Abstract
Purpose
Gonadotropins, interacting with their gonadal receptors, play a key role in sexual development, reproductive functions and metabolism. In this study we performed the genetic analysis of FSHR and LHR and semen investigation in 14 infertile men with normal level of T and elevated levels of FSH and/or LH in the absence of other causes of infertility.
Methods
Sperm parameters were analysed following WHO (2010) guidelines and sperm morphology by Transmission Electron Microscopy (TEM) analysis mathematically elaborated. FSHR and LHR gene mutations have been searched by PCR technique, followed by DHPLC analysis and direct sequencing.
Results
In FSHR, we found no difference in the frequency between Ala or Thr at position 307, Ser was at codon 680 in all subjects. Three patients had an heterozygous mutation at codon 419. Three intronic polymorphisms (rs2091787, rs6708637, rs1922464) were significantly found compared to controls; the single allele frequency and the odds ratio were calculated. Two new variants: the Cys338Arg and the Gln123Glu were detected in two different patients. Regarding LHR, three patients were heterozygous for the known variant Glu354Lys and two for Ile374Thr. Intronic polymorphisms were not identified. A new variant, the Val144Ile was found. By the routine semen analysis, variable seminal conditions in this group of patients was observed, on the contrary TEM data mathematically elaborated showed a homogeneous decrease in fertility index and increase in sperm pathologies such as apoptosis and immaturity.
Conclusions
The obtained results suggest that a deeper examination of spermatozoa, achieved by the use of more powerful tools such as TEM or molecular analysis, are advisable in patients with hypergonadotropic hypogonadism.
Keyword: FSHR, LHR, Male infertility, Sperm evaluation, Transmission electron microscopy
Introduction
Gonadotropins, the actions of which are mediated at the level of their receptors, play a key role in sexual development, reproductive functions and metabolism. In particular, spermatogenesis is regulated by FSH acting on Sertoli cells and LH acting on Leydig cells [1]. Knowledge of the structure of gonadotropin receptors and their mode of action has rapidly advanced [2]. FSH receptor (FSHR) and LH receptor (LHR) are members of a novel family of G-protein-coupled receptors and their genes are located on chromosome 2p21 [2]. Recently, significant new information has emerged about the structure, mechanisms of activation, expression and genetic mutations of these receptors [3]. Mutations can be activating or inactivating, and descriptions of only 10 FSHR-inactivating mutations and one activating mutation have been published. In men, inactivating LHR mutations are associated with severe phenotypes, such as the presence of a 46,XY karyotype correlated with disorders of sex development [4]; in addition, an LH receptor splice mutation responsible for male hypogonadism with subnormal sperm production has been reported [5].
Studies have demonstrated that naturally occurring mutations of FSHR are very rare, however several single nucleotide polymorphisms (SNP), responsible for FSH level alterations, have been associated with risk of impaired gametogenesis. There are three common FSHR gene polymorphisms: one in the promoter region (at nucleotide position 29) and two in exon 10 [6]; the first SNP in exon 10 is located in the extracellular domain at position 307 (nucleotide 919) which can be occupied either by alanine (Ala) or by threonine (Thr), the second is located in the intracellular domain at position 680 (nucleotide 2039) and has either asparagine (Asn) or serine (Ser). A decrease in testicular FSHR binding ability has been reported in infertile men, but investigations on the distribution of the above mentioned variants gave conflicting results [7], showing the importance of ethnic differences [8–10]. Although no significant correlation between serum FSH levels and semen characteristics and FSHR gene polymorphisms was found, in some studies the combination of heterozygous Thr/Ala + Asn/Ser genotypes has been shown to increase the risk of male infertility [7, 11, 12]. However, a published study of 364 idiopathic infertile patients and 281 fertile controls in the Han-Chinese population [13] and a subsequent meta-analysis of the literature [14] did not show an association between male infertility and SNPs at positions 307 and 680.
Nevertheless, analysis of the FSHR gene may represent a valid pharmacogenetic approach for the treatment of male infertilit, while also confirming the importance of strict criteria for the selection of patients to be administered FSH [15]. Tuttelmann et al [16]. reported on the combined effects of the FSHB -211G >T and FSHR 2039A >G variants on male reproductive parameters, demonstrating that oligozoospermic patients carrying unfavorable variants affecting FSH action may benefit from FSH treatment.
In this paper, semen quality in a group of selected patients with hypergonadotropic hypogonadism without obvious testicular noxa was evaluated by optical microscopy and sperm morphology was observed by transmission electron microscopy (TEM). At the same time, the genetic analysis of known/unknown SNPs in FSHR and LHR genes was carried out.
Materials and methods
Patients
Among patients with elevated levels of FSH and/or LH and normal testosterone (T) at the Interdepartmental Centre for Research and Therapy of Male Infertility and at the Endocrinology Unit of the University of Siena from 2010 to 2012, we selected 14 individuals (aged 34–78 years) who did not show or report other pathologies potentially responsible for hypergonadotropic hypogonadism.. Patients were interviewed about their case histories, reproductive problems and family backgrounds. They had a normal 46, XY karyotype evaluated by conventional cytogenetic analysis, no history of diabetes, radiotherapy, chemotherapy, chronic illness or medications. Microbiological investigation of semen specimens and urethral fluids excluded the presence of genitourinary infections. Clinical and physical examinations and scrotal Eco-color Doppler were performed in all patients to detect the possible presence of varicocele or other anatomical problems. All patients showed a BMI <25 kg/m2. Serum hormone levels were evaluated using chemiluminescence performed with a commercial kit (Beckman Coulter Access for FSH, LH and Testosterone). Normal ranges were: 0.7–11.00 mU/ml for FSH (sensitivity 0.2 mUI/ml, intra- and inter-assay coefficient of variation <10 %), 0.8–8.0 mU/L for LH (sensitivity 0.2 mUI/ml, intra- and inter-assay coefficient of variation <10 % and 2.7–10.9 mg/ml) for testosterone (sensitivity 0.1 ng/ml, intra- and inter-assay coefficient of variation <10 %). The control group for allelic frequencies was represented by 40 fertile men (aged 30–51), <25 kg/m2, with normal karyotype and without endocrine pathologies examined at the blood donor centre of Siena Hospital. All patients and controls provided written informed consent before inclusion in this study.
