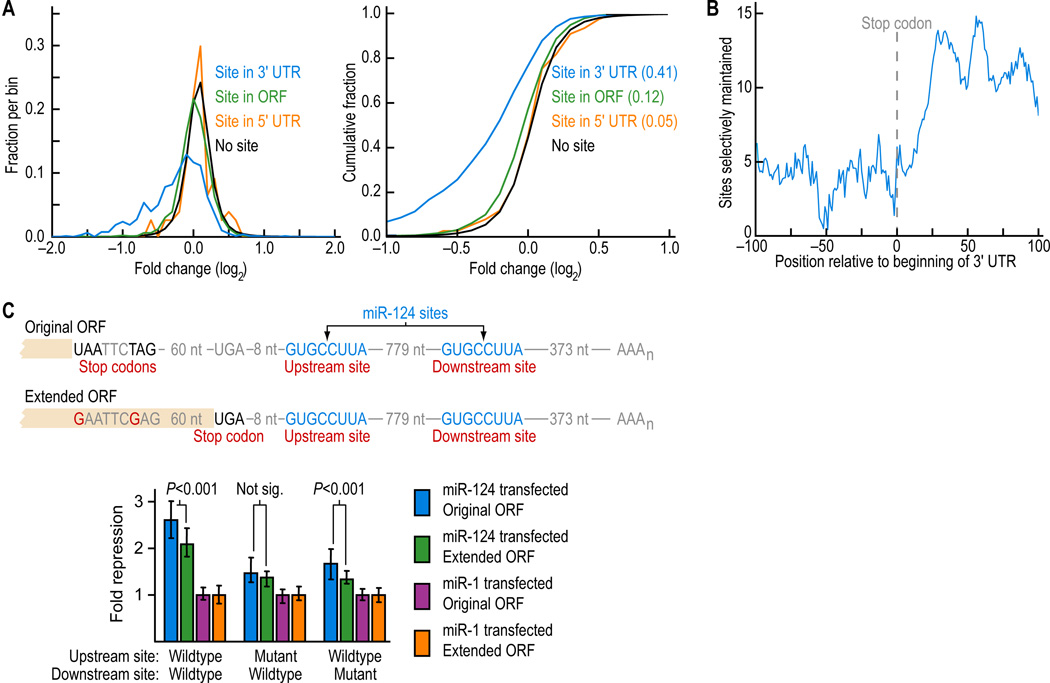
Effective Sites Preferentially Reside in the 3'UTR but not too Close to the Stop Codon
Although the majority of investigation into miRNA function has been for sites located in 3'UTRs, the 5'UTRs and open reading frames (ORFs) of mammalian genes contain, on average, twice the sequence length as 3'UTRs, and artificial siRNAs can direct cleavage at perfectly complementary sites throughout the message. For single 8mer sites, we observed no detectable efficacy in 5'UTRs, detectable but marginal efficacy in ORFs, and high efficacy in 3'UTRs (Figure 4A). These results mirrored those from previous site-conservation analysis, expression arrays, and site-depletion analysis (Farh et al., 2005; Lewis et al., 2005; Lim et al., 2005).
Figure 4. Poor efficacy of sites within 15 nt of the stop codon.
(A) Distributions (left) and cumulative distributions (right) of changes to the levels of messages with one 8mer site following miRNA transfection, analyzed and displayed as in Figure 1B. Messages with a site in their 3'UTR or ORF were significantly more repressed than those with no site (P < 10−126 and < 10−16, respectively), whereas those in 5'UTRs were not (P = 0.181).
(B) Conservation of 7-8mer sites in the region near the stop codon. Plotted are the number of sites conserved above background per nucleotide in the ±5-nt window centered on the indicated mRNA position, with position 0 being the first nucleotide of the 3'UTR. A site was scored as within the window if the 5'-most nucleotide of the 7-8mer was within the window.
(C) MicroRNA-mediated repression of luciferase reporter genes fused to 3'UTR fragments containing miR-124 sites or mutant derivatives. After normalizing for transfection, luciferase activity from HeLa cells cotransfected with each reporter construct and its cognate miRNA (green and blue bars) was normalized to that from co-transfection of each reporter with its non-cognate miRNA (purple and orange bars). Error bars represent the fourth largest and smallest values among 18 replicates. P values (Wilcoxon rank-sum test) indicate whether repression from reporters containing the original ORF (blue) was significantly greater than that with a reporter with an extended ORF that stopped 9 nt from the upstream site (green).
When plotting the number of sites conserved above chance, we found that this number, which was low in the ORF, remained low in the first ~15 nt following the stop codon, and thus the transition between ORF targeting and 3'UTR targeting did not occur precisely at the stop codon, but was offset ~15 nt downstream (Figure 4B). Concurring with this finding, 3'UTR sites within 15 nt of the stop codon were less effective on the array, compared to sites elsewhere in the 3'UTR (P <0.01 for a 1-sided K-S test comparing expression values for messages with an 8mer, data not shown). Further supporting these findings were results from reporters that were identical except at two nucleotides that disrupted two stop codons, thereby extending the ORF by 69 residues and bringing into service a stop codon falling within 8 nt of a miR-124 site (Figure 4C). These two point substitutions, both more than 70 nt upstream of the site, specifically abrogated function of the site, while having no effect on the function of a downstream site (Figure 4C). These results confirmed that the segment immediately following the stop codon was inhospitable for targeting.
