Abstract
Cardiovascular drugs are the most commonly prescribed medications. Some prior assays successfully detect cardiovascular drugs among multiple classes using a single sample. Here, we develop an assay to detect a broad range of cardiovascular drug classes to include commonly used cardiovascular drugs and evaluate the assay’s analytical and statistical properties in a clinical setting. We describe a protocol for drug detection that encompasses 34 commonly prescribed cardiovascular drugs or drug metabolites with a single LC-MS/MS method using 100µl of serum or plasma. Drug classes monitored by this assay include: anticoagulants, angiotensin converting enzyme inhibitors (ACEI), angiotensin II receptor blockers (ARB), beta blockers, calcium channel blockers, diuretics, statins, and vasodilators, as well as digoxin, fenofibrate, and niacin.
Analytical accuracy and precision for each drug was evaluated by repeating the assay on spiked samples at low, medium, and high concentrations. In 294 clinical samples obtained from hospitalized patients for whom medication administration was recorded, we evaluated the assay’s statistical sensitivity, specificity, and accuracy. For the 34 drugs or drug metabolites, the assay was statistically sensitive (>0.90) for all drugs except captopril (0.25), isosorbide (0.81), and niacin (0.89). The assay was statistically specific for all drugs, with a minimum specificity of 0.94 (aspirin). To our knowledge, this method is the first method of simultaneous analysis of 34 cardiovascular drugs or drug metabolites from nine drug classes evaluated using clinical samples from hospitalized patients.
Keywords: cardiovascular drugs, drug monitoring, selectivity, mass spectrometry, liquid chromatography, clinical samples
1. Introduction
Cardiovascular disease causes more than 800,000 deaths in the United States each year [1]. Pharmacological treatment can reduce the risk of cardiovascular disease, but most patients require more than one medication to achieve risk reduction [2]; cardiovascular medications comprise the most commonly prescribed medication class in the United States [3].
Prior assays developed to detect cardiovascular medications have generally quantified medications from the same drug class or with similar structure (e.g., diuretics, angiotensin II receptor antagonists, and beta blockers) [4–9]. When cardiovascular drugs from multiple classes have been included in a single assay, commonly prescribed medications such as hydralazine, isosorbide, methyldopa, aliskiren, clonidine, digoxin, fenofibrate, and niacin were not included in the assay, and the assay was tested in a small number of clinical samples [10]. Many patients take cardiovascular medications from different drug classes, and there is a need for a rapid assay that can detect the range of cardiovascular medications that a patient may be taking using a single small blood sample.
Drug effectiveness is related to drug concentration. Individual variation in drug metabolism and drug-drug interactions impacts the concentration and, therefore, the effectiveness of cardiovascular medications. Pharmacogenomics research is identifying factors that affect drug concentration and effect, but studies can be confounded by unknown medication adherence. Medication adherence also affects patient outcomes, but the lack of readily available, objective measures of adherence such as therapeutic drug screening limits the development of effective interventions to improve adherence [11–14].
We describe a rapid, high-throughput mass spectrometry (MS) assay that detects 34 cardiovascular drugs or their drug metabolites in nine drug classes. The medications targeted by the assay were selected based on the 200 most commonly prescribed medications and regional prescribing patterns of clinicians [15]. The assay uses a single, small volume sample and was designed for detection of drug, to assist physicians and researchers in determining whether drugs are present. In addition, the assay was evaluated using samples obtained from patients for whom the administration of cardiovascular medications was documented during hospitalization.
2. Material and methods
2.1 Reagents and chemicals
Liquid chromatography (LC)-MS-grade acetonitrile, methanol, and water were purchased from Fisher Scientific (Suwanee, GA, USA). Formic acid >99%, 1.5 mL high performance liquid chromatography (HPLC) autosampler vials, inserts, caps, 1.5 mL eppendorf tubes, and pipette tips were obtained from Sigma-Aldrich (St. Louis, MO, USA). The internal standard, sulfameter, and analytical standards were obtained through Sigma-Aldrich (St. Louis, MO, USA), with the following exceptions: L-methyldopa, losartan, lisinopril, and valsartan were obtained from AK Scientific (Union City, CA, USA). Drug-free human plasma was purchased from Innovative Research, Inc. (Novi, MI, USA).
2.2 Preparation of standard solutions
A 1mg/mL primary stock solution was made for each analyte. Secondary stocks of 250 µg/mL or 10 µg/mL were made, as required to reach the approximate expected concentration. When an expected concentration range was known from prior published data, appropriate calculated amounts of analyte standards were added to create a single stock solution that contained each analyte at an expected concentration (Table 1) [16–33]. A 1 µg/mL working internal standard solution was made from the 1mg/mL primary stock of sulfameter. Sulfameter, a veterinary antibiotic that is chemically distinct from all 34 cardiovascular drugs included on the assay, was used as the internal standard to control for shifts in retention time.
Table 1.
Analyzed compounds
| Drug Class | Compound | Calibration curve range (µg/mL) |
Published Drug Concentrations (µg/mL) |
Regression Equation | R2 |
|---|---|---|---|---|---|
| Anticoagulants | Acetylsalicylic acid/salicylic acid | 0.1–50 | 44–330 [27] | =0.96x−37.70 | 0.97 |
| Clopidogrel | 0.0002–0.1 | 0.0145 [30] | =42.53x−2.53 | 0.98 | |
| Warfarin | 0.1–50 | 3.1 [27] | =−0.003x2+300.08x+40934.95 | 0.97 | |
| Angiotensin converting enzyme inhibitors | Captopril | 0.005–2.5 | 0.82–0.88 [27] | =113.88x−578.64 | 0.95 |
| Enalapril | 0.001–0.5 | 0.045–0.07 [27] | =1652.23x −1455.89 | 0.99 | |
| Enalaprilat | 0.001–0.5 | 0.045–0.07 [27] | =415.28x−304.59 | 0.93 | |
| Lisinopril | 0.001–0.5 | 0.082 [27] | =69.73x−95.16 | 0.94 | |
| Ramipril | 0.001–0.5 | 0.023 [37] | =2196.82x−2230.51 | 0.98 | |
| Losartan | 0.0005–0.25 | 0.0845–1.3949 [20] | =557.90x−95.55 | 0.91 | |
| Losartan | 0.0005–0.25 | 0.0845–1.3949 [20] | =557.90x−95.55 | 0.91 | |
| Angiotensin II receptor blockers | Telmisartan Valsartan | 0.001–0.5 | 0.084±0.029 [25] | =200.57x−43.99 | 0.97 |
| 2.22±1.622 [25] | =178.88x−198.07 | 0.97 | |||
| Beta blockers | Atenolol | 0.01–5 | 1.48–2.75 [27] | =157.53x+729.22 | 0.97 |
| Carvedilol | 0.001–0.5 | 0.0265–.205 [17] | =426.37x+142.57 | 0.98 | |
| Metoprolol | 0.001–0.5 | 116 [27] | =201.61x−26.52 | 0.96 | |
| Propranolol | 0.001–0.5 | 335 [27] | =306.89x−65.16 | 0.96 | |
| Amlodipine | 0.0005–0.25 | 0.024 [27] | =456.72x−227.47 | 0.99 | |
| Diltiazem | 0.001–0.5 | 0.1–0.2 [27] | =2463.22x − 1740.23 | 0.99 | |
| Nifedipine | 0.001–0.5 | 0.115 [27] | =281.82x+26.31 | 0.95 | |
| Verapamil | 0.001–0.5 | 355 [27] | =2035.92x−827.44 | 0.99 | |
| Diuretics | Canrenone (Spironolactone) | 0.005–2.5 | 0.1–0.5 [26] | =43.77x+28.69 | 0.96 |
| Furosemide | 0.005–2.5 | 1–2 [19] | =10.79x−12.37 | 0.97 | |
| Hydrochlorothiazide | 0.005–2.5 | 0.202 [27] | =3.40x+20.58 | 0.98 | |
| Triamterene | 0.0002–0.1 | 0.0157–0.0446 [24] | =232.95x+0.37 | 0.97 | |
| Statins | Atorvastatin | 0.0001–0.05 | 0.0054–0.0078 [16] | =179.89x−25.49 | 0.90 |
| Lovastatin | 0.0002–0.1 | 0.00977 [21] | =53.06x+1.38 | 0.97 | |
| Pravastatin | 0.002–1 | 0.0274±0.0107 [22] | =4.37x+1.15 | 0.97 | |
| Simvastatin | 0.001–0.5 | 0.0073–0.0285 [23] | =29.37x−12.85 | 0.96 | |
| Vasodilators | Hydralazine | 0.01–5 | 0.3–0.5 [27] | =211.23x−4261.64 | 0.99 |
| Isosorbide | 0.001–0.5 | 0.0258 [27] | =11.45x+65 | 0.96 | |
| Methyldopa | 0.01–5 | 2.2 [27] | =120.39x−936.79 | 0.96 | |
| Other | Aliskiren | 0.001–0.5 | 0.2–0.4 [29] | =92.89x−152.09 | 0.97 |
| Clonidine | 0.0001–0.05 | 0.0013 [27] | =140.04x+7.57 | 0.95 | |
| Digoxin | 0.0001–0.05 | 0.0014 [27] | =19.63x−1.08 | 0.99 | |
| Fenofibrate/ic acid | 0.1–50 | 5–15 [18] | =−0.001x2+170.76x+11465.34 | 0.95 | |
| Niacin | 0.0002–0.1 | 0.0182–0.109 [21] | =46.28x+9.09 | 0.97 |
2.3 Extraction of test and clinical samples
Prior to each assay, fresh standards were prepared to provide all analytes at expected concentrations: 90µL of drug-free human plasma was transferred to an eppendorf tube, and 10µL of a stock solution containing all 34 cardiovascular drugs and their metabolites, each at the appropriate concentration to produce the appropriate concentration in the biological matrix after 1 to 10 dilution (Table 1).
