SUMMARY
Nuclear shape and size vary between species, during development and in many tissue pathologies, but the causes and effects of these differences remain poorly understood. During fertilization, sperm nuclei undergo a dramatic conversion from a heavily compacted form into decondensed, spherical pronuclei, accompanied by rapid nucleation of microtubules from centrosomes. Here we report that the assembly of the spherical nucleus depends on a critical balance of microtubule dynamics, which is regulated by the chromatin-binding protein Developmental pluripotency-associated 2 (Dppa2). While microtubules normally promote sperm pronuclear expansion, in Dppa2-depleted Xenopus egg extracts excess microtubules cause pronuclear assembly defects leading to abnormal morphology and disorganized DNA replication. Dppa2 inhibits microtubule polymerization in vitro, and Dppa2 activity is needed at a precise time and location during nascent pronuclear formation. This demonstrates a strict spatiotemporal requirement for local suppression of microtubules during nuclear formation, fulfilled by chromatin-bound microtubule regulators.
INTRODUCTION
Eukaryotic nuclei vary widely in size and shape between species and during development (Brandt et al., 2006; Levy and Heald, 2010; Solovei et al., 2013), facilitating specific mechanical functions and gene expression programs (Solovei et al., 2009; Wang et al., 2009). Altered nuclear shape is also linked to many tissue dystrophies and is a chief diagnostic feature of metastatic cancer (Zink et al., 2004; Webster et al., 2009), but the regulation of these changes and their functional implications are poorly understood.
The nucleus is dismantled and reassembled over the course of open mitosis, during which chromosomes are segregated by the microtubule spindle. Both nuclear and microtubule dynamics are regulated by intrinsic, cell cycle-dependent signals from the chromosomes themselves. During M phase, chromatin stimulates microtubule polymerization for spindle assembly (Heald et al., 1996). This is mediated firstly by generation of RanGTP by chromosome-bound RCC1, which liberates microtubule assembly factors from importins around chromatin (Gruss et al., 2001; Nachury et al., 2001; Wiese et al., 2001; Kalab et al., 2002). Secondly, Aurora B, the kinase subunit of the chromosomal passenger complex (CPC), is activated on chromatin and inhibits microtubule-destabilizing factors (Sampath et al., 2004; Gadea and Ruderman, 2006; Kelly et al., 2007; Maresca et al., 2009; Tseng et al., 2010). In interphase, DNA templates switch to driving nuclear envelope assembly (Forbes et al., 1983; Newport, 1987). This process also relies on RCC1 and RanGTP hydrolysis (Zhang and Clarke, 2000; Hetzer et al., 2000; Walther et al., 2003; Harel et al., 2003). In contrast, the CPC inhibits nuclear formation and must be removed from chromatin upon entry into interphase for nuclear assembly (Ramadan et al., 2007; Kelly et al., 2010). The switch from chromatin-driven microtubule assembly to nuclear formation has not been extensively studied. Imaging studies have suggested that remnants of spindle microtubules may physically impede nuclear membrane closure (Haraguchi et al., 2008; Lu et al., 2011), but the spatial and temporal coupling of these large-scale cellular rearrangements remains unclear.
The events of nuclear formation are dramatically illustrated by the changes that occur during normal spermatogenesis and subsequent fertilization. Mature sperm are genetically inert and highly compact, with bulk histones replaced by protamines, and adopt distinct nuclear shapes, ranging from paddle-shaped in humans, hook-shaped in rodents to crescent-shaped in Xenopus (Wright, 1999). Upon exposure to egg cytoplasm, these modifications are reversed as sperm chromatin is decompacted and protamines are exchanged for histones (Philpott et al., 1991). This is followed by recruitment of nuclear envelope proteins and a dramatic shape conversion into a spherical pronucleus that expands with nuclear import (Wright, 1999). Concomitant with pronuclear formation, sperm-associated centrosomes nucleate long astral microtubules, which capture both sperm and egg pronuclei and transport them over millimeter-scale distances to meet in preparation for the first zygotic mitosis (Wühr et al., 2009). Pronuclear assembly, migration and fusion are each essential for fertility, but few modulators of these processes have been described.
Here we report that a poorly-described chromatin-binding protein, Dppa2, couples microtubule disassembly to nuclear formation and is critical for nuclear function. Dppa2 is specifically required to inhibit local microtubule polymerization around chromatin during early nuclear formation. In the absence of Dppa2, excess microtubules lead to distorted nuclear shape and slower, disorganized DNA replication. The activity of Dppa2 opposes the CPC, and normal nuclear morphology is rescued by CPC depletion or depolymerizing microtubules. However, the complete abolition of microtubules delays nuclear expansion, indicating that microtubule dynamics are carefully balanced for proper nuclear formation. At later time points, nuclear assembly is no longer sensitive to microtubule perturbations. Our study therefore reveals a unique, spatially and temporally-restricted regulation of microtubule dynamics, balanced by chromatin-associated factors to ensure proper nuclear formation and function.
RESULTS
The Xenopus chromatin-binding protein Dppa2 is required for nuclear assembly and replication
Cell-free extracts of Xenopus laevis cytoplasm provide a biochemically-accessible system for investigating nuclear assembly and microtubule dynamics (Newport, 1987; Philpott et al., 1991; Murray, 1991). Like most vertebrates, Xenopus eggs are naturally arrested at meiotic metaphase II, and released into interphase upon sperm entry by an intracellular calcium wave. This is mimicked by adding demembranated sperm together with calcium to metaphase-arrested egg extracts, which recapitulates the dramatic conversion of crescent-shaped sperm nuclei into spherical pronuclei and subsequent nuclear expansion (Figure S1A).
We initially identified Dppa2 as a regulator of this process from a proteomics screen for novel chromatin-binding proteins (Figure S1B). Dppa2 (also known as XDppa2/4) is the Xenopus homolog of mammalian pluripotency-associated Dppa2 and Dppa4 (also known as ECAT15-2 and ECAT15-1), and is indispensible for Xenopus embryogenesis (Maldonado-Saldivia et al., 2007; Siegel et al., 2009). To investigate the molecular function of Dppa2, we raised polyclonal antibodies against recombinant Dppa2 protein (Figure S1C). These antibodies recognized a single major protein in Xenopus egg extracts (Figures S1D and S1E).
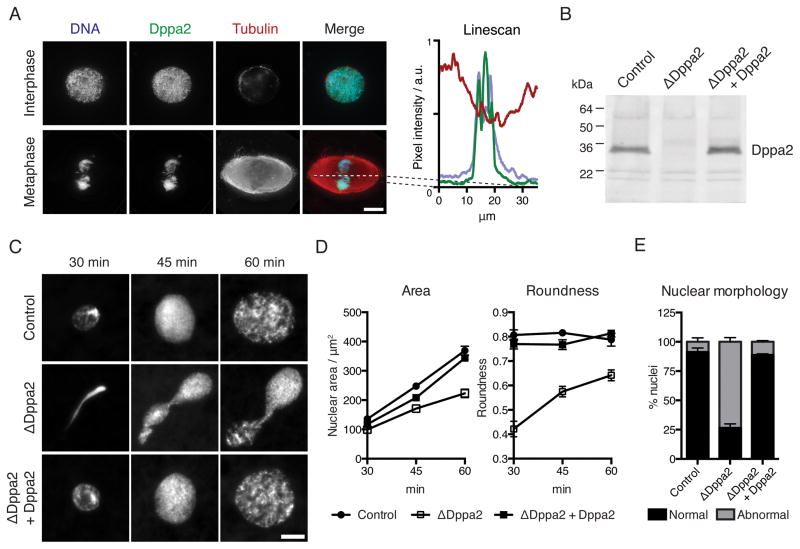
Dppa2 localized uniformly to chromatin in both interphase and metaphase (Figure 1A). We used anti-Dppa2 antibodies to immunodeplete Dppa2 from Xenopus egg extracts (Figure 1B). When sperm nuclei were added to mock-depleted control extracts and released into interphase, they assembled spherical nuclei within 30 min and then continued to expand. In contrast, in Dppa2-depleted extracts (ΔDppa2), sperm formed abnormal, stretched structures that expanded slowly (Figure 1C). We quantified these defects in nuclear size and shape as reductions in nuclear cross-sectional area and roundness (Figure 1D; roundness is defined as the ratio of minor to major axes of a best-fit ellipse). This phenotype was specific to Dppa2 depletion, since it was rescued by adding back recombinant Dppa2 protein to ΔDppa2 extracts (Figures 1C – 1E).