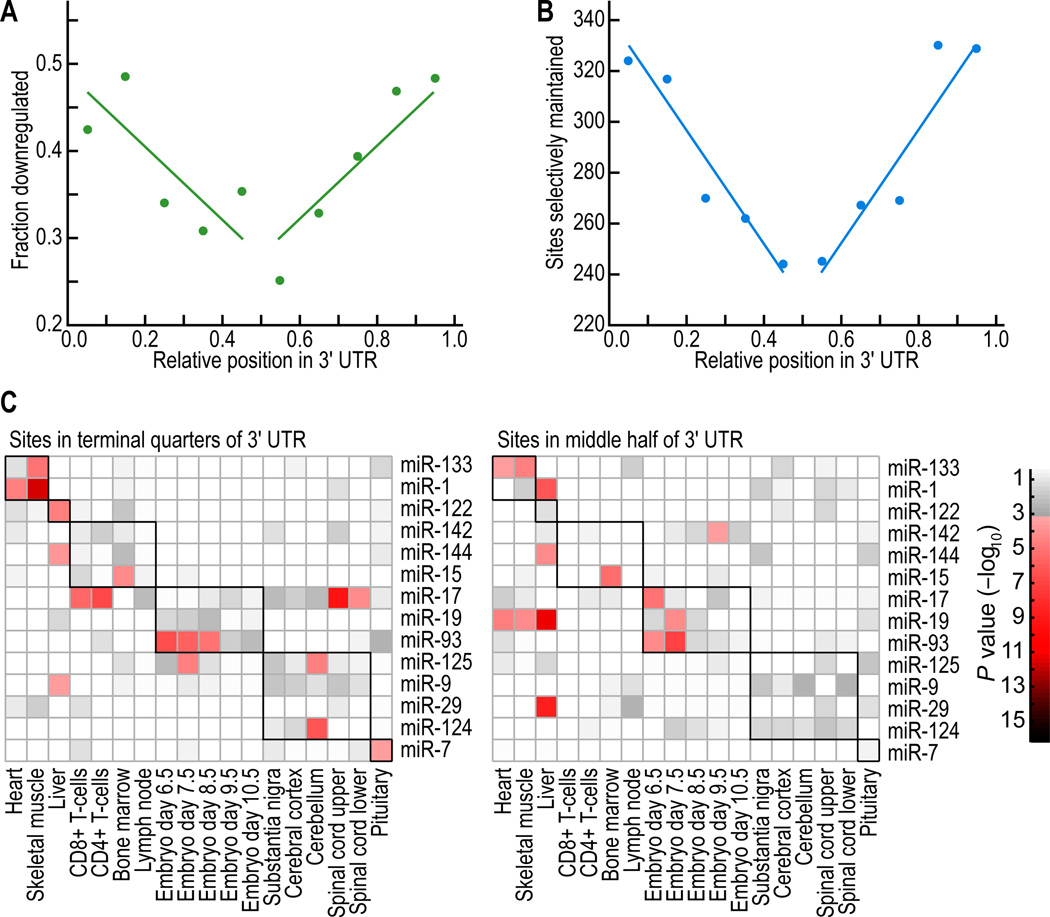
Effective Sites Preferentially Reside Near Both Ends of the 3'UTR
When examining whether the location of the site within the remainder of the 3'UTR influenced performance, we found that sites residing near the two ends of long UTRs generally were more effective than those near the center (Figures 5A; P = 0.0011, Pearson correlation). Moreover, site-conservation analysis revealed that more sites were selectively maintained near the ends than in the central region (Figure 5B; P < 10−4, Pearson correlation). Site-depletion in messages preferentially co-expressed with miRNAs was also more severe near the ends of long UTRs than near the center, indicating that newly emergent sites are more likely to be functional in the animal if they fall near the ends (Figure 5C, P = 0.032, 1-sided K-S test). Thus, all three lines of evidence—the experimental approach, which monitored mRNA destabilization, plus the two computational approaches, which addressed protein output in the animal—indicated that the UTR quartiles near the ORF and near the poly(A) tail were more hospitable for effective targeting than were the two central quartiles. This effect was most pronounced for longer UTRs (>1300 nt).
Figure 5. Poorer performance of sites near the middle of long 3'UTRs.
(A) Fraction of sites associated with repression for 8mers residing at different positions in 3'UTRs. UTRs of at least 1300 nt with single 8mer sites were split into 10 equally spaced bins based on the relative position of the site (distance from stop codon divided by the UTR length). For each bin, the point is plotted that corresponded to the mean site position and the fraction of messages downregulated at a threshold P < 0.05 on the microarray. The lines are the least-squares fit to all the data, using a model assuming equal effects from both ends of the UTR.
(B) Number of sites conserved above background for 8mers residing at different positions in 3'UTRs. UTRs of at least 1300 nt were divided into 10 equally spaced bins, and each bin, the point is plotted that corresponded to the mean site position and the number of sites conserved above the background. The lines are the least-squares fit to the binned data, using a model assuming equal effects from both ends of the UTR.
(C) Site-depletion analysis implying greater efficacy of sites near the ends of UTRs. To evaluate the efficacy of sites in different UTR regions, total 3'UTR sequence for mouse UTRs of at least 1300 nt was divided into the middle half (right panel) and the two remaining terminal quarters (left), and the depletion analysis was performed for each group as in Figure 3D.