2.4 Clinical samples
Patients provided informed consent, and the study was approved by the Vanderbilt University institutional review board. Serum or plasma was collected from patients who were hospitalized at Vanderbilt University Medical Center. Verification of the medications, doses, routes, frequency, and timing of medication administration was performed by manual review of the medication administration record, the electronic medical record, and outpatient pharmacy records. Samples were extracted as described below, and 100µL of serum or plasma from each patient was analyzed.
2.5 Preparation of test and clinical samples
One hundred microliters of either a known test sample or a clinical sample were aliquoted into eppendorf tubes and then extracted using a protein precipitation procedure: 10µL of the working internal standard was added to 90µL of 9:1 0.1% formic acid in water:MeOH. Proteins were precipitated by adding 600µL of cold acetonitrile. Samples were then vortexed and centrifuged at 2300 G for 15 minutes at 4°C. After centrifugation, supernatant was transferred to new eppendorf tubes and evaporated to dryness using a Fischer Scientific Savant SpeedVac concentrator (Suwanee, GA, USA). The residue was then reconstituted in 50µL 9:1 Water:MeOH, vortexed, and transferred to autosampler vials for analysis by LC-MS/MS.
2.6 LC-MS/MS analyses
The chromatographic system consisted of two 1260 binary pumps (Agilent Technologies Santa Clara, CA, USA), a 1260 μ-degasser, a thermostatted column compartment (TCC) SL temperature-controlled column compartment, and a temperature-controlled 1260 HiP-ALS auto-sampler, kept at 4°C. Separation of a 10µL aliquot of the sample was achieved using an Agilent Zorbax Eclipse Plus C-18 (rapid resolution high throughput [RRHT] 2.1 × 50mm 1.8 micron) column (Agilent Technologies Santa Clara, CA, USA). The temperature of the column compartment was set to 40°C. The injection needle was repeatedly rinsed with a 1:1 Water:MeOH solution after each injection. A gradient of 0.1% formic acid in water (mobile phase A) and 0.1% formic acid in acetonitrile (mobile phase B) was used. The second binary pump was used to re-equilibrate the column while a second equilibrated column was switched into line for the next injection. The dwell time was 20 ms; a minimum of 10 data points over each HPLC peak were used. The dead volume of the HPLC was minimized by bypassing the pulse damper and using a smaller mixer, resulting in a final dead volume of 200 µL.
LC–MS/MS analyses were performed on two instrument platforms to demonstrate the ability to transfer the method to another platform and evaluate the need to alter sample preparation or LC conditions. The first was a 6430 triple quadrupole mass spectrometer (Agilent Technologies, Santa Clara, CA, USA), with an API-Electrospray source operated in both positive and negative ion mode switching between the two as necessary. The mass spectrometer was operated in multiple-reaction monitoring (MRM) mode. The source conditions were chosen to give satisfactory signal for all analytes and are as follows: gas temperature: 325°C; gas flow: 10 L/min; nebulizer: 40 psi; capillary voltage: positive 4000, negative 3500. This instrument was operated with the Mass Hunter data acquisition software, and data was processed using Quantitative Analysis B.04.00/Build 4.0.225.19 (Agilent Technologies, Santa Clara, CA, USA).
The second instrument, a Waters TQD with an Aquity UPLC system consisting of a Binary Solvent manager and a temperature controlled Sample manager (Waters, Milford, MA, USA), was operated in MRM mode, switching between positive and negative ion monitoring as necessary. The source conditions that were determined to be optimal were as follows: source temperature, 150°C; desolvation temperature, 450°C; and desolvation gas flow, 800L/hr. This instrument was operated through the Mass Lynx software v4.1, and the samples were processed using TagetLynx v4.1 (Waters, Milford, MA, USA). Results of assays performed on the Agilent Technologies instrument platform are reported in this manuscript.
2.7 Quantitative Data
Data were processed using Agilent MassHunter Quantitative Analysis B.04.00/Build 4.0.225.19 Software (Agilent Technologies Santa Clara, CA, USA). Linear regression with 1/x weighting best fit the curves for all analytes except warfarin, fenofibrate, and losartan. Quadratric regression was used to model warfarin and fenofibrate over the quantitation range; losartan was modeled using 1/x2 weighting to achieve a functional curve over the quantitation range (Table 1). Plasma samples were spiked with low (0.25%), medium (2.5%), and high (25%) concentrations of drugs. Inter-day analytical accuracy was calculated as the average of five replicates at each concentration per day and over three days, compared against the theoretical value. Analytic selectivity, or the extent to which the assay identified each analyte without interferences from other compounds, was evaluated based on its level of detection (LOD) and level of quantitation (LOQ). The lowest concentration for each of the 34 cardiovascular drugs with a consistent signal-to-noise ratio of three or more during the process of developing and testing the assay was used as the level of detection (LOD). Analytic accuracy was determined from spiked standards and was computed by dividing the calculated concentration by the theoretical concentration. Percentage of relative standard deviation (%RSD), or the absolute value of the coefficient of variation, was computed by dividing the standard deviation by the average concentration. Validation criteria were based on FDA guidance for drug analysis,[34] modified for multi-class drug analysis to require that a minimum of two thirds of quality control samples be within 25% of their nominal value. A minimum of six matrix-based, standard calibration samples were required, in addition to a minimum of duplicate quality control samples at three concentrations. Similarly, because strict quantitation was not the goal of this assay, samples outside quantitation limits were not diluted. Instead, note was made of the sample outside the limits of quantitation for potential future analysis. Where available, published values were used to evaluate whether concentrations were within therapeutic range.
2.8 Statistical Analysis
The primary outcome of this assay was binary: detection or lack of detection of drugs administered to patients using the LC-MS/MS assay. To evaluate the test characteristics of the assay when used in clinical samples, the medication administration record and manual, detailed review of the electronic medical record and outpatient pharmacy records served as the reference standard to determine the true drug presence status. Using this reference standard, we computed the assay’s statistical sensitivity, specificity, and accuracy for detecting the presence or absence of each of the 34 cardiovascular drugs or drug metabolites, compared against the reference standard of recorded medication administration history. Statistical sensitivity was calculated as the proportion of patients who correctly had drug detected within therapeutic range by the LC-MS/MS assay (true positives) among all patients classified as taking drug based on the recorded medication administration history (true positives and false negatives). Statistical specificity was calculated as the proportion of patients who correctly had no detected drug within therapeutic range on the LC-MS/MS assay (true negatives) among those for whom there was no recorded history of drug administration (true negatives and false positives). For each drug, statistical accuracy was calculated as the proportion of patients correctly categorized as having or not having detectable drug out of total number of tested patient samples. Statistical sensitivity and specificity were computed for serum and plasma samples separately; because the results were similar, we present the results of all samples together. Bootstrapping was used to compute the 95% confidence intervals (CIs) around statistical sensitivity, specificity, and accuracy.[35]
3. Results
3.1 LC and MS conditions
Chromatographic conditions were optimized and mobile phase was chosen to maximize conditions across all 34 drugs and metabolites. The optimized gradients are found in Table 2. The fragmentor voltage, collision energy, and cell accelerator voltages were optimized as in the Table 3. Assays were performed using 7V for the cell accelerator. Two product ions for the LC-MS/MS analysis were selected for each compound, except simvastatin where three product ions were selected to increase the reliability of the assay.