Figure 1. The chromosome-binding protein Dppa2 is essential for sperm pronuclear assembly in Xenopus egg extracts.
(A) Chromosomal localization of Dppa2 protein in interphase and metaphase visualized by immunofluorescence. Right: linescan of fluorescence intensity across the metaphase spindle axis.
(B) Dppa2 protein levels in mock-depleted (Control), Dppa2-depleted (ΔDppa2) and Dppa2-depleted extracts reconstituted with recombinant Dppa2 protein (ΔDppa2 + Dppa2) were analyzed by Western blotting.
(C) Demembranated sperm nuclei were added to metaphase extracts together with calcium to release into interphase and initiate pronuclear assembly. Samples were fixed and stained with Hoechst 33342.
(D) Quantification of nuclear cross-sectional area and roundness from (C). Roundness is defined as the ratio of minor to major axes of a best-fit ellipse. Data shown are mean and standard error from > 30 nuclei per sample in a single representative experiment.
(E) Scoring of abnormal nuclear morphology from 3 independent experiments as performed in (C). Nuclei were fixed at 60 min after calcium addition. Bars indicate mean and standard error, 200 nuclei scored per sample per experiment.
Scale bars, 10 μm.
See also Figure S1.
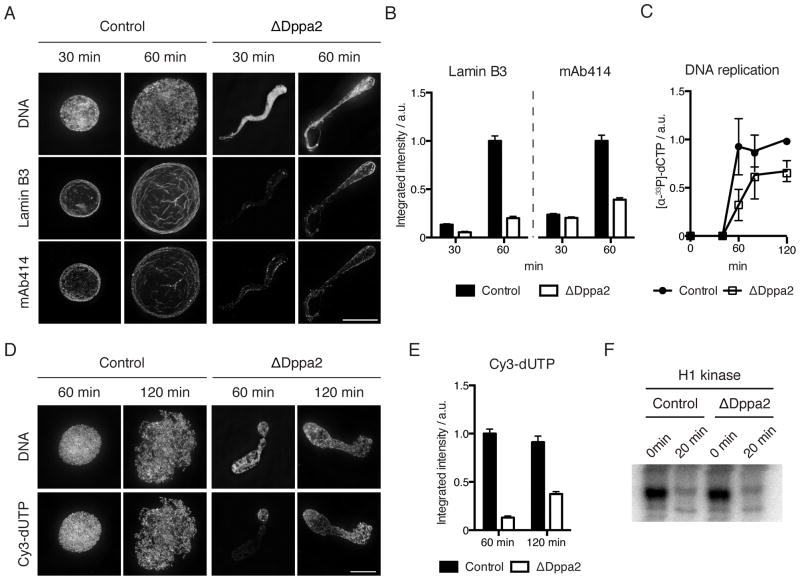
This apparent failure of nuclear assembly was accompanied by reduced incorporation of nuclear lamin and nuclear pore complexes (Figure 2A and 2B) and slower DNA replication (Figures 2C – E). Strikingly, we observed uneven DNA synthesis in ΔDppa2 extracts with regions of DNA failing to incorporate nucleotides (Figure 2D) despite bulk nuclear import not being affected (see Figure 5A). Replication origins are normally uniformly spaced in Xenopus embryos (Blow et al., 2001), suggesting that a nuclear organization defect in ΔDppa2 extracts led to uneven origin firing.
Figure 2. Dppa2 depletion compromises nuclear envelope integrity and leads to disorganized DNA replication.
(A) Visualization of lamin B3 and nuclear pore complexes (mAb414) by immunofluorescence.
(B) Quantification of integrated fluorescence intensity from (A). Bars represent mean and standard error from > 30 nuclei per sample and are representative of 3 independent experiments.
(C) DNA replication assayed by total incorporation of [α-33P]dCTP at the indicated time points. Data shown are mean and standard error from 3 independent experiments.
(D) DNA replication visualized by incorporation of Cy3-dUTP.
(E) Quantification of integrated fluorescence intensity from (D). Bars represent mean and standard error from > 30 nuclei per sample and are representative of 3 independent experiments.
(F) Histone H1 kinase assay was performed at 0 and 20 min after calcium addition.
Scale bars, 10 μm.
See also Figure S2.
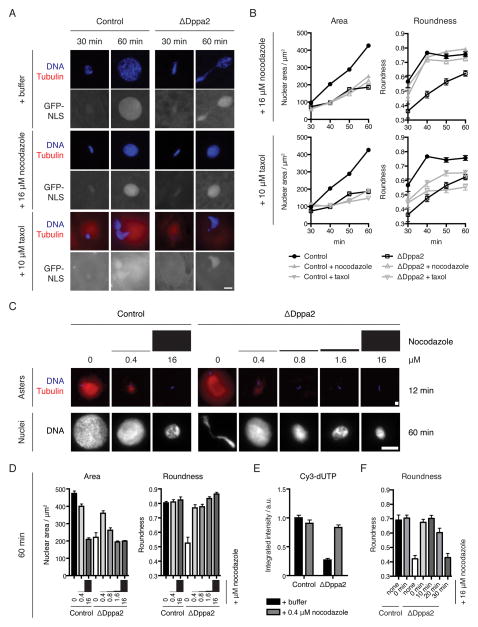
Figure 5. Pronuclear formation requires timely microtubule disassembly and is compromised by persistent microtubules.
(A) The nuclear shape defect in ΔDppa2 extracts is reversed by nocodazole treatment and mimicked by taxol treatment. Demembranated sperm nuclei were added to metaphase extracts together with calcium and 16 μM nocodazole or 10 μM taxol, as well as rhodamine-labeled tubulin (red) and GST-GFP-NLS to monitor nuclear import. Samples were fixed and stained with Hoechst 33342 (blue).
(B) Quantification of nuclear size and shape from (A). Data from nocodazole and taxol-treated samples are shown on separate axes together with untreated samples. Each point represents mean and standard error from > 30 nuclei and is representative of 3 independent experiments.
(C) Low dose nocodazole does not eliminate microtubules and rescues both nuclear expansion and morphology in ΔDppa2 extracts. Nuclei were assembled in extracts treated with the indicated doses of nocodazole. Asters were visualized at 12 min after calcium addition using rhodamine-labeled tubulin (red) and Hoechst 33342 (blue). Nuclei were visualized at 60 min after calcium addition using Hoechst 33342.
(D) Quantification of nuclear size and shape at 60 min from (C). Each bar represents mean and standard error from > 30 nuclei.
(E) Low-dose nocodazole rescues DNA replication in ΔDppa2 extracts. DNA replication was visualized using Cy3-dUTP and quantified as integrated nuclear fluorescence intensity. Each bar represents mean and standard error from > 30 nuclei.
(F) Delayed nocodazole treatment no longer bypasses the requirement for Dppa2. Nuclear assembly was initiated in control and ΔDppa2 extracts, and nocodazole added at the indicated timepoints. Nuclei were fixed and stained at 60 min after calcium addition. Bars indicate mean and standard error from > 30 nuclei per sample.
Scale bars, 10 μm.
See also Figure S5.
These replication defects were not caused by delayed cell cycle exit from metaphase, since both histone H1 kinase activity (Figure 2F) and M phase-specific phosphorylation of histone H3 at threonine 3 (H3T3ph) were downregulated with normal timing (Figure S2A). Sperm remodeling, which involves replacement of sperm protamines with histones upon exposure to egg cytoplasm, was not impaired, since histone H2B was loaded equally in control and ΔDppa2 extracts (Figures S2B and S2C). Similarly, chromosomal loading of RCC1 and the CPC, both regulators of nuclear formation in Xenopus egg extracts (Zhang and Clarke, 2000; Hetzer et al., 2000; Ramadan et al., 2007), was also not affected by Dppa2 depletion (Figure S2D). The nuclear morphology defects observed in ΔDppa2 extracts were not a consequence of impaired DNA replication, since inhibiting replication using recombinant non-degradable geminin (McGarry and Kirschner, 1998) or aphidicolin did not lead to comparable nuclear defects (Figures S2E and S2F).