A Quantitative Model for Site Efficacy
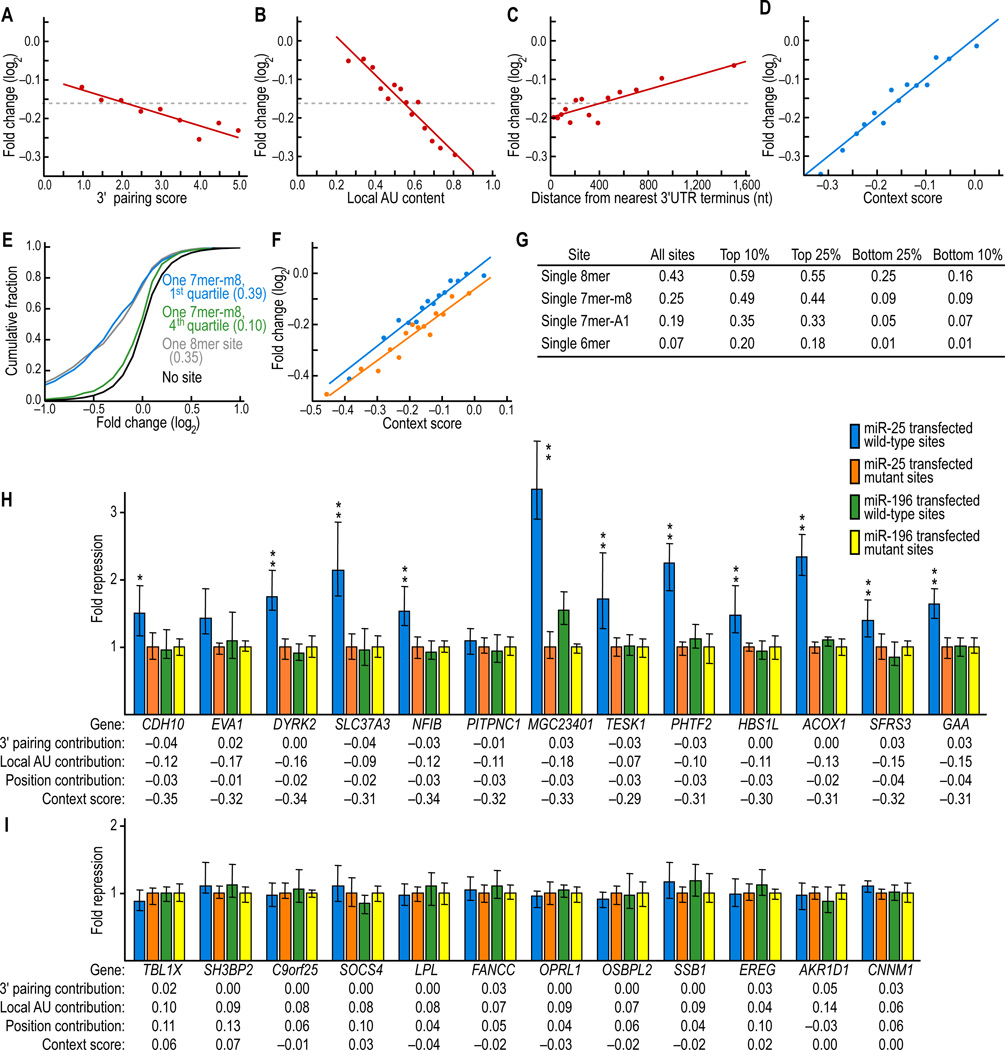
To develop a quantitative tool for predicting the efficacy of single sites, irrespective of their evolutionary conservation, we used linear regression to model the relationship between downregulation on the array and the context of single 7mer-m8 sites. To build a model general to various cell types, proximity to sites for co-expressed miRNAs was not considered. Sites in 5'UTRs, in ORFs, or within 15 nt of the stop codon were excluded, in accord with our results showing that such sites were generally not effective (Figure 4). For additional pairing to the 3' region of the miRNA, scores were calculated as described in Figure 2F, with the score equal to the greatest number of contiguously paired bases, weighted towards pairing at nucleotides 13–16 (Figure 6A). For local AU composition, scores were calculated as described in Figure 3B, with a higher score indicating greater local AU composition in the region flanking the site (Figure 6B). For position effects, scores were based on the distance in nucleotides between the site and the closest end of the 3'UTR (Figure 6C).
Figure 6. A quantitative model that considers site context to predict site efficacy.
(A) Increased efficacy of 7mer-m8 sites with more 3' pairing. Messages with a single site were scored as described in Figure 2F, and for each score, the mean downregulation on the microarray is plotted. The regression line is the best least-squares fit to the full data and represents the relationship between score and downregulation on the array (P < 10−3, r = −0.07, Pearson correlation). The average log2 fold change (−0.161) is indicated (dashed line). The few sites with pairing score <1 or >5 were folded into the first and last bins, respectively.
(B) Increased efficacy of 7mer-m8 sites within higher local AU content. Messages with a single site were split into 14 equally sized bins based on scoring described in Figure 3B, where 1.0 equaled the maximum possible score. For each bin, the point is plotted that corresponded to the mean score and the mean downregulation on the microarray, and the line was fit as in (A) (P < 10−32, r = −0.21, Pearson correlation).