Table 2.
Optimized gradient for LC-MS/MS analysis
| Time (min) | Organic phase (%) | Aqueous phase (%) |
|---|---|---|
| 0.00 | 4.00 | 96.00 |
| 1.00 | 10.00 | 90.00 |
| 3.00 | 20.00 | 80.00 |
| 10.00 | 98.00 | 2.00 |
| 11.00 | 98.00 | 2.00 |
| 11.10 | 4.00 | 96.00 |
| 13.50 | 4.00 | 96.00 |
Table 3.
Optimal MS/MS conditions and quantitation limits for analyzed compounds.
| Compound | Precursor Ion (m/z) |
Product Ion 1 (m/z) |
Product Ion 2 (m/z) |
Fragmentor Voltage |
Collision Energy |
Polarity | Limit of Detection* (ng/mL) |
Limit of Quantitation * (ng/mL) |
|---|---|---|---|---|---|---|---|---|
| Acetylsalicylic Acid | 179.1 | 137 | 93 | 50 | 4 | − | 100 | 250 |
| Aliskiren | 552.8 | 436.3 | 418.2 | 155 | 13 | + | 0.5 | 1 |
| Amlodipine | 409.2 | 294.1 | 238.2 | 90 | 9 | + | 0.25 | 0.5 |
| Atenolol | 267.2 | 190.1 | 145.1 | 116 | 14 | + | 1 | 10 |
| Atorvastatin | 559.6 | 440.2 | 250.1 | 140 | 20 | + | 0.1 | 0.25 |
| Canrenone (Spironolactone) | 341.2 | 107.1 | 91.1 | 148 | 30 | + | 2.5 | 5 |
| Captopril | 218.3 | 116.1 | 70.1 | 80 | 9 | + | 500 | NQ |
| Carvedilol | 407.5 | 222.1 | 100.1 | 140 | 24 | + | 0.5 | 1 |
| Clonidine | 230.2 | 160 | 44.2 | 148 | 38 | + | 0.2 | 1 |
| Clopidogrel | 322.8 | 184 | 155 | 110 | 20 | + | 1 | 5 |
| Digoxin | 651.3 | 131.1 | 97.1 | 160 | 21 | + | 0.25 | 1 |
| Diltiazem | 415.3 | 178 | 150 | 140 | 21 | + | 0.5 | 1 |
| Enalapril | 377.2 | 234.1 | 91.1 | 116 | 14 | + | 0.25 | 1 |
| Enalaprilat | 349.4 | 206 | 117 | 120 | 13 | + | 1 | 1 |
| Fenofibrate | 361.1 | 233 | 139 | 130 | 13 | + | 5 | 100 |
| Fenofibric Acid | 319.08 | 233 | 139 | 110 | 13 | + | 5 | 250 |
| Furosemide | 329.3 | 285 | 204.9 | 110 | 9 | + | 5 | 12.5 |
| Hydrochlorothiazide | 296.2 | 268.9 | 204.9 | 140 | 13 | + | 5 | 5 |
| Hydralazine | 161.1 | 89.1 | 63.1 | 100 | 22 | + | 5 | 25 |
| Isosorbide | 147.1 | 88.1 | 69.1 | 50 | 5 | + | 10 | NQ |
| Lisinopril | 406.2 | 246.1 | 84.1 | 132 | 22 | + | 1 | 5 |
| Losartan | 423.2 | 207.1 | 180 | 116 | 22 | + | 0.1 | 0.5 |
| Lovastatin | 405.6 | 225.2 | 199.2 | 80 | 13 | + | 2 | 5 |
| Methyldopa | 212.2 | 166.1 | 139 | 80 | 12 | + | 100 | 500 |
| Metoprolol | 268.3 | 133.1 | 116.1 | 116 | 26 | + | 0.25 | 1 |
| Metoprolol acid | 268.3 | 133.1 | 116.1 | 116 | 26 | + | ** | NQ |
| Niacin | 124.1 | 80.1 | 78.1 | 110 | 21 | + | NQ | NQ |
| Nifedipine | 347.3 | 315.1 | 194.1 | 80 | 5 | + | 1 | 1 |
| Pravastatin | 423.5 | 321.2 | 101.1 | 140 | 9 | − | 2 | 10 |
| Propranolol | 260.4 | 116.1 | 56.2 | 110 | 16 | + | 0.5 | 1 |
| Ramipril | 417.3 | 234.1 | 117.1 | 140 | 17 | + | 0.2 | 1 |
| Ramiprilat | 389.3 | 206.2 | 156.2 | 140 | 17 | + | ** | ** |
| Salicylic acid | 137.1 | 93 | 65.1 | 80 | 12 | − | 50 | 250 |
| Simvastatin | 419.6 | 285.1 | 225.1 | 80 | 4 | + | 0.5 | 1 |
| Simvastatin | 419.6 | 199.1 | 80 | 4 | + | 0.5 | 1 | |
| Sulfameter (IS) | 281.3 | 108.1 | 92.1 | 110 | 25 | + | N/A | N/A |
| Sulfameter (IS) | 279.3 | 264 | 196 | 110 | 5 | − | N/A | N/A |
| Telmisartan | 515.2 | 497.3 | 276.1 | 160 | 40 | + | 0.2 | 1 |
| Triamterene | 254.3 | 237.1 | 104.1 | 165 | 25 | + | 0.2 | 0.2 |
| Valsartan | 436.2 | 291.1 | 207.1 | 100 | 14 | + | 0.5 | 10 |
| Verapamil | 455.2 | 165.1 | 150.1 | 164 | 26 | + | 0.2 | 1 |
| Warfarin | 309.3 | 251.1 | 163 | 110 | 17 | + | 0.2 | 100 |
Where significant interference prevented quantification below a set LOD, LOD and LOQ are identical
when the authenticated standard was unavailable, we used the estimated metabolite mass, parameters of the parent compound, and fragment ion based on literature review
Abbreviation: LOD, limit of detection; LOQ, limit of quantitation; N/A, not applicable; NQ, not quantifiable
3.2 Extraction procedure
The extraction method was developed based on published protocols and refined by an iterative process aimed at optimizing extraction of all 34 drugs and metabolites. The parameters chosen provided the simplest, fastest, and optimal extraction method based on the entire drug panel. Solid phase extraction was used initially, but this resulted in the need for multiple runs over multiple columns. Liquid-liquid phase and precipitations resulted in optimization for the majority of the drugs, although this method was not ideal for a few individual drugs.
An acidified extraction was used to prevent loss of hydralazine and also to improve the recovery of captopril. Hydralazine reacts with pyruvic acid present in plasma and serum to form a metabolite [36], but this reaction is reversible by lowering the pH of the sample. The effects on analyte recovery were evaluated after the addition of HCl 0.25N, 0.5N, and 1N and were compared against the results obtained after adding formic acid 0.05%, 0.1%, 0.2%, and 0.4%. Addition of HCl improved the recovery of hydralazine, but the signal for several other analytes was adversely affected even at low HCl concentrations. Addition of formic acid showed improved recovery of hydralazine and captopril and no significant deleterious effect on other analytes. Thus, formic acid was used during the extraction procedure.
3.4 Level of quantitation and working range
Figure 1 shows the chromatogram of the test sample containing 34 cardiovascular drugs or drug metabolites. Figure 2 shows a representative calibration curve for atenolol superimposed on individual concentrations for each clinical sample analyzed. Three patients had received their last dose more than 24 hours before the sample was collected; one had a value below the expected 20ng/mL minimum efficacious concentration [37]. The limit of detection and limit of quantitation for each drug or drug metabolite are reported in Table 3. Although this assay is not intended to be a quantitative assay, calculations for analytical accuracy and precision of test samples run in triplicate at low, medium, and high concentrations on different days, are reported in Table 4.
