Abstract
The Southern house mosquito, Culex quinquefasciatus - a vector of West Nile virus - is equipped with 130 odorant receptors (ORs), which enable young females to locate plants and blood-meal sources and older females to find suitable sites for oviposition. In our attempts to de-orphanize ORs expressed in female antennae, we identified CquiOR37 and CquiOR99, which were narrowly tuned to two phenolic compounds, 4-methylphenol and 4-ethylphenol. When tested in the Xenopus oocyte recording system the observed EC50s for 4-methylphenol and 4-ethylphenol were 6.4 and 18.2 µM for CquiOR37 and 14.4 and 0.74 µM for CquiOR99 (goodness of fit, R2 =0.88–0.99), respectively. Indoor behavioral assays demonstrated that gravid female mosquitoes laid significantly more eggs in water trays spiked with these compounds than in control water trays. Field studies with gravid traps corroborated that 4-ethylphenol is active in a wide range of doses from 0.1 to 10 µg/l, as required for practical applications. A dsRNA construct based on the two genes, CquiOR37/99-dsRNA was stable in pupa hemolymph for up to 3 h. Pupae injected with CquiOR37/99-dsRNA, β-galactosidasedsRNA or water had more than 40% survival rate at the peak of oviposition (day-9). qPCR analysis showed individual variation, but significant mean reduction in CquiOR37 and CquiOR99 transcript levels in CquiOR37/99-dsRNA-treated mosquitoes. Water-injected females and those treated with the control gene laid significantly more eggs in trays containing 4-ethylphenol than in water trays, whereas CquiOR37/99-dsRNA-treated mosquitoes laid normal number of eggs, but could not discriminate treatment from control. This study linked for the first time specific receptors for 4-ethylphenol with increased oviposition in the important vector Cx. quinquefasciatus.
Keywords: Culex quinquefasciatus, CquiOR37, CquiOR99, 4-methylphenol, 4-ethylphenol, RNAi
1. Introduction
Culex mosquitoes are vectors for human diseases, including filariasis and various types of encephalitis, throughout the world (Nasci and Miller, 1996). In the United States, they spread West Nile Virus while feeding on birds and humans (Syed and Leal, 2009). The Southern house mosquito, Culex quinquefasciatus Say, may possess one of the most, if not the most, acute olfactory system in mosquitoes for the reception of host-derived compounds, such as nonanal (Syed and Leal, 2009), and has the largest repertoire of odorant receptors (ORs) of all dipteran species whose genomes have been hitherto sequenced (Arensburger et al., 2010; Pelletier et al., 2010b; Xu et al., 2013). De-orphanization of these ORs gives us better insights on how mosquitoes perceive the world through small, signal-carrying molecules (semiochemicals), and may lead to the discovery of novel repellents for reducing bites and disease transmission as well as “green chemicals” for monitoring and controlling mosquito populations. Monitoring mosquito populations and mosquito-borne virus activity are the cornerstones of surveillance programs. Sampling female mosquitoes with gravid traps is in principle more effective as they collect mosquitoes that have taken blood meals and therefore had the opportunity to become horizontally infected, but there is limited availability of lures for these traps (Leal et al., 2008). Previously, we have identified and de-orphanized two ORs expressed in Cx. quinquefasciatus female antennae and sensitive to oviposition attractants (Hughes et al., 2010; Pelletier et al., 2010b), which were named CquiOR2 and CquiOR10 and now renamed CquiOR121 and CquiOR21, respectively. In our additional attempts to de-orphanize Culex ORs expressed in female antennae, we have identified silent, generic, and plant kairomone sensitive ORs (Xu et al., 2013). Envisioning that OR(s) for the mosquito oviposition pheromone (MOP), (5R,6S)-6-acetoxy-5-hexadecanolide (Laurence and Pickett, 1982b) may lead us to more effective oviposition attractants, we attempted to identify MOP-sensitive OR(s). To narrow down from 130 ORs in the genome of Cx. quinquefasciatus (Arensburger et al., 2010; Pelletier et al., 2010b; Xu et al., 2013), we reasoned that as MOP structure contains a hexadecanolide moiety, its receptors might be orthologs of lactone-sensitive ORs. Previously, two ORs from the malaria mosquito Anopheles gambiae have been demonstrated to respond to lactones (Carey et al., 2010; Wang et al., 2010), namely, AgamOR30 and AgamOR48, which showed small and robust responses (Pask et al., 2013), respectively. With phylogenetic analysis by the neighbor-joining method (Tamura et al., 2011) we found no orthologs of AgamOR30, but there are putative orthologs of AgamOR48 in Culex genome, i.e., CquiOR37 (VectorBase ID=CPIJ004163), CquiOR38 (CPIJ004164), and CquiOR99 (CPIJ011787), which are clustered with AgamOR48 into one group (94–100% bootstrap support) (Pelletier et al., 2010b; Xu et al., 2013). We successfully cloned CquiOR37 and CquiOR99, but CquiOR38 is not expressed in adult female antennae and may be a pseudogene. We accidentally discovered that these two ORs, albeit insensitive to MOP and other lactones, responded specifically to phenols, i.e., 4-methylphenol (p-cresol) and 4-ethylphenol. Indoor (cage) oviposition assays and field experiments demonstrated that indeed these semiochemicals are oviposition attractants and that 4-ethylphenol at various doses could be used for trapping gravid female Culex mosquitoes. Furthermore, RNAi experiments showed that dsRNA-treated phenotypes had a long-lasting reduction of OR transcript levels and defect of oviposition response to 4-ethylphenol.
2. Materials and Methods
2.1 Insects
Culex quinquefasciatus mosquitoes used in this study were originated from a stock laboratory colony, which in turn started from adult mosquitoes collected in Merced, CA in the 1950s and maintained by Dr. Anthony Cornel in the Kearney Agricultural Center, University of California. In Davis, mosquitoes were maintained in an insectary at 27±1°C, under a photoperiod of 16:8 hr (L:D) for the last 3 years.
2.2 RNA Extraction and Cloning of CqOr37 and CqOr99
Antennae were dissected from 3-5-day-old female mosquitoes. Total RNA extraction and cDNA synthesis were performed as previously described (Pelletier and Leal, 2009). To amplify full-length coding sequences of CquiOR37 and CquiOR99, the following gene specific primers were designed on the basis of gene annotations for Cx. quinquefasciatus (Arensburger et al., 2010) with restriction site adaptors at ends of primers: CquiOr37-F: (XmaI) 5’-CCCGGGATGTCCCCAAACATCCCACC-3’; CquiOr37-R: (HindIII) 5’-AAGCTTTCAAAAGCGCTTCTTCAGTATC-3’; CquiOr38-F: (XmaI) 5’-CCCGGGATGTTCCTGCAGCGCTTCTTCC-3' CquiOr38-R: (XbaI) 5’-TCTAGATCAAGCGTTGAACCGTTCCTTC-3' CquiOr99-F: (XmaI) 5’-CCCGGGATGCACTTCCTCAAAAGATCGCTCC-3’; CquiOr99-R: (XbaI) 5’-TCTAGATTAAAAGCTGTTCTTCAGTATCAAATA-3’. PCR reactions were carried out using Pfu Ultra II polymerase (Stratagene), and products were purified using QIAquick Gel Extraction kit (Qiagen, Valencia, CA) and ligated into pBlueScript SK (+) (Stratagene). Three attempts to clone CquiOR38 with the following primers were unsuccessful: CquiOr38F: (Xma I) 5’-CCCGGGATGTTCCTGCAGCGCTTCTTCC-3’ and CquiOr38R: (Xba I) 5’-TCTAGATCAAGCGTTGAACCGTTCCTTC-3’. We concluded that this gene may not be expressed in adult antennae or might be a pseudogene. Ligation products of CquiOR37 and CquiOR99 were used to transform One Shot OmniMAX competent cells (Invitrogen, Carlsbad, CA). White clones were selected on LB medium plates containing ampicillin and grown in LB liquid media. Plasmids were purified using QIAprep Spin Miniprep kit (Qiagen) and sequenced (Davis Sequencing Inc, Davis, CA). Subsequently, inserts were subcloned into Xenopus oocyte expression plasmid pGEMHE (Liman et al., 1992) with restriction sites, XmaI and HindIII/XbaI at both ends (see underlined sequences). The subcloned sequences were validated by sequencing (Davis Sequencing Inc, Davis, CA). CquiOrco-pGEMHE was available from a previous project (Pelletier et al., 2010b).
