Abstract
Purpose
To find out whether the MTHFR rs1801133 polymorphism is a risk factor for male infertility in the Spanish population. To determine if a pattern of sperm DNA hypomethylation at the paternally imprinted loci H19-ICR and/or IG-DMR is related to the MTHFR rs1801133 polymorphism and/or CTCFL mutations.
Methods
One hundred and seven samples from individuals who sought consultation for fertility problems and twenty-five semen samples from sperm donors were analyzed. The MTHFR rs1801133 SNP was analyzed in all samples by the PCR-RFLP method. We compared the distribution of the genotypes between control and infertile populations and among the groups of patients with altered seminal parameters. In those patients with the most severe hypomethylation pattern (n = 12) we also analyzed the CTCFL protein-coding exons by sequencing.
Results
There were no significant differences in the distribution of the genotypes among the control and infertile populations. Moreover, none of the genotypes were associated, neither to the characteristics of the seminogram, nor to the presence of sperm DNA hypomethylation. We did not identify frameshift, nonsense or missense mutations of the CTCFL gene.
Conclusions
The MTHFR rs1801133 polymorphism is not associated with male infertility in the Spanish population. Neither the MTHFR polymorphism, nor CTCFL mutations explain a pattern of sperm hypomethylation at paternally imprinting loci.
Electronic supplementary material
The online version of this article (doi:10.1007/s10815-013-0013-2) contains supplementary material, which is available to authorized users.
Keywords: MTHFR polymorphism, CTCFL, DNA methylation, Male infertility, Imprinting errors
Introduction
In previous years, several reports have described an association between male infertility and methylation abnormalities at imprinted loci in spermatozoa [4, 8, 12, 14, 15, 17–20, 23]. Imprinting is a specific epigenetic mark consisting of parental-specific DNA methylation at CpG dinucleotides and histone post-translational modifications. This mechanism controls the expression of a gene on the basis of its parental origin resulting in monoallelic expression. In order to achieve a specific gender epigenetic mark, imprinting is erased in primordial germ cells and established de novo during gametogenesis in a sex-specific manner. There are several histone post-translational modifications leading to a specific expression profile. One of these is methylation at lysines or arginines catalyzed by histone methyltransferases (HMT). On the other hand, DNA methyltransferases (DNMT) are the enzymes that add the methyl groups to CpG dinucleotides. DNMT3A and DNMT3B are those implicated in de novo DNA methylation, and DNMT3L is a co-factor specifically expressed in the germline [2].
In the testis, the specific factor CCCTC-binding factor-like protein (CTCFL/BORIS) cooperates with Protein Arginine Methyltransferase 7 (PRMT7) in establishing symmetrical dimethyl modification of arginine residues in histones H2A and H4. This action happens close to the H19-Imprinting Control Region (H19-ICR) leading to the DNA methylation of this ICR [13]. Thus, CTCFL-deficient activity could cause imprinting errors at H19/IGF2 as well as in other paternally methylated ICR, such as the one that controls MEG3/DLK1.
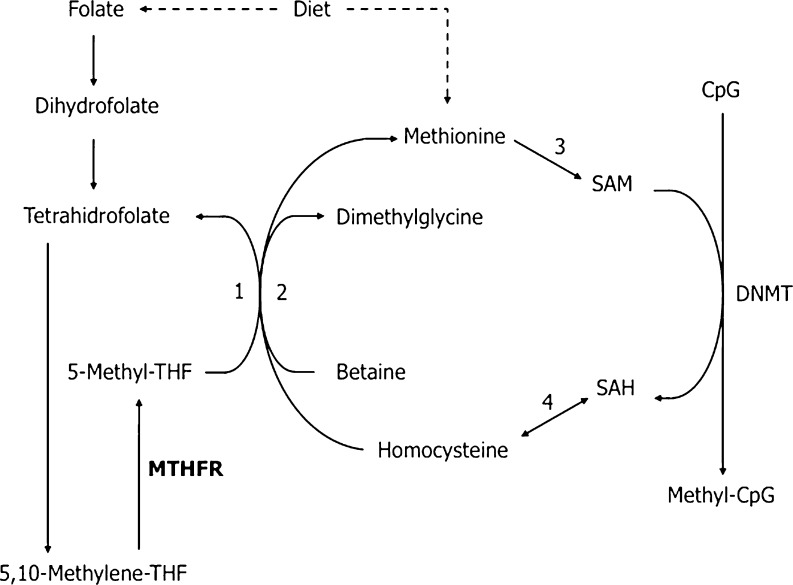
Methyl groups needed by methyltransferases are provided by S-adenosyl-L-methionine (SAM) through the folate pathway. The availability of 5,10-methylenetetrahydrofolate (5,10-methylene-THF) influences the pool of methyl groups and it is derived from folic acid. The 5,10-methylenetetrahydrofolate reductase (MTHFR) is a key regulatory enzyme that plays an important role in the metabolic cycle of methionine, since it catalyzes the reduction of 5,10-methylene-THF to 5-methylenetetrahydrofolate (5-methyl-THF). The 5-methylene-THF is the methyl donor for homocysteine (Hcy) methylation to methionine by the methionine synthase. Methionine is the precursor of SAM, which is the direct methyl donor for the methylation of histones and DNA by HMT and DNMT. The cycle is completed by the conversion of the product resulting from the methyl donation of SAM, the S-adenosylhomocysteine (SAH) to Hcy (Fig. 1).