Light microscopy of semen
Semen samples were available from 13 patients. One patient refused to participate due to religious beliefs. Semen was collected by masturbation after 4 days of sexual abstinence and examined after liquefaction for 30 min at 37 °C. Volume, pH, sperm concentration and motility were evaluated according to WHO [17] guidelines. In patients with a sperm concentration of less than 10 million/ml, PCR analysis on DNA extracted from blood lymphocytes (QIAamp DNA Blood kit, QIAGEN, Milan, Italy) was carried out to exclude the presence of Y chromosome microdeletions.
Transmission electron microscopy of semen
For electron microscopy, sperm samples were fixed in cold Karnovsky fixative and maintained at 4 °C for 2 h. Fixed semen was washed in 0.1 mol/l cacodylate buffer (pH 7.2) for 12 h, postfixed in 1 % buffered osmium tetroxide for 1 h at 4 °C, then dehydrated and embedded in Epon Araldite. Ultra-thin sections were cut with a Supernova ultramicrotome (Reickert Jung, Vienna, Austria), mounted on copper grids, stained with uranyl acetate and lead citrate and then observed and photographed with a Philips EM208 electron microscope (Philips Scientifics, Eindhoven, The Netherlands).
For each patient, three hundred ultra-thin sperm sections were analysed. Major submicroscopic characteristics were recorded by two trained examiners who were blind to the experiment. TEM data were evaluated using the statistical mathematical formula by Baccetti et al [18]. This formula considers 16 selected submicroscopic characteristics of sperm organelles able to define sperm function and calculates the number of spermatozoa probably free of structural defects (fertility index) and the percentages of three main phenotypic sperm pathologies: immaturity, necrosis and apoptosis [19], each one characterised by typical alterations of organelles. Reduced acrosomes, round or elliptical nuclei with uncondensed chromatin and the presence of cytoplasmic droplets were the examined characteristics for immaturity. Marginated chromatin and altered shaped nuclei were considered as main ultrastructural markers of apoptosis, whereas spermatozoa with broken plasma membranes, reacted acrosomes and disrupted chromatin are affected by necrosis. Moreover, in Table 1 we reported the sperm characteristics from 25 men of proven fertility as reference values, previously published by Collodel and Moretti [19].
Table 1.
Primer sequences for FSHR and LHR
| FSHR | LHR | ||
|---|---|---|---|
| Primer name | Sequence | Primer name | Sequence |
| FSHR1F | CTGCTCCTGGTCTCTTTGC | LHR1F | GTGTGTGGAAGGGCAGC |
| FSHR1R | ATATCAGCCTAATGTAAA | LHR1R | CTGGGCTTCTGCGGCTT |
| FSHR2F | AGCACTTAGTATATGGTA | LHR2F | TGACAATCACTTTTACCC |
| FSHR2R | CATGTCAGTTCGGCTAC | LHR2R | CAGAACACAAATCTGGGAG |
| FSHR3F | GACACAAGGCTCTATGCTG | LHR3F | CCATTCCTAAACCCTCTC |
| FSHR3R | GACAATCTAGGGATCTGAG | LHR3R | GATCTTGTAGCCAGAGTA |
| FSHR4F | TACCATCAAGATCACTAGC | LHR4F | GAGCTACGAATGCTTACACA |
| FSHR4R | CTCTCCACCTCCACTTCTGC | LHR4R | GCCTGGAATAGATGCTT |
| FSHR5F | CCCAGTTAGTTTCCTTCATT | LHR5F | GAACAAAGCATAAATAGAC |
| FSHR5R | CCTCATTTCTCATTTACC | LHR5R | CCGTAACCAAGACTTGTA |
| FSHR6F | GGAAATGTTACCCTTGAG | LHR6F | AATCCCTTACCTCAAGCC |
| FSHR6R | TCGTGGAGTTAGATGGCAG | LHR6R | CAGTCTATGGCATGGTTAT |
| FSHR7F | CACTCACTCTCATTTTG | LHR7F | CTGTATTCGCAAGTGAAC |
| FSHR7R | TGGAGGATGTAGACTA | LHR7R | GACCTTCATGTAATTGCTG |
| FSHR8F | GAGTAGAGCTGATGTAAG | LHR8F | CAGCAATTACATGAAGGTC |
| FSHR8R | GTGGAGAAATGCCAAAG | LHR8R | TATAAAGTTCAGCCCGACG |
| FSHR9F | ACTAACCTGTGCTGTCTGCC | ||
| FSHR9R | GACTTCTAACTTACACAAAC | ||
| FSHR10F | AGGTCCATAGTAATCCTG | ||
| FSHR10R | AAGGGCTTGAGACAGGGAA | ||
| FSHR11F | TACTGGATCTGAGATGTTG | ||
| FSHR11R | CATAGTTGTGATATTGGCTC | ||
| FSHR12F | GAGCCAATATCACAACTATG | ||
| FSHR12R | GTGAAACAGAACCAGCAGAATC | ||
| FSHR13F | GATTCTGCTGGTTCTGTTTCAC | ||
| FSHR13R | CAGCTTCCTAATGTATCAC | ||
| FSHR14F | GTGATACATTAGGAAGCTG | ||
| FSHR14R | TTGGAGACCCAGCCGTTGC | ||
DNA extraction