Figure 1. Chromatogram of known test sample, spiked with 34 cardiovascular drugs or drug metabolites (each at 5% of the high standard reported in Table 4).
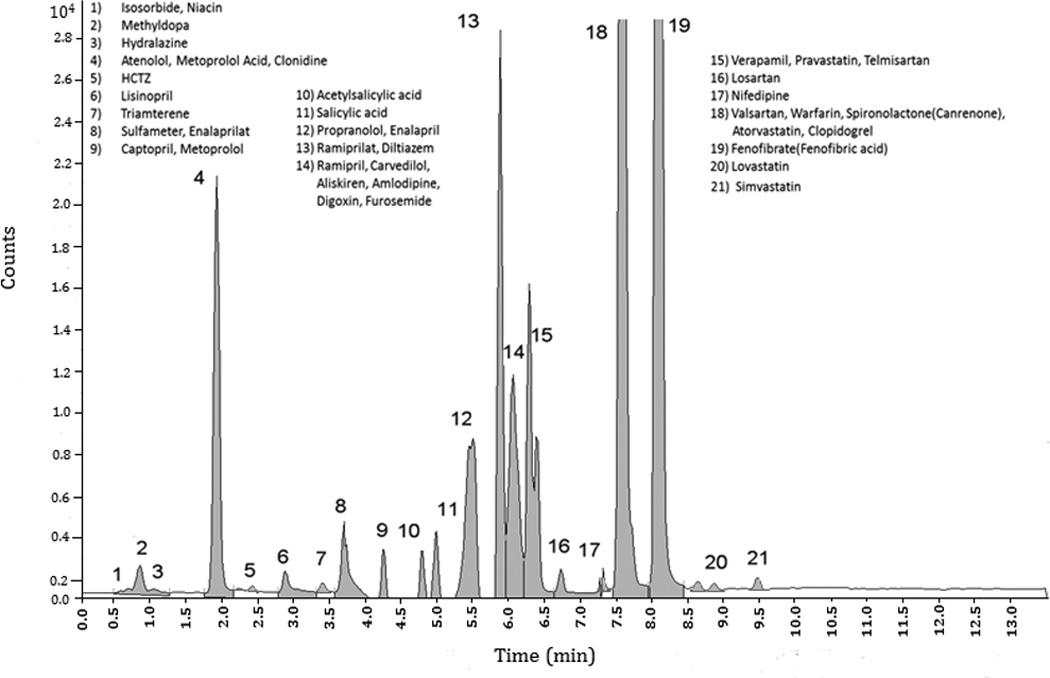
Abbreviations: HCTZ, hydrochlorothiazide; min, minutes
Figure 2. Calibration curve for atenolol, superimposed on individual clinical sample results.
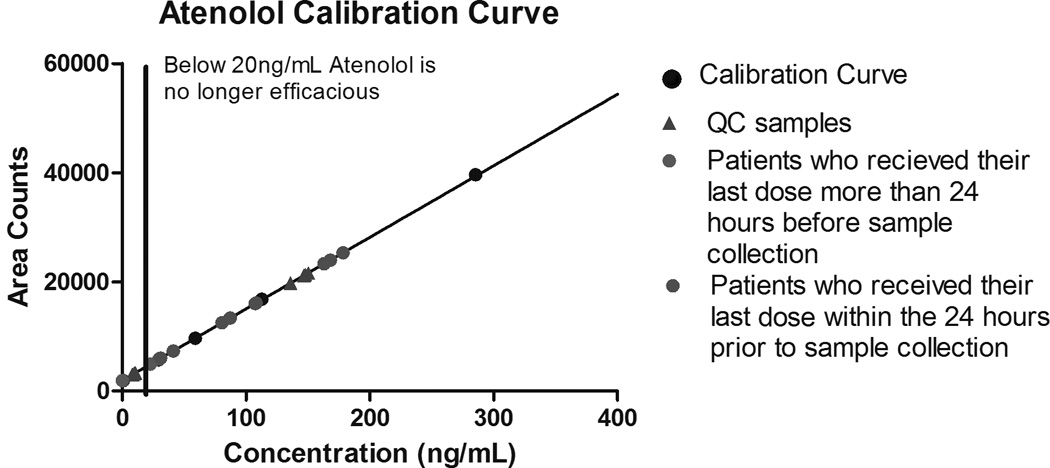
Abbreviations: QC: quality control
Table 4.
Analytic accuracy and precision of spiked samples
| Drug Name | Low Concentration | Medium Concentration | High Concentration | Meets FDA quantification recommendations** |
|||
|---|---|---|---|---|---|---|---|
| Accuracy (%) |
Precision (%RSD) |
Accuracy (%) |
Precision (%RSD) |
Accuracy (%) |
Precision (%RSD) |
||
| Acetylsalicylic acid (0.125, 1.25, 12.5) | 110.96 | 8.65 | 143.72 | 10.29 | 111.33 | 17.70 | No |
| Aliskiren (0.00125, 0.0125, 0.125) | 89.44 | 14.03 | 101.99 | 6.90 | 95.88 | 6.02 | Yes |
| Amlodipine (0.000625, 0.00625, 0.0625) | 72.13 | 22.72 | 86.86 | 26.12 | 109.29 | 15.47 | No |
| Atenolol (0.0125, 0.125, 1.25) | 72.26 | 11.36 | 106.42 | 4.06 | 91.76 | 4.29 | Yes |
| Atorvastatin (0.000125, 0.00125, 0.0125) | 95.14 | 16.03 | 80.11 | 12.80 | 92.62 | 5.79 | No |
| Captopril (0.00625, 0.0625, 0.625) | 277.19 | 5.79 | 135.29 | 10.84 | 114.46 | 29.50 | No |
| Carvedilol (0.00125, 0.0125, 0.125) | 86.49 | 27.67 | 98.15 | 26.95 | 109.83 | 20.92 | Yes |
| Clonidine (0.000125, 0.00125, 0.0125) | 99.18 | 27.18 | 88.79 | 8.34 | 96.51 | 5.16 | Yes |
| Clopidogrel (0.00025, 0.0025, 0.025) | 81.22 | 63.45 | 79.74 | 31.56 | 111.09 | 14.30 | Yes |
| Digoxin (0.000125, 0.00125, 0.0125) | 116.57 | 52.38 | 104.22 | 16.22 | 111.35 | 5.33 | No |
| Diltiazem (0.00125, 0.0125, 0.125) | 93.22 | 11.36 | 82.40 | 17.01 | 80.23 | 22.30 | Yes |
| Enalapril (0.00125, 0.0125, 0.125) | 76.22 | 8.71 | 95.72 | 5.49 | 101.37 | 4.85 | No |
| Enalaprilat (0.00125, 0.0125, 0.125) | 79.71 | 7.37 | 90.82 | 5.09 | 98.43 | 4.44 | No |
| Fenofibric acid (0.125, 1.25, 12.5) | 47.22 | 15.12 | 113.89 | 5.21 | 85.46 | 3.70 | No |
| Furosemide (0.00625, 0.0625, 0.625) | 77.35 | 14.57 | 92.58 | 6.89 | 91.00 | 5.64 | Yes |
| Hydrochlorothiazide (0.00625, 0.0625, 0.625) | 69.32 | 58.20 | 109.34 | 26.50 | 89.58 | 15.66 | No |
| Hydralazine (0.0125, 0.125, 1.25) | 58.40 | 86.34 | 118.91 | 17.13 | 549.15 | 12.93 | No |
| Isosorbide (0.00125, 0.0125, 0.125) | 1122.04 | 200 | 87.14 | 196.64 | 127.37 | 24.63 | No |
| Lisinopril (0.00125, 0.0125, 0.125) | 160.00 | 3.83 | 98.36 | 4.74 | 97.96 | 4.63 | No |
| Losartan (0.000625, 0.00625, 0.0625) | 83.12 | 16.89 | 88.62 | 10.32 | 96.66 | 8.39 | Yes |
| Lovastatin (0.00025, 0.0025, 0.025) | 119.53 | 54.01 | 96.52 | 29.53 | 105.84 | 27.36 | No |
| Methyldopa (0.0125, 0.125, 1.25) | 73.63 | 14.14 | 79.02 | 15.51 | 80.27 | 22.11 | Yes |
| Metoprolol (0.00125, 0.0125, 0.125) | 55.86 | 14.59 | 87.24 | 7.28 | 88.16 | 4.66 | No |
| Niacin (0.00025, 0.0025, 0.025) | 106.89 | 137.67 | 79.98 | 38.77 | 87.18 | 7.35 | No |
| Nifedipine (0.00125, 0.0125, 0.125) | 73.13 | 19.77 | 94.43 | 10.68 | 94.02 | 12.09 | Yes |
| Pravastatin (0.0025, 0.025,0.25) | 44.87 | 40.85 | 84.27 | 6.66 | 82.79 | 7.09 | No |
| Propranolol (0.00125, 0.0125, 0.125) | 116.22 | 11.40 | 105.30 | 15.97 | 99.54 | 15.96 | Yes |
| Ramipril (0.00125, 0.0125, 0.125) | 53.18 | 6.76 | 82.92 | 5.67 | 100.91 | 4.08 | Yes |
| Salicylic acid (0.125, 1.25, 12.5) | 46.03 | 49.82 | 103.26 | 3.91 | 81.75 | 7.54 | No |
| Simvastatin (0.00125, 0.0125, 0.125) | 85.12 | 34.90 | 77.83 | 26.52 | 96.47 | 20.24 | No |
| Spironolactone (0.00625, 0.0625, 0.625) | 77.91 | 12.93 | 92.24 | 8.05 | 96.49 | 6.76 | Yes |
| Telmisartan (0.00125, 0.0125, 0.125) | 82.54 | 9.75 | 105.58 | 19.29 | 97.80 | 8.75 | No |
| Triamterene (0.00025, 0.0025, 0.025) | 85.57 | 19.52 | 92.80 | 12.51 | 91.95 | 9.74 | Yes |
| Valsartan (0.0125, 0.125, 1.25) | 74.14 | 9.09 | 83.20 | 5.72 | 89.52 | 5.80 | No |
| Verapamil (0.00125, 0.0125, 0.125) | 67.51 | 15.53 | 88.22 | 14.17 | 91.45 | 10.67 | No |
| Warfarin (0.125, 1.25, 12.5) | 99.50 | 9.91 | 144.19 | 4.09 | 91.86 | 3.55 | No |
Abbreviations: % RSD, percentage of relative standard deviation
Low, medium, and high concentrations, in µg/mL
FDA quantification recommendations for acceptance criteria: “At least 67% (4 out of 6) of QC samples should be within 15% of their respective nominal valude, 33% of the QC samples (not all replicates at the same concentration) may be outside 15% of nominal value. In certain situations, wider acceptance criteria may be justified.” [40]
3.5 Matrix effects
Matrix effects were assessed by comparing the area of the drug peak for a spiked sample immediately following extraction and prior to evaporation against that of a standard prepared in a neat solution [10, 38]. As expected given their extreme hydrophobic or hydrophilic properties, the following compounds were susceptible to matrix effects: isosorbide, niacin, methyldopa, hydralazine, clopidogrel, atorvastatin, lovastatin, and simvastatin. Matrix effects were also significant for salicylic acid, aliskiren, and telmisartan due to co-eluting drugs.
