Abstract
Osteogenesis imperfecta (OI) is typically caused by mutations in type 1 collagen genes, but in recent years new recessive and dominant forms caused by mutations in a plethora of different genes have been characterized. OI type V is a dominant form caused by the recurrent (c.-14C>T) mutation in the 5′UTR of the IFITM5 gene. The mutation adds 5 residues to the N-terminus of the IFITM5 but the pathophysiology of the disease still remains to be elucidated. Typical clinical features present in the majority of OI type V patients include interosseous membrane calcification between the radius and ulna, and the tibia and fibula, radial head dislocation and significant hyperplastic callus formation at the site of fractures. We report a 5 year-old child with clinical features of OI type III or severe OI type IV (characteristic facies, grey sclerae, typical fractures) and absence of classical features of OI type V with a de novo recurrent IFITM5 mutation (c.-14C>T), now typical of OI type V. This highlights the variability of OI caused by IFITM5 mutations and suggests screening for mutations in this gene in most cases of OI where type 1 collagen mutations are absent.
Keywords: Osteogenesis Imperfecta, Type V, IFITM5, hyperplastic callus, interosseous membrane calcification, whole exome sequencing
Introduction
Osteogenesis Imperfecta (OI; OMIM 166200, 1666210, 259420, and 166220) is a genetically and clinically heterogeneous disorder of bone and connective tissue characterized by low bone mass, density, and strength leading to fragility fractures and deformities. Originally, Sillence et al. classified OI into four distinct phenotypes (1). Identification of new genetic causes of OI over the last decade has resulted in an expanded classification, which includes eight phenotypes based on the involved genes (2,3). In the year 2000, Glorieux et al. described OI type V as a clinically distinct entity distinguished by the frequent occurrence of hyperplastic callus following fracture or surgery, the presence of bilateral radial head dislocation, ossification of the interosseous membranes between both the radius and ulna and the tibia and fibula, and radiodense metaphyseal bands adjacent to growth plates. Type V patients typically do not have discoloration of sclerae or dentinogenesis imperfecta (DI) (4). Recently, a single recurrent heterozygous mutation (c.-14C>T) in the 5′ UTR of the IFITM5 gene has been shown to be the cause of OI type V in four different cohorts, but the patients demonstrate high phenotypic variability (5–8). The role of IFITM5 in bone development and maintenance is still only beginning to be uncovered (9–11). We now report a case of OI which phenotypically resembled OI type III or a severe form of type IV, but upon exome sequencing, the recurrent IFITM5 mutation of OI type V was uncovered.
Case Report
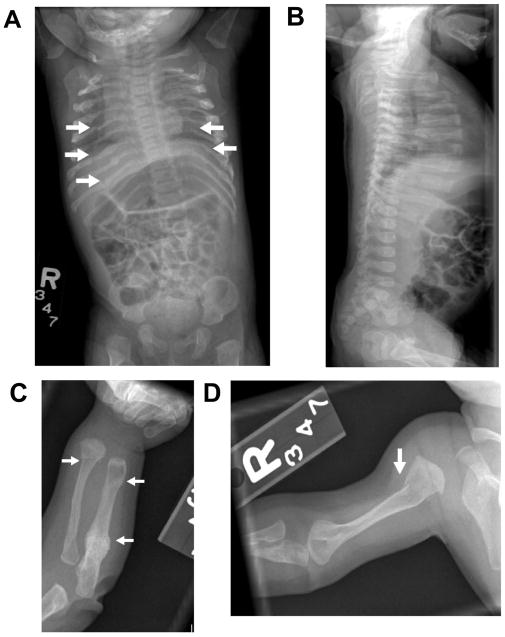
We report a 5 ½ year old Hispanic female with OI originally classified as type III/IV. She was born to a 28 year old female by full term spontaneous vaginal delivery. The mother has had one miscarriage in the past. Her birth weight was 5 lbs. 13 oz. Prenatally, there was concern that she was small for gestational age or had intrauterine growth restriction. Her mother underwent amniocentesis that showed normal karyotype of the fetus. At birth, her weight was at the 5th percentile, length below the 5th percentile, and head circumference was at the 25th percentile. Shortly after birth, bilateral ulnar deviation of the wrists and prominence of the right radial styloid process with an abnormal junction between the hand and the ulna/radius were noted. There was normal range of motion at elbows, wrists, knees, or ankles. She had normal carrying angles at the elbows. Her muscle tone, bulk and deep tendon reflexes were normal at birth in all four extremities. The rest of her examination was unremarkable and she passed the neonatal hearing screen. Given the history of fussiness on handling and the deformity on examination, a skeletal survey (Figure 1) was performed which revealed multiple bilateral posterior rib fractures, bowing deformities of both forearms, and non-displaced fractures at the left ulna and proximal right humeral shaft. There was no evidence of any vertebral body abnormalities.