(C) Decreased efficacy of 7mer-m8 sites further from the 3'UTR ends. Messages with a single site were split into 14 equally sized bins based on their distance from the closest UTR ends, and the points and regression line were plotted as in (B) (P < 10−8, r = 0.11, Pearson correlation).
(D) Correspondence between the predicted and observed efficacy of single 7mer-m8 sites. Messages with a single site were split into 14 equally sized bins based on their the context score, calculated for each message by predicting the offsets from the mean for the three context determinants (panels A–C) and then adding these three contributions to the average log2 fold change for 7mer-m8 sites (Figure S6). Points correspond to the mean score and mean downregulation for each bin. The regression line is the best least-squares fit to the full data (P < 10−45, r = 0.25, Pearson correlation).
(E) Performance of the combined model when applied to a dataset that was not used to derive the model. Shown are the cumulative distributions of changes in mRNA levels following siRNA transfection (Jackson et al. 2003) after first predicting the top and bottom quartiles using context scores calculated as in (D) for messages with single 7mer-m8 sites to the siRNAs (Figure S5); otherwise as shown for Figure 1B (P < 10−31, 2-sided KS test, comparing top and bottom quartiles).
(F) A unified model that considers the differential efficacy of the 7-8mer canonical sites and the influence of context determinants. Human messages with a single 7mer-A1, 7mer-m8, or 8mer site were split into two sets based on whether the site was conserved in orthologous UTRs of mouse, rat and dog. For each site, a context score was calculated using the regressions of panels A–C and analogous ones for the other two types of sites (Table S6). For conserved (orange) and nonconserved (blue) sets, sites were divided into 14 bins based on context score, and the mean score and mean repression was plotted for each bin. The regression lines were fit to the full data (conserved: P < 10−24, r = 0.25, nonconserved: P < 10−50, r = 0.32, Pearson correlations).
(G) Performance of different types of sites predicted to be in favorable or unfavorable contexts, as ranked using the context scores of (F). Shown are the minimum number of downregulated messages, as calculated using cumulative distributions like that of (E).
(H) MicroRNA-mediated repression of luciferase reporter genes fused to 3'UTR fragments containing a single 7mer-m8 miR-25 site located in favorable context. After normalizing to the transfection control, luciferase activity from HEK293 cells cotransfected with each reporter construct and its cognate miRNA (miR-25) was normalized to that from cotransfection of a reporter with a mutated miR-25 site and cognate miRNA. Similarly, luciferase acitivity from cotransfected wild-type reporter constructs and non-cognate miRNA (miR-196) was normalized to that from cotransfection of mutant reporter constructs and non-cognate miRNA. Plotted are the normalized values, with error bars representing the third largest and third smallest values among 12 replicates, with significant repression when comparing results for the cognate and noncognate miRNA indicated (*, P < 0.01; **, P < 0.001; Wilcoxon rank-sum test). Below each gene name is the context score for the miR-25 site and the three context contributions used to calculate, as in (D), this score.
(I) MicroRNA-mediated repression of luciferase reporter genes fused to 3'UTR fragments containing a single 7mer-m8 miR-25 target site located in unfavorable context, otherwise as in (H).
The scores for each context feature were not significantly co-correlated (Figure S4), indicating that their effects could be considered independently and combined into a single model (Figure 6D). When tested on an independent series of siRNA transfections this model accurately anticipated the messages destabilized in response to the siRNAs (Figure 6E), and the individual features of the model were each effective on their own (Figure S5). This result showed that our model was predictive for data that had not been used in its derivation and demonstrated that it applied to siRNA off-targeting as well as miRNA targeting.
Using the same approach, we considered the 8mer, 7mer-A1, and 6mer sites and combined the models for these sites into a single unified model. To address the question of how evolutionary conservation impinged on the model, the effects were plotted separately for conserved and nonconserved sites (Figure 6F). For both conserved and nonconserved sites the unified model yielded a clear correspondence between the predicted and observed effects on message stability. Nonetheless, conserved sites with the same score were somewhat more effective than were the nonconserved sites (P < 10−11, 2-sided KS test). Perhaps additional, uncharacterized context features remain to be discovered and have been under selective pressure for optimization, leading to the greater observed efficacy of the conserved sites. Alternatively, our model might not be parameterized to fully capture the effects of known features, or for conserved sites evolution might have combined known factors into more optimal, synergistic arrangements.
Despite the difference in magnitude of mRNA destabilization for conserved and nonconserved sites, correspondence between predicted and observed effects for these two classes of sites was indistinguishable in terms of its slope. The strong correspondence when considering only the nonconserved sites alleviated concern that our context features might have merely pointed to conserved sites and thereby artifactually predicted repression primarily through unknown determinants associated with conserved sites. Such a concern would have been particularly relevant for the AU-content and UTR-position features because these features correlated with all nucleotide conservation, not just that matching miRNAs (Supplementary discussion). Further alleviating this concern was the performance with siRNA off-targets, which proved that the model as well its component features functioned well for sites that have not been under any selective pressure for targeting efficacy (Figures 6E and S5). Moreover, local AU content and UTR position were each supported by site-depletion analysis, which was also independent of conservation and had the added benefit of speaking to targeting efficacy in the animal (Figures 3D and 5C).