Blood samples were obtained from 14 patients and 40 healthy controls. Genomic DNA was extracted using salting out procedures. Briefly, 5 ml of blood were lysed with 5 ml of TKM1 (10 mM Tris–HCl, 10 mM KCl, 10 mM MgCl2 and 2 mM EDTA) with Igepal for 7 min. Samples were then centrifuged at 2,400 rpm for 10 min at 18–20 °C. Pellets were washed twice with 10 ml of TKMI without Igepal. Finally, pellets were resuspended in 800 μl of TKM2 (10 mM Tris–HCl, 10 mM KCl, 10 mM MgCl2, 2 mM EDTA and 40 mM MaCl) and 10 % SDS and incubated at 55 °C for 15 min. At the end of the incubation period, 600 μl of 3 M NaCl were added and samples were centrifuged at 1,400 rpm for 15 min (room temperature). A supernatant was then added to 4 ml of 100 % EtOH and mixed by inversion until DNA appeared. DNA was then washed with 70 % EtOH by centrifugation at 1,400 rpm for 10 min (room temperature). The pellets were then resuspended with 100 μl of water and left O/N at 37 °C. For each sample DNA concentration was assessed by spettrophotometry and stock solutions of 200 ng/50 μl were prepared and used for PCR analysis.
FSHR and LHR gene analysis
Primers (Table 1) were designed to cover all FSHR and LHR gene exons and the intron/exon junction using Primer 3 (version 0.4.0). For FSHR1, FSHR2, FSHR5, FSHR7, FSHR11, FSHR12, LHR2, LHR3, LHR4, LHR5, LHR6, LHR7 and LHR8 conditions were 2.5 mM MgCl2 and 50 °C (35 cycles); for FSHR8, FSHR9 and FSHR14 conditions were 2.5 mM MgCl2 and 56 °C (35 cycles); for FSHR13 conditions were conditions were 2 mM MgCl2 and 50 °C (35 cycles); for FSHR4, FSHR6 and FSHR10 conditions were conditions were 1.5 mM MgCl2 and 55 °C (35 cycles) and for LHR1 conditions were 1.5 mM MgCl2, 65 °C in GC-rich buffer (35 cycles). PCR products were run in agarose gel (1.5 %) and visualized with ethidium bromide. Mutations/SNPs that were searched for in the FSHR included: Ala419Thr, Ala307Thr, Asn112Thr, Asp224Val, Pro346Arg, Arg573Cys, Pro519Thr, Leu601Val, Ile160Thr, Ala189Val, Ans191Ile, Thr666Ala, Asn680Ser, Cys338Arg, Gln123Glu, rs114573531, rs6708637, rs2091787, rs115961828, and 1922464. Mutations/SNPs that were searched for in the LHR included: LLKLLLLLQ insertion, Val144Ile, Cys133Arg, Phe194Val, Val144Phe, Cys343Ser, Glu354Lys, Ile374Thr, Thr392Ile, Trp491Stp, Cys543Arg, Cys545Stop, Arg554Stp, 589insTfs, Ala593Pro, Leu608/Val609 deletion, Ser616Tyr, Tyr623Ser, Ile625Lys, exon 8 and 10 deletions.
DHPLC and sequencing
PCR products were denatured at 95 °C for 10 min and then left at 56 °C for 10 min to allow hybridization of the PCR products and formation of a mixture of homo- or heteroduplexes. Then the samples were analyzed with the Denaturing High Performance Liquid Chromatography (DHPLC) technique (Transgenomic) at specific temperatures calculated by NavigatorTM Software for each amplicon to confirm the presence/absence of mutations/polymorphisms. The temperature was calculated in order to allow the partial denaturation of DNA heteroduplexes. The melted heteroduplexes were then resolved from the corresponding homoduplexes by reversed-phase ion-pair liquid chromatography using their different retention times on the DNASep Cartridge matrix. The WAVETM Low-Range Mutation and the WAVETM High-Range Mutation were used as the standard. Samples with different DHPLC chromatograms were subjected to direct sequencing (Genechron, Rome, Italy). In particular, for all samples, when a chromatogram suggested the presence of a wild type sample, the latter was mixed with a known control to find possible homozygous mutations. All samples with an heteroduplex chromatogram were sequenced.
Analysis of variants
When applicable, new variants were analyzed with specific software to predict a possible pathogenetic role. In particular, we used the Swiss-model to insert the variant into the protein sequence, the Pymol (version 1.5) to simulate the three-dimensional structure of the protein and PolyPhen-2 to analyze the potential pathogenetic role. Every intronic polymorphism was analyzed with NetGene2 Server Splicing to predict a site of splicing.
Statistical analysis
Frequencies of polymorphisms were calculated using the SPSS software package version 13.0 (IBM Company, Chicago, Illinois). Interaction with SNPs was tested by χ2 analysis at the genotypic and allelic levels. P < 0.05 was considered statistically significant.