Fig. 1.
Folate and methionine pathway. DNMT DNA methyltransferase; MTHFR 5,10-methylenetetrahydrofolate reductase; 1 Methionine-synthase (vitamin B12 and zinc dependent); 2 Betaine-homocysteine-methyltransferase (zinc dependent); 3 Methionine-adenosyltransferase; 4 S-adenosylhomocysteine hydrolase. Adapted from Ebisch et al. [7]
In humans, only two studies have tried to relate mutations in genes involved in DNA methylation to the presence of imprinting errors in spermatozoa. Kobayashi et al. [14] reported DNMT3A (GeneID: 1788) and DNMT3L (GeneID: 29947) variations in some infertile patients with sperm DNA methylation anomalies at specific imprinted loci. In relation to the CTCFL gene (GeneID: 140690), Poplinski et al. [23], they did not identify mutations in somatic DNA from infertile patients that showed sperm DNA hypomethylation at IGF2/H19-ICR or severe MEST hypermethylation.
On the other hand, the common single nucleotide polymorphism (SNP) rs1801133 of the MTHFR gene (GeneID: 4524), which corresponds to a C > T transition causing an alanine-to-valine substitution at position 222 (p.A222V), compromises the enzyme activity [9], and it has been described as associated with male infertility [1, 3, 10, 16, 21, 22, 25, 26]. Nevertheless, to our best knowledge, sperm DNA methylation analysis of infertile men together with rs1801133 genotyping has not yet been investigated.
In this report, we have explored the possibility that DNA hypomethylation at the paternally imprinted ICR’s H19-ICR and IG-DMR is the consequence of deficient MTHFR activity because of the presence of the rs1801133 polymorphism or due to mutations in the CTCFL gene. We have also compared the frequency of MTHFR rs1801133 genotypes between the control and the infertile populations. Finally, we have analyzed if there is any relationship between the distribution of the genotypes and specific alterations of the seminal parameters.
Material and methods
Sperm samples
Twenty-five semen samples from sperm donors were selected as a control population. All controls met the following characteristics: 1) Normal karyotype, 2) Proven fertility, 3) More than 90 × 106 of total motile progressive sperm in the raw ejaculates, 4) More than 14 % of normal forms (strict criteria), and 5) More than 10 × 106 of progressive forms per ml after a post-thawing survival test. The average age of the control group was 26 ± 6.15 years of age (range: 19–45).
One hundred and seven samples from individuals who sought consultation for fertility problems were analysed (Supplemental Table 1): 15 normozoospermic (N), 1 oligozoospermic (O), 8 asthenozoospermic (A), 30 teratozoospermic (T), 1 oligoastenozoospermic (OA), 5 oligoteratozoospermic (OT), 31 asthenoteratozoospermic (AT) and 16 oligoasthenoteratozoospermic (OAT). The average age of the infertile individuals was 36 ± 5.50 years of age (range: 26–53). Seminal classification was carried out following the criteria of the World Health Organisation [29].
Donors and patients gave their informed consent to participate in the study, which was approved by our Institutional Ethics Committee.
The spermatozoa were separated from the rest of the cells of the ejaculate by the direct swim-up technique [27]. The extraction of genomic DNA from the spermatozoa was performed with the commercial extraction kit PUREGEN (Gentra Systems; Minneapolis, MN, USA).
In a previous report by our group, the sperm DNA samples from the infertility patients (n = 107) were classified according to the presence of normal methylation or hypomethylation at the paternally imprinted ICR H19-ICR and IG-DMR [5]. Among the group of 12 cases with hypomethylation at H19-ICR and the group of 40 cases with hypomethylation at IG-DMR (Table 1), we have selected those samples with the most severe pattern of hypomethylation (Tables 2 and 3).