To illustrate the utility of our model for predicting which sites are more likely to mediate repression, we examined the destabilization of messages with single sites predicted to be in favorable or unfavorable contexts (Figure 6G). For messages with 7mer-m8 sites, the minimal fraction of messages downregulated on the array was 0.49 for the sites in favorable contexts, a substantial discrimination when compared to the 0.09 value observed for sites in unfavorable contexts. Analogous discrimination was observed for the other types of canonical sites.
To evaluate the model further, we constructed reporters designed to test the efficacy of single 7mer-m8 miR-25 sites in either a favorable or an unfavorable context, as predicted by the model. miR-25, which was not among the miRNAs used in the array experiments, was chosen to confirm that the model extended to other miRNAs, and assays were performed in HEK293 cells to confirm that the model extended to cell types other than HeLa cells. UTRs were selected without regard to site conservation. In retrospect, more sites in favorable contexts were conserved, as was expected (6 of 13, compared to 1 of 12).
Nearly all of the sites in favorable contexts yielded significant repression (Figure 6H), whereas none of the sites in unfavorable contexts yielded detectable repression (Figure 6I). Importantly, of those 7mer-m8 sites predicted to be in favorable contexts, the fraction yielding repression in the reporter assay substantially exceeded the 0.49 value derived from the expression array data alone (Figure 6G,6H). Two factors explained this observation. The first was that the arrays underestimated the number of downregulated messages because of their large measurement noise. For example, in a scenario in which every message with a 7mer site was downregulated by 15%, the 7mer expression distribution would shift to the left, but because of the noise of the array experiment (as indicated by the spread of the no-site distribution), only 53% of the sites would be scored as effective. The second factor was that the array monitored only mRNA destabilization, whereas the reporter experiments monitored protein output. Therefore, messages that were repressed translationally with little or no change in mRNA level would yield detectable repression with the reporters but not the arrays. Regardless of the relative contributions of these two factors, the larger extent of repression observed at the protein level with the reporters compared to that observed at the RNA level with the arrays indicated that the context features uncovered in our study were general—they successfully predicted sites that mediated repression at the mRNA level and those that mediated it at the protein level. Their general relevance was expected because these context features were supported by analyses of site conservation and depletion, which are evolutionary consequences of modulating protein output.
Because our approaches searched for general context determinants, we could not exclude the existence of additional, unrelated context determinants specific to targets repressed only at the protein level. However, we have no reason to suspect that such specific determinants exist. Indeed, the absence of detectable targeting for all the assayed sites predicted to fall in non-favorable contexts (Figure 6I) indicated that if such unknown determinants specific to translational repression exist, they either are rarely present or have negligible impact.
In addition to the striking difference in performance between sites in favorable and unfavorable contexts, we observed quantitative differences among sites with indistinguishable context scores (Figure 6H), which showed that some aspect of reporter readout was not yet captured in our model. Such unexplained variability could contribute to the low (though significant) Pearson-correlation r values observed when fitting the array data (e.g., Figure 6D). However, the variability of array data extended to poorly scoring sites, which were nevertheless uniformly ineffective in reporter assays (Figure 6I), indicating that array-measurement noise or effects downstream of actual targeting were major causes of variability. Thus, the low r values would persist even for an improved model that captured all variability attributable to targeting.
Specificity Determinants apply to endogenous miRNA-message interactions
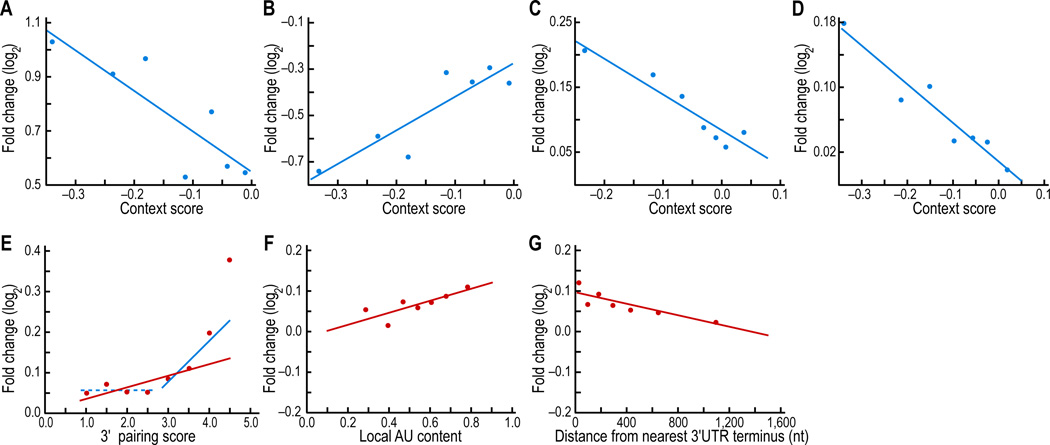
From the results of our conservation and depletion analyses, which examined the evolutionary effects of in vivo miRNA targeting, we anticipated that our model would apply not only to exogenously supplied miRNAs and siRNAs but also to endogenous miRNA-message interactions (Figures 2–6). To confirm this relevance to targeting in vivo, we used the model to predict the effects on endogenous messages after perturbing three miRNAs: knockout and rescue of miR-430 in the zebrafish embryo, inhibition of miR-122 in the adult mouse liver, and ablation of miR-155 in murine T cells (Krutzfeldt et al., 2005; Giraldez et al., 2006; Rodriguez et al., 2007). In each case, context scores of messages with single sites to the relevant miRNA corresponded significantly to the in vivo response, thereby confirming the predictive power of our model for miRNA targeting in vivo (Figure 7A–D; P < 10−7 for each panel, r = −0.26, 0.27, −0.14, −.19, respectively, Pearson correlations). Moreover, for each component feature of the combined model, the predicted repression for messages with single miR-155 sites corresponded significantly to the in vivo response, thereby confirming the relevance of 3' pairing, local AU content and site position for endogenous targeting in the mouse (Figure 7E–G). These data also supported the idea that 3' pairing was consequential for only the small minority of sites with high scores (Figure 7E and S5A).