2.3 In vitro Transcription and Oocyte Microinjection
Capped cRNAs were prepared using a mMESSAGE mMACHINE T7 Kit (Ambion) as previously described (Xu et al., 2012). Purified OR cRNAs were re-suspended in nuclease-free water at 200 ng/µl and microinjected with the same amount of CquiOrco into Xenopus laevis oocytes on stage V or VI (purchased from EcoCyte Bioscience, Austin, TX). Injected oocytes were incubated at 18°C for 3–7 days in modified Barth's solution [NaCl 88 mM, KCl 1 mM, NaHCO3 2.4 mM, MgSO4 0.82 mM, Ca(NO3)2 0.33 mM, CaCl2 0.41 mM, HEPES 10 mM, pH 7.4] supplemented with 10 µg/ml of gentamycin, 10 µg/ml of streptomycin and 1.8 mM sodium pyruvate. Two-electrode voltage-clamp technique (TEVC) was employed to observe odorant-induced currents at holding potential of −80 mV. Signals were amplified with an OC-725C amplifier (Warner Instruments, Hamden, CT), low-pass filtered at 50 Hz and digitized at 1 kHz. Data acquisition and analysis were carried out with Digidata 1440A and software pCLAMP 10 (Molecular Devices, LLC, Sunnyvale, CA).
The following panel of compounds were used to de-orphanize ORs: 1-hexanol, 1-octanol, 2,3-butanediol, (E)-2-hexen-1-ol, (Z)-2-hexen-1-ol, 1-hexen-3-ol, 1-heptene-3-ol, 3-octanol, 1-octen-3-ol, guaiacol, 1-hexadecanol, geraniol, linalool, ethyl acetate, butyl acetate, octyl acetate, decyl acetate, methyl propionate, ethyl propionate, methyl butyrate, ethyl 3-hydroxyhexanoate, (E)-2-hexenyl acetate, (Z)-3-hexenyl acetate, ethyl lactate, methylsalicylate, geranyl acetate, octadecyl acetate, 2-heptanone, fenchone, cyclohexanone, acetophenone, 6-methyl-5-hepten-2-one, 2-butoxylacetone, 2-tridecanone, thujone, ethyl stearate, methyl myristate, MOP, γ-valerolactone, γ-hexalactone, γ-octalactone, γ-decalactone, 2-undecanone, propanal, penatanal, heptanal, octanal, nonanal, decanal, undecanal, phenylacetaldehyde, furfural, trans-2-methyl-2-butenal, benzaldehyde, phenol, 2-methylphenol, 3-methylphenol, 4-methylphenol, 4-ethylphenol, 3,5-dimethylphenol, 2,3-dimethylphenol, 2-methoxy-4-propylphenol, indole, 1-methylindole, 2-methylindole, 3-methylindole, 4-methylindole, 5-methylindole, 6-methylindole, 7-methylindole, butylamine, heptylamine, octylamine, trimethylamine, lactic acid, and nonanoic acid. They were acquired from Sigma-Aldrich (Milwaukee, MO), except for 4-methylindole (Tokyo Chemical Industry, Tokyo, Japan) and MOP, which was a synthetic sample from a previous work (Leal et al., 2008). All compounds were of purity 95% or higher, except for trimetylamine, which was a 45% water solution.
2.4 Indoor and Field Behavior Assays
Oviposition bioassay was performed in Bioquip’s Collapsible Field Cages (30.5 × 30.5 × 30.5 cm); cage covers were extensively washed with hot water before and after tests. In each cage two plastic trays (Falcon, Specimen Container #354014) filled with 100 ml of distilled water were placed in opposite corners. One microliter of an ethanol solution of the test compound was added into treatment, whereas the same volume of ethanol was added into control tray. Four cages were used per treatment per day. Forty gravid females per cage (5 days after the first of two blood meals) were used for oviposition assay during scotophase. Egg rafts in treated and control trays were counted in the following morning, cages were changed, and trays rotated for the next replicate with a fresh group of females. Data were analyzed, as previously performed (Braks et al., 2007), using Wilcoxon Rank Sign Test (KaleidaGraph, Synergy Software, Reading, PA) and student t-test (http://studentsttest.com/).
Field tests were conducted from June to November, 2012 in Recife, Brazil, a city with abundant populations of Cx. quinquefasciatus that breed throughout the year (Leal et al., 2008). Tests were conducted in the backyards of residences and in the campus of the Universidade Federal de Pernambuco. Gravid mosquito traps (Bioquip, Rancho Dominguez, CA) were deployed in duplicate per location (2 control and 2 treatments), except for dose comparison when three traps were deployed per location. Traps were modified to operate with a power supply as to minimize variation in fan speed. Traps were filled with 5L of water plus 100 µl of an ethanol solution of 4-ethylphenol to generate the final concentrations of 0.1, 1, or 10 µg/l. They were placed on the ground with an intertrap distance of at least 5 m, inspected and rotated every morning. Collecting chambers were replaced, trapped mosquito were frozen and counted. Data were analyzed using Wilcoxon Rank Sign Test for paired data or transformed to log (x+1) and analyzed by ANOVA and Tukey honestly significant differences (KaleidaGraph, Synergy Software, Reading, PA).
2.5 dsRNA Synthesis
Double-strand RNA (dsRNA) of CquiOR37/99 and β-galactosidase were synthesized by in vitro transcription from PCR product using the MEGAscript T7 transcription kit (Ambion, Austin, TX). PCR was performed using plasmids containing the target genes as DNA template with the following gene specific primers that included T7 promoter sequence (underlined); CquiOR37/99-F: 5’-TAATACGACTCACTATAGGGAGACGGACCTGCAAGTGCTCAAGT-3’ and CquiOR37/99-R: 5’-TAATACGACTCACTATAGGGAGAAAACACCACCTTCACACAGGT-3’; Cquiβ-gal-F: 5’-TAATACGACTCACTATAGGGAATGGTTCAGGTCGAAAACG-3’ and Cquiβ-gal-R: 5’-TAATACGACTCACTATAGGGCCGCCTCGTACAAAACAAGT-3’. Large scale dsRNAs were purified with MEGAclear kit (Ambion, Austin, TX) and precipitated with 5 M ammonium acetate to yield 13~14 µg/µl of CquiOR37/99-dsRNA.