3.6 Limit of Detection and Limit of Quantitation
The limit of detection for each analyte was determined by serial dilution and was defined as the lowest concentration with a signal-to-noise ratio ≥3. The limit of quantitation was determined using calculated concentrations to estimate a range of concentrations that would be likely to occur in patients (Table 1); the lowest calibration standard with a signal-to-noise ratio ≥10 was used as the limit of quantitation. With the exception of warfarin, the low end of the calibration curves were near the limit of detection for all analytes. Warfarin had a 500-fold difference between its limit of detection and the lowest calibration standard; because concentrations below the standard curve are not biologically meaningful, no effort was made to determine an accurate limit of quantitation for warfarin.
3.7 Clinical samples: statistical sensitivity, specificity, and accuracy
Two hundred ninety four clinical samples were obtained and analyzed, including 243 serum samples from hospitalized patients and 51 plasma samples from patients participating in cardiovascular clinical trials. Results were interpreted as false negative if medical records indicated the drug had been administered but was not detected. Results were interpreted as false positive if a drug was detected but review of medical and pharmacy records did not reveal evidence of drug administration. Supplemental Figure A shows the results for each of the 294 clinical samples. Table 5 reports the number of samples obtained in which the medication administration record indicated the patient had been administered drug, as well as the assay’s statistical sensitivity, specificity, and accuracy and their 95% CI’s for each of the 34 cardiovascular drugs or drug metabolite. Figure 3 shows the statistical sensitivity and specificity of the assay for each of the cardiovascular drugs or drug metabolites, as well as their 95% CIs. Thresholds to detect the presence of 34 drugs or drug metabolites were determined as shown in Table 6.
Table 5.
Test characteristics of mass spectrometry assay for 34 cardiovascular drugs or drug metabolites, in 294 clinical samples: Statistical sensitivity, specificity, and accuracy
| Drug Name | TN | TP | FN1 | FP | Sensitivity | Specificity | Accuracy | |
|---|---|---|---|---|---|---|---|---|
| 1 | Acetylsalicylic Acid | 108 | 171 | 8 | 7 | 0.96 | 0.94 | 0.95 |
| 2 | Aliskiren | 292 | 2 | 0 | 0 | 1.00 | 1.00 | 1.00 |
| 3 | Amlodipine | 241 | 49 | 1 | 3 | 0.98 | 0.99 | 0.99 |
| 4 | Atenolol | 264 | 30 | 0 | 0 | 1.00 | 1.00 | 1.00 |
| 5 | Atorvastatin | 239 | 52 | 2 | 1 | 0.96 | 1.00 | 0.99 |
| 6 | Canrenone (Spironolactone) | 232 | 60 | 0 | 2 | 1.00 | 0.99 | 0.99 |
| 7 | Captopril | 290 | 1 | 3 | 0 | 0.25 | 1.00 | 0.99 |
| 8 | Carvedilol | 225 | 69 | 0 | 0 | 1.00 | 1.00 | 1.00 |
| 9 | Clonidine | 266 | 25 | 0 | 3 | 1.00 | 0.99 | 0.99 |
| 10 | Clopidogrel | 219 | 68 | 7 | 0 | 0.91 | 1.00 | 0.98 |
| 11 | Digoxin | 264 | 28 | 1 | 1 | 0.97 | 1.00 | 0.99 |
| 12 | Diltiazem | 261 | 30 | 0 | 3 | 1.00 | 0.99 | 0.99 |
| 13 | Enalapril | 279 | 15 | 0 | 0 | 1.00 | 1.00 | 1.00 |
| 14 | Fenofibrate | 268 | 25 | 1 | 0 | 0.96 | 1.00 | 1.00 |
| 15 | Furosemide | 174 | 119 | 1 | 0 | 0.99 | 1.00 | 1.00 |
| 16 | HCTZ | 247 | 46 | 0 | 1 | 1.00 | 1.00 | 1.00 |
| 17 | Hydralazine | 259 | 34 | 1 | 0 | 0.97 | 1.00 | 1.00 |
| 18 | Isosorbide | 252 | 26 | 6 | 10 | 0.81 | 0.96 | 0.95 |
| 19 | Lisinopril | 203 | 88 | 0 | 3 | 1.00 | 0.99 | 0.99 |
| 20 | Losartan | 262 | 32 | 0 | 0 | 1.00 | 1.00 | 1.00 |
| 21 | Lovastatin | 293 | 1 | 0 | 0 | 1.00 | 1.00 | 1.00 |
| 22 | Methyldopa | 293 | 1 | 0 | 0 | 1.00 | 1.00 | 1.00 |
| 23 | Metoprolol | 182 | 111 | 1 | 0 | 0.99 | 1.00 | 1.00 |
| 24 | Niacin | 285 | 8 | 1 | 0 | 0.89 | 1.00 | 1.00 |
| 25 | Nifedipine | 270 | 24 | 0 | 0 | 1.00 | 1.00 | 1.00 |
| 26 | Pravastatin | 266 | 28 | 0 | 0 | 1.00 | 1.00 | 1.00 |
| 27 | Propranolol | 287 | 7 | 0 | 0 | 1.00 | 1.00 | 1.00 |
| 28 | Ramipril | 286 | 8 | 0 | 0 | 1.00 | 1.00 | 1.00 |
| 29 | Simvastatin | 225 | 63 | 4 | 2 | 0.94 | 0.99 | 0.98 |
| 30 | Triamterene | 287 | 7 | 0 | 0 | 1.00 | 1.00 | 1.00 |
| 31 | Telmisartan | 292 | 2 | 0 | 0 | 1.00 | 1.00 | 1.00 |
| 32 | Valsartan | 255 | 35 | 0 | 4 | 1.00 | 0.98 | 0.99 |
| 33 | Verapamil | 275 | 17 | 0 | 2 | 1.00 | 0.99 | 0.99 |
| 34 | Warfarin | 223 | 71 | 0 | 0 | 1.00 | 1.00 | 1.00 |
Abbreviations: TN, true negative; TP, true positive; FN1, false negative (drug not detected, drug reported in patient records); FP2, false positive (drug detected, drug not reported in patient records); HCTZ, hydrochlorothiazide.