Figure 1.
A: Anteroposterior views of the chest and abdomen from skeletal survey demonstrates multiple bilateral posterior rib fractures B: A lateral view of the thoracolumbar spine reveals no vertebral compression fractures C: Anteroposterior view of the left forearm showing a healing proximal ulnar fracture, an acute distal radial fracture and expansion of the distal ulnar metaphyses consistent with previous fracture. D: Anteroposterior view of the right humerus reveals an acute proximal metaphyseal fracture.
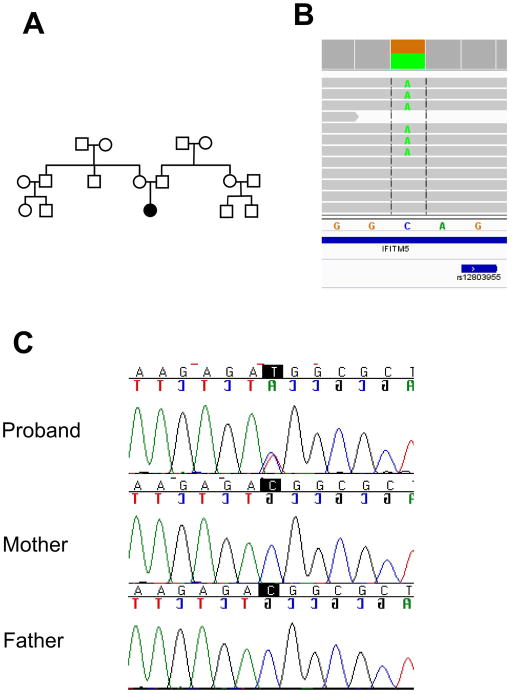
There is no family history of OI or other skeletal diseases, and the parents are not consanguineous by report (Figure 2A).
Figure 2.
A Pedigree. B Alignment of the next-generation sequencing reads as viewed in the IGV viewer from the Broad Institute. Grey lines represent the sequence reads matching to the reference sequence, while the green letter “A” represents the recurrent IFITM5 5′UTR mutation. C Sanger sequencing chromatograms shows the validation of the mutation identified by next-generation sequencing.
Currently, at 5 ½ years, her height and weight are both below the 3rd percentiles. She has the classic triangular facies seen in OI, grey sclerae, midfacial hypoplasia and hypotonia of the limbs. There is absence of rhizomelia (as characterized in OI type VII) (12) or problems with supination/pronation (as characterized in OI type V) (4), and she has normal hearing. She has had multiple dental caries but no evidence of dentinogenesis imperfecta.
Her past medical history is significant for eczema and multiple low trauma/no trauma fractures of long bones. Given the early onset of fractures and the other clinical features, she was clinically diagnosed as OI type III or a severe form of OI type IV.
Over the years, she has received physical therapy, DXA scans, pulmonary function tests and hearing screen as part of the Linked Clinical Research Center (LCRC) - longitudinal study of osteogenesis imperfecta.
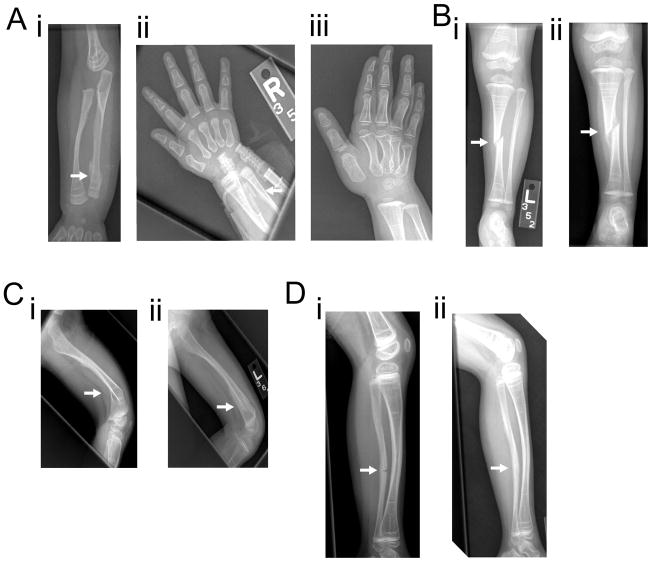
Radiological examination (Figure 3) shows generalized low bone density and rows of retained calcified cartilage at metaphyses due to bisphosphonate therapy. There has been no evidence of hyperplastic callus formation despite multiple fractures over the years, interosseous membrane calcification or radial head dislocation. There have been no vertebral compression fractures noted.