Table 1.
Number and percentage of infertile patients with hypomethylation or normal methylation at H19-ICR and IG-DMR [5]
| Region | Hypomethylation | Normal methylation |
|---|---|---|
| H19-ICR | 12/107 (11.21 %) | 95/107 (88.78 %) |
| IG-DMR | 40/107 (37.38 %) | 67/107 (62.61 %) |
Table 2.
Percentage of methylation of each analyzed CpG at the H19-ICR locus in the six infertile patients showing the most severe pattern of hypomethylation
| Case | CpG 1 | CpG 2 | CpG 3 | CpG 4 | CpG 6 | CpG 7 | CpG 8 | CpG 9 | CpG 10 | CpG 11 | CpG 12 | CpG 13 | CpG 14 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 37 | 84 | 84 | 78 | 67 | 77 | 85 | 82 | 83 | 74 | 82 | 79 | 89 | 80 |
| 50 | 83 | 84 | 74 | 71 | 73 | 81 | 80 | 83 | 74 | 76 | 75 | 85 | 83 |
| 80 | 86 | 92 | 85 | 56 | 82 | 88 | 79 | 81 | 83 | 84 | 73 | 84 | 88 |
| 91 | 83 | 88 | 81 | 70 | 74 | 90 | 87 | 84 | 74 | 81 | 73 | 88 | 80 |
| 95 | 9 | 8 | 11 | 10 | 10 | 8 | 7 | 8 | 7 | 11 | 10 | 10 | 12 |
| 102 | 70 | 73 | 68 | 64 | 66 | 74 | 71 | 75 | 67 | 74 | 70 | 78 | 76 |
| Reference range | 86–96 | 87–97 | 84–93 | 61–88 | 82–88 | 83–100 | 85–97 | 85–97 | 74–92 | 83–92 | 73–95 | 89–99 | 84–96 |
Hypomethylated CpGs are indicated in bold. All patients showed hypomethylation affecting more than 50 % of the CpGs
Table 3.
Percentage of methylation of each analyzed CpG at the IG-DMR locus in the eight infertile patients showing the most severe pattern of hypomethylation
| Case | CpG 1 | CpG 2 | CpG 3 | CpG 4 | CpG 5 |
|---|---|---|---|---|---|
| 6 | 91 | 93 | 57 | 79 | 63 |
| 18 | 94 | 93 | 56 | 77 | 60 |
| 37 | 85 | 82 | 48 | 62 | 55 |
| 39 | 93 | 90 | 58 | 78 | 64 |
| 59 | 94 | 93 | 57 | 80 | 63 |
| 79 | 90 | 93 | 56 | 77 | 61 |
| 104 | 92 | 93 | 56 | 79 | 64 |
| Range of reference | 97–83 | 100–83 | 69–55 | 89–76 | 72–59 |
Hypomethylated CpGs are indicated in bold. CpGs with methylation values close to the lowest threshold of the normal referred range are indicated in bold/italics
MTHFR rs1801133 analysis
The MTHFR rs1801133 SNP was analyzed in all samples (25 from sperm donors and 107 from infertile patients) by the PCR-RFLP method using primers and conditions described by Frosst et al. [9]. Briefly, the C > T transition creates a HinfI recognition sequence. The digested PCR product yields two fragments of 175 and 23 bp for the T allele whereas the wild C allele remains undigested and yields a single 198-bp amplicon.
To evaluate whether MTHFR rs1801133 SNP genotypes CC, CT and/or TT showed different frequencies between the control (n = 25) and the infertile (n = 107) populations, a contingency analysis was performed. This analysis was carried out considering whether the control and infertile individuals presented the genotype: CC, non-CC; CT, non-CT and TT, non-TT.
A second contingency analysis was performed to determine whether some rs1801133 SNP genotypes are preferably associated with normozoospermia, oligozoospermia, asthenozoospermia or teratozoospermia in the infertile population (n = 107). For this purpose, the analysis was done considering whether the infertile individuals presented the variables of: normozoospermia, non-normozoospermia; oligozoospermia, non-oligozoospermia; asthenozoospermia, non-asthenozoospermia; teratozoospermia, non-teratozoospermia. The finality of this separation allowed us to determine if a specific component of the spermiogram was preferentially linked to any genotype.
We also checked in the infertile population (n = 107) a possible relationship between the presence of hypomethylation at imprinted loci and rs1801133 SNP genotypes. Again, a contingency table was performed contrasting the presence of a normal or hypomethylation pattern at H19-ICR and IG-DMR with every single genotype.