2.6 Ex vivo Determination of dsRNA Stability
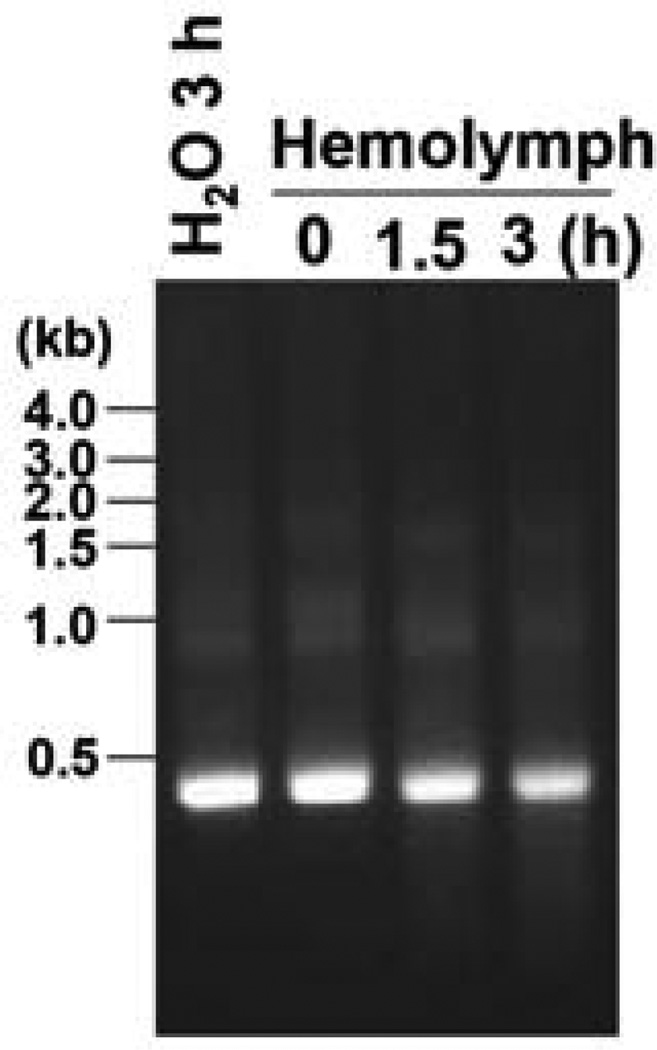
Hemolymph was collected from day-0 pupae, combined in chilled 1.5 ml microcentrifuge tube containing phenylthiourea (PTU) to prevent melanization (Arakawa, 1995). Pooled hemolymph was centrifuged at 1,000×g for 10 min at 4°C to separate cell-free plasma from hemocytes. CquiOR37 dsRNA (1 µg) was mixed with 3 µl cell-free hemolymph plasma and incubated at room temperature for 0–3 h, as reported for M. sexta (Garbutt et al., 2013). dsRNA in DEPC-water was used as control. After incubation, dsRNA samples were recovered with 30 µl nuclease-free water using an RNeasy Mini Kit (Qiagen, Valencia, CA), according to RNA Cleanup instruction. Recovered dsRNA (10 µl) was separated on 1% agarose EtBr gel. dsRNA on gel was visualized using a UV transilluminator (Bio-Rad, Hercules, CA).
2.7 dsRNA Microinjection and Blood Feeding
Day-0 female pupae were collected in a plastic cup filled with distilled water and kept on ice for 15 min. The sharp end of a yellow pipette tip was cut diagonally to make a stage to hold a pupa. Twenty-two nanograms of dsRNAs in 10 nl volume were injected in the thorax membrane close to the base of the trumpet using a NanoLiter 2000 inject (World Precision Instruments, Sarasota, FL). The injected pupae were put in new plastic cup with distilled water and kept at 27°C. The newly emerged adults were transferred into mesh cages with sugar water (10%). The same number of newly emerged males was released into the cage for mating. Three blood meals were provided to females on the morning of day 4, 5, and 6. Five days after the first blood meal the blood-fed females were used for oviposition bioassay.
2.8 Quantitative analysis of receptor transcription levels
For qRT-PCR, one pair or twenty pairs of antennae were dissected and collected in DEPC-water on ice using a stereo microscope (Zeiss, Stemi DR 1663, Germany). Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA). cDNA were synthesized using RT-for-PCR kit according to the manufacturer’s instructions (Clontech, Mountain View, CA). Real-time quantitative PCR (qPCR) was carried out by using a CFX96 TouchTM Real-Time PCR Detection System (Bio-Rad, Hercules, CA) and iQTM SYBR Green Supermix (Bio-Rad, Hercules, CA). CquiOrco gene was used as reference. We designed two pairs of primer for each gene by Primer 3 program (http://frodo.wi.mit.edu/), IDT (http://www.idtdna.com/scitools/Applications/RealTimePCR/), and OligoPerfect™ Designer (http://tools.invitrogen.com/content.cfm?pageid=9716). CquiOr37 forward and reverse: (A1 pair), 5’- GGTTCTTATGGGCGAGATGA-3’ and 5’-TACGAGTACGACGCTTGCAC-3’; (A2 pair), 5’-GTGGATTGGGTTTTGGGTTGTG-3’ and 5’-AGCAGAACACAAGGTAGATCG-3’; CquiOr99 forward and reverse: (B1 pair), 5’-GTCATCCAGTACGGGGCTAA-3’ and 5’-AGCAAACGGTCTCTTCCAGA-3’; (B2 pair), 5’-AAGTTGTATGTTGATTTCGTGCC-3’ and 5’- GCATTGTTCCGTTGAAGCAG-3’; CquiOrco forward and reverse (C1 pair), 5’-TTCCTGCTGTTCTCGATGTG-3’and 5’-CGCTCAGGTCAAAGTCCTTC-3’; (C2 pair), 5’-GCCGGATACGTTTTCTCCTTC-3’ and 5’- GCGCATAATTCCCTTCAGATG-3’. qPCR data analysis was performed using the 2−ΔΔCT method.
3. Results and Discussion
3.1 Cloning of CquiOR37 and CquiOR99
Using cDNA from adult female antennae as template, we cloned CquiOR37 and CquiOR99, which encode proteins with amino acid sequences differing slightly from those in VectorBase. They may be isoforms caused by single nucleotide polymorphism (SNPs) differences, and their sequences have been deposited in GenBank: CquiOR37, KF049489 and CquiOR99, KF049490. While the Cx. quinquefasciatus genome was sequenced from the Johannesburg strain (Arensburger et al., 2010), we cloned the genes using cDNA from a California strain. CquiOR37 and CquiOR99 differed from the annotated proteins in one and three amino acid residues, respectively: Arg- vs His-175 in CquiOR37; Asp- vs. Glu-114, Leu- vs. Val-210, and Ala- vs. Val-21 in CquiOR99. These differences are not surprising given that Cx. quinquefasciatus and related Culex pipiens complex mosquitoes have very high densities of SNPs, in fact more than any other mosquito thus far studied (Lee et al., 2012).