Dppa2 is a direct inhibitor of microtubule assembly
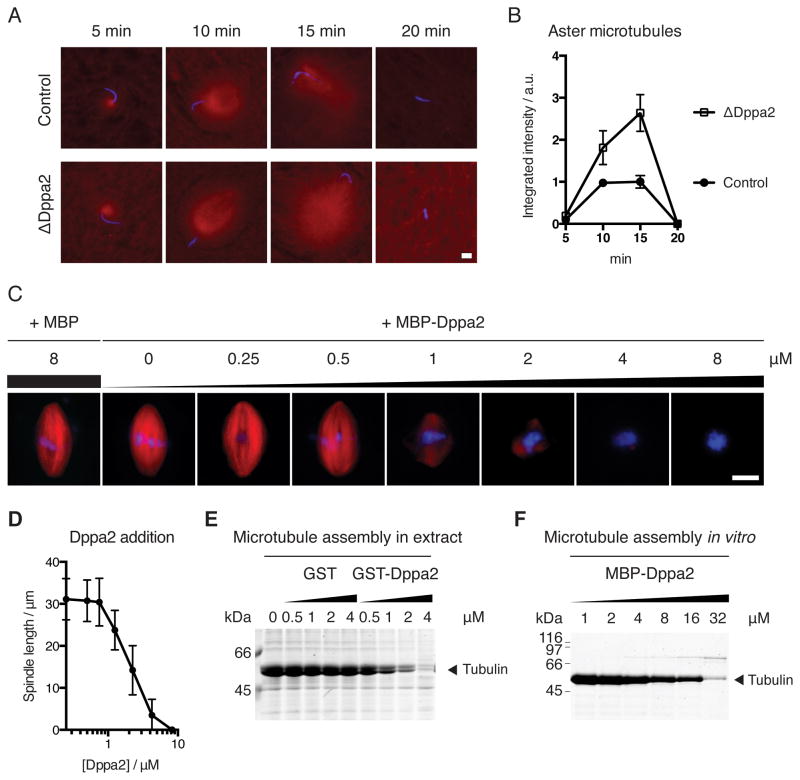
During fertilization, microtubule asters are rapidly nucleated from sperm centrosomes, which eventually capture both sperm and egg nuclei and transport them to the center of the egg for zygotic fusion (Wühr et al., 2009). However, as the egg is simultaneously released from meiotic metaphase into interphase, microtubules become less dynamic due to decreased Cdk1 activity (Verde et al., 1990; Belmont et al., 1990; Verde et al., 1992; Niethammer et al., 2007). We observed this transition in microtubule behavior after adding sperm to egg extracts together with calcium. Microtubules were rapidly nucleated from sperm centrosomes, peaking within 15 min but then diminishing by 20 min (Figure 3A, top). At later time points, interphase microtubules were longer but less dense, to the extent that they were not visible under our usual fixation conditions (compare Figure 3A with Figure S3A).
Figure 3. Dppa2 inhibits microtubule assembly around chromatin and in vitro.
(A) Dppa2 inhibits sperm aster microtubule assembly. Demembranated sperm nuclei were added together with calcium to metaphase extracts supplemented with rhodamine-labeled tubulin (red). Samples were fixed and stained with Hoechst 33342 (blue).
(B) Quantification of tubulin fluorescence intensity from (A). Data shown indicate mean and standard error from > 30 asters per sample and are representative of 3 independent experiments.
(C) Dppa2 inhibits spindle assembly in a dose-dependent manner. Metaphase spindles were assembled in extracts supplemented with MBP-Dppa2 fusion proteins and rhodamine-labeled tubulin.
(D) Quantification of spindle length from (C). Data shown are mean and standard deviation from 30 spindles per sample.
(E) Chromatin-independent microtubule assembly in Xenopus egg extracts. Recombinant GST-Dppa2 protein was added to metaphase extracts together with 0.5 % DMSO. Polymerized microtubules were recovered by pelleting and analyzed by Coomassie staining.
(F) Dppa2 inhibits microtubule polymerization in vitro. MBP-Dppa2 was added to purified bovine tubulin together with 0.5 % DMSO. Polymerized microtubules were recovered by pelleting and analyzed by Coomassie staining.
Scale bars, 10 μm.
See also Figure S3.
During this time, Dppa2 was localized exclusively to chromatin (Figures S3B and S3C) just as in metaphase (Figure 1A). However, in ΔDppa2 extracts we observed greater microtubule growth during the initial nucleation phase (Figures 3A and 3B), suggesting that Dppa2 inhibits microtubule assembly. In support of this idea, addition of recombinant Dppa2 fused to maltose-binding protein (MBP-Dppa2; Figure S3D) to metaphase extracts inhibited spindle assembly in a dose-dependent manner (Figure 3C). We estimated the concentration of endogenous Dppa2 to be around 400 nM, and addition of 4 μM MBP-Dppa2 or more essentially abolished spindle assembly (Figures 3C and 3D). This activity was more potent than that of Op18/stathmin, an established inhibitor of microtubule assembly, which requires greater than 15 μM for comparable inhibition of spindle formation (Houghtaling et al., 2009). Both untagged recombinant Dppa2 and GST-tagged Dppa2 proteins also showed the same inhibition of spindle assembly as MBP-Dppa2 (Figures S3E, S3F and data not shown).
The RanGTP and CPC microtubule assembly pathways are restricted to chromatin in order to prevent ectopic spindle formation in the absence of chromosomes (Kalab et al., 2002; Caudron et al., 2005; Athale et al., 2008; Kelly et al., 2007; Tseng et al., 2010). We asked therefore whether the activity of Dppa2 is similarly limited to chromatin-dependent microtubule assembly. Microtubule polymerization can be stimulated without chromatin by adding dimethyl sulfoxide (DMSO) to Xenopus egg extracts or purified tubulin (Budde et al., 2006). We found that recombinant Dppa2 inhibited DMSO-induced microtubule polymerization both in egg extracts (Figure 3E; see also Figure 6D) and from purified bovine tubulin in vitro (Figure 3F) in a dose-dependent manner. Higher concentrations of recombinant Dppa2 were required to suppress polymerization of purified tubulin compared to tubulin in Xenopus egg extracts (Figure 3C – 3E), which may reflect the more dynamic behavior of microtubules in extracts compared to purified systems (Kinoshita et al., 2001), and/or the contribution of additional cofactors or modifications to Dppa2 in egg extracts.
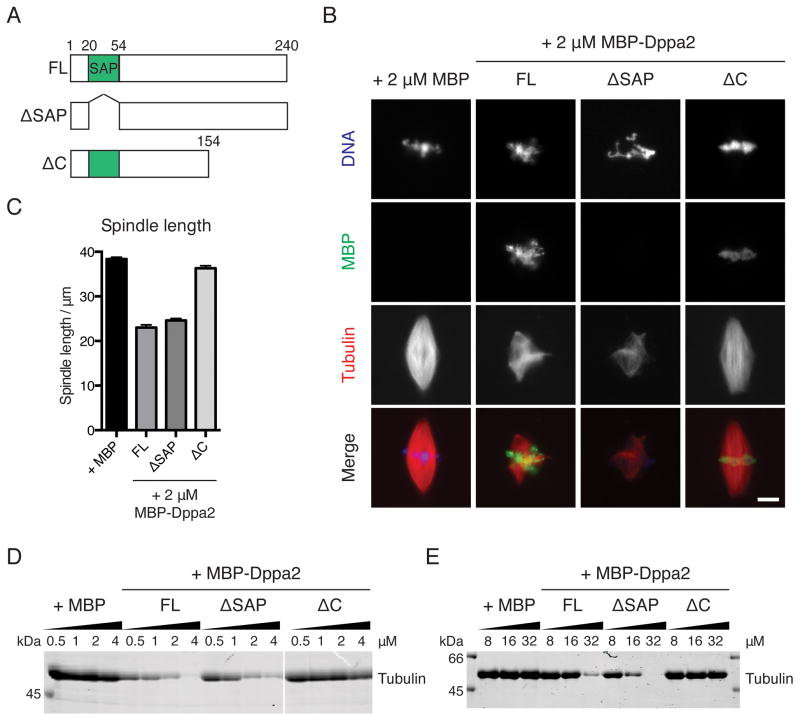
Figure 6. Dppa2 requires its C terminus but not DNA-binding to inhibit microtubule assembly.
(A) Schematic of Dppa2 deletion constructs used.
(B) Inhibition of spindle assembly requires the Dppa2 C terminus but not DNA-binding. Metaphase spindles were assembled in extracts supplemented with 2 μM MBP-Dppa2 fusion proteins. MBP-Dppa2 localization was visualized by immunofluorescence using an anti-MBP antibody. Scale bar, 10 μm.
(C) Quantification of spindle length in (B). Bars represent mean and standard error from 30 spindles and are representative of 3 independent experiments.