Results
The results of semen parameters analyzed by light microscopy, sperm morphology examined by TEM and serum gonadotropin levels are reported in Table 2. One patient refused semen analysis. Serum FSH was elevated in all (13/14) but one patient (n°1) who showed elevated levels of LH. Serum LH was elevated in 6/14 patients and an increase of both FSH and LH was observed in 5/14 patients. Serum testosterone was within normal range in all patients.
Table 2.
Semen parameters detected by light microscopy (volume, Sperm/ml × 106and motility), ultrastructural sperm analysis by transmission electron microscopy (fertility index and necrosis, apoptosis and immaturity percentages) and blood gonadotropin levels in 14 selected patients
| Semen parameters | TEM evaluation of sperm | Gonadotropin levels | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Case | Vol/ml | Sperm/ml × 106 | Motility % | Necrosis% | Apoptosis% | Immaturity% | Fertility index (FI) n° | FSH mU/ml | LH mU/ml |
| 1 | 2.5 | 0.32 | 0 | 87.51 | 11.90 | 72.85 | 0 | 7.5 | 10.5 |
| 2 | 4.5 | 38.25 | 12 | 60.02 | 28.92 | 67.35 | 4864 | 12.5 | 5.4 |
| 3 | 2 | 4 | 13 | 68.04 | 9.26 | 69.76 | 63 | 13.1 | 3.3 |
| 4 | 4 | 14.25 | 44 | 34.50 | 6.67 | 59.56 | 63733 | 12.9 | 2.1 |
| 5 | 3 | 39.75 | 30 | 19.59 | 5.80 | 89.84 | 354652 | 23.8 | 11 |
| 6 | 7.5 | 11 | 36 | 32.57 | 23.29 | 56.08 | 100582 | 41.8 | 16.5 |
| 7 | 2 | 57 | 54 | 27.94 | 7.05 | 57.24 | 560837 | 16.1 | 5.5 |
| 8 | 1.4 | 5 | 31 | 70.81 | 18.86 | 78.22 | 3004 | 15.8 | 2.1 |
| 9 | 7.5 | 14.25 | 35 | 28.64 | 6.50 | 51.33 | 26125 | 13.6 | 7.3 |
| 10 | 2.3 | 24 | 13 | 30.40 | 9.54 | 66.57 | 10860 | 12.1 | 2.06 |
| 11 | 3 | 1.95 | 11 | 12.6 | 7.2 | ||||
| 12 | 2 | Azo | 20.5 | 8.3 | |||||
| 13 | 2.8 | Azo | 45.3 | 27.7 | |||||
| 14 | Refused | 56 | 29 | ||||||
| Reference range | WHO [17] | WHO [17] | WHO [17] | 32.13 [19] | 4.05 [19] | 48.83 [19] | >1,691,122 [19] | 0.7–11 | 0.8–8 |
Semen evaluation
Two out of 13 patients were azoospermic. Sperm concentration was lower than the 2.5 centile [17] in 4 out of 11 patients (n° 1, 3, 8 11), comprised between the 2.5 centile and the 5 centile in 3 out of 11 men (n° 4, 6, 9), comprised between the 10 centile and rhe 25 centile in 3 out of 11 patients (n° 2, 5, 10), and comprised between the 25 centile and the 50 centile in only 1 patient (n° 7).