Figure 7. Relevance of the model and its component features for endogenous miRNA targeting.
(A) Correspondence between context score and in vivo efficacy for endogenous miR-430-target interactions in zebrafish. Messages containing 6mer, 7mer-A1, 7mer-m8, or 8mer sites were analyzed as in Figure 6F, using published array data (Giraldez et al. 2006) monitoring the stabilization of endogenous messages after removing miR-430 and other miRNAs.
(B) Correspondence between context score and in vivo efficacy for miR-430-target interactions after restoring miR-430 to embryos missing all miRNAs (Giraldez et al. 2006), analyzed as in (A).
(C) Correspondence between context score and in vivo efficacy for endogenous miR-122-target interactions in mouse liver. Analyzed as in (A), using published array data (Krutzfeldt et al. 2005) monitoring the stabilization of endogenous messages after inhibiting endogenous miR-122.
(D) Correspondence between context score and in vivo efficacy for endogenous miR-155-target interactions in mouse T cells. Analyzed as in (A), using published array data (Rodriguez et al. 2007) monitoring the stabilization of endogenous messages after deleting miR-155.
(E) Confirmation that the 3' pairing score correlates with endogenous targeting efficacy. Messages containing 6mer, 7mer-A1, 7mer-m8, or 8mer sites were analyzed as in Figure 6A, using published array data (Rodriguez et al. 2007) monitoring the stabilization of endogenous T-cell messages after deleting miR-155. To enable data for all four site types to be considered in aggregate, values were normalized by the average log2 fold change for each site type. Linear regression on all the data (red line), as in Figure 6A, yielded a significant correlation (P = 0.003, r = .10, Pearson correlation). As discussed for the siRNA transfection dataset (Figure S5A), this linear model appeared too simple because messages with sites scoring < 3 displayed no discernable trend (dashed blue line), whereas the handful of messages with high-scoring sites (<3% of sites have scores ≥ 4) were associated with strong de-repression. Linear regression on only the data with scores ≥ 3 (blue line) yielded a significant correlation (P = 0.004, Pearson correlation).
(F) Confirmation that local AU content correlates with endogenous targeting efficacy. Analyzed as in (A), except sites were scored for local AU content, as in Figure 6B (P < 10−3, r = 0.12, Pearson correlation).
(G) Confirmation that site position correlates with endogenous targeting efficacy. Analyzed as in (A), except sites were scored for position, as in Figure 6C (P < 10−4, r = −0.14, Pearson correlation).
Mechanistic Implications
The mechanism of specificity can be explained at least partly in terms of site accessibility and site affinity, which influence the association and dissociation of the silencing complex. For example, the differential efficacy of 8mer, 7mer, and 6mer sites presumably reflects differences in binding affinity. Supplemental pairing outside of the seed region, particularly to nucleotides 13–16 of the miRNA, could further decrease the dissociation rate of the bound silencing complex. In contrast, the local AU context determinant might be associated with weaker mRNA secondary structure in the vicinity of the site and thus increased accessibility to the seed site.
Increased repression with multiple sites could be explained by an increased likelihood of any one site being bound or by a beneficial effect of having more than one silencing complex bound simultaneously. Our observation that overlapping or near-overlapping sites for two different miRNAs yielded less downregulation than did more distantly spaced sites favored the latter possibility. A beneficial effect of having multiple silencing complexes bound simultaneously implied that interactions with the downstream repressive machinery are limiting, either in quantity or duration, for repression. The finding that cooperativity extended to different sites for co-expressed miRNAs ruled out as the mechanism of cooperative action a local-concentration effect in which the second, nearby site merely re-captures a dissociating complex. Instead, the cooperative function of optimally spaced sites might be explained by cooperative contacts with the repressive machinery. Alternatively, it might be explained by binding at one site increasing binding at the other site, either through favorable contacts between silencing complexes or by displacing occlusive mRNA structure.
Additional evidence that miRNA sites must remain bound in order to confer repression came from the reduced site effectiveness in the 5'UTR and ORF, which presumably results from displacement of silencing complexes as the ribosome translocates from the cap-binding complex to the stop codon. We found that this effect extended beyond the stop codon and into the first ~15 nt of the 3'UTR. The length of ~15 nt was in agreement with the observation that mRNA enters the ribosome ~15 nt downstream of the decoding site (Yusupova et al., 2001; Takyar et al., 2005), which would presumably strip off any silencing complex and block rebinding until the ribosome dissociated from the message. The apparent interference by the ribosome along its entire path of translation strongly implied that messages under miRNA control experience at least one complete round of translation prior to or concurrently with their repression, thereby disfavoring models in which an appreciable fraction of messages are sequestered prior to translation of a full-length protein. In contrast to natural miRNA sites, sites perfectly complementary to artificial siRNAs function well in ORFs and thus do not appear subject to ribosome interference. Perhaps the ribosome has more difficulty disrupting the more extensive pairing. Moreover, extensively paired sites might not need to remain associated because they result in catalytic cleavage, whereas 7-8mer sites might require longer periods of association to confer appreciable repression.