Data of the three contingency tables were analyzed using a Chi-squared test. The level of statistical significance was established as 0.05. The statistical analysis was performed with the specialized support of Servei d’Estadística Aplicada (Applied Statistics Service) of the Universitat Autònoma de Barcelona.
CTCFL sequence analysis
According to GenBank accession number DQ778111, the protein-coding exons of the CTCFL gene (exons 3 to 13) were analyzed in sperm DNA samples. This analysis was done exclusively in 12 infertile individuals selected for showing the most severe pattern of hypomethylation in the paternal germline/primary differentially methylated regions; 50 % of the analyzed CpGs hypomethylated or with methylation values close to the lowest threshold of the normal referred range [5], (H19-ICR n = 6, IG-DMR n = 7).
Due to limitations in sperm DNA availability, the analysis was performed by nested PCR using 15 pg of the sperm isolated DNA in a total volume of 25 μl for the first round, and 4 ul of the first purified PCR reaction was used to perform the second round also in 25 μl. The intronic primers and conditions are described in Table 4. First and second round PCR purification was done using ExoSAP-IT™ (Affymetrix Inc/USB products, High Wycombe, United Kingdom) and the final products were sequenced with the forward and reverse primer using the BigDye™ Terminator Sequencing Kit v3.1 (Applied Biosystems, Foster City, USA) on an ABIPRISM 3100.
Table 4.
Primers and amplification conditions used for CTCFL sequence analysis. Exon numbering according to the GenBank accession number DQ778111
| EXON | Primer | Sequence (5′–3′) | Amplicon length | Annealing |
|---|---|---|---|---|
| E3 | CTCFL E3F | ggccagaccttgtttcaactcc | 717 bp | 64 °C |
| CTCFL E3R | gcaactgcaaaataccttgtcc | |||
| nCTCFL E3Fa | ctcataccacccccttctcc | 676 bp | 58 °C | |
| nCTCFL E3Ra | ttgcataaaagccgacttga | |||
| E4 | CTCFL E4Fa | gcttcatccaggcaaaagtc | 355 bp | 64 °C |
| CTCFL E4Ra | accatcacctgcccataaag | |||
| nCTCFL E4F | caggcaaaagtctaattatac | 338 bp | 58 °C | |
| nCTCFL E4R | tgcccataaagaaaaacagca | |||
| E5 | CTCFL E5Fa | ttgaaaggacgatgtgctga | 396 bp | 58 °C |
| CTCFL E5Ra | tggaaattcaagaaaaagaagaca | |||
| nCTCFL E5F | gatttactttcaagtattta | 318 bp | 58 °C | |
| nCTCFL E5R | gaaacaggtggattcacatt | |||
| E6 | CTCFL E6Fa | gaattccttggaatgtttctgg | 303 bp | 58 °C |
| CTCFL E6Ra | aaaaagcctgtttgtaacagattc | |||
| nCTCFL E6F | ggttaagtcttgttttgaaa | 266 bp | 58 °C | |
| nCTCFL E6R | cagattctactgttagttac | |||
| E7 | CTCFL E7Fa | aatttaaatctcttttacaatgctga | 252 bp | 58 °C |
| CTCFL E7Ra | aacactgtaggtatcaggccttc | |||
| nCTCFL E7F | ttaatggaagactctgccat | 217 bp | 58 °C | |
| nCTCFL E7R | ggtatcaggccttcagcacc | |||
| E8 | CTCFL E8Fa | ggtgagaagggggttgataa | 332 bp | 58 °C |
| CTCFL E8Ra | ataggaccacgctccaaaga | |||
| nCTCFL E8F | aacactttcagcacaggtg | 277 bp | 58 °C | |
| nCTCFL E8R | ctccaaagagccagcaaatg | |||
| E9 | CTCFL E9Fa | gaaaccccggtttagaggag | 300 bp | 58 °C |
| CTCFL E9Ra | cagccctccattcttccata | |||
| nCTCFL E9F | ggagaggggaagcaggctaa | 267 bp | 58 °C | |
| nCTCFL E9R | atacaccactgcctctccaa | |||
| E10 | CTCFL E10F | gccttgttcagaatgtgttt | 463 bp | 58 °C |
| CTCFL E10R | aggcatgacagatgctcctgaag | |||
| nCTCFL E10Fa | ttggggagggaaataaaagg | 323 bp | 58 °C | |
| nCTCFL E10Ra | gatctttccatgggggattt | |||
| E11 | CTCFL E11Fa | attgccctcgaaagaactca | 397 bp | 64 °C |
| CTCFL E11Ra | caaataggggctctggacac | |||
| nCTCFL E11F | tttctttccatgcgtggggtcc | 356 bp | 58 °C | |