Sperm progressive motility [17] appeared lower than the 2.5 centile in 5 out of 11 patients (n° 1, 2, 3, 9, 10), comprised between the 2.5 centile and thw 5 centile in 3 out of 11 analyzed samples (n° 5, 8, 11), comprised between the 5 centile and the 10 centile in one case (n° 6) and between the 10 centile and the 25 centile in another (n° 4), and finally between the 25 centile and the 50 centile in the last sample (n° 7).
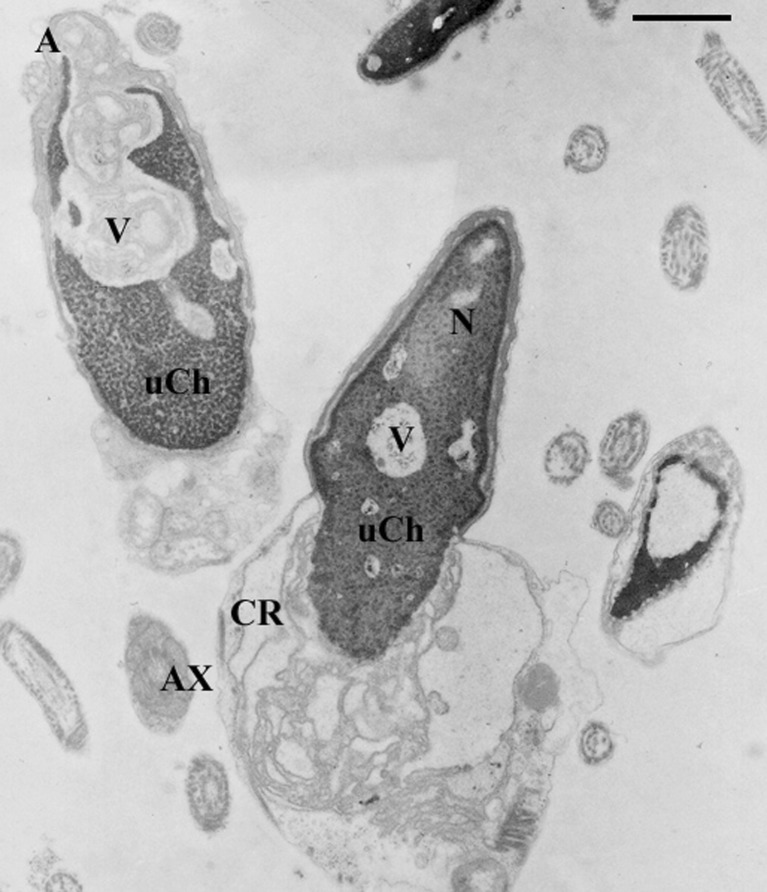
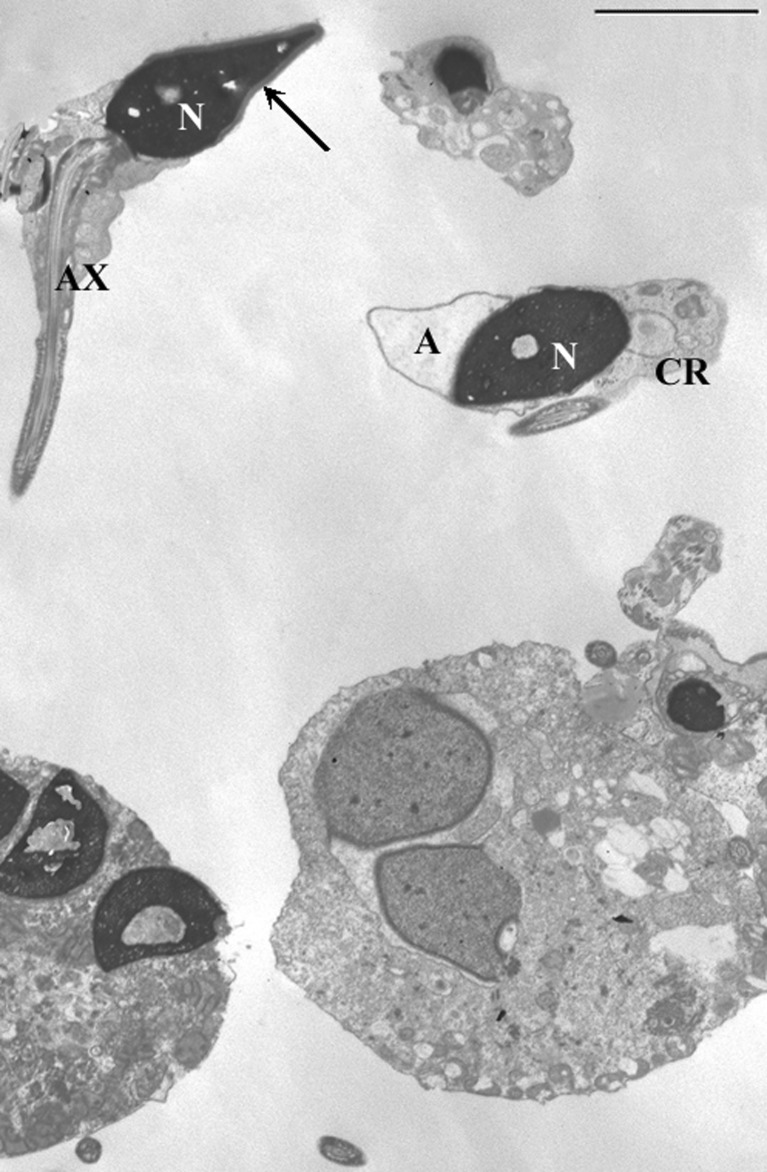
TEM analysis was possible in 10 patients with a sufficient number of sperm/ml. In all specimens the fertility index (number of spermatozoa devoid of ultrastructural defects) resulted clearly lower than that reported for fertile men. To understand the presumptive effect of FSH and LH levels on sperm morphology, we considered the incidence of sperm pathologies that directly came from the mathematical elaboration of TEM data [18, 19]. The necrosis percentage was increased in 4 out of 10 patients (n°1, 2, 3, 8) compared with the reference value obtained from semen analysis of proven fertile men. In all analyzed specimens, the percentages of apoptosis and immaturity (Figs. 1 and 2) were higher than the reference values (Table 2).
Fig. 1.
TEM micrograph of sections of sperm with altered nuclei (N) and acrosomes (a). The chromatin appears uncondensed (uCh) and marginated, vacuoles (V) are present. Cytoplasmic residue (CR) is evident. All these alterations are characteristic of sperm pathologies such as immaturity and apoptosis. Axoneme (Ax). Bar 1 μm
Fig. 2.
TEM micrograph of sections of sperm. Up—a normal sperm is shown (arrow), down—two multiple germinal cells are detected: on the right a young double spermatid, on the left an apoptotic multiple mature sperm. Nucleus (N), acrosome (A), axoneme (Ax), cytoplasmic residue (CR). Bar 3 μm
In addition, a common characteristic of the analyzed semen samples was the presence of spermatocytes and round spermatids. In some cases, multiple spermatozoa and spermatids were observed (Fig. 2).