The increased efficacy of sites falling near the ends of long UTRs might be attributed either to proximity with the translation machinery or to increased site accessibility. Within the circularized structures of mRNAs, with the poly(A) tail interacting with the 5' cap, sites located in the middle of long 3'UTRs would be furthest from the translational machinery, whereas sites closer to the ORF and 5'UTR might be better situated to interact with the translation machinery and hence induce repression. Proximity would be greater within the 5'UTR or ORF, but such sites would face ribosome interference, leaving sites near the ends of the 3'UTR as the most optimal. Alternatively, if a long UTR is visualized as a cloud of interconverting structures, bound by the ribosome on one end and the polyA-binding proteins on the other, the regions in the middle of the UTR would be less accessible because they would have opportunities to form occlusive interactions with segments from either side, whereas residues near the ends would not.
The strong influence of local AU content was another context determinant suggesting a role for occlusive UTR structure. We followed up on this possibility using predicted local secondary structure with published rubrics to score site accessibility (Robins et al., 2005; Zhao et al., 2005; Long et al., 2007; Zhao et al., 2007). The method of Long et al. (2007) is not yet available for large-scale analyses, but we were able to evaluate the sites of Figure 6H using STarMir (sfold.wadsworth.org), which implements this algorithm. Of the thirteen sites we predicted to be in optimal context, STarMir predicted only two to be functional. When implementing the method of Zhao et al. in a genome-wide analysis, a correlation was observed between downregulation and weaker secondary structure in the vicinity of the site, but secondary-structure prediction was less informative than was local AU content and had no utility after accounting for local AU content (Figure S3). Perhaps direct recognition of A’s and U’s flanking the seed sites is a component of target recognition, in which case scoring local AU content would provide a more reliable measure of this recognition feature than would secondary-structure predictions. Or perhaps because of RNA-binding proteins, RNA tertiary structure, and the compact but multiple competing conformations of arbitrary RNA sequence (Schultes et al., 2005), the details of intracellular UTR structures differ substantially from those of the predicted structures, such that scoring local AU content is significantly more reliable for predicting site accessibility.
To the extent that the context features revealed in our analyses reflected the negative effects of occlusive secondary structure and ribosome interference, we anticipate that they will apply not only to miRNA regulatory sites but also to a range of other elements, including binding sites for regulatory proteins. For instance, messages with many conserved regulatory elements would be associated with higher overall AU content, whereas messages selectively avoiding any regulatory targeting their 3'UTR would benefit from increased GC content that makes any newly emergent fortuitous sites less likely to function, thereby helping to explain the strong correlation between high nucleotide conservation and high global AU composition in 3'UTRs. Likewise, poor accessibility of elements within 15 nt of the stop codon and near the center of long UTRs would explain why other 3'UTR regions have higher nucleotide conservation.
A Resource for Prioritizing Conserved Sites and Predicting Functional Nonconserved Sites
In addition to providing insights and constraints for mechanistic models, the context features described here provide valuable information for selecting which of the many mammalian miRNA:target relationships are most promising for experimental follow-up. Accordingly, for all conserved and nonconserved 7-8mer sites matching known miRNAs, each of these determinants is evaluated and reported (Figure S6; targetscan.org), with the goal of providing a resource for enabling even more rapid progress in understanding this recently appreciated mode of gene regulation.
Experimental Procedures
MicroRNA and mRNA Sequence Data
MicroRNAs conserved in human, mouse, rat, dog, and zebrafish were clustered into 73 families based on miRNA nucleotides 2–8 (Table S1). Human annotated 5'UTR, ORF, and 3'UTR sequences were obtained from RefSeq, and orthologous sequences in mouse, rat, and dog were derived from the UCSC genome browser multiZ multiple genome alignments (Blanchette et al., 2004). When multiple RefSeq identifiers mapped to a single Entrez Gene entry, the RefSeq annotation with the longest UTR was used.
Conservation Analysis
Site-conservation analysis was as in Lewis et al. (2005). In addition, to account for the observation that the four nucleotides are conserved at different rates in mammalian 3'UTRs, control sequences were also limited to those that had identical or nearly identical (within one nucleotide) composition as the authentic sites. The number of conserved control sites for each miRNA was normalized by dividing by the number of control sites in human 3' UTRs and multiplying by the number of miRNA sites in human 3' UTRs.
Array Experiments
HeLa cells were transfected with synthetic miRNA duplexes (Table S2) as described (Lim et al., 2005). Probes were mapped to their representative mRNA sequence through Entrez Gene. Only probes exceeding median intensity in at least half of all experiments were considered for the expression analysis, which limited the analysis to genes expressed at a level sufficient for down-regulation to be readily detectable. When multiple probes matched a single gene, their geometric mean and most significant P value were used. Array data from siRNA transfections was processed analogously.