| nCTCFL E11R | ctggacacatcccctggaca | |||
| E12 | CTCFL E12Fa | tgcacagtttataaattccaattcc | 492 bp | 58 °C |
| CTCFL E12Ra | acctgcaatgtttctttgaaat | |||
| nCTCFL E12F | ttagctttctgataaatttgct | 439 bp | 58 °C | |
| nCTCFL E12R | ctacaaacaggagaacaattcta | |||
| E13 | CTCFL E13F | gagttctggagatcagtcatgg | 351 bp | 58 °C |
| CTCFL E13Rb | GGTCGTTCAGAGGAGTGTGGb | |||
| nCTCFL E13F | tatgttgtgttagtcctttct | 235 bp | 58 °C | |
| nCTCFL E13Rb | TGTGGCGCCTCCCCCTCTCTb |
Results
MTHFR rs1801133 analysis
In the control group (n = 25), 8 individuals (32 %) were homozygous CC, 15 (60 %) were heterozygous CT and 2 (8 %) were homozygous TT. In the infertile population (n = 107), 47 subjects (44 %) were homozygous CC, 43 (40 %) were heterozygous CT and 17 (15 %) were homozygous TT. The contingency analysis results showed no significant differences (p > 0.05) in the distribution of the three genotypes among the control and the infertile populations (Table 5).
Table 5.
MTHFR rs1801133 results in control and infertile populations
| CC | CT | TT | Total | |
|---|---|---|---|---|
| Control | 8 | 15 | 2 | 25 |
| Infertile | 47 | 43 | 17 | 107 |
| Total | 55 | 58 | 19 | 132 |
Likewise, none of the three rs1801133 genotypes were preferentially associated (p > 0.05), neither to the characteristics of the seminogram (Table 6) nor to the presence of an abnormal methylation pattern in paternally methylated H19-ICR or IG-DMR (Table 7).
Table 6.
MTHFR rs1801133 results in the infertile population classified according to the presence or absence of normozoospermia, asthenozoospermia, oligozoospermia and teratozoospermia
| CC | CT | TT | Total | |
|---|---|---|---|---|
| Normozoospermia | 5 | 7 | 3 | 15 |
| Non-Normozoospermia | 42 | 36 | 14 | 92 |
| Asthenozoospermia | 21 | 26 | 9 | 56 |
| Non-Asthenozoospermia | 26 | 17 | 8 | 51 |
| Oligozoospermia | 12 | 9 | 2 | 23 |
| Non-Oligozoospermia | 35 | 34 | 15 | 84 |
| Teratozoospermia | 40 | 29 | 13 | 82 |
| Non-Teratozoospermia | 7 | 14 | 4 | 25 |
Table 7.
MTHFR rs1801133 results in the infertile population according to the methylation pattern at H19-ICR and IG-DMR
| CC | CT | TT | Total | ||
|---|---|---|---|---|---|
| H19-ICR | Normal Methylation | 41 | 38 | 16 | 95 |
| Hypomethylation | 6 | 5 | 1 | 12 | |
| IG-DMR | Normal Methylation | 34 | 23 | 10 | 67 |
| Hypomethylation | 13 | 20 | 7 | 40 | |
CTCFL sequence analysis
We did not identify nonsense or missense mutations in any case. In addition to several referred single nucleotide polymorphisms, two novel synonymous sequence variations not listed in the dbSNP database were identified. One was present in homozygosis in one patient (Case 18) with IG-DMR slight hypomethylation (c.288G > A;pLeu69Leu), the other one was present in heterozygosis in a patient (Case 102) with H19-ICR hypomethylation (c.1308G > A;pGln409Gln).
Discussion
The results of MTHFR rs1801133 SNP genotyping showed no significant association of the polymorphism neither to the condition of infertility nor to the presence of a hypomethylation pattern at paternally imprinted loci. These results point out that this polymorphism is not a risk factor for male infertility in the Spanish population analyzed. Moreover, the presence of the T allele is not the cause of the lower levels of sperm DNA methylation at the loci that showed hypomethylation. Importantly, even the distribution of the genotypes among the 12 patients with a more marked hypomethylation pattern fits to the values observed in the control population (8 homozygous CC and 4 heterozygous CT).