FSHR gene analysis
All patients and controls were analysed for known and unknown mutations/SNPs in the FSHR gene associated with infertility. Among the already described SNPs (Thr307 and Asn680), we found no difference in the frequency between Ala or Thr at position 307 and all patients presented Ser at codon 680. Three patients had a heterozygous mutation at codon 419 (Ala419Thr). No other known mutations were present, in patients or in the control group. Since FSHR mutations are known to be pathogenetic only when they occur in homozygosis, we also searched for other genetic alterations in the intronic regions of the gene and we observed several intronic polymorphisms. The rs114573531 (nucleotide 157890, Ensembl database) and the rs115961828 (nucleotide 157891, Ensembl database) were found in heterozygosis in 1 patient out of 14 (7.2 %) but in none of controls. For three polymorphisms, the genotype frequencies were statistically different for patients compared to controls (Table 3). For polymorphism rs6708637 (nucleotide 140428, Ensembl database), 14.3 % of patients but none of controls were CC, 71.4 % of patients and 47.5 % of controls were CT, and finally 14.3 % of patients and 52.5 % of controls were TT (P = 0.006, Pearson chi-square test). For polymorphism rs2091787 (nucleotide 137718, Ensembl database), we found that 42.9 % of patients and 95 % of controls were TT, 42.9 % of patients and 5 % of controls were TC and 14.3 % of patients but none of controls were CC (P < 0.001, Pearson chi-square test). The third polymorphism was the rs1922464 (nucleotide 157847, Ensembl database). For this variant we observed that 57.1 % of patients and 75 % of controls were CC, none of patients but 25 % of controls were CG and 42.9 % of patients but none of controls were GG with a P < 0.001 with the Pearson chi-square. For these three polymorphisms, we analyzed the single allele frequency and the odds ratio in order to evaluate a possible estimation risk (Table 4). For rs6708637 the frequencies of allele C were 50 % and 23.8 % in patients and controls respectively and the frequencies of T were 50 % and 76.3 % in patients and in controls respectively (P = 0.016, odds ratio 0.311). For rs2091787 the frequencies of allele T were 64.3 % and 97.5 % in patients and controls respectively and the frequencies of C were 35.7 % and 2.5 % in patients and controls respectively (P < 0.001, odds ratio 4.36). For rs1922464, the frequencies of allele C were 57.1 % and 87.5 % for patients and controls respectively and the frequencies of G were 42.9 % and 12.5 % in patients and controls respectively (P < 0.01, odds ratio 5.25). Every polymorphism was analyzed with NetGene2 Server Splicing software and none produced new splice sites. We found two new variants that were not previously described (Ensembl database): the Cys338Arg (patient n°9) and the Gln123Glu (patient n°11).
Table 3.
Frequency of FSHR polymorphisms in patients and controls
| CRL frequency (%) | Patients frequency (%) | P | |
|---|---|---|---|
| rs6708637 | |||
| CC | 0 | 14.3 | 0.006 |
| CT | 47.5 | 71.4 | |
| TT | 52.5 | 14.3 | |
| rs2091787 | |||
| TT | 95 | 42.9 | <0.001 |
| TC | 5 | 42.9 | |
| CC | 0 | 14.3 | |
| rs1922464 | |||
| CC | 75 | 57.1 | <0.001 |
| CG | 25 | 0 | |
| GG | 0 | 42.9 | |
Table 4.
Single allele frequency for FSHR polymorphisms in patients and controls
| CRL frequency (%) | Patients frequency (%) | P | Risk estimation (Odds ratio) | |
|---|---|---|---|---|
| rs6708637 | ||||
| C | 23.8 | 50 | 0.016 | 0.311 |
| T | 76.3 | 50 | ||
| rs2091787 | ||||
| T | 97.5 | 64.3 | <0.001 | 4.36 |
| C | 2.5 | 35.7 | ||
| rs1922464 | ||||
| C | 87.5 | 57.1 | <0.01 | 5.25 |
| G | 12.5 | 42.9 | ||
LHR gene analysis
Patients and controls were analyzed for LHR gene mutations. We identified three patients (21.4 %) heterozygous for the known variant Glu354Lys and two patients (14.2 %) heterozygous for the already described mutation Ile374Thr. None of controls were positive for LHR mutations. Again, these variants are known to exert a pathogenetic role when found in homozygosis. We extended our analysis to intronic regions, but we did not identify intronic polymorphisms in patients nor in controls. However, patient 11, who showed a new variant in FSHR, was also heterozygous for a new LHR variant, the Val144Ile.
Discussion
Gonadotropins, interacting with their gonadal receptors, play a key role in sexual development, reproductive functions and metabolism [20]. In this study we performed an analysis of FSHR and LHR genes and semen investigation in men with normal levels of T and elevated FSH and/or LH levels.
FSHR contains two common linked polymorphisms, Thr307Ala (rs6165) and Asn680Ser (rs6166), which have been shown to modulate ovarian function in women but inconclusive results have been reported on male fertility and reproductive parameters. Recently, the FSHR Asn680Ser variation was searched for in 1790 Baltic men demonstrating a significant association among the FSHR Ser680 allele, lower total testes volume, high serum FSH, lower inhibin B and total testosterone [21].
In our group of patients, we observed the same frequency of Ala or Thr at codon 307 and all patients had Ser at position 680. Moreover, we searched for the described pathogenetic alterations in both FSHR and LHR genes, but we did not find homozygous patients for these mutations, and only three patients were heterozygous for Ala419Thr. None of the mutations were present in the control group. However, we observed several intronic polymorphisms, three of which were significantly more frequent in patients compared to controls. For these three polymorphisms, we statistically analyzed the single allele frequency and we obtained significant values for rs2091787 and for rs1922464 (by SPSS version 13.0). Nevertheless, these variants were not implicated with new splice site production (NetGene2 Server Splicing), they appeared to be involved with a higher risk of developing infertility as indicated by the odds ratio (4.36 and 5.25 for rs2091787 and rs1922464, respectively by SPSS version 13.0).
We found two new FSHR gene variants, not previously described, the Cys338Arg and the Gln123Glu. Both variants are located in the extracellular domain of the protein. For the Gln123Glu we were able to predict possible changes in protein structure using the Swiss-model program, however, this variant did not produce any significant functional abnormalities. Accordingly, both variations were not considered pathogenetic using PolyPhen-2.