3.2 De-orphnization of CquiOR37 and CquiOR99
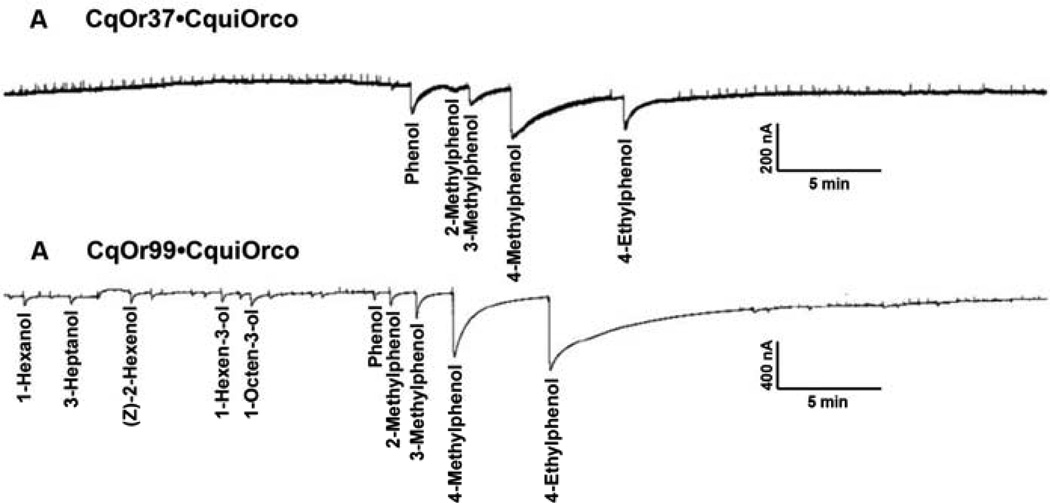
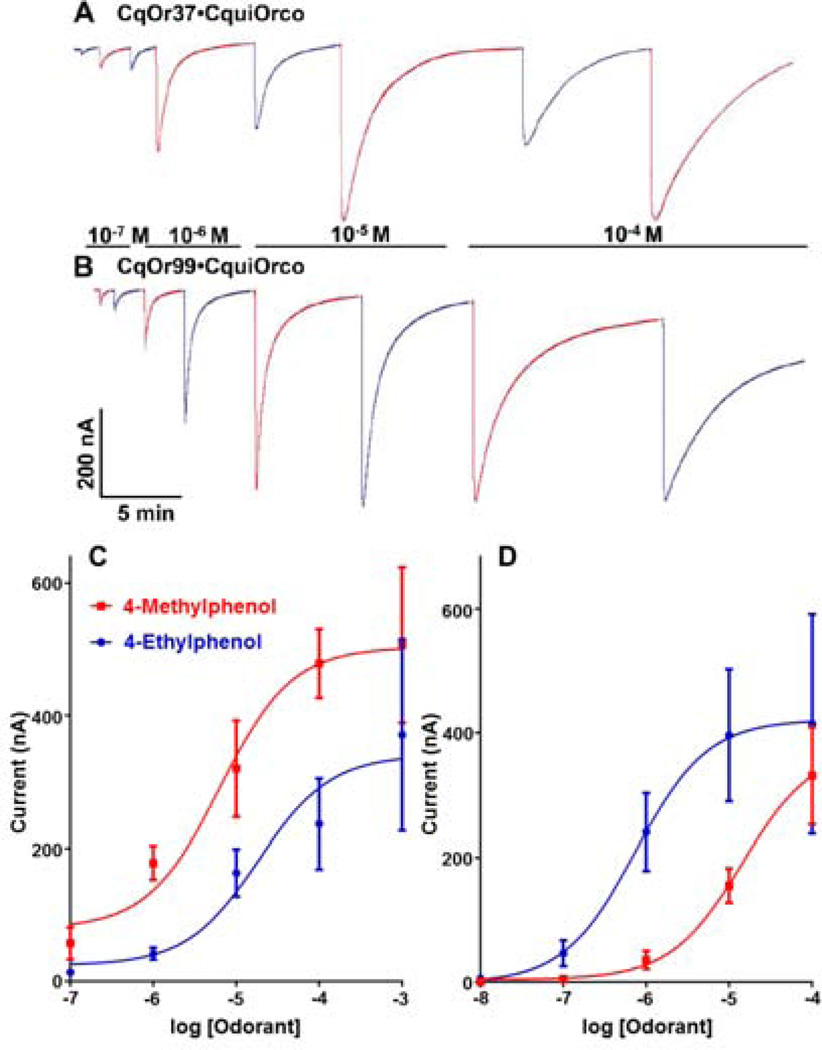
To de-orphanize these receptors, we screened CquiOR37●CquiOrco- and CquiOR99●CquiOrco-expressing oocytes with a panel of more than 70 compounds, including known oviposition attractants, such as, trimethylamine (Leal et al., 2008), nonanal (Leal et al., 2008), and skatole (Blackwell et al., 1993; Leal et al., 2008; Millar et al., 1992), and various other compounds of physiological importance for mosquitoes. Interestingly, oocytes expressing these receptors (n=20 eggs from 3 batches) were insensitive to the mosquito oviposition pheromone (MOP) (Laurence and Pickett, 1982a) and to other known oviposition attractants as well as to the largest majority of the compounds tested at 1 mM doses (Fig. 1). They all responded, however, to phenolic compounds with reverse profiles. 4-Methylphenol and 4-ethylphenol elicited, respectively, higher and lower current responses in CquiOR37●CquiOrco-expressing oocytes. By contrast, CquiOR99●CquiOrco-oocytes showed stronger responses to 4-ethylphenol than to 4-methylphenol (Fig. 1). These response profiles were systematically compared in dose-dependent relationships (Fig. 2). To avoid desensitization, the expressed receptors were challenged from low to high concentrations of ligands, and starting each dose tested with the odorant that elicits lower responses. Clearly, 4-methylphenol was the most potent of these two compounds for CquiOR37 (Fig 2A), whereas 4-ethylphenol was the best ligand for CquiOR99 (Fig 2B). In-depth concentration-response analysis confirmed that 4-methylphenol and 4-ethylphenol are the best ligands for CquiOR37 (Fig. 2C) and CuqiOr99 (Fig. 2D), respectively. 4-Methylphenol and 4-ethylphenol activated CquiOR37●CquiOrco with EC50 of 6.4 µM and 18.2 µM, respectively. By contrast, the EC50 for 4-ethylphenol and 4-methylphenol with CquiOR99●CquiOrco were 0.74 µM and 14.4 µM, respectively. The goodness of fit for the data (R2) ranged from 0.88 to 0.99.
Fig. 1.
Screening of putative CquiORs with a panel of odorants. Electrophysiological recordings from (A) CquiOR37●CquiOrco and (B) CquiOR39●CquiOrco-expressing oocytes. Each odorant was applied at concentration of 1 mM for 1 s, with washing between applications until baseline recovery. In these representative traces oocytes were challenged with the largest majority of the test ligands: 73 and 72, respectively. Only the names of compounds generating reproducible responses above baseline levels were displayed. See Materials & Methods, for the full list of test compounds.
Fig. 2.
Current responses recorded from CquiOR●CquiOrco-expressing oocytes challenged with phenolic compounds and dose-dependent relationships. While (A) CquiOR37 showed stronger responses to 4-methylphenol (red traces), (B) CquiOR99 was more sensitive to 4-ethylphenol (blue traces). (C) CquiOR37●CquiOrco-expressing oocytes and (D) CquiOR99●CquiOrco-expressing oocytes were stimulated with increasing doses of 4-methylphenol (red curves) and 4-ethylphenol (blue curves). CquiOR99 was saturated at doses higher than 10−4M. n=3–5, error bars in all figures represent SEM.