Figure 3.
Figure 3A: i Right forearm radiograph at 8 months of age showing distal right ulnar fracture. Retained calcified cartilage at the metaphyses is noted, likely from bisphosphonate therapy. ii Right hand radiograph from 6 months later showing interval healing. iii Right hand radiograph 12 months after the occurrence of fracture shows healing at the fracture site.
Figure 3B: i Anteroposterior left leg radiograph at 2 years of age showing an acute oblique fracture of the mid-tibial diaphysis with one-half shaft-width lateral displacement. ii Repeat radiograph 1 month later shows an appropriate amount of callus has developed at the tibial fracture site.
Figure 3C: i Lateral left femoral radiograph at 2.5 years of age shows an acute fracture through the distal diaphysis of the left femur which is associated with approximately 40 degrees of posterior angulation. ii Follow up radiograph a month later shows interval callus formation and improved angulation.
Figure 3D: i Lateral left leg radiograph at 5 years of age demonstrates an acute non-displaced fracture involving the anterior cortex of the midshaft of the left fibula with associated bowing deformity. ii Repeat radiograph 6 months later shows healing at the fracture site.
DXA scan (Hologic Delphi-A version12.1) shows a total body bone mineral density (BMD) z-score of −1.8 at 4 years and −1.5 at 5 years of age. We do not have DXA scans prior to the start of treatment as bisphosphonate therapy was started at 1 month of age and at the time the patient was unable to lie still for the duration of the scan without sedation. Patient’s 2nd metacarpal cortical thickness was within 1 standard deviation at ages 1, 2 and 4 years.
Laboratory analyses were done at regular intervals from 1 month to 5 ½ years of age which showed normal serum calcium (10.3± 0.25 mg/dl), phosphorus (4.4 ± 0.9 mg/dl), intact parathyroid hormone (23.8 ± 10.6 pg/ml), 25 OH-Vitamin D (44 ± 13 ng/ml), 1, 25 (OH)2 Vitamin D (56 ± 11 ng/ml) and osteocalcin (27.8 ±3.8 ng/ml) levels. She had elevated serum alkaline phosphatase (598 ± 200 U/L) and urine N-telopeptide/creatinine ratio (544± 228 nM bone collagen equivalent/mM creatinine) at the time of healing of an acute fracture.
The patient was started on bisphosphonate therapy at 1 month of age. She has been treated with IV pamidronate from 1 month to 3 years of age (9 mg/kg/year) followed by IV zoledronic acid (0.10 mg/kg/year) after 3 years of age. She has also received supplemental calcium and vitamin D orally. She has tolerated the therapy well.
Materials and Methods
Exome Sequencing
Exome sequencing and analysis were conducted as previously described (13). Briefly, exomes were captured on Nimblegen’s SeqCap EZ V2.0 library, and sequencing was conducted on Illumina HiSeq2000. Reads were aligned with BWA (Burrows-Wheeler Aligner) to human reference genome GRCh37/hg19 and locally realigned with GATK (Genome Analysis Toolkit) around potential INDELs. SNPs were called using Samtools Pileup (version 0.1.17) (14) and short indels were called using Samtools (14), Atlas-INDEL (15), and GATK (16). Variants were annotated with ANNOVAR (17); protein-impacting variants that were rare (minor allele frequency <5%) or novel were preferentially explored.
Sanger sequencing
Amplicons were generated using 1 ng/microliter of genomic DNA using TaqMan polymerase (ABI, Life Technologies, Carlsbad, CA) using the manufacturer’s protocol with an annealing temperature of 60 °C and an amplification of 1 minute. Primers used were forward: 5′-AGGGCGACAGGGCTATAAGTGAG-3′ and reverse: 5′-GAAGCCGAGGCAACACAGATTCAGGTAG-3′. Products were verified by agarose gel then sequenced by Sanger sequencing using the same primers at Beckman Coulter Genomics.