The association between male infertility and the T allele has mainly been described in Asian population studies [1, 16, 22, 26]. This association has also been described in a population from Brazil [10] and Iran [25]. In European studies, the male infertility-MTHFR SNP association is less conclusive. Bezold et al. [3] clearly found this association comparing a large series of infertile and control patients. Paracchini et al. [21] reported a prospective study on men seeking care at the infertility clinic in Milano (Italy), concluding that those subjects carrying the TT genotype were at increased risk of being infertile after the 1-year follow-up. Nevertheless, the rest of the reports from European groups [7, 24, 28] and one from an Indian group [6] did not find any association between male infertility and the MTHFR SNP.
Two recent meta-analysis support the results obtained in our infertile population [11, 30]. First, because both studies conclude that the T allele is a risk factor for infertility particularly related to azoospermia; none of infertile patients analyzed in our series were azoospermic. Second, the article from Wu et al. [30] supports that the rs1801133 polymorphism is capable of causing male infertility susceptibility only in Asians, but not in Caucasians infertile patients.
What is still a matter of debate is the origin of this difference according to the geographical localization of the populations. Results suggest that the presence of the T allele affects the reproductive phenotype according to the nutritional characteristics of the population. Thus, in populations without nutritional deficiencies in vegetables, green leafy vegetables, grains and nuts, which are rich in folate, the polymorphism does not represent a risk factor for DNA methylation nor for fertility. However, the effects of the SNP would be detectable in populations where folate nutritional contributions are deficient. Nevertheless, it has to be considered that the presence of other MTHFR SNPs together with rs1801133, as well as in other enzymes of the folate pathway, could increase the deleterious effect on infertility and methylation by methyltransferases [7, 21]. That is, the relation between genotype and diet, and the interactions among polymorphisms, has been shown to affect the impact on fertility.
Concerning the CTCFL gene sequence analysis, we were unable to relate the pattern of hypomethylation with the presence of CTCFL mutations. The imprinting mark is a very complex mechanism that regulates gene expression through the participation of a huge number of molecules in a highly coordinated way. Some of the most important genes involved in this mechanism are the following: DNMT1, DNMT3A, DNMT3B, DNMT3L, HAT, HDAC, HMT, HDM, HP1, CTCFL, Zfp57, CTCF, and PRMT7.
Like us, other authors have tried to identify the origin of sperm DNA imprinting errors through the analysis of single genes using PCR-based techniques. Overall, the results have not been encouraging enough; Poplinski et al. [23] did not identify any mutation in CTCFL while Kobayashi et al. [14] reported the DNMT3A and DNMT3L variations. However, the imprinting abnormalities were not present in all of the regions analyzed and were related to hypo-, hypermethylation errors and also found in cases with normal sperm methylation patterns. Moreover, the most stand out sequence variations in DNMT3L present in homozygosis in patients with hypomethylation at paternally imprinted loci (H19-ICR and/or IG-DMR) correspond to a referred SNP (rs75396112) and to a synonymous sequence variation (c.788C > T; pIle98Ile; GenBank accession number AF194032). Accordingly, we do believe that the complexity of the process requires the analysis of the Exome through the application of next-generation sequencing techniques. Unfortunately, we were unable to perform this kind of large-scale analysis in our population due to limitations in the DNA availability.
In conclusion, the MTHFR rs1801133 polymorphism is not a risk factor for male infertility in the Spanish population analyzed. Concerning the analysis of the MTHFR rs1801133 polymorphism and CTCFL mutations as the cause of sperm DNA hypomethylation, the lack of association in patients with the most severe pattern of hypomethylation suggest the implication of other factors.
Electronic supplementary material
Age and spermiogram parameters of the infertile patients. (DOC 175 kb)
Acknowledgments
The authors wish to thank Dr. Javier Nadal and the embryologist from the Unidad de Reproducción Asistida of the Centro Médico Teknon (Barcelona, Spain) and the Laboratorio de Andrología y Banco de Semen of the Instituto Universitario IVI Valencia (Valencia, Spain) for providing the semen samples. This work was supported by Projects PS09/00330 (Fondo de Investigación Sanitaria, Instituto de Salud Carlos III, Ministerio de Ciencia e Innovación, Spain) and SGR2009–282 (Agència de Gestió d’Ajuts Universitaris i de Recerca, Generalitat de Catalunya, Spain). Marta Pladevall was the recipient of the grant number UAB2006–00213 from the Universitat Autònoma de Barcelona.
Footnotes
Capsule The MTHFR rs1801133 polymorphism is not associated with male infertility in the Spanish population. Neither the MTHFR polymorphism, nor CTCFL mutations explain a pattern of sperm hypomethylation at paternally imprinting loci.
Cristina Camprubí and Marta Pladevall contributed equally to this work.