(D) Chromatin-independent microtubule assembly in Xenopus egg extracts. MBP-Dppa2 proteins were added to metaphase extracts together with 0.5 % DMSO. Polymerized microtubules were recovered by pelleting and analyzed by Coomassie staining.
(E) Polymerization of purified tubulin in vitro. Purified bovine tubulin was treated with MBP-Dppa2 proteins and 0.5 % DMSO, and polymerized microtubules analyzed by Coomassie staining.
Dppa2 functionally opposes the CPC
Our data indicated that Dppa2 promotes pronuclear formation while inhibiting microtubule assembly. These activities contrast with the known functions of the CPC, which promotes spindle assembly but inhibits nuclear formation in Xenopus egg extracts (Sampath et al., 2004; Ramadan et al., 2007). We discovered that the CPC functionally opposes Dppa2, since the increase in microtubule assembly observed in ΔDppa2 extracts was reduced to control levels following co-depletion of the CPC (Figures 4A – 4C). Depletion of the CPC also rescued the nuclear roundness defect of Dppa2 depletion, although it did not restore nuclear size (Figures 4D and 4E). However, Dppa2 is not simply an upstream repressor of the CPC, since excess Dppa2 did not affect CPC localization or Aurora B substrate phosphorylation (Figures S4A and S4B), nor centromeric localization of MCAK (Figure S4C) which is CPC-dependent (Lan et al., 2004). Furthermore, Dppa2 was still able to inhibit microtubule assembly in the absence of the CPC (Figure S4D). Thus, Dppa2 suppresses microtubules independently of the CPC, but does so to an equal and opposite extent as CPC-induced microtubule assembly.
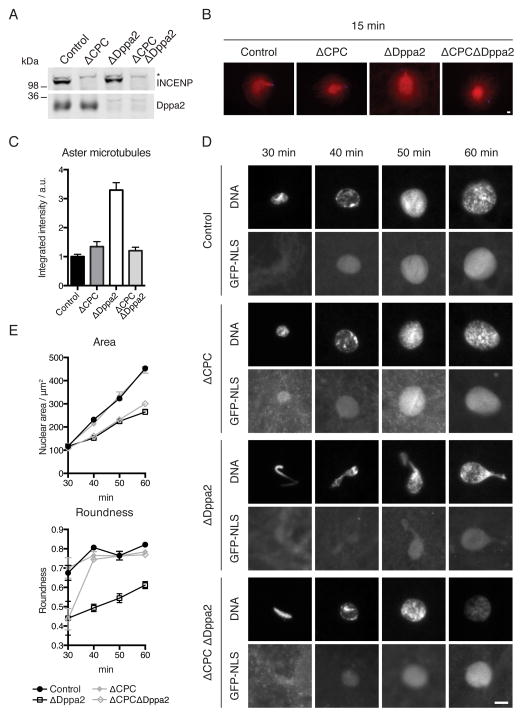
Figure 4. CPC depletion rescues ΔDppa2 phenotypes.
(A) CPC was depleted from extracts using anti-INCENP antibodies (Sampath et al., 2004) and depletion efficiency assessed by Western blotting. * indicates non-specific reactivity.
(B) Sperm nuclei were added together with calcium to metaphase extracts containing rhodamine-labeled tubulin (red). Sperm-associated asters were fixed after 15 min and stained with Hoechst 33342 (blue).
(C) Quantification of tubulin intensity from (B). Bars indicate mean and standard error of > 30 asters from each sample and are representative of 3 independent experiments.
(D) Sperm nuclei were added together with calcium to metaphase extracts supplemented with GST-GFP-NLS. Nuclei were fixed and visualized with Hoechst 33342.
(E) Quantification of nuclear area and roundness from (D). Bars indicate mean and standard error of > 30 nuclei from each sample and are representative of 3 independent experiments.
Scale bars, 10 μm.
See also Figure S4.
Nuclear formation requires timely microtubule disassembly by Dppa2
Since CPC depletion rescued both the presence of additional microtubules and abnormal nuclear morphology in ΔDppa2 extracts (Figure 4), we reasoned that excess microtubules in ΔDppa2 extracts might have perturbed concomitant nuclear formation. We therefore asked whether this nuclear morphology defect could be reversed by ectopically depolymerizing microtubules. Indeed, in the presence of 16 μM nocodazole, sperm assembled into spherical nuclei in ΔDppa2 extracts with no measurable shape defect, demonstrating that nuclear distortion in the absence of Dppa2 depends on microtubules (Figures 5A and 5B). Conversely, stabilizing excess microtubules by treatment with 10 μM taxol mimicked the effect of Dppa2 depletion, leading to nuclear size and shape defects in control extracts that were comparable to ΔDppa2 extracts (Figures 5A and 5B).
However, we noted that treatment with 16 μM nocodazole resulted in decreased nuclear size in control extracts (Figures 5A and 5B). Abolishing microtubules using colcemid or with a triple alanine mutant and active form of Op18 (Op18AAA; (Budde et al., 2001) had the same effect (Figure S5A). This delay in nuclear expansion following microtubule depolymerization was not due to activation of the spindle assembly checkpoint (SAC), since much higher concentrations of sperm are typically required to activate the SAC (Minshull et al., 1994), and we confirmed that microtubule depolymerization did not delay dephosphorylation of M phase-specific H3T3ph in our assays (Figure S5B). Similarly, both nocodazole and taxol treatment led to decreased nuclear size even in ΔCPC extracts (Figure S5C) where the SAC is inactivated (Vigneron et al., 2004). We titrated down the dosage of nocodazole and found similar effects until we reached 0.4 μM, which no longer fully eliminated microtubules but largely did not affect nuclear expansion (Figure 5C). Unlike higher doses, this low concentration was able to rescue both nuclear expansion and roundness in ΔDppa2 extracts (Figures 5C and 5D). The same concentration also restored normal DNA replication in ΔDppa2 extracts (Figure 5E). Thus we conclude that the precise balance of microtubule dynamics is critical to nuclear assembly, and even slight deviations towards excessive or insufficient microtubule polymerization can prevent formation of normal, spherical nuclei.
We reasoned that suppression of microtubule assembly by Dppa2 might be particularly important for maintaining this balance during the early, rapid and highly dynamic phase of aster growth that we observed at the metaphase-interphase transition (Figure 3A). Indeed, we found that nocodazole could no longer bypass the requirement for Dppa2 in maintaining nuclear shape if treatment was delayed until 30 min (Figure 5F). Similarly, inducing ectopic microtubules with taxol at 30 min did not significantly perturb subsequent pronuclear assembly (Figure S5D). Taken together, these data suggest that Dppa2 activity is required during an early time window when microtubule structures undergo dynamic rearrangements and impinge on nuclear assembly, after which nuclei are committed to normal or abnormal expansion and morphology irrespective of microtubule status.
Nuclear assembly requires local inhibition of microtubules by chromatin-bound Dppa2
The exclusive localization of Dppa2 on chromatin rather than microtubules (Figures 1A and S3B) suggested that the spatial distribution of Dppa2 activity might be important for its function. To test this hypothesis, we carried out domain deletion mutagenesis on Dppa2. Dppa2 contains a conserved SAF-A/B, Acinus and PIAS (SAP) DNA-binding domain (Figure 6A; Siegel et al., 2009). Deletion of the SAP domain (MBP-Dppa2 ΔSAP) abrogated binding to chromatin, but this mutant inhibited spindle assembly equally effectively as full-length Dppa2 (MBP-Dppa2FL). In contrast, deletion of the C-terminal 86 amino acids (MBP-Dppa2ΔC) retained chromatin binding, but no longer inhibited spindle assembly (Figures 6B and 6C).
We observed the same dependencies in the absence of chromatin both in egg extracts and in vitro. When microtubule assembly was stimulated in egg extracts without chromatin by adding DMSO, MBP-Dppa2FL and MBP-Dppa2ΔSAP, but not MBP-Dppa2ΔC, inhibited this assembly in a dose-dependent manner (Figure 6D). Similarly, MBP-Dppa2 also inhibited polymerization of purified bovine tubulin in a manner dependent on its C terminus (Figure 6E).