3.3 Indoor and Field Behavioral Assays
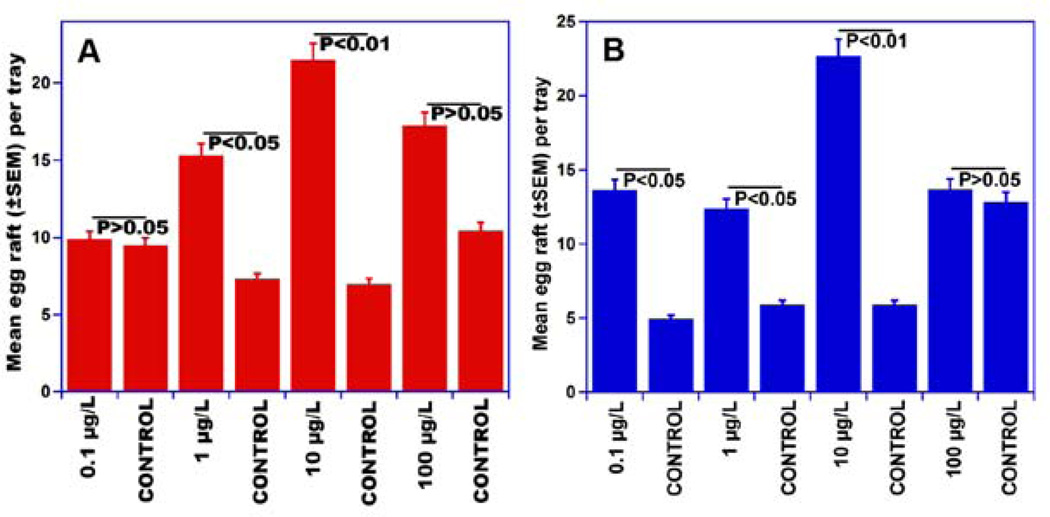
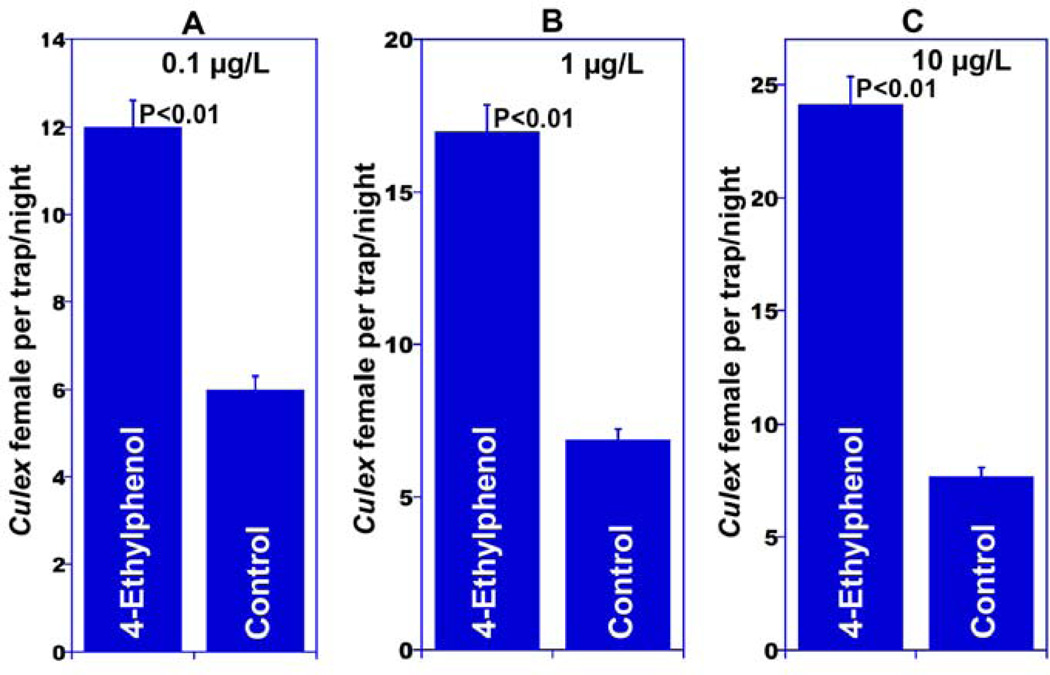
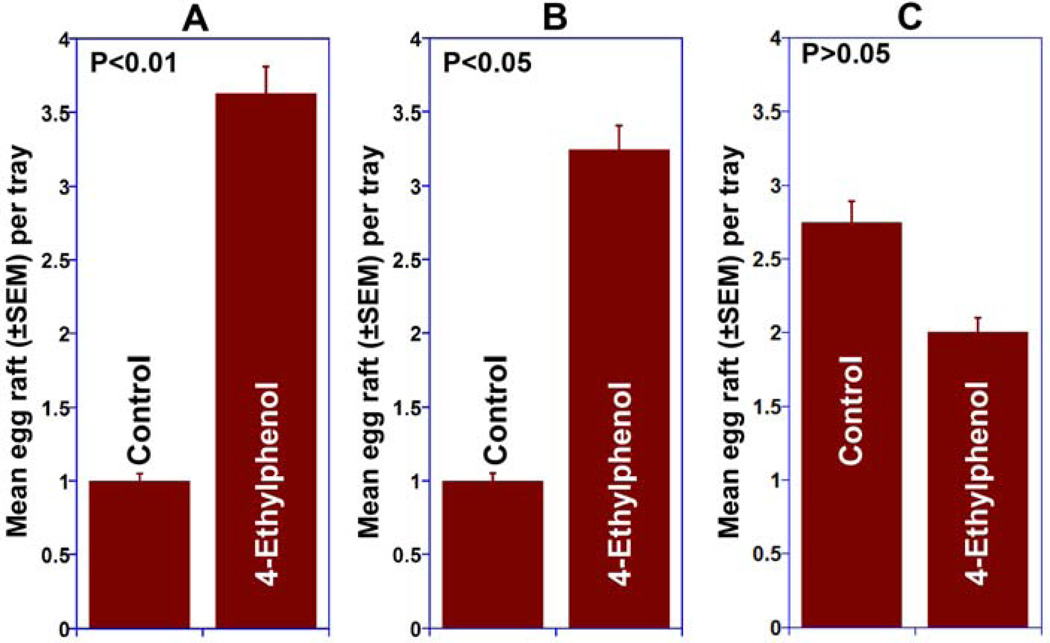
4-Methylphenol and 4-ethylphenol have been previously isolated from fermented Bermuda grass infusion along with skatole (Millar et al., 1992), but the same cage bioassays that led to the first characterization of skatole as an oviposition attractant (Millar et al., 1992) were negative or inconsistent for these phenolic compounds (Blackwell et al., 1993; Millar et al., 1992). Intrigued by the fact that two genes in the genome of Cx. quinquefasciatus express OR specifically tuned to these compounds and responding in a way, we decided to perform meticulous cage behavioral assays. To avoid inconsistencies, we controlled the physiological status of the tested mosquitoes and used only gravid females five days after the first of three blood meals, and performed bioassays in an insectary with controlled temperature, humidity and photoperiod (see Material and Methods). To minimize possible positional effects, treatment and control trays were rotated daily. The results demonstrate that gravid Cx. quinquefasciatus mosquitoes laid significantly more eggs in trays treated with 4-methylphenol or 4-ethylphenol at various doses than in controls (Fig. 3). 4-Ethylphenol was active at three doses, i.e. 0.1, 1 and 10 µg/l (Fig 3B), whereas 4-methylphenol was active in a narrower range, i.e., 1 to 10 µg/l (Fig 3A). The numbers of egg rafts laid in traps baited with either 4-methylphenol or 4-ethylphenol at a higher dose, 100 µg/l, were not significantly different from those laid in water (control) trays (Fig. 3). Oviposition attractants that have a wide range of active doses like 4-ethylphenol are highly desirable for practical applications given that under field conditions water evaporation from gravid traps and odorant losses may render formulations of narrow range compounds inactive with these changes in concentration. We then subjected 4-ethyphenol to tests under field conditions. Gravid traps baited with 4-ethylphenol and control traps were deployed in an urban area in Recife, Brazil for five consecutive months. Trapping field data mirrored the cage observation for oviposition, with 4-ethylphenol-treated traps capturing significantly more female Culex mosquitoes than control traps at three doses (Fig. 4A, B, and C). It is worth mentioning, however, that gravid traps capture female mosquitoes, whereas cage behavioral assays are based on the number of egg rafts laid. Although it may appear that higher doses of 4-ethylphenol led to higher catches, it is worth mentioning that these results are not statistically comparable as each experiment was performed independently, i.e., comparing only treatment and control, thus the experimental design does not allow comparison between treatments. Multiple comparisons are very simple in agricultural setting, but very challenging in medical entomology. First, mosquito populations in human dwellings are heterogeneous.