References
- 1.A ZC, Yang Y, Zhang SZ, Li N, Zhang W. Single nucleotide polymorphism C677T in the methylenetetrahydrofolate reductase gene might be a genetic risk factor for infertility for Chinese men with azoospermia or severe oligozoospermia. Asian J Androl. 2007;9:57–62. doi: 10.1111/j.1745-7262.2007.00225.x. [DOI] [PubMed] [Google Scholar]
- 2.Aapola U, Kawasaki K, Scott HS, Ollila J, Vihinen M, Heino M, et al. Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics. 2000;65:293–298. doi: 10.1006/geno.2000.6168. [DOI] [PubMed] [Google Scholar]
- 3.Bezold G, Lange M, Peter RU. Homozygous methylenetetrahydrofolate reductase C677T mutation and male infertility. New Engl J Med. 2001;344:1172–1173. doi: 10.1056/NEJM200104123441517. [DOI] [PubMed] [Google Scholar]
- 4.Boissonnas CC, Abdalaoui HE, Haelewyn V, Fauque P, Dupont JM, Gut I, et al. Specific epigenetic alterations of IGF2-H19 locus in spermatozoa from infertile men. Eur J Hum Genet. 2010;18:73–80. doi: 10.1038/ejhg.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camprubí C, Pladevall M, Grossmann M, Garrido N, Pons M, Blanco J. Semen samples showing an increased rate of spermatozoa with imprinting errors have a negligible effect in the outcome of assisted reproduction techniques. Epigenetics. 2012;7:1115–1124. [DOI] [PMC free article] [PubMed]
- 6.Dhillon VS, Shahid M, Husain SA. Associations of MTHFR DNMT3b 4977 bp deletion in mtDNA and GSTM1 deletion, and aberrant CpG island hypermethylation of GSTM1 in non-obstructive infertility in Indian men. Mol Hum Reprod. 2007;13:213–222. doi: 10.1093/molehr/gal118. [DOI] [PubMed] [Google Scholar]
- 7.Ebisch IM, van Heerde WL, Thomas CM, van der Put N, Wong WY, Steegers-Theunissen RP. C677T methylenetetrahydrofolate reductase polymorphism interferes with the effects of folic acid and zinc sulfate on sperm concentration. Fertil Steril. 2003;80:1190–1194. doi: 10.1016/S0015-0282(03)02157-5. [DOI] [PubMed] [Google Scholar]
- 8.El Hajj N, Zechner U, Schneider E, Tresch A, Gromoll J, Hahn T, et al. Methylation status of imprinted genes and repetitive elements in sperm DNA from infertile males. Sex Dev. 2011;5:60–69. doi: 10.1159/000323806. [DOI] [PubMed] [Google Scholar]
- 9.Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, Matthews RG, et al. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet. 1995;10:111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- 10.Gava MM, Chagas Ede O, Bianco B, Christofolini DM, Pompeo AC, Glina S, et al. Methylenetetrahydrofolate reductase polymorphisms are related to male infertility in Brazilian men. Genet Test Mol Biomarkers. 2011;15:153–157. doi: 10.1089/gtmb.2010.0128. [DOI] [PubMed] [Google Scholar]
- 11.Gupta N, Gupta S, Dama M, David A, Khanna G, Khanna A, et al. Strong association of 677 C > T substitution in the MTHFR gene with male infertility—a study on an indian population and a meta-analysis. PLoS One. 2011;6:e22277. doi: 10.1371/journal.pone.0022277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammoud SS, Purwar J, Pflueger C, Cairns BR, Carrell DT. Alterations in sperm DNA methylation patterns at imprinted loci in two classes of infertility. Fertil Steril. 2010;94:1728–1733. doi: 10.1016/j.fertnstert.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 13.Jelinic P, Stehle JC, Shaw P. The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation. PLoS Biol. 2006;4:e355. doi: 10.1371/journal.pbio.0040355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi H, Hiura H, John RM, Sato A, Otsu E, Kobayashi N, et al. DNA methylation errors at imprinted loci after assisted conception originate in the parental sperm. Eur J Hum Genet. 2009;17:1582–1591. doi: 10.1038/ejhg.2009.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi H, Sato A, Otsu E, Hiura H, Tomatsu C, Utsunomiya T, et al. Aberrant DNA methylation of imprinted loci in sperm from oligospermic patients. Hum Mol Genet. 2007;16:2542–2551. doi: 10.1093/hmg/ddm187. [DOI] [PubMed] [Google Scholar]