Results
DNA sample from the patient was obtained after obtaining informed consent. Investigation was approved by the local ethics committee. Genomic DNA was extracted from peripheral blood by standard techniques. We performed a SNP array which did not identify any regions of homozygosity suggestive of absence of consanguinity. This was consistent with the family history. Genomic DNA was sequenced for the known genes of OI, and no mutations were identified in the coding regions and exon-intron boundaries of the following genes: COL1A1, COL1A2, CRTAP, LEPRE1, PPIB, FKBP10, SERPINH1, SERPINF1, PLOD2 and SP7. Collagen studies were performed on fibroblasts and were also negative. Hence, next we performed whole exome sequencing. We identified a heterozygous change in 5′UTR of IFITM5 (Figure 2B) which we and others have previously reported to be a causative gene for OI type V. This was confirmed by Sanger sequencing with a new genomic DNA sample. The parents’ DNA was also sequenced for this gene, and we did not identify the same mutation. Hence, we concluded that it is a de novo mutation (Figure 2C).
Discussion
Variable expressivity, both within and between affected families, is characteristic of OI type V (5–8). This case is unusual as this mutation has been recently described in four cohorts of OI type V patients who are characterized by hyperplastic callus formation, calcification of interosseous membrane and radial head dislocation. Our patient lacked all of these classical features. This fact raises several questions: Is it possible that the classical physical and radiological features of type V OI develop later in childhood? We believe this is unlikely as there are reports of patients as young as 2 months who have classical features of OI type V (8). Could development of these features be prevented by an early start of bisphosphonate therapy? We would need case control studies to evaluate this question but this appears to be unlikely as the described cohorts include patients who were started on treatment early and have still developed the hyperplastic callus or calcification of interosseous membranes (8). This leads us to the third question i.e., whether this mutation is specific to OI type V. Is it possible that this case is another example of marked phenotypic variability and that there is no genotype-phenotype correlation in OI? It suggests that there are other factors that may modify the expressivity of this mutation, such as polymorphisms in other genes. Given the high phenotypic variability of OI type V and the fact that all patients have the same IFITM5 mutation, this condition would be ideal to search for modifier genes in the age of next-generation sequencing. Based on our experience with this case, and the fact that there is no feature present in 100% of patients in the previous reports, we suggest that all unsolved OI patients, in spite of absence of classical type V phenotype, should be screened for this IFITM5 gene mutation. It also remains to be determined whether mutations elsewhere in IFITM5 might cause a skeletal phenotype, arguing for sequencing of the rest of the gene in unsolved OI cases as a second or third tier molecular testing strategy.
Acknowledgments
We thank Alyssa Tran for patient enrollment and Yuqing Chen for technical assistance. This research was supported by NIH grants PO1 HD22657 and PO1 HD070394, and the BCM Intellectual and Developmental Disabilities Research Center (HD024064) from the Eunice Kennedy Shriver National Institute of Child Health & Human Development. PC is supported by a CIHR clinician-scientist training award and the O’Malley Foundation. MG was supported by The Pediatric Endocrine Society Research Fellowship award. Authors’ roles: Whole exome sequencing analysis: JL, RG, PC and MG Drafting manuscript: MG. Revising manuscript content: MG, PC, CL, AS, and BL.
Footnotes
Disclosure:
All authors state they have no conflicts of interest.