Fig. 3.
Indoor (cage) oviposition assays. In these two-choice assays gravid female Culex mosquitoes laid significantly more eggs in trays baited with (A) 4-methylphenol and (B) 4-ethylphenol than in control trays at the doses of 1 (P<0.01) and 10 µg/l (P<0.05) and 0.1 (P<0.05), 1 (P<0.01) and 10 µg/l (P<0.05), respectively. There were no significant differences in the number of eggs laid in trays loaded with a higher dose (100 µg/l) and their respective controls. n=20 (with 40 gravid females per trial).
Fig. 4.
Field data comparing captures of female Cx. quinquefasciatus. Catches in gravid traps baited with 0.1, 1 and 10 µg/l of 4-ethylphenol were significantly higher than in their respective control traps. n=13–15 repetitions per dose.
Secondly, there are limited physical spaces in single residences that restrict the number of traps deployed per residence. Thus, it may be misleading to compare trap captures if these traps are deployed in different residences and/or at different times, i.e., tested under different conditions. To be able to compare different doses directly and considering the limitations for intertrap distance, we omitted control traps and ran triplicate experiments with traps baited with 0.1, 1 and 10 µg/l of 4-ethylphenol being rotated over the duration of the experiment. Although there were smaller captures in traps baited with 0.1 µg/l of 4-ethylphenol (5.7±0.98 Culex female per trap/night) than 1 µg/l (8±0.73) and 10 µg/l (8±0.9), these differences were not statistically significant (Tukey honestly significant difference at 5%).
3.4 Gene silencing
That 4-ethylphenol and 4-methylphenol activated CquiOR99 and CquiOR37 and elicited oviposition behavior, prompted us to attempt to silence these OR genes and test the hypothesis that they are directly involved in the reception of these oviposition attractants. Given that no OR genes have been successfully silenced in insect, except for the co-receptor Orco (e.g: (Howlett et al., 2012; Zhao et al., 2011)) and a larval OR gene from Anopheles gambiae (Liu et al., 2010), we started a systematic approach to knock down CquiOR37 and CquiOR99 in adult mosquitoes. It has been suggested that failures to silence insect genes might be related to rapid degradation of dsRNA in insect hemolymph, as shown for Manduca sexta (Garbutt et al., 2013). Thus, we first performed an ex vivo experiments to determine if CquiOR-dsRNA is degraded in Culex hemolymph. As expression of adult mosquito olfactory genes starts in the pupal stage (Xia and Zwiebel, 2006), we surmised that pupae would be the most suitable stage for treatment. Our ex vivo experiments showed that dsRNA is stable in pupal hemolymph, with a residence time of at least 3 h (Fig. 5). These results were encouraging as we expect the residence time for a dsRNA in the hemolymph (before reaching the target olfactory cells) to be much shorter than 3 h. This assumption is supported by the previous success in knocking down odorant-binding proteins from Cx. quinquefasciatus (Pelletier et al., 2010a) and the malaria mosquito Anopheles gambiae (Biessmann et al., 2010). Given that CquiOR37 and CquiOR99 share sequence identity as high as 62.5% at nucleotide level, we chose 400nt sequence from CquiOr37 as dsRNA template to construct the dsRNA, which we named CquiOR37/99-dsRNA. First, we blasted the DNA template in VectorBase and found no other significant match than CquiOR37 (E-value, 0), CquiOR99 (E-value, 4e−23) and CquiOR38 (E-value, 2 e−7). We then attempted to clone CquiOR38 using multiple conditions and were unsuccessful, probably because this gene is not expressed in antennae or may be a pseudogene.
Fig. 5.
Ex vivo determination of dsRNA stability. RT-PCR data showing that there is no significant degradation of CquiOR37/99-dsRNA when incubated with Culex mosquito hemolymph at room temperature as compared to incubation in DEPC-treated water up to 3 h.
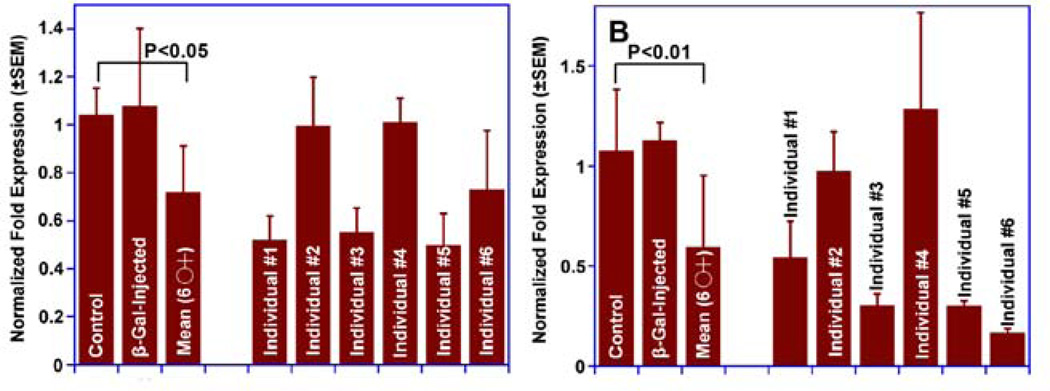
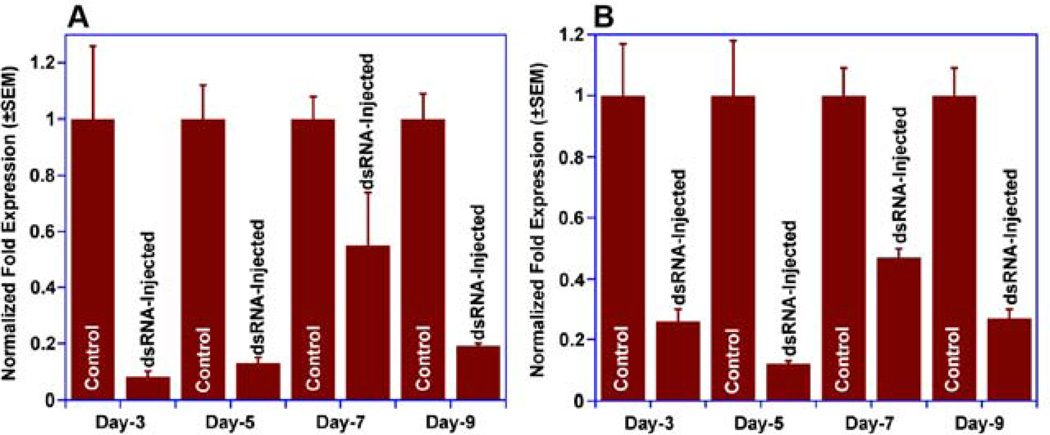
Having determined that there are no off-target genes in Culex genome, we next evaluated the effect of injection of CquiOR37/99-dsRNA in pupae on the level of transcripts of CquiOR37 and CquiOR99 by sampling antennae of individual day-9 adults. As previously observed with CquiOBP1 transcript levels (Pelletier et al., 2010a), even though there was considerable individual variation in the level of transcripts of OR genes (Fig. 6), there was significant reduction on average of levels of CquiOR37 (Fig. 6A) and CquiOR99 transcripts (Fig. 6B) when compared with water-treated and β-galactosidase controls. To minimize these individual variations and considering that a larger sample of gravid female mosquitoes is needed to test the phenotypes, hereafter we utilized a large pool (40 antennae per treatment), starting with the analysis of the time course of OR level of transcripts from day-3 to day-9. qPCR experiments showed that levels of CquiOR37 (Fig. 7A) and CquiOR99 transcripts (Fig. 7B) were significantly reduced from day-3 to day-9. These results show that by the time gravid females are tested in cage bioassays (day-9, i.e., 5-days after the first blood meal) level of transcripts of both OR genes were still low. Such long-lasting gene silencing in Culex has also been observed with transcripts of a heat-shock protein 9-day after larval uptake of dsRNA (Lopez-Martinez et al., 2012).