Different studies reported no significant correlation among serum FSH levels, semen characteristics or fertility status and FSHR gene polymorphisms [7, 10, 14, 22]. However, the hypothesis that the combination of heterozygous FSHR (codon 307 and 607) can be responsible for male infertility was reported by Shimoda et al [11]. Recently, the synergic effect of FSHR 307 and FSHR 680 polymorphisms and androgen receptors on semen quality was described by Lazaros et al [23]. Similarly, the polymorphisms found in our patients might, in combination with each other, predispose for infertility although they are not directly involved in the production of a non-functioning receptor.
Regarding LHR gene mutations, we did not identify any homozygous patients, however three patients were heterozygous for the known variant Glu354Lys and two for the already described mutation Ile374Thr. No patients or controls were positive for LHR mutations or any intronic polymorphisms. Moreover, for LHR we found one patient who was heterozygous for a new variant: the Val144Ile. This aminoacidic substitution is located in the extracellular domain of the protein, but it does not significantly modify protein structure or exert a pathogenetic role according to the Swiss-model or the PolyPhen-2 software. The semen characteristics of patients carrying the new FSHR (Cys338Arg and Gln123Glu) or LHR (Val144Ile) variants, although reduced with respect to controls, were similar to those observed in the other cases. FSH levels were elevated in both patients (numbers 9 and 11, Table 2) with the new FSHR alterations. Patient number 11 (Table 2), carrying the new LHR variant, had a normal level of LH.
It is known that LHR mutations may represent an underestimated cause of infertility in women and it can be responsible for male hypogonadism with reduced spermatogenesis [5]; these data are concordant with our results that showed LHR polymorphisms associated with a reduced number of ejaculated sperm.
Of particular interest was the analysis of semen parameters performed at light microscopy and TEM levels. By routine semen analysis, carried out with an optical microscope, sperm concentrations and motility percentages were included in a wide range of values, indicating variable seminal conditions in our group of patients. On the contrary, with TEM we observed a homogeneous sperm pattern with a decreased fertility index, as well as increased apoptosis and immaturity in agreement with a mathematical formula based on probability calculation [18, 19].
The obtained results support the evidence of possible common sperm alterations in patients with hypergonadotropic hypogonadism undetectable by conventional semen analysis; for this reason a deeper examination of spermatozoa, achieved by the use of more powerful tools, such as TEM or molecular analysis, are advisable.
In addition, these data seem to confirm the important role of FSH during the spermatogenetic process, as reported in previous studies [24], since modification in the action of this gonadotropins at the testicular level increases the incidence of sperm pathologies such as apoptosis and immaturity.
We are aware that the patient group is small; however, it should be considered that the patients were enrolled following stringent selection criteria to exclude the presence of other pathologies involved in male infertility. Nevertheless, an accurate haplotype analysis to confirm the role of the association of multiple receptor polymorphisms in infertility would require a greater number of patients. In conclusion, a variable seminal pattern at the light microscopy level was detected in men with hypergonadotropic hypogonadism and with different polymorphisms in LHR and FSHR. All patients showed sperm with ultrastructural characteristics of apopotosis and immaturity, however these observations deserve further confirmation using different methods to detect such sperm pathologies in the presence of LHR and FSHR polymorphisms.
Footnotes
Capsule
To perform the genetic analysis of FSHR and LHR and semen investigation in men with normal level of T and elevated levels of FSH and/or LH. The use of more powerful tools such as TEM or molecular analysis, are advisable in patients with hypergonadotropic hypogonadism.
Collodel and Cantara equally contributed to the work.
References
- 1.O’Shaughnessy PJ, Monteiro A, Verhoeven G, De Gendt K, Abel MH. Effect of FSH on testicular morphology and spermatogenesis in gonadotrophin-deficient hypogonadal mice lacking androgen receptors. Reproduction. 2010;139:177–184. doi: 10.1530/REP-09-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huhtaniemi I, Alevizaki M. Gonadotrophin resistance. Best Pract Res Clin Endocrinol Metab. 2006;20:561–576. doi: 10.1016/j.beem.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Menon KM, Menon B. Structure, function and regulation of gonadotropin receptors—a perspective. Mol Cell Endocrinol. 2012;356:88–97. doi: 10.1016/j.mce.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Themmen APN, Huhtaniemi IT. Mutations of gonadotropins and gonadotropin receptors: elucidating the physiology and pathophysiology of pituitary-gonadal function. Endocr Rev. 2000;21:551–583. doi: 10.1210/er.21.5.551. [DOI] [PubMed] [Google Scholar]