Figure 3. Statistical sensitivity and specificity of mass spectrometry assay for the detection of each of 34 cardiovascular drugs or drug metabolites.
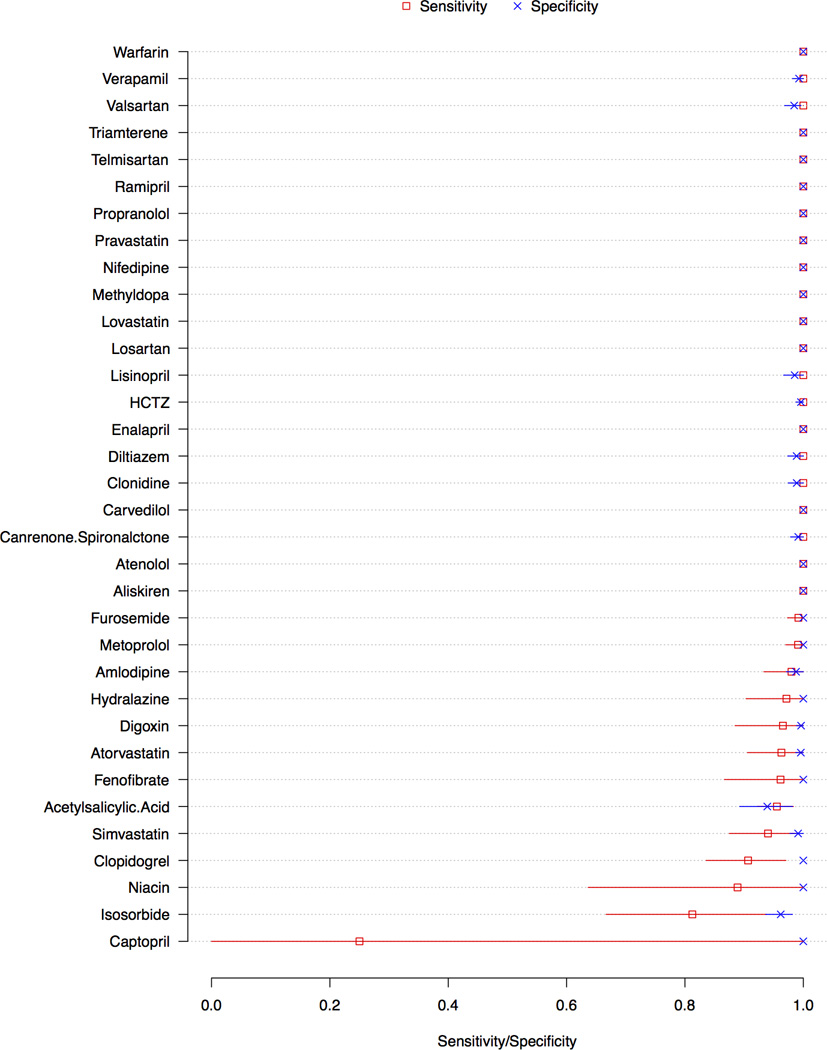
Red and blue lines indicate 95% confidence intervals for sensitivity and specificity, respectively
Abbreviation: HCTZ, hydrochlorothiazide
Table 6.
Determination of detection threshold
| Drug | Retention Time (minutes) |
% of High Standard |
Ratio | Comment |
|---|---|---|---|---|
| Acetylsalicylic acid | 4.79–4.95 | 0.07 | 30–50 | The presence of salicylic acid may indicate the patient received aspirin |
| Aliskiren | 6.1–6.2 | 0.06 | 35–55 | |
| Amlodipine | 6.15-6.4 | 0.3 | 60–90 | |
| Atenolol | 1.9–2.1 | 0.08 | 70 | |
| Atorvastatin | 8.04–8.15 | 0.4 | 29–50 | |
| Captopril | 4 | 3.5 | 80–160 | There is crosstalk between metoprolol and captopril; use a narrow retention time range when considering positives |
| Carvedilol | 6.05–6.35 | 0.1 | 45–55 | |
| Clonidine | 2.2 | 2 | 5–11 | |
| Clopidogrel | 7.85–8.05 | 0.8 | 55–125 | |
| Digoxin | 6.2–6.4 | 14 | 9–30 | Very low intensity and low abundance make this difficult to detect. |
| Diltiazem | 5.85–6.25 | 0.9 | 27–47 | Unknown interferences have produced peaks at the correct retention time with the correct ratio but below the threshold. |
| Enalapril | 5.5–5.7 | 0.02 | 38–70 | Large, obvious tailing easily distinguishes this analyte from potential false positives. |
| Enalaprilat | 3.7–3.85 | 0.02 | 30–50 | The large, obvious fronting for this analyte can be used as an additional confirmation of drug presence. |
| Fenofibric Acid (Fenofibrate) | 8.05–8.15 | 0.4 | 60–75 | |
| Furosemide | 6.2–6.4 | 0.07 | 30–75 | |
| Hydralazine | 1–1.1 | 0.06 | 35 | Adding formic acid allows measurement of total hydralazine rather than apparent hydralazine |
| Hydrochlorothiazide | 2.4 | 0.4 | 40–80 | Symmetric peak with a lower response suggests the patient received the drug more than 24 hours ago |
| Isosorbide | 0.6–0.68 | 20 | 10–70 | |
| Lisinopril | 2.87–3.1 | 0.06 | 15–30 | There is significant tailing on lisinopril peaks |
| Losartan | 6.7–6.85 | 0.04 | 20–30 | |
| Lovastatin | 9.05 | 0.3 | 50–75 | |
| Methyldopa | 0.86 | 0.3 | 85–94 | L-dopa interferes with a retention time at 1 minute; compare the retention time of the peak with the retention time of the standard. |
| Metoprolol | 4.3 | 0.06 | 35–60 | A metoprolol acid metabolite has a retention time of 2.9 with the same m/z transitions |
| Niacin | 0.61 | 0.4 | 75–150 | A low area count may indicate niacin or other supplemental vitamins. |
| Nifedipine | 7.31 | 0.03 | 15–35 | Symmetric peak with a lower response suggests the patient received the drug more than 24 hours ago. |
| Pravastatin | 6.24–6.37 | 0.07 | 55–140 | Patient samples that are positive for Pravastatin show two peaks in the ion trace about 0.2 minutes apart with baseline separation, this is clearly apparent in low abundant samples. Since this is only observed in patient samples it is theorized that this is a metabolite. |
| Propranolol | 5.4–5.5 | 1.2 | 65–80 | |
| Ramipril | 6–6.15 | 0.003 | 1–5 | Significant fronting distinguishes this analyte from false positives |
| Ramiprilat | 5.4–5.5 | N/A | 20–75 | Unique peak shape distinguishes true positives from false positives |
| Salicylic acid | 5 | 0.3 | 5–7 | P-hydroxybenzoic acid is suspected to interfere in the salicylic acid determination, and there are multiple sources of salicylic acid. The presence of salicylic acid does not prove the presence of aspirin. |
| Simvastatin | 9.4–9.5 | N/A | N/A | Even in −80°C, simvastatin converts to its acid metabolite; over time, the reliability of detection and quantitation decreases. |
| Spironolactone (Canrenone) | 7.72–7.96 | 0.08 | 50–150 | |
| Telmisartan | 6.39-6.5 | 0.1 | 60–70 | |
| Triamterene | 3.4 | 0.7 | 60 | |
| Valsartan | 7.55-7.7 | 0.5 | 45–80 | Most true positives are significantly higher than the minimum |
| Verapamil | 6.27-6.5 | 0.09 | 25–40 | |
| Warfarin | 7.59-7.7 | 0.7 | 40–50 | Because warfarin is detectable several days after the treatment has stopped, peaks with area less than 0.5% of the Standard are not considered positive. |
Abbreviation: %, percentage
4. Discussion
This mass spectrometry method was developed with the intention of simultaneously detecting multiple, broad classes of 34 cardiovascular drugs/drug metabolites using a single sample. Our work is unique because the assay includes many of the most commonly prescribed cardiovascular drugs, the assay was evaluated in a large population of clinical patient samples and because the assay was developed for detection of 34 cardiovascular drugs or drug metabolites using a single, small volume sample (Table 1).