Fig. 6.
qPCR data showing individual variation in RNAi treatment. Although there was no difference between water- and β-galactosidase dsRNA-injected mosquitoes, in average there was significant reduction in (A) CquiOR37 and (B) CquiOR99 transcript levels in 9-day-old gravid female mosquitoes. However, in both cases there were significant individual variations. n=3 (a pair of antennae per individual, 6 mosquitoes per treatment, three replicates per quantitative analysis).
Fig. 7.
Time dependence of RNAi treatment as observed with a large pool of mosquito antennae. (A) CquiOR37 and (B) CquiOR99 transcript levels in day-3, 5, 7, and 9 gravid female mosquitoes treated with dsRNA and water-injected controls. n=3 (40 antennae per age and per trial). In all treatments the levels of transcripts were significantly lower (P<0.01) than in the respective controls.
Lastly, we treated mosquitoes in groups of 100 pupae to evaluate the effect of CquiOR37/99-dsRNA on the oviposition behavior as compared to two controls. Seventy-six to 82% of the female pupae injected at day 0 with CquiOR37/99-dsRNA, β-galactosidase-dsRNA, and water emerged as adults 1 day later. Equal numbers of males were added to mosquito cages, and three blood meals were provided at days 4, 5, and 6. Forty-two to 47% of the treated mosquitoes survived to day 9. Their oviposition behavior was evaluated comparatively by using 4-ethylphenol as lure at the lowest active dose (0.1 µg/l) (Fig. 3B), with 4 replicates and using 10 gravid female mosquito per cage. Oviposition by β-galactosidase-treated and water-injected mosquitoes were significantly higher in trays baited with 4-ethylphenol than in control trays (Fig. 8A and B). By contrast, there was no significant difference in the number of eggs laid in control and 4-ethylphenol trays by CquiOR37/99-dsRNA-treated mosquitoes (Fig. 8C). These results demonstrate that these receptors are required for the reception of 4-ethylphenol. Additionally, the number of eggs laid by these phenotypes suggests that dsRNA-treatment did not interfere with egg laying behavior, except for the ability to identify the 4-ethylphenol treated trays.
Fig. 8.
Indoor oviposition assay with RNAi treated mosquitoes. (A) Water-injected mosquitoes, (B) β-galactosidase dsRNA-injected, and (C) CquiOR37/99-ds RNA-injected. Oviposition in traps baited with 0.1 µg/l of 4-ethylphenol was significantly higher in treatment with (A) water-and (B) β-galactosidase dsRNA-treated mosquitoes, but (C) CquiOR37/99-dsRNA-treated mosquitoes showed no significant preference for treatment or control. n=8 (10 gravid female per cage).
4. Conclusions
We demonstrated that two ORs in the genome of Cx. quinquefasciatus, CquiOR37 and CquiOR99, are narrowly tuned to two oviposition attractants, 4-methylphenol and 4-ethylphenol. Field tests corroborate that 4-ethylphenol has a wide range of active doses and, consequently, potential practical application as a lure for trapping Culex mosquitoes. Injection of dsRNA in pupae led to significant reduction of CquiOR37 and CquiOR99 transcript levels in adult females at the peak of egg-laying activity, i.e., 9-day after the first blood meal. Indoor behavioral assay showed that water-injected and β-galactosidase-treated controls preferred 4-ethylphenol-treated water for egg-laying, whereas the phenotype with reduced CquiOR37 and CquiOR99 transcript levels laid eggs indiscriminately, thus, suggesting that these receptors are required for the reception of 4-ethylphenol in female Culex mosquitoes.
HIGHLIGHTS.
Identification and Characterization of Culex Odorant Receptors Narrowly Tuned to Phenolic Compounds
Evaluation of Oviposition Attractants by Indoor Assays and Field Tests
Silencing of Odorant Receptor Genes and Behavioral Evaluation of the Phenotype
Acknowledgments
We thank Dr. Anthony Cornel (Kearney Agricultural Center, University of California) for providing mosquitoes that allowed us to duplicate his colony at the Davis campus. Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R01AI095514. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH. FZ sabbatical leave at UC Davis was supported in part by the China Scholarship Council.