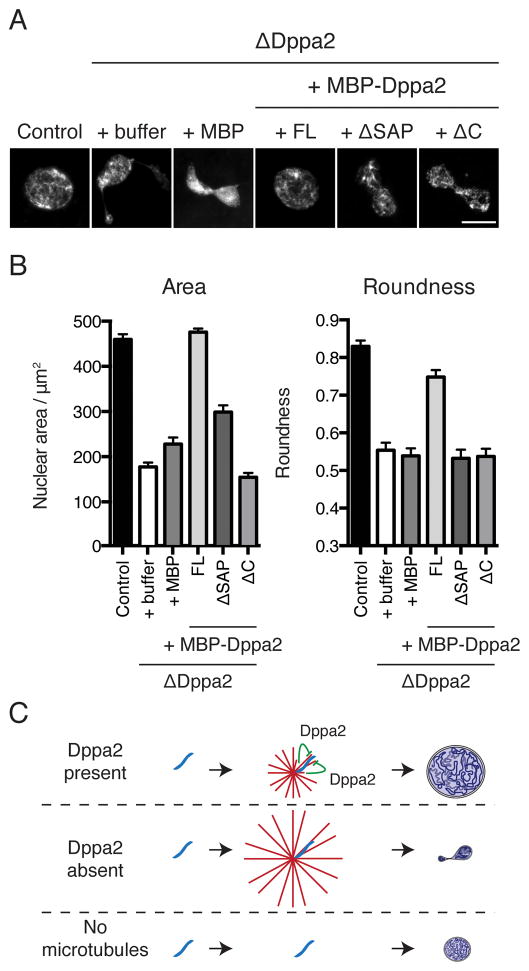
Although DNA-binding was dispensable for inhibition of microtubule assembly by Dppa2 (Figure 6), we discovered that it was critical for nuclear formation. Adding back endogenous levels of MBP-Dppa2ΔSAP, which inhibited microtubule polymerization but did not localize to chromosomes, could not recover the nuclear assembly defects of Dppa2 depletion, nor could MBP-Dppa2ΔC, which bound chromosomes but did not inhibit microtubule polymerization (Figures 7A and 7B). This observation is consistent with the hypothesis that both chromatin-binding and inhibition of microtubule assembly must be coupled to support nuclear formation. Taken together, we conclude that normal nuclear assembly requires temporally and spatially-restricted microtubule assembly, mediated by Dppa2 localized on chromatin (Figure 7C).
Figure 7. Pronuclear assembly requires localized microtubule disassembly by chromatin-bound Dppa2.
(A) Both DNA-binding and microtubule-inhibitory domains of Dppa2 are indispensable for pronuclear formation. Nuclear assembly was initiated in control and ΔDppa2 extracts reconstituted with MBP-Dppa2 fusion proteins to endogenous levels (400 nM). Nuclei were fixed at 60 min after release into interphase and stained with Hoechst 33342. Scale bar, 10 μm.
(B) Quantification of nuclei from (A). Bars indicate mean and standard error from > 30 nuclei per sample and are representative of 3 independent experiments.
(C) Dppa2 is required during nascent pronuclear formation to inhibit local microtubule assembly on chromosomes. Excess microtubules compromise nuclear shape, while completely abolishing microtubules impairs nuclear expansion.
DISCUSSION
In this study we identified Dppa2 as a regulator of microtubule dynamics that inhibited microtubule polymerization in vitro, whose activity was required for nuclear assembly and organized DNA replication. Dppa2 regulated microtubules during an early time window, when we found nuclear formation to be uniquely sensitive to altered microtubule dynamics. Stabilization of microtubules by Dppa2 depletion or taxol treatment during this window may cause defects by physically obstructing nuclear envelope closure (Haraguchi et al., 2008; Lu et al., 2011). However, this cannot fully explain the nuclear morphology defect that we observed, since abolishing microtubules at a later time point, thereby removing such obstacles, was insufficient to restore normal nuclear shape (Figure 5C). It is possible that, instead, these nuclei were irreversibly damaged by microtubule-dependent shear stress, given the reduced lamin assembly and weakening of the nuclear envelope in ΔDppa2 extracts (Figure 2A). During entry into mitosis, microtubules and cytoplasmic dynein bind and tear the nuclear envelope to facilitate nuclear envelope breakdown (Beaudouin et al., 2002; Salina et al., 2002), and excess microtubules at mitotic exit may allow similar forces to disrupt the partially assembled nuclear envelope. Once nuclear envelope assembly is complete, the nuclear lamina structure may acquire enough strength to resist microtubule-dependent forces.
Meanwhile, although completely abolishing microtubules with nocodazole did not interfere with the shape conversion of sperm into spherical nuclei, nuclear expansion was delayed (Figures 5A and 5B). This may be explained by microtubules helping to deliver membrane vesicles and nuclear pore complexes to the nascent nucleus (Waterman-Storer et al., 1995; Ewald et al., 2001), underscoring both positive and negative roles of microtubules and hence a need for tight regulation of microtubule dynamics during nuclear assembly.
Dppa2ΔSAP, which maintained the capacity to inhibit microtubule assembly but did not localize to chromatin, failed to support proper pronuclear assembly (Figure 7), suggesting that Dppa2 is needed to suppress microtubules in the immediate vicinity of nascent nuclei. The spatial restriction of Dppa2 activity to chromosomes may prevent perturbation of global microtubule dynamics, which could otherwise compromise nuclear expansion and nuclear positioning. This suppression of microtubules by a chromosomal factor contrasts with the known regulators of the RanGTP and CPC pathways, which stabilize microtubules around chromatin (Gruss et al., 2001; Nachury et al., 2001; Wiese et al., 2001; Sampath et al., 2004; Gadea and Ruderman, 2006; Maresca et al., 2009). At the exit from M phase, the CPC is inactivated by two mechanisms. First, the CPC is recruited to mitotic chromatin by H3T3ph, and this mark is rapidly dephosphorylated upon entering interphase (Wang et al., 2010; Kelly et al., 2010; Yamagishi et al., 2010). Second, CPC subunits are ubiquitylated and removed from chromatin by the Cdc48/p97 ATPase (Ramadan et al., 2007; Dobrynin et al., 2011). We demonstrate that these inactivation mechanisms alone are not sufficient to support nuclear formation; Dppa2 is required for additional suppression of microtubules, since Dppa2 depletion caused excess microtubules in a CPC-dependent manner (Figure 4A). Conversely, CPC depletion rescued nuclear morphology but not the nuclear size defect observed in ΔDppa2 extracts, indicating that Dppa2 must play additional roles in nuclear formation beyond simply opposing the CPC.
Dppa2 is highly expressed in Xenopus eggs and the early embryo until the mid-blastula transition, and antisense morpholino-mediated knockdown of Dppa2 leads to defective gastrulation (Siegel et al., 2009). This suggests that Dppa2 may be specifically required for early embryonic events, including fertilization and the embryo’s first mitotic divisions. Our experiments recapitulate the events of fertilization, when sperm chromatin is decompacted and assembled into a spherical pronucleus. In the absence of Dppa2, we observed nuclear morphology defects and disorganized replication, and decreased pronuclear size is known to result in fertilization failure (Meyerzon et al., 2009). This underscores the importance of nuclear formation processes, especially since infertility remains a poorly understood phenomenon. Dppa2 may also assist in accelerating nuclear formation during the rapid, 30-minute cell division cycles of early cleavage stage embryos. In particular, early embryos of Xenopus and zebrafish assemble nuclear envelopes around individual anaphase chromosomes, forming micronuclei known as karyomeres, in order to initiate DNA synthesis prior to completion of mitosis (Lemaitre et al., 1998; Schoft et al., 2003). This may place additional demands on the nuclear assembly machinery, necessitating specialized mitotic mechanisms.
Our study unveils a specific window of time during the formation of nascent nuclei when dynamic microtubules can leave critical impacts on the size and shape of the nucleus. Organized DNA replication depends on proper regulation of these processes. To this end, chromatin-bound Dppa2 mediates local microtubule disassembly in a temporally-restricted manner. Further studies should examine nuclear formation and address how microtubule and nuclear dynamics are spatially and temporally coupled in other systems, since altered nuclear structure is linked to many diseases (Webster et al., 2009; Zink et al., 2004), and accumulated defects in nuclear reassembly may contribute to age-related loss of nuclear integrity (Haithcock et al., 2005).
EXPERIMENTAL PROCEDURES
Xenopus Egg Extracts and Identification of Dppa2
CSF-arrested Xenopus laevis egg extracts were prepared as previously described (Murray, 1991). DNA beads without DNA ends were prepared as previously described (Heald et al., 1996; Postow et al., 2008). Chromatin was assembled on DNA beads in metaphase and interphase egg extracts, and chromatin-bound proteins analyzed by mass spectrometry. See Supplemental Experimental Procedures for further details.