Unless indicated otherwise, only messages with a single site to the cognate miRNA were considered. For instance, when evaluating single 8mer sites, only genes with a single 8mer in the UTR and no additional 7mers or 6mers were considered. Likewise, when a single 6mer was being considered, genes were excluded in which that 6mer was part of a canonical 7-8mer. Analyses for multiple sites (Figures 1C–1E, 1J) and additional specificity determinants (Figures 2–7) were performed in the same way, requiring the exact numbers of the motif being examined and no other canonical sites, except that additional 6mers were allowed in order to increase the sample sizes, based on the observation that the presence of additional 6mers had marginal effect in the context of stronger sites.
Calculating the Fraction of Genes Downregulated
To determine for each type of site the minimal fraction of genes down-regulated on the array, we compared the cumulative distribution of expression changes for messages with the site versus those with no canonical site, calculating the maximum positive cumulative difference between the two distributions. To correct for bumpiness in the cumulative distributions, we performed 100 permutations in which the down-regulation values for all genes were randomly shuffled, while maintaining the size of the original distributions, and the median maximum positive deviation for these 100 control permutations was subtracted from the value obtained from the real distributions.
Depletion Analysis
Site-depletion analyses were as in Farh et al. (2005) but focused on a subset of tissues and tissue-specific miRNAs with very robust depletion signatures and employed only mouse UTR data. To test the hypothesis that sites falling in one context were more depleted than in the other context, direct comparisons were performed between sites in the two contexts by aggregating the relative expression values for targeted genes in each tissue expressing the cognate miRNA (boxed in the figures).
Simulating Repression from Dual Sites
The simulated dual-site distributions (green lines, Figure 1D–E) were derived by selecting two genes randomly from the single 7mer site distribution and summing their log2 expression changes. This procedure was repeated 100 times for each gene in the dual-site distribution. For comparison to the simulated distributions, the observed dual-site distributions were modified by selecting one gene randomly from the dual site distribution and one gene randomly from the no-site distribution and summing their log2 expression changes (blue lines, Figure 1D–E). This modification was necessary for comparison because by summing two distributions the variance of the resulting distribution greatly increased because microarray measurement noise was compounded. The distributions for single site plus no site and for no site plus no site are also shown for comparison. Genes with dual sites typically had longer 3'UTRs than those with one or no sites; to control for possible length effects, pairs of randomly sampled genes were required to have UTR lengths within 30% of each other.
Regression analyses
For each analysis, significance was determined using both Pearson (parametric) and Spearman (nonparametric) correlation tests. Pearson statistics are reported. With the exception of correlations in Figure 7E and S5A, the significance of the correlations did not substantially differ when using the two tests. Correlations between 3' pairing score and repression (Figure 7E and S5A) were not significant using the Spearman test because this test was insensitive to the influence of a small number of high-scoring sites.
Reporter assays
Assays were as in Farh et al. (2005), except for Figure 1I, in which 25 nM miR-1 and 1 nM miR-133 were co-transfected. Because co-transfection of one miRNA titrated repression by the other, relative miRNA concentrations were adjusted such that both were similarly active when combined. For each construct assayed, multiple independent experiments, each comprising three replicate values, were combined. To combine replicate values from independent experiments, firefly-normalized Renilla values were normalized to the geometric mean of values from otherwise identical constructs in which all sites were disrupted. Values plotted for each construct are the geometric mean of normalized Renilla values from transfection with the cognate miRNA divided by that value from transfection with the noncognate miRNA (Figures 1G–I, 4C). Construct and miRNA sequences are provided (Tables S3–S5). Assays utilizing HEK293 cells were performed identically, except transfections contained 100 ng firefly control reporter, 100 ng TK-Renilla reporter, 1 µg pUC19 and 25 nM miRNA duplex.
Supplementary Material
Acknowledgments
We are grateful to J. Schelter and P. Linsley for providing the microarray data for the miR-133 transfection and to the Rosetta Gene Expression Laboratory for microarray work. Supported by an NIH grant (D.B.) and NIH postdoctoral fellowship (A.G.). D.B. is an HHMI investigator.
References
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Lai EC, Tam B, Rubin GM. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev. 2005;19:1067–1080. doi: 10.1101/gad.1291905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Long D, Lee R, Williams P, Chan CY, Ambros V, Ding Y. Potent effect of target structure on microRNA function. Nat Struct Mol Biol. 2007;14:287–294. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Robins H, Li Y, Padgett RW. Incorporating structure to predict microRNA targets. Proc Natl Acad Sci U S A. 2005;102:4006–4009. doi: 10.1073/pnas.0500775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins H, Press WH. Human microRNAs target a functionally distinct population of genes with AT-rich 3' UTRs. Proc Natl Acad Sci U S A. 2005;102:15557–15562. doi: 10.1073/pnas.0507443102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, Vetrie D, Okkenhaug K, Enright AJ, Dougan G, Turner M, Bradley A. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saetrom P, Heale BS, Snove O, Jr, Aagaard L, Alluin J, Rossi JJ. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucl Acids Res. 2007;35:2333–2342. doi: 10.1093/nar/gkm133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultes EA, Spasic A, Mohanty U, Bartel DP. Compact and ordered collapse of randomly generated RNA sequences. Nat Struct Mol Biol. 2005;12:1130–1136. doi: 10.1038/nsmb1014. [DOI] [PubMed] [Google Scholar]
- Takyar S, Hickerson RP, Noller HF. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- Yusupova GZ, Yusupov MM, Cate JH, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.