The developed LC-MS/MS assay was fully evaluated using test samples injected known concentrations of each of the 34 cardiovascular drugs or drug metabolites, as well as clinical samples obtained from hospitalized patients (Table 4). Detection of multiple cardiovascular medications from varied drug classes shows acceptable statistical specificity, sensitivity, and precision when applied to clinical samples obtained from hospitalized patients. This is particularly important given that clinical samples were obtained at variable time intervals after drug administration. The assay was successfully designed for use in a wide variety of clinical and research settings.
Because the analytical conditions were chosen to optimize detection of all 34 cardiovascular drugs or drug metabolites using a single clinical sample and assay, the detection of a few drugs was not maximized. For example, isosorbide has very poor retention on columns, making it susceptible to matrix effects, which may result in unreliable quantitation and/or detection. Because aspirin is rapidly metabolized to salicylic acid, we measured the metabolite. Other drugs can be metabolized to salicylic acid, and a common preservative, p-hydroxybenzoic acid, can interfere with detection of salicylic acid, reducing assay selectivity. In this case it is possible to minimize the risks of false positive detection by setting a high threshold and monitoring acetylsalicylic acid as well as the metabolite. With respect to captopril, the disulfide bonds that are formed prevent detection without reducing the disulfide bond and derivatizing the thiol group to prevent the reformation of the disulfide. In order to attempt to detect captopril, we adjusted the pH to 3, protonating the thiol group and preventing disulfide bond formation. This avoided the need to derivatize the sample but it did not hydrolyze the bonds that may have already formed. By our methods, captopril was detectable within four hours of the last dose, but if the blood was drawn more than six hours after drug administration captopril was not detected. Finally, we did not explore optimal sample storage conditions. Although we analyzed many samples within 1 week, we found that the assay detected most drugs in samples that had been stored for more than three years.
For clinical samples, the medical administration record was used to determine whether drug should have been present. Interestingly, we discovered that the LC-MS/MS cardiovascular drug assay is so sensitive that it detected medications that were administered before arrival to our hospital or that were administered during procedures such as cardiac catheterizations, where medication administration records were not initially available. The assay also detected medication regimen changes that occurred shortly before hospitalization. In cases where drug was detected by the assay but was not present on review of medication or pharmacy records, results were labeled false positive. It is possible, however, that drug was truly present but that we lacked proper documentation of their administration.
Prior assays developed to detect cardiovascular medications have sometimes been limited to the measurement of medications from the same drug class or with similar structure, e.g., diuretics, angiotensin II receptor antagonists, and beta blockers [4–9]. Gergov et al. developed an assay to monitor 238 therapeutic and illegal drugs using autopsy blood samples [9]; their methods were developed for screening and establishing limits of detection. In addition, this method used a single ion, which increases the potential for interference from co-eluting compounds. Clinical samples for each drug included in the assay were not available to establish test characteristics. Mueller et al. developed a method for detecting 301 drugs using a QTrap liquid chromatography/tandem mass spectrometry system and automated searching [39]. Their extraction methods were not optimized for the entirety of the group of drugs, and the assay was not evaluated using clinical samples. Remane et al. developed a method to monitor 136 analytes, but it required multiple analyses of the same sample [8]. Gonzalez et al. developed a method that detected ten cardiovascular drugs from seven classes [4]; this work was followed by an assay that detects 57 antihypertensive medications, 12 anti-diabetic medications, 2 anti-coagulants, 5 hypo-lipemiants, and 2 anti-platelet medications [10]. Unlike the assay presented here, the assay developed by Gozalez et al. does not include the commonly used medications hydralazine, isosorbide, methyldopa, aliskiren, clonidine, digoxin, fenofibrate, or niacin. Our validation clinical sample size of 294 patients is significantly larger than any of the prior studies
5. Conclusions
To our knowledge, this method is the first method of simultaneous detection of 34 commonly prescribed cardiovascular drugs or drug metabolites from multiple drug classes to be evaluated in samples obtained from patients.
Supplementary Material
Figure 4. Chromatogram of patient clinical sample. The patient was administered metoprolol, aspirin, furosemide, and losartan while hospitalized.
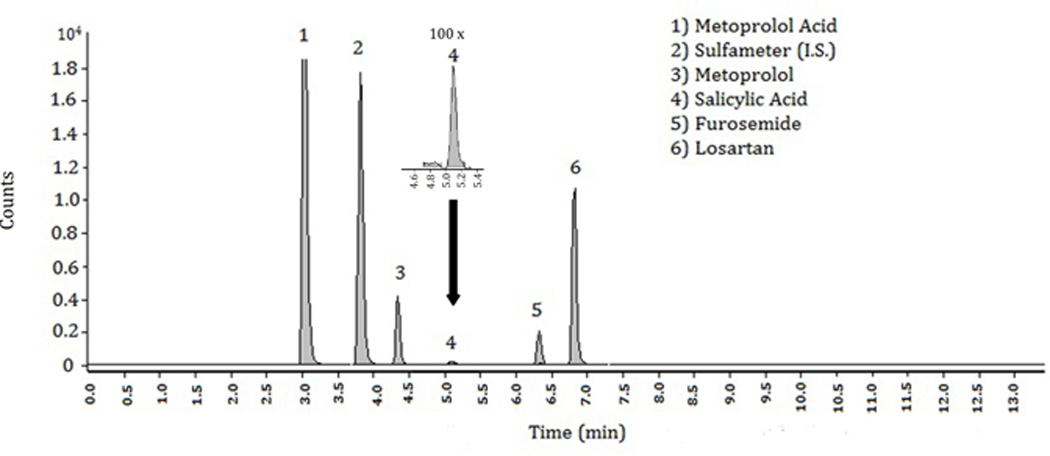
Abbreviations: min, minutes; I.S., internal standard
Highlights.
Development of an LC-MS assay for 34 cardiovascular drugs from nine drug classes.
The assay’s performance was evaluated using samples from 294 patients.
Specificity was ≥0.94 and sensitivity ≥0.90 except for captopril, isosorbide, niacin.