Recombinant Proteins and Anti-Dppa2 Antibodies
GST-tagged Dppa2 was expressed in E. coli from pGEX-6p-1, and purified using glutathione sepharose 4b resin (GE Healthcare). The GST tag was removed using Prescission protease (GE Healthcare). Untagged protein was used to immunize rabbits for polyclonal antibody production (Covance). MBP-tagged Dppa2 was expressed in E. coli and purified using amylose resin (NEB). See Supplemental Experimental Procedures for further details.
Immunodepletion
Egg extracts containing 100 μg/ml cycloheximide were immunodepleted by incubation at 4 °C with antibodies pre-bound to magnetic protein A beads (Invitrogen; see Supplemental Experimental Procedures). To deplete 100 μl extract, 100 μl beads were coupled to 10 μg rabbit IgG or polyclonal anti-Dppa2 antibodies, and 2 rounds of depletion performed using 50 μl beads and 1 h incubation per round. To deplete the CPC from 100 μl extract, 2 rounds of 100 μl beads per round were used.
Western blots
Immunoblots were blocked with 4 % nonfat dry milk in PBS for 1 h at room temperature. Primary antibodies were diluted in Odyssey Blocking Buffer (Li-Cor) as follows: 5 μg/ml anti-Dppa2; 1:1,000 anti-H3 (Abcam ab1791), 5 μg/ml anti-INCENP (Sampath et al., 2004); 1 μg/ml anti-RCC1 (gift of Rebecca Heald), 3 μg/ml anti-Xkid (Funabiki and Murray, 2000). IRDye 800CW and 680LT secondary antibodies were used at 50 ng/ml and detected on an Odyssey infrared imaging system (Li-Cor).
Nuclear and Spindle Assembly
Demembranated sperm were added to metaphase extracts at 500/μl together with 0.3 mM CaCl2 to induce release into interphase and nuclear assembly. For subsequent spindle assembly, 3 volumes of additional metaphase extracts were added after 1 h 30 min, and incubated at 20 °C for 1 h to cycle back into metaphase. Chromosomes were visualized by squashing 1 μl extract with 3 μl fixative (5 mM HEPES, 100 mM NaCl, 50 % glycerol, 10 % formaldehyde, 1 μg/ml Hoechst 33342, pH 7.7) under an 18 × 18 mm square coverslip. Nuclear import was visualized by supplementing extracts with 1 μM GST-GFP-NLS (gift of Cristina Ghenoiu), expressed and purified from E. coli using pMD49 (gift of Satoru Mochida and Mary Dasso). Spindles were visualized by supplementing extracts with 0.2 μM bovine tubulin labeled with rhodamine succinimidyl ester (Invitrogen C-1309). Nocodazole (Sigma M1404) was used at 0.4 to 16 μM, taxol (Sigma T7402) at 10 μM and colcemid (Fisher A430033M001) at 13 μM. GST-Op18AAA (gift of Jessica Rosenberg; Budde et al., 2001) was used at 4 μM.
Sperm Centrosome Aster Assembly
Sperm were added together with calcium to metaphase extracts containing 0.2 μM rhodamine-labeled tubulin and incubated at 20 °C. Samples were withdrawn and fixed at regular time points.
Immunofluorescence Microscopy
Nuclei and spindles were processed for immunofluorescence as described (Funabiki and Murray, 2000). Briefly, 10 μl extract samples were fixed in 2 % formaldehyde and spun down onto glass coverslips. Primary antibodies were diluted in AbDil (10 mM Tris, 150 mM NaCl, 2 % BSA, 0.1% Triton X-100, pH 7.4) as follows: 6 μg/ml anti-Dasra A (Sampath et al., 2004); 0.2 μg/ml anti-Dppa2; 2 μg/ml anti-GFP (Roche 11814460001); 1 μg/ml anti-H3S10ph (mAb 7G1G7, gift of Hiroshi Kimura); 1:200 anti-lamin B3 (gift of Dale K. Shumaker); 0.2 μg/ml mAb414 (Covance MMS-120P); 0.2 μg/ml anti-MCAK (gift of Ryoma Ohi); 1 μg/ml anti-MBP (NEB E8032), 1:1,000 anti-α-tubulin (Sigma T9026). Alexa Fluor 488 and Cy3-conjugated secondary antibodies (Jackson ImmunoResearch) were used at 1.5 μg/ml, DNA was stained with 1 μg/ml Hoechst 33342 (Sigma B2261).
Quantification of Microscope Images
Nuclei and spindles were identified from images by global thresholding in the DNA or tubulin channels respectively. Nuclear cross-sectional area and roundness were measured using ImageJ (NIH). Spindle length was defined as the Feret diameter (the longest distance between 2 points on the perimeter) of thresholded spindles.
Sperm Replication
Nuclei were assembled in extracts supplemented with 1 kBq/μl [α-33P]dCTP (PerkinElmer) or 10 μM Cy3-dUTP (GE Healthcare). To quantify incorporation of radiolabeled nucleotides, 15 μl extract samples were taken at various time points and added to 200 μl stop buffer (20 mM Tris, 20 mM EDTA, 0.5 % SDS, pH 8) containing 50 μg/ml RNase A and incubated at 37 °C for 15 min. A further 200 μl stop buffer containing 1 mg/ml proteinase K was then added, and the reaction incubated at 37 °C for 1 h. Samples were then extracted twice with 400 μl phenol-chloroform and once with 400 μl chloroform. DNA was precipitated by addition of 40 μl 3 M sodium acetate and 1 ml ethanol and incubation at 4 °C for 15 min, and pelleted by centrifugation at 16,000 g for 30 min at 4 °C. DNA was then resuspended in 15 μl TE buffer (10 mM Tris, 1 mM EDTA, pH 8) containing 50 μg/ml RNase A. 10 μl of each sample was loaded on an agarose gel, which was dried and exposed to a PhosphorImager screen (Fujifilm). To quantify incorporation of Cy3-dUTP, nuclei were spun down for immunofluorescence as described above. Replication was inhibited by addition of 8 μM recombinant non-degradable geminin (gift of Christian Zierhut; McGarry and Kirschner, 1998) or 40 μM aphidicolin (Fisher BP615).
Histone H1 Kinase Assay
1 μl samples of metaphase and interphase Xenopus egg extracts were snap-frozen in liquid nitrogen and stored at −80 °C. Samples were then thawed by bringing up to 10 μl in kinase buffer (15 mM MgCl2, 20 mM EGTA, 80 mM β-glycerophosphate, 0.1 % Igepal CA-630, 1 mM DTT, 50 μM ATP, 10 μg/ml leupeptin, 10 μg/ml pepstatin, 10 μg/ml chymostatin, 125 μg/ml histone H1, 60 μCi/ml [γ-32P]ATP). The reaction mixture was incubated at 30 °C for 10 min and stopped by addition of 30 μl SDS sample buffer. 10 μl was analyzed on a 15 % polyacrylamide gel, dried and exposed to a PhosphorImager screen.
DMSO-Induced Microtubule Assembly in Egg Extracts
DMSO-induced microtubule assembly in CSF extracts was performed as described (Budde et al., 2006). Briefly, 5 % DMSO was added to 20 μl metaphase egg extracts, and incubated at 20 °C for 30 min. The extract was diluted in 0.4 ml BRB80 (80 mM PIPES, 1 mM MgCl2, 1 mM EGTA, pH 6.9) + 30 % glycerol + 1 % Triton X-100, and layered over 0.8 ml BRB80/40 % glycerol in microcentrifuge tubes. Microtubules were pelleted by centrifugation at 16,000 g for 15 min at room temperature. The supernatant was removed and the cushion interface washed with 2 × 0.4 ml ddH2O. The cushion was then removed and the pellet resuspended in 40 μl SDS sample buffer.
DMSO-Induced Assembly of Purified Tubulin
For in vitro microtubule assembly, MBP or MBP-Dppa2 proteins were added to 20 μM bovine tubulin in 10 μl volume of BRB80/1 mM DTT/1 mM GTP on ice. 5 % DMSO was then added and the mixture warmed to 37 °C for 30 min. The mixture was layered over 0.5 ml warm BRB80/40 % glycerol and pelleted at 90,000 rpm (14.5 k-factor) in a TLA120.1 rotor (Beckman) for 5 min at 37 °C. The supernatant was removed and the cushion interface washed with 2 × 0.2 ml ddH2O. The cushion was removed and the pellet resuspended in 30 μl SDS sample buffer.