Abbreviations
- MOP
mosquito oviposition pheromone
- OR
odorant receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arakawa T. Phenylthiourea, an effective inhibitor of the insect hemolymph melanization reaction, interferes with the detection of lipoprotein hydroperoxide. Japanese Journal of Applied Entomology and Zoology. 1995;30:443–449. [Google Scholar]
- Arensburger P, Megy K, Waterhouse RM, Abrudan J, Amedeo P, Antelo B, Bartholomay L, Bidwell S, Caler E, Camara F, Campbell CL, Campbell KS, Casola C, Castro MT, Chandramouliswaran I, Chapman SB, Christley S, Costas J, Eisenstadt E, Feschotte C, Fraser-Liggett C, Guigo R, Haas B, Hammond M, Hansson BS, Hemingway J, Hill SR, Howarth C, Ignell R, Kennedy RC, Kodira CD, Lobo NF, Mao C, Mayhew G, Michel K, Mori A, Liu N, Naveira H, Nene V, Nguyen N, Pearson MD, Pritham EJ, Puiu D, Qi Y, Ranson H, Ribeiro JM, Roberston HM, Severson DW, Shumway M, Stanke M, Strausberg RL, Sun C, Sutton G, Tu ZJ, Tubio JM, Unger MF, Vanlandingham DL, Vilella AJ, White O, White JR, Wondji CS, Wortman J, Zdobnov EM, Birren B, Christensen BM, Collins FH, Cornel A, Dimopoulos G, Hannick LI, Higgs S, Lanzaro GC, Lawson D, Lee NH, Muskavitch MA, Raikhel AS, Atkinson PW. Sequencing of Culex quinquefasciatus establishes a platform for mosquito comparative genomics. Science. 2010;330:86–88. doi: 10.1126/science.1191864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biessmann H, Andronopoulou E, Biessmann MR, Douris V, Dimitratos SD, Eliopoulos E, Guerin PM, Iatrou K, Justice RW, Krober T, Marinotti O, Tsitoura P, Woods DF, Walter MF. The Anopheles gambiae odorant binding protein 1 (AgamOBP1) mediates indole recognition in the antennae of female mosquitoes. PloS one. 2010;5:e9471. doi: 10.1371/journal.pone.0009471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell A, Mordue AJ, Hansson BS, Wadhams LJ, Pickett JA. A behavioral and electrophysiological study of oviposition cues for Culex quinquefasciatus. Physiol Entomol. 1993;18:343–348. [Google Scholar]
- Braks MA, Leal WS, Carde RT. Oviposition responses of gravid female Culex quinquefasciatus to egg rafts and low doses of oviposition pheromone under semifield conditions. Journal of chemical ecology. 2007;33:567–578. doi: 10.1007/s10886-006-9223-8. [DOI] [PubMed] [Google Scholar]
- Carey AF, Wang G, Su CY, Zwiebel LJ, Carlson JR. Odorant reception in the malaria mosquito Anopheles gambiae. Nature. 2010;464:66–71. doi: 10.1038/nature08834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbutt JS, Belles X, Richards EH, Reynolds SE. Persistence of double-stranded RNA in insect hemolymph as a potential determiner of RNA interference success: Evidence from Manduca sexta and Blattella germanica. Journal of insect physiology. 2013;59:171–178. doi: 10.1016/j.jinsphys.2012.05.013. [DOI] [PubMed] [Google Scholar]
- Howlett N, Dauber KL, Shukla A, Morton B, Glendinning JI, Brent E, Gleason C, Islam F, Izquierdo D, Sanghavi S, Afroz A, Aslam A, Barbaro M, Blutstein R, Borovka M, Desire B, Elikhis A, Fan Q, Hoffman K, Huang A, Keefe D, Lopatin S, Miller S, Patel P, Rizzini D, Robinson A, Rokins K, Turlik A, Mansfield JH. Identification of chemosensory receptor genes in Manduca sexta and knockdown by RNA interference. BMC genomics. 2012;13:211. doi: 10.1186/1471-2164-13-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes DT, Pelletier J, Luetje CW, Leal WS. Odorant receptor from the southern house mosquito narrowly tuned to the oviposition attractant skatole. Journal of chemical ecology. 2010;36:797–800. doi: 10.1007/s10886-010-9828-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurence BR, Pickett JA. erythro-6-Acetoxy-5-hexadecanolide, the major component of a mosquito attractant pheromone. Journal of the Chemical Society, Chemical Communications. 1982a:59–60. [Google Scholar]
- Laurence BR, Pickett JA. erythro-6-Acetoxy-5-hexadecanolide, the major compound of a mosquito attractant pheromone. J. Chem. Soc., Chem. Commun. 1982b:59–60. [Google Scholar]
- Leal WS, Barbosa RM, Xu W, Ishida Y, Syed Z, Latte N, Chen AM, Morgan TI, Cornel AJ, Furtado A. Reverse and conventional chemical ecology approaches for the development of oviposition attractants for Culex mosquitoes. PloS one. 2008;3:e3045. doi: 10.1371/journal.pone.0003045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Seifert SN, Nieman CC, McAbee RD, Goodell P, Fryxell RT, Lanzaro GC, Cornel AJ. High degree of single nucleotide polymorphisms in California Culex pipiens (Diptera: Culicidae) sensu lato. Journal of medical entomology. 2012;49:299–306. doi: 10.1603/me11108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liman ER, Tytgat J, Hess P. Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron. 1992;9:861–871. doi: 10.1016/0896-6273(92)90239-a. [DOI] [PubMed] [Google Scholar]
- Liu C, Pitts RJ, Bohbot JD, Jones PL, Wang G, Zwiebel LJ. Distinct olfactory signaling mechanisms in the malaria vector mosquito Anopheles gambiae. PLoS biology. 2010;8 doi: 10.1371/journal.pbio.1000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Martinez G, Meuti M, Denlinger DL. Rehydration driven RNAi: a novel approach for effectively delivering dsRNA to mosquito larvae. Journal of medical entomology. 2012;49:215–218. doi: 10.1603/me11122. [DOI] [PubMed] [Google Scholar]
- Millar JG, Chaney JD, Mulla MS. Identification of oviposition attractants for Culex quinquefasciatus from fermented Bermuda grass infusions. Journal of the American Mosquito Control Association. 1992;8:11–17. [PubMed] [Google Scholar]
- Nasci RS, Miller BR. Culicine mosquitoes and the agents they transmit. In: Beaty BJ, Marquardt WC, editors. The biology of disease vectors. University Press of Colorado: Niwot; 1996. [Google Scholar]
- Pask GM, Romaine IM, Zwiebel LJ. The molecular receptive range of a lactone receptor in Anopheles gambiae. Chemical senses. 2013;38:19–25. doi: 10.1093/chemse/bjs074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Guidolin A, Syed Z, Cornel AJ, Leal WS. Knockdown of a mosquito odorant-binding protein involved in the sensitive detection of oviposition attractants. J Chem Ecol. 2010a;36:245–248. doi: 10.1007/s10886-010-9762-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Hughes DT, Luetje CW, Leal WS. An odorant receptor from the southern house mosquito Culex pipiens quinquefasciatus sensitive to oviposition attractants. PloS one. 2010b;5:e10090. doi: 10.1371/journal.pone.0010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Leal WS. Genome analysis and expression patterns of odorant-binding proteins from the Southern House mosquito Culex pipiens quinquefasciatus. PloS one. 2009;4:e6237. doi: 10.1371/journal.pone.0006237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed Z, Leal WS. Acute olfactory response of Culex mosquitoes to a human- and bird-derived attractant. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18803–18808. doi: 10.1073/pnas.0906932106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular biology and evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Carey AF, Carlson JR, Zwiebel LJ. Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:4418–4423. doi: 10.1073/pnas.0913392107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Zwiebel LJ. Identification and characterization of an odorant receptor from the West Nile virus mosquito, Culex quinquefasciatus. Insect biochemistry and molecular biology. 2006;36:169–176. doi: 10.1016/j.ibmb.2005.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Choo YM, Pelletier J, Sujimoto FR, Hughes DT, Atungulu E, Cornel JA, Luetje CW, Leal WS. Silent, generic, and plant kairomone sensitive odorant receptors from the Southern house mosquito. J. Insect Physiol. 2013 doi: 10.1016/j.jinsphys.2013.07.004. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Garczynski SF, Atungulu E, Syed Z, Choo YM, Vidal DM, Zitelli CH, Leal WS. Moth sex pheromone receptors and deceitful parapheromones. PloS one. 2012;7:e41653. doi: 10.1371/journal.pone.0041653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YY, Liu F, Yang G, You MS. PsOr1, a potential target for RNA interference-based pest management. Insect molecular biology. 2011;20:97–104. doi: 10.1111/j.1365-2583.2010.01049.x. [DOI] [PubMed] [Google Scholar]