- 5.Bruysters M, Christin-Maitre S, Verhoef-Post M, Sultan C, Auger J, Faugeron I, et al. A new LH receptor splice mutation responsible for male hypogonadism with subnormal sperm production in the propositus, and infertility with regular cycles in an affected sister. Hum Reprod. 2008;23:1917–1923. doi: 10.1093/humrep/den180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simoni M, Nieschlag E, Gromoll J. Isoforms and single nucleotide polymorphisms of the FSH receptor gene: implications for human reproduction. Hum Reprod Update. 2002;8:413–421. doi: 10.1093/humupd/8.5.413. [DOI] [PubMed] [Google Scholar]
- 7.Safarinejad MR, Shafiei N, Safarinejad S. Evaluating the role of the FSH receptor gene Thr307-Ala and Asn680-Ser polymorphisms in male infertility and their association with semen quality and reproductive hormones. BJU Int. 2011;108:E117–E125. doi: 10.1111/j.1464-410X.2010.09890.x. [DOI] [PubMed] [Google Scholar]
- 8.Pengo M, Ferlin A, Arredi B, Ganz F, Selice R, Garolla A, et al. FSH receptor gene polymorphisms in fertile and infertile Italian men. Reprod Biomed Online. 2006;13:795–800. doi: 10.1016/S1472-6483(10)61026-7. [DOI] [PubMed] [Google Scholar]
- 9.Kuijper EA, Blankenstein MA, Luttikhof LJ, Roek SJ, Overbeek A, Hompes PG, et al. Frequency distribution of polymorphisms in the FSH receptor gene in infertility patients of different ethnicity. Reprod Biomed Online. 2010;20:588–593. doi: 10.1016/j.rbmo.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 10.Ghirelli-Filho M, Peluso C, Christofolini DM, Gava MM, Glina S, Barbosa CP, et al. Variants in follicle-stimulating hormone receptor gene in infertile Brazilian men and the correlation to FSH serum levels and sperm count. Reprod Sci. 2012;19:733–739. doi: 10.1177/1933719111432872. [DOI] [PubMed] [Google Scholar]
- 11.Shimoda C, Koh E, Yamamoto K, Matsui F, Sugimoto K, Sin HS, et al. Single nucleotide polymorphism analysis of the follicle-stimulating hormone (FSH) receptor in Japanese with male infertility: identification of codon combination with heterozygous variations of the two discrete FSH receptor gene. Endocr J. 2009;56:859–865. doi: 10.1507/endocrj.K09E-130. [DOI] [PubMed] [Google Scholar]
- 12.Balkan M, Gedik A, Akkoc H, Izci Ay O, Erdal ME, Isi H, et al. FSHR single nucleotide polymorphism frequencies in proven fathers and infertile men in Southeast Turkey. J Biomed Biotechnol. 2010;2010:640318. doi: 10.1155/2010/640318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Gu A, Yang H, Ding X, Ji G, Lu C, et al. FSH receptor gene polymorphisms in fertile and infertile Han-Chinese males. Clin Chim Acta. 2011;412:1048–1052. doi: 10.1016/j.cca.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 14.Wu W, Cai H, Sun H, Lu J, Zhao D, Qin Y, et al. Follicle stimulating hormone receptor G-29A, 919A>G, 2039A>G polymorphism and the risk of male infertility: a meta-analysis. Gene. 2012;505:388–392. doi: 10.1016/j.gene.2012.02.023. [DOI] [PubMed] [Google Scholar]
- 15.Selice R, Garolla A, Pengo M, Caretta N, Ferlin A, Foresta C. The response to FSH treatment in oligozoospermic men depends on FSH receptor gene polymorphisms. Int J Androl. 2011;34:306–312. doi: 10.1111/j.1365-2605.2010.01086.x. [DOI] [PubMed] [Google Scholar]
- 16.Tüttelmann F, Laan M, Grigorova M, Punab M, Sõber S, Gromoll J. Combined effects of the variants FSHB -211G>T and FSHR 2039A>G on male reproductive parameters. J Clin Endocrinol Metab. 2012;97:3639–3647. doi: 10.1210/jc.2012-1761. [DOI] [PubMed] [Google Scholar]
- 17.World Health Organization. WHO laboratory manual for the examination and processing of human semen. 5th ed. WHO Press; 2010.
- 18.Baccetti B, Bernieri G, Burrini AG, Collodel G, Crisa N, Mirolli M, et al. Notulae seminologicae. 5. Mathematical evaluation of interdependent submicroscopic sperm alterations. J Androl. 1995;16:356–371. [PubMed] [Google Scholar]
- 19.Collodel G, Moretti E. Morphology and meiotic segregation in spermatozoa from men of proven fertility. J Androl. 2008;29:106–114. doi: 10.2164/jandrol.107.002998. [DOI] [PubMed] [Google Scholar]
- 20.Casarini L, Pignatti E, Simoni M. Effects of polymorphisms in gonadotropin and gonadotropin receptor genes on reproductive function. Rev Endocr Metab Disord. 2011;12:303–321. doi: 10.1007/s11154-011-9192-2. [DOI] [PubMed] [Google Scholar]
- 21.Grigorova M, Punab M, Poolamets O, Sõber S, Vihljajev V, Zilaitienė B, et al. Study in 1790 Baltic men: FSHR Asn680Ser polymorphism affects total testes volume. Andrology. 2013;1:293–300. doi: 10.1111/j.2047-2927.2012.00028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zalata AA, Hassan AH, Nada HA, Bragais FM, Agarwal A, Mostafa T. Follicle-stimulating hormone receptor polymorphism and seminal anti-Müllerian hormone in fertile and infertile men. Andrologia. 2008;40:392–397. doi: 10.1111/j.1439-0272.2008.00877.x. [DOI] [PubMed] [Google Scholar]
- 23.Lazaros L, Xita N, Takenaka A, Sofikitis N, Makrydimas G, Stefos T, et al. Synergistic effect of follicle-stimulating hormone receptor and androgen receptor gene variants on semen quality. Andrologia. 2012 doi: 10.1111/and.12021. [DOI] [PubMed] [Google Scholar]
- 24.Baccetti B, Strehler E, Capitani S, Collodel G, De Santo M, Moretti E, et al. The effect of follicle stimulating hormone therapy on human sperm structure (Notulae seminologicae 11) Hum Reprod. 1997;12:1955–1968. doi: 10.1093/humrep/12.9.1955. [DOI] [PubMed] [Google Scholar]