Supplementary Material
HIGHLIGHTS.
Nuclear formation in Xenopus depends on the chromatin-binding protein Dppa2
Dppa2 directly inhibits microtubule polymerization
Perturbation of microtubules compromises nuclear morphology and replication
Microtubule dynamics must be balanced spatially and temporally for nuclear formation
Acknowledgments
We thank members of the Funabiki lab for discussions; A. Clark and L. Fish for technical assistance; M. Dasso, C. Ghenoiu, R. Heald, C. Jenness, H. Kimura, S. Mochida, R. Ohi, J. Rosenberg, D. Shumaker and C. Zierhut for reagents. Microscopy was performed at the Rockefeller University Bio-Imaging Resource Center. This work was supported by the NIH (GM075249 to H.F., RR00862 to B.T.C., CA09673 to L.P.), HHMI (E.M.W.) and the Leukemia and Lymphoma Society (L.P.). H.F. was supported by a Searle Scholarship, the Alexandrine and Alexander Sinsheimer Fund and the Irma T. Hirschl/Monique Weill-Caulier Trust.
Footnotes
AUTHOR CONTRIBUTIONS
J.Z.X., E.M.W., B.T.C. and H.F. conceived experiments. E.M.W. and L.P. independently identified Dppa2. J.Z.X. and E.M.W. performed experiments. J.Z.X., E.M.W. and H.F. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Athale C, Dinarina A, Mora-Coral M, Pugieux C, Nédélec FJ, Karsenti E. Regulation of microtubule dynamics by reaction cascades around chromosomes. Science. 2008;322:1243–1247. doi: 10.1126/science.1161820. [DOI] [PubMed] [Google Scholar]
- Beaudouin J, Gerlich D, Daigle N, Eils R, Ellenberg J. Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell. 2002;108:83–96. doi: 10.1016/s0092-8674(01)00627-4. [DOI] [PubMed] [Google Scholar]
- Belmont LD, Hyman AA, Sawin KE, Mitchison TJ. Real-time visualization of cell cycle-dependent changes in microtubule dynamics in cytoplasmic extracts. Cell. 1990;62:579–589. doi: 10.1016/0092-8674(90)90022-7. [DOI] [PubMed] [Google Scholar]
- Blow JJ, Gillespie PJ, Francis D, Jackson DA. Replication origins in Xenopus egg extract are 5–15 kilobases apart and are activated in clusters that fire at different times. J Cell Biol. 2001;152:15–25. doi: 10.1083/jcb.152.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt A, Papagiannouli F, Wagner N, Wilsch-Bräuninger M, Braun M, Furlong EEM, Loserth S, Wenzl C, Pilot F, Vogt N, et al. Developmental control of nuclear size and shape by kugelkern and kurzkern. Curr Biol. 2006;16:543–552. doi: 10.1016/j.cub.2006.01.051. [DOI] [PubMed] [Google Scholar]
- Budde PP, Desai A, Heald R. Analysis of microtubule polymerization in vitro and during the cell cycle in Xenopus egg extracts. Methods. 2006;38:29–34. doi: 10.1016/j.ymeth.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Budde PP, Kumagai A, Dunphy WG, Heald R. Regulation of Op18 during spindle assembly in Xenopus egg extracts. J Cell Biol. 2001;153:149–158. doi: 10.1083/jcb.153.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudron M, Bunt G, Bastiaens PIH, Karsenti E. Spatial coordination of spindle assembly by chromosome-mediated signaling gradients. Science. 2005;309:1373–1376. doi: 10.1126/science.1115964. [DOI] [PubMed] [Google Scholar]
- Dobrynin G, Popp O, Romer T, Bremer S, Schmitz MHA, Gerlich DW, Meyer HH. Cdc48/p97-Ufd1-Npl4 antagonizes Aurora B during chromosome segregation in HeLa cells. J Cell Sci. 2011;124:1571–1580. doi: 10.1242/jcs.069500. [DOI] [PubMed] [Google Scholar]
- Ewald A, Zünkler C, Lourim D, Dabauvalle MC. Microtubule-dependent assembly of the nuclear envelope in Xenopus laevis egg extract. Eur J Cell Biol. 2001;80:678–691. doi: 10.1078/0171-9335-00207. [DOI] [PubMed] [Google Scholar]
- Forbes DJ, Kirschner MW, Newport JW. Spontaneous formation of nucleus-like structures around bacteriophage DNA microinjected into Xenopus eggs. Cell. 1983;34:13–23. doi: 10.1016/0092-8674(83)90132-0. [DOI] [PubMed] [Google Scholar]
- Funabiki H, Murray AW. The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell. 2000;102:411–424. doi: 10.1016/s0092-8674(00)00047-7. [DOI] [PubMed] [Google Scholar]
- Gadea BB, Ruderman JV. Aurora B is required for mitotic chromatin-induced phosphorylation of Op18/Stathmin. Proc Natl Acad Sci USA. 2006;103:4493–4498. doi: 10.1073/pnas.0600702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss OJ, Carazo-Salas RE, Schatz CA, Guarguaglini G, Kast J, Wilm M, Le Bot N, Vernos I, Karsenti E, Mattaj IW. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell. 2001;104:83–93. doi: 10.1016/s0092-8674(01)00193-3. [DOI] [PubMed] [Google Scholar]
- Haithcock E, Dayani Y, Neufeld E, Zahand AJ, Feinstein N, Mattout A, Gruenbaum Y, Liu J. Age-related changes of nuclear architecture in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2005;102:16690–16695. doi: 10.1073/pnas.0506955102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi T, Kojidani T, Koujin T, Shimi T, Osakada H, Mori C, Yamamoto A, Hiraoka Y. Live cell imaging and electron microscopy reveal dynamic processes of BAF-directed nuclear envelope assembly. J Cell Sci. 2008;121:2540–2554. doi: 10.1242/jcs.033597. [DOI] [PubMed] [Google Scholar]
- Harel A, Chan RC, Lachish-Zalait A, Zimmerman E, Elbaum M, Forbes DJ. Importin β negatively regulates nuclear membrane fusion and nuclear pore complex assembly. Mol Biol Cell. 2003;14:4387–4396. doi: 10.1091/mbc.E03-05-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker PB, Hyman AA, Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Hetzer MW, Bilbao-Cortes D, Walther TC, Gruss OJ, Mattaj IW. GTP hydrolysis by Ran is required for nuclear envelope assembly. Mol Cell. 2000;5:1013–1024. doi: 10.1016/s1097-2765(00)80266-x. [DOI] [PubMed] [Google Scholar]
- Houghtaling BR, Yang G, Matov A, Danuser G, Kapoor TM. Op18 reveals the contribution of nonkinetochore microtubules to the dynamic organization of the vertebrate meiotic spindle. Proc Natl Acad Sci USA. 2009;106:15338–15343. doi: 10.1073/pnas.0902317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P, Weis K, Heald R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 2002;295:2452–2456. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- Kelly AE, Ghenoiu C, Xue JZ, Zierhut C, Kimura H, Funabiki H. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science. 2010;330:235–239. doi: 10.1126/science.1189505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AE, Sampath SC, Maniar TA, Woo EM, Chait BT, Funabiki H. Chromosomal enrichment and activation of the Aurora B pathway are coupled to spatially regulate spindle assembly. Dev Cell. 2007;12:31–43. doi: 10.1016/j.devcel.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita K, Arnal I, Desai A, Drechsel DN, Hyman AA. Reconstitution of physiological microtubule dynamics using purified components. Science. 2001;294:1340–1343. doi: 10.1126/science.1064629. [DOI] [PubMed] [Google Scholar]
- Lan W, Zhang X, Kline-Smith SL, Rosasco-Nitcher SE, Barrett-Wilt GA, Shabanowitz J, Hunt DF, Walczak CE, Stukenberg PT. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr Biol. 2004;14:273–286. doi: 10.1016/j.cub.2004.01.055. [DOI] [PubMed] [Google Scholar]
- Lemaitre JM, Géraud G, Méchali M. Dynamics of the genome during early Xenopus laevis development: karyomeres as independent units of replication. J Cell Biol. 1998;142:1159–1166. doi: 10.1083/jcb.142.5.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DL, Heald R. Nuclear size is regulated by importin α and Ntf2 inXenopus. Cell. 2010;143:288–298. doi: 10.1016/j.cell.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Ladinsky MS, Kirchhausen T. Formation of the postmitotic nuclear envelope from extended ER cisternae precedes nuclear pore assembly. J Cell Biol. 2011;194:425–440. doi: 10.1083/jcb.201012063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado-Saldivia J, van den Bergen J, Krouskos M, Gilchrist MJ, Lee C, Li R, Sinclair AH, Surani MA, Western PS. Dppa2 and Dppa4 are closely linked SAP motif genes restricted to pluripotent cells and the germ line. Stem Cells. 2007;25:19–28. doi: 10.1634/stemcells.2006-0269. [DOI] [PubMed] [Google Scholar]