References
- 1.Sillence DO, Senn A, Danks DM. Genetic heterogeneity in osteogenesis imperfecta. J Med Genet. 1979;16(2):101–116. doi: 10.1136/jmg.16.2.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rauch F, Glorieux FH. Osteogenesis imperfecta Lancet. Vol. 363. England: 2004. pp. 1377–1385. [DOI] [PubMed] [Google Scholar]
- 3.Van Dijk FS, Pals G, Van Rijn RR, Nikkels PG, Cobben JM. Classification of Osteogenesis Imperfecta revisited. European journal of medical genetics. 2010;53(1):1–5. doi: 10.1016/j.ejmg.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 4.Glorieux FH, Rauch F, Plotkin H, Ward L, Travers R, Roughley P, Lalic L, Glorieux DF, Fassier F, Bishop NJ. Type V osteogenesis imperfecta: a new form of brittle bone disease. Journal of bone and mineral research: the official journal of the American Society for Bone and Mineral Research. 2000;15(9):1650–1658. doi: 10.1359/jbmr.2000.15.9.1650. [DOI] [PubMed] [Google Scholar]
- 5.Semler O, Garbes L, Keupp K, Swan D, Zimmermann K, Becker J, Iden S, Wirth B, Eysel P, Koerber F, Schoenau E, Bohlander SK, Wollnik B, Netzer C. A mutation in the 5′-UTR of IFITM5 creates an in-frame start codon and causes autosomal-dominant osteogenesis imperfecta type V with hyperplastic callus. American journal of human genetics. 2012;91(2):349–357. doi: 10.1016/j.ajhg.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rauch F, Moffatt P, Cheung M, Roughley P, Lalic L, Lund AM, Ramirez N, Fahiminiya S, Majewski J, Glorieux FH. Osteogenesis imperfecta type V: marked phenotypic variability despite the presence of the IFITM5 c.-14C>T mutation in all patients. J Med Genet. 2013;50(1):21–24. doi: 10.1136/jmedgenet-2012-101307. [DOI] [PubMed] [Google Scholar]
- 7.Cho TJ, Lee KE, Lee SK, Song SJ, Kim KJ, Jeon D, Lee G, Kim HN, Lee HR, Eom HH, Lee ZH, Kim OH, Park WY, Park SS, Ikegawa S, Yoo WJ, Choi IH, Kim JW. A single recurrent mutation in the 5′-UTR of IFITM5 causes osteogenesis imperfecta type V. American journal of human genetics. 2012;91(2):343–348. doi: 10.1016/j.ajhg.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shapiro JR, Lietman C, Grover M, Lu JT, Nagamani SC, Dawson BC, Baldridge DM, Bainbridge MN, Cohn DH, Blazo M, Roberts TT, Brennen FS, Wu Y, Gibbs RA, Melvin P, Campeau PM, Lee BH. Phenotypic variability of osteogenesis imperfecta type V caused by an IFITM5 mutation. J Bone Miner Res. 2013 doi: 10.1002/jbmr.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanagata N, Li X. Osteoblast-enriched membrane protein IFITM5 regulates the association of CD9 with an FKBP11-CD81-FPRP complex and stimulates expression of interferon-induced genes. Biochemical and biophysical research communications. 2011;409(3):378–384. doi: 10.1016/j.bbrc.2011.04.136. [DOI] [PubMed] [Google Scholar]
- 10.Hanagata N, Li X, Morita H, Takemura T, Li J, Minowa T. Characterization of the osteoblast-specific transmembrane protein IFITM5 and analysis of IFITM5-deficient mice. Journal of bone and mineral metabolism. 2011;29(3):279–290. doi: 10.1007/s00774-010-0221-0. [DOI] [PubMed] [Google Scholar]
- 11.Moffatt P, Gaumond MH, Salois P, Sellin K, Bessette MC, Godin E, de Oliveira PT, Atkins GJ, Nanci A, Thomas G. Bril: a novel bone-specific modulator of mineralization. Journal of bone and mineral research: the official journal of the American Society for Bone and Mineral Research. 2008;23(9):1497–1508. doi: 10.1359/jbmr.080412. [DOI] [PubMed] [Google Scholar]
- 12.Ward LM, Rauch F, Travers R, Chabot G, Azouz EM, Lalic L, Roughley PJ, Glorieux FH. Osteogenesis imperfecta type VII: an autosomal recessive form of brittle bone disease Bone. Vol. 31. United States: 2002. pp. 12–18. [DOI] [PubMed] [Google Scholar]
- 13.Campeau PM, Kim JC, Lu JT, Schwartzentruber JA, Abdul-Rahman OA, Schlaubitz S, Murdock DM, Jiang MM, Lammer EJ, Enns GM, Rhead WJ, Rowland J, Robertson SP, Cormier-Daire V, Bainbridge MN, Yang XJ, Gingras MC, Gibbs RA, Rosenblatt DS, Majewski J, Lee BH. Mutations in KAT6B, encoding a histone acetyltransferase, cause Genitopatellar syndrome. Am J Hum Genet. 2012;90(2):282–289. doi: 10.1016/j.ajhg.2011.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics (Oxford, England) 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Havlak P, Chen R, Durbin KJ, Egan A, Ren Y, Song XZ, Weinstock GM, Gibbs RA. The Atlas genome assembly system. Genome research. 2004;14(4):721–732. doi: 10.1101/gr.2264004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]