Acknowledgements
The project described was supported by an Innovation and Discovery in Engineering And Science (IDEAS) grant from Vanderbilt University and the National Center for Research Resources, Grant UL1 RR024975-01, which is now at the National Center for Advancing Translational Science Grant 2 UL1 TR00045-06. Dr. McNaughton’s time was supported by the Office of Academic Affiliations, Department of Veterans Affairs, VA National Quality Scholars and by the Vanderbilt Emergency Medicine Research Training (VEMRT) Program (HL 1K12HL109019). The funding sources played no role in the conduction, analysis, analysis, or publication of these findings; the content is solely the responsibility of the authors and does not represent the opinions of the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Roger VL, Go AS, Lloyd-Jones DM, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, Fullerton HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Makuc DM, Marcus GM, Marelli A, Matchar DB, Moy CS, Mozaffarian D, Mussolino ME, Nichol G, Paynter NP, Soliman EZ, Sorlie PD, Sotoodehnia N, Turan TN, Virani SS, Wong ND, Woo D, Turner MB. Circulation. 2012;125:e2–e220. doi: 10.1161/CIR.0b013e31823ac046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heidenreich PA, Trogdon JG, Khavjou OA, Butler J, Dracup K, Ezekowitz MD, Finkelstein EA, Hong Y, Johnston SC, Khera A, Lloyd-Jones DM, Nelson SA, Nichol G, Orenstein D, Wilson PW, Woo YJ. Circulation. 2011;123:933–944. doi: 10.1161/CIR.0b013e31820a55f5. [DOI] [PubMed] [Google Scholar]
- 3.Wright JD, Hughes JP, Ostchega Y, Yoon SS, Nwankwo T. National health statistics reports. 2011:1–22. 24. [PubMed] [Google Scholar]
- 4.Gonzalez O, Iriarte G, Rico E, Ferreiros N, Maguregui MI, Alonso RM, Jimenez RM. J Chromatogr B Analyt Technol Biomed Life Sci. 2010;878:2685–2692. doi: 10.1016/j.jchromb.2010.07.026. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez O, Iriarte G, Ferreiros N, Maguregui MI, Alonso RM, Jimenez RM. J Pharm Biomed Anal. 2009;50:630–639. doi: 10.1016/j.jpba.2008.10.037. [DOI] [PubMed] [Google Scholar]
- 6.Iriarte G, Gonzalez O, Ferreiros N, Maguregui MI, Alonso RM, Jimenez RM. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:3045–3053. doi: 10.1016/j.jchromb.2009.07.018. [DOI] [PubMed] [Google Scholar]
- 7.Kristoffersen L, Oiestad EL, Opdal MS, Krogh M, Lundanes E, Christophersen AS. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;850:147–160. doi: 10.1016/j.jchromb.2006.11.030. [DOI] [PubMed] [Google Scholar]
- 8.Remane D, Meyer MR, Peters FT, Wissenbach DK, Maurer HH. Anal Bioanal Chem. 2010;397:2303–2314. doi: 10.1007/s00216-010-3820-7. [DOI] [PubMed] [Google Scholar]
- 9.Gergov M, Ojanpera I, Vuori E. J Chromatogr B Analyt Technol Biomed Life Sci. 2003;795:41–53. doi: 10.1016/s1570-0232(03)00498-7. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez O, Alonso RM, Ferreiros N, Weinmann W, Zimmermann R, Dresen S. J Chromatogr B Analyt Technol Biomed Life Sci. 2011;879:243–252. doi: 10.1016/j.jchromb.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 11.Jackevicius CA, Li P, Tu JV. Circulation. 2008;117:1028–1036. doi: 10.1161/CIRCULATIONAHA.107.706820. [DOI] [PubMed] [Google Scholar]
- 12.Osterberg L, Blaschke T. N Engl J Med. 2005;353:487–497. doi: 10.1056/NEJMra050100. [DOI] [PubMed] [Google Scholar]
- 13.Ho PM, Bryson CL, Rumsfeld JS. Circulation. 2009;119:3028–3035. doi: 10.1161/CIRCULATIONAHA.108.768986. [DOI] [PubMed] [Google Scholar]
- 14.Heidenreich PA. Am J Med. 2004;117:130–132. doi: 10.1016/j.amjmed.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Bartholow MP. Pharmacy Times. 2011 [Google Scholar]
- 16.Reddy P, Ellington D, Zhu Y, Zdrojewski I, Parent SJ, Harmatz JS, Derendorf H, Greenblatt DJ, Browne K., Jr Br J Clin Pharmacol. 2011;72:434–441. doi: 10.1111/j.1365-2125.2011.03996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tenero D, Boike S, Boyle D, Ilson B, Fesniak HF, Brozena S, Jorkasky D. J Clin Pharmacol. 2000;40:844–853. doi: 10.1177/00912700022009576. [DOI] [PubMed] [Google Scholar]
- 18.Bhavesh D, Shah S. Biomed Chromatogr. 2009;23:922–928. doi: 10.1002/bmc.1203. [DOI] [PubMed] [Google Scholar]
- 19.Lee WI, Yoon WH, Shin WG, Song IS, Lee MG. Biopharm Drug Dispos. 1997;18:753–767. doi: 10.1002/(sici)1099-081x(199712)18:9<753::aid-bdd63>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 20.Ohtawa M, Takayama F, Saitoh K, Yoshinaga T, Nakashima M. Br J Clin Pharmacol. 1993;35:290–297. doi: 10.1111/j.1365-2125.1993.tb05696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menon R, Tolbert D, Cefali E. Biopharm Drug Dispos. 2007;28:297–306. doi: 10.1002/bdd.555. [DOI] [PubMed] [Google Scholar]
- 22.Singhvi SM, Pan HY, Morrison RA, Willard DA. Br J Clin Pharmacol. 1990;29:239–243. doi: 10.1111/j.1365-2125.1990.tb03626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lilja JJ, Neuvonen M, Neuvonen PJ. Br J Clin Pharmacol. 2004;58:56–60. doi: 10.1111/j.1365-2125.2004.02095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brodie RR, Chasseaud LF, Darragh A, Taylor T, Walmsley LM. Biopharm Drug Dispos. 1982;3:361–370. doi: 10.1002/bdd.2510030409. [DOI] [PubMed] [Google Scholar]
- 25.Miura M, Satoh S, Kagaya H, Saito M, Inoue T, Ohkubo T, Habuchi T, Suzuki T. J Clin Pharm Ther. 2009;34:683–692. doi: 10.1111/j.1365-2710.2009.01053.x. [DOI] [PubMed] [Google Scholar]
- 26.Karim A, Zagarella J, Hutsell TC, Dooley M. Clin Pharmacol Ther. 1976;19:177–182. doi: 10.1002/cpt1976192177. [DOI] [PubMed] [Google Scholar]
- 27.Baselt RC. Disposition of Toxic Drugs and Chemicals in Man, 3rd Ed. 6th ed. Foster City: Biomedical Publications; 2002. [Google Scholar]
- 28.Karim A, et al. Clin Pharmacol Ther. 1976;19 doi: 10.1002/cpt1976192158. [DOI] [PubMed] [Google Scholar]
- 29.Vaidyanathan S, Jarugula V, Dieterich HA, Howard D, Dole WP. Clin Pharmacokinet. 2008;47:515–531. doi: 10.2165/00003088-200847080-00002. [DOI] [PubMed] [Google Scholar]
- 30.Mani H, Toennes SW, Linnemann B, Urbanek DA, Schwonberg J, Kauert GF, Lindhoff-Last E. Ther Drug Monit. 2008;30:84–89. doi: 10.1097/FTD.0b013e31815c13fd. [DOI] [PubMed] [Google Scholar]
- 31.Lee MG, Li T, Chiou WL. Biopharm Drug Dispos. 1986;7:537–547. doi: 10.1002/bdd.2510070603. [DOI] [PubMed] [Google Scholar]
- 32.DiPalma JR, Thayer WS. Annu Rev Nutr. 1991;11:169–187. doi: 10.1146/annurev.nu.11.070191.001125. [DOI] [PubMed] [Google Scholar]
- 33.Benge CD, Muldowney JA., 3rd Expert Opin Drug Metab Toxicol. 2012 doi: 10.1517/17425255.2012.725721. [DOI] [PubMed] [Google Scholar]
- 34.U.S.N.L.o. Medicine, editor. LiverTox: Clinical and Research Information on Drug-Induced Liver Injury. Bethesda: [PubMed] [Google Scholar]
- 35.Efron B. Ann Statist. 1979;7:1–26. [Google Scholar]
- 36.Iwaki M, Ogiso T, Ito Y. J Pharm Sci. 1988;77:280–283. doi: 10.1002/jps.2600770320. [DOI] [PubMed] [Google Scholar]
- 37.Israili ZH. Annu Rev Pharmacol Toxicol. 1979;19:25–52. doi: 10.1146/annurev.pa.19.040179.000325. [DOI] [PubMed] [Google Scholar]
- 38.Matuszewski BK, Constanzer ML, Chavez-Eng CM. Anal Chem. 2003;75:3019–3030. doi: 10.1021/ac020361s. [DOI] [PubMed] [Google Scholar]
- 39.Mueller CA, Weinmann W, Dresen S, Schreiber A, Gergov M. Rapid Commun Mass Spectrom. 2005;19:1332–1338. doi: 10.1002/rcm.1934. [DOI] [PubMed] [Google Scholar]
- 40.2001 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