- Maresca TJ, Groen AC, Gatlin JC, Ohi R, Mitchison TJ, Salmon ED. Spindle assembly in the absence of a RanGTP gradient requires localized CPC activity. Curr Biol. 2009;19:1210–1215. doi: 10.1016/j.cub.2009.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- Meyerzon M, Gao Z, Liu J, Wu JC, Malone CJ, Starr DA. Centrosome attachment to the C. elegans male pronucleus is dependent on the surface area of the nuclear envelope. Developmental biology. 2009;327:433–446. doi: 10.1016/j.ydbio.2008.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshull J, Sun H, Tonks NK, Murray AW. A MAP kinase-dependent spindle assembly checkpoint in Xenopus egg extracts. Cell. 1994;79:475–486. doi: 10.1016/0092-8674(94)90256-9. [DOI] [PubMed] [Google Scholar]
- Murray AW. Cell cycle extracts. Meth Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- Nachury MV, Maresca TJ, Salmon WC, Waterman-Storer CM, Heald R, Weis K. Importin β is a mitotic target of the small GTPase Ran in spindle assembly. Cell. 2001;104:95–106. doi: 10.1016/s0092-8674(01)00194-5. [DOI] [PubMed] [Google Scholar]
- Newport JW. Nuclear reconstitution in vitro: stages of assembly around protein-free DNA. Cell. 1987;48:205–217. doi: 10.1016/0092-8674(87)90424-7. [DOI] [PubMed] [Google Scholar]
- Niethammer P, Kronja I, Kandels-Lewis S, Rybina S, Bastiaens P, Karsenti E. Discrete states of a protein interaction network govern interphase and mitotic microtubule dynamics. PLoS Biology. 2007;5:e29. doi: 10.1371/journal.pbio.0050029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott A, Leno GH, Laskey RA. Sperm decondensation in Xenopus egg cytoplasm is mediated by nucleoplasmin. Cell. 1991;65:569–578. doi: 10.1016/0092-8674(91)90089-h. [DOI] [PubMed] [Google Scholar]
- Postow L, Ghenoiu C, Woo EM, Krutchinsky AN, Chait BT, Funabiki H. Ku80 removal from DNA through double strand break-induced ubiquitylation. J Cell Biol. 2008;182:467–479. doi: 10.1083/jcb.200802146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadan K, Bruderer R, Spiga FM, Popp O, Baur T, Gotta M, Meyer HH. Cdc48/p97 promotes reformation of the nucleus by extracting the kinase Aurora B from chromatin. Nature. 2007;450:1258–1262. doi: 10.1038/nature06388. [DOI] [PubMed] [Google Scholar]
- Salina D, Bodoor K, Eckley DM, Schroer TA, Rattner JB, Burke B. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell. 2002;108:97–107. doi: 10.1016/s0092-8674(01)00628-6. [DOI] [PubMed] [Google Scholar]
- Sampath SC, Ohi R, Leismann O, Salic A, Pozniakovsky A, Funabiki H. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell. 2004;118:187–202. doi: 10.1016/j.cell.2004.06.026. [DOI] [PubMed] [Google Scholar]
- Schoft VK, Beauvais AJ, Lang C, Gajewski A, Prüfert K, Winkler C, Akimenko MA, Paulin-Levasseur M, Krohne G. The lamina-associated polypeptide 2 (LAP2) isoforms β, γ and ω of zebrafish: developmental expression and behavior during the cell cycle. J Cell Sci. 2003;116:2505–2517. doi: 10.1242/jcs.00450. [DOI] [PubMed] [Google Scholar]
- Siegel D, Schuff M, Oswald F, Cao Y, Knöchel W. Functional dissection of XDppa2/4 structural domains in Xenopus development. Mech Dev. 2009;126:974–989. doi: 10.1016/j.mod.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Solovei I, Kreysing M, Lanctôt C, Kösem S, Peichl L, Cremer T, Guck J, Joffe B. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 2009;137:356–368. doi: 10.1016/j.cell.2009.01.052. [DOI] [PubMed] [Google Scholar]
- Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, Zwerger M, Cohen TV, Devys D, Foisner R, Peichl L, et al. LBR and Lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell. 2013;152:584–598. doi: 10.1016/j.cell.2013.01.009. [DOI] [PubMed] [Google Scholar]
- Tseng BS, Tan L, Kapoor TM, Funabiki H. Dual detection of chromosomes and microtubules by the chromosomal passenger complex drives spindle assembly. Dev Cell. 2010;18:903–912. doi: 10.1016/j.devcel.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde F, Dogterom M, Stelzer E, Karsenti E, Leibler S. Control of microtubule dynamics and length by cyclin A- and cyclin B-dependent kinases in Xenopus egg extracts. J Cell Biol. 1992;118:1097–1108. doi: 10.1083/jcb.118.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde F, Labbé JC, Dorée M, Karsenti E. Regulation of microtubule dynamics by cdc2 protein kinase in cell-free extracts of Xenopus eggs. Nature. 1990;343:233–238. doi: 10.1038/343233a0. [DOI] [PubMed] [Google Scholar]
- Vigneron S, Prieto S, Bernis C, Labbé JC, Castro A, Lorca T. Kinetochore localization of spindle checkpoint proteins: who controls whom? Mol Biol Cell. 2004;15:4584–4596. doi: 10.1091/mbc.E04-01-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Askjaer P, Gentzel M, Habermann A, Griffiths G, Wilm M, Mattaj IW, Hetzer MW. RanGTP mediates nuclear pore complex assembly. Nature. 2003;424:689–694. doi: 10.1038/nature01898. [DOI] [PubMed] [Google Scholar]
- Wang F, Dai J, Daum JR, Niedzialkowska E, Banerjee B, Stukenberg PT, Gorbsky GJ, Higgins JMG. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science. 2010;330:231–235. doi: 10.1126/science.1189435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Tytell JD, Ingber DE. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat Rev Mol Cell Biol. 2009;10:75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer CM, Gregory J, Parsons SF, Salmon ED. Membrane/microtubule tip attachment complexes (TACs) allow the assembly dynamics of plus ends to push and pull membranes into tubulovesicular networks in interphase Xenopus egg extracts. J Cell Biol. 1995;130:1161–1169. doi: 10.1083/jcb.130.5.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster MT, Witkin KL, Cohen-Fix O. Sizing up the nucleus: nuclear shape, size and nuclear-envelope assembly. J Cell Sci. 2009;122:1477–1486. doi: 10.1242/jcs.037333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C, Wilde A, Moore MS, Adam SA, Merdes A, Zheng Y. Role of importin-β in coupling Ran to downstream targets in microtubule assembly. Science. 2001;291:653–656. doi: 10.1126/science.1057661. [DOI] [PubMed] [Google Scholar]
- Wright SJ. Sperm nuclear activation during fertilization. Curr Top Dev Biol. 1999;46:133–178. doi: 10.1016/s0070-2153(08)60328-2. [DOI] [PubMed] [Google Scholar]
- Wühr M, Dumont S, Groen AC, Needleman DJ, Mitchison TJ. How does a millimeter-sized cell find its center? Cell Cycle. 2009;8:1115–1121. doi: 10.4161/cc.8.8.8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi Y, Honda T, Tanno Y, Watanabe Y. Two histone marks establish the inner centromere and chromosome bi-orientation. Science. 2010;330:239–243. doi: 10.1126/science.1194498. [DOI] [PubMed] [Google Scholar]
- Zhang C, Clarke PR. Chromatin-independent nuclear envelope assembly induced by Ran GTPase in Xenopus egg extracts. Science. 2000;288:1429–1432. doi: 10.1126/science.288.5470.1429. [DOI] [PubMed] [Google Scholar]
- Zink D, Fischer AH, Nickerson JA. Nuclear structure in cancer cells. Nat Rev Cancer. 2004;4:677–687. doi: 10.1038/nrc1430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.