- 16.Lee HC, Jeong YM, Lee SH, Cha KY, Song SH, Kim NK, et al. Association study of four polymorphisms in three folate-related enzyme genes with non-obstructive male infertility. Hum Reprod. 2006;21:3162–3170. doi: 10.1093/humrep/del280. [DOI] [PubMed] [Google Scholar]
- 17.Marques CJ, Carvalho F, Sousa M, Barros A. Genomic imprinting in disruptive spermatogenesis. Lancet. 2004;363:1700–1702. doi: 10.1016/S0140-6736(04)16256-9. [DOI] [PubMed] [Google Scholar]
- 18.Marques CJ, Costa P, Vaz B, Carvalho F, Fernandes S, Barros A, et al. Abnormal methylation of imprinted genes in human sperm is associated with oligozoospermia. Mol Hum Reprod. 2008;14:67–74. doi: 10.1093/molehr/gam093. [DOI] [PubMed] [Google Scholar]
- 19.Marques CJ, Francisco T, Sousa S, Carvalho F, Barros A, Sousa M. Methylation defects of imprinted genes in human testicular spermatozoa. Fertil Steril. 2010;94:585–594. doi: 10.1016/j.fertnstert.2009.02.051. [DOI] [PubMed] [Google Scholar]
- 20.Minor A, Chow V, Ma S. Aberrant DNA methylation at imprinted genes in testicular sperm retrieved from men with obstructive azoospermia and undergoing vasectomy reversal. Reproduction. 2011;141:749–757. doi: 10.1530/REP-11-0008. [DOI] [PubMed] [Google Scholar]
- 21.Paracchini V, Garte S, Taioli E. MTHFR C677T polymorphism, GSTM1 deletion and male infertility: a possible suggestion of a gene-gene interaction? Biomarkers. 2006;11:53–60. doi: 10.1080/13547500500442050. [DOI] [PubMed] [Google Scholar]
- 22.Park JH, Lee HC, Jeong YM, Chung TG, Kim HJ, Kim NK, et al. MTHFR C677T polymorphism associates with unexplained infertile male factors. J Assist Reprod Genet. 2005;22:361–368. doi: 10.1007/s10815-005-6795-0. [DOI] [PubMed] [Google Scholar]
- 23.Poplinski A, Tüttelmann F, Kanber D, Horsthemke B, Gromoll J. Idiopathic male infertility is strongly associated with aberrant methylation of MEST and IGF2/H19 ICR1. Int J Androl. 2010;33:642–649. doi: 10.1111/j.1365-2605.2009.01000.x. [DOI] [PubMed] [Google Scholar]
- 24.Ravel C, Chantot-Bastaraud S, Chalmey C, Barreiro L, Aknin-Seifer I, Pfeffer J, et al. Lack of association between genetic polymorphisms in enzymes associated with folate metabolism and unexplained reduced sperm counts. PLoS One. 2009;4:e6540. doi: 10.1371/journal.pone.0006540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Safarinejad MR, Shafiei N, Safarinejad S. Relationship between genetic polymorphisms of methylenetetrahydrofolate reductase (C677T, A1298C, and G1793A) as risk factors for idiopathic male infertility. Reprod Sci. 2011;18:304–315. doi: 10.1177/1933719110385135. [DOI] [PubMed] [Google Scholar]
- 26.Singh K, Singh SK, Sah R, Singh I, Raman R. Mutation C677T in the methylenetetrahydrofolate reductase gene is associated with male infertility in an Indian population. Int J Androl. 2005;28:115–119. doi: 10.1111/j.1365-2605.2004.00513.x. [DOI] [PubMed] [Google Scholar]
- 27.Solvas I, Grossmann M, Santaló J, Pons MC. Estudio comparativo entre dos métodos de swim-up. Revista ASEBIR. 2002;7:28–32.
- 28.Stuppia L, Gatta V, Scarciolla O, Colosimo A, Guanciali-Franchi P, Calabrese G, et al. The methylenetethrahydrofolate reductase (MTHFR) C677T polymorphism and male infertility in Italy. J Endocrinol Invest. 2003;26:620–622. doi: 10.1007/BF03347018. [DOI] [PubMed] [Google Scholar]
- 29.WHO laboratory manual for the examination of human semen and sperm-cervical mucus interaction. 4. New York: Cambridge University Press; 1999. [Google Scholar]
- 30.Wu W, Shen O, Qin Y, Lu J, Niu X, Zhou Z, et al. Methylenetetrahydrofolate reductase C677T polymorphism and the risk of male infertility: a meta-analysis. Int J Androl. 2012;35:18–24. doi: 10.1111/j.1365-2605.2011.01147.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Age and spermiogram parameters of the infertile patients. (DOC 175 kb)