Summary
Innate immune recognition of foreign nucleic acids induces protective interferon responses. Detection of cytosolic DNA triggers downstream immune signaling through activation of cyclic GMP–AMP synthase (cGAS). We report here the crystal structure of human cGAS, revealing an unanticipated zinc-ribbon DNA-binding domain appended to a core enzymatic nucleotidyl transferase scaffold. The catalytic core of cGAS is structurally homologous to the RNA sensing enzyme, 2′–5′ oligo-adenylate synthase (OAS), and divergent C-terminal domains account for specific ligand-activation requirements of each enzyme. We show that the cGAS zinc-ribbon is essential for STING-dependent induction of the interferon response, and conserved amino acids displayed within the intervening loops are required for efficient cytosolic DNA recognition. These results demonstrate that cGAS and OAS define a new family of innate immunity sensors, and that structural divergence from a core nucleotidyl transferase enables second-messenger responses to distinct foreign nucleic acids.
Introduction
The human innate immune system deploys cellular sensors to detect and respond to the presence of pathogens. Many of these sensors activate innate immunity by recognizing aberrant nucleic acid localization within the cell (Holm et al., 2013; Kagan, 2012; Medzhitov, 2007). Foreign RNA detection by toll-like receptors and RIG-I has been studied in some detail, but the mechanistic basis of DNA detection and signal initiation within the cytoplasm has remained enigmatic. Recently, the enzyme cyclic GMP–AMP synthase (cGAS) was identified as requisite for DNA detection, and cyclic GMP–AMP (cGAMP) was shown to function as a second messenger that stimulates innate immunity through the endoplasmic reticulum receptor STING (Sun et al., 2013; Wu et al., 2013). Identification of cGAS explains the potent immune response to cytosolic DNA and reveals a major source of ligands responsible for STING activation, but does not show how cGAS responds selectively to DNA and how it relates to other nucleic acid receptors.
Results and Discussion
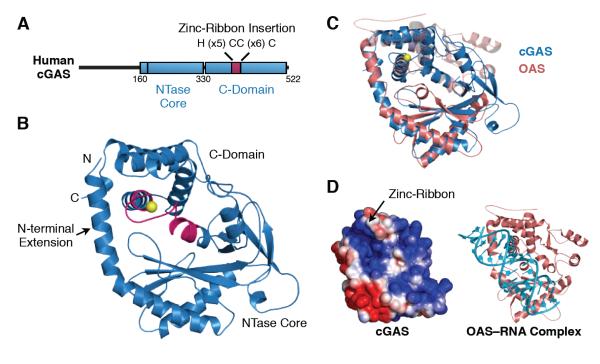
To investigate the mechanism and evolution of cytosolic DNA recognition, we determined the 2.5 Å crystal structure of human cGAS. Analysis of purified human cGAS by partial proteolytic digestion revealed a protease-sensitive ~150 amino-acid N-terminus attached to a protease-resistant fragment containing all regions previously determined to be required for cytosolic DNA detection (fig. S1) (Sun et al., 2013). A fluorescence scan of crystallized cGAS (amino acids 157–522) detected zinc, and a single bound zinc ion provided anomalous X-ray diffraction data sufficient for initial phase determination (table S1, fig. S2). Human cGAS adopts the overall fold of other template-independent nucleotidyl transferase (NTase) enzymes, including tRNA CCA-adding enzymes and the RNA sensor 2′–5′ oligo-adenylate synthase (OAS) (Donovan et al., 2013; Hartmann et al., 2003; Xiong and Steitz, 2004). Appended to the NTase core scaffold is an unanticipated zinc-ribbon domain resulting from a unique sequence insertion conserved in the carboxy-terminal domain (C-domain) of all vertebrate cGAS enzymes (Fig. 1A, B; fig. S3).
Fig. 1. Structure of Human cGAS.
(A) Cartoon schematic of the human cGAS primary sequence. (B) Overall structure of human cGAS with the N-terminal helical extension, NTase core scaffold, and carboxy-terminal domain (C-domain) shown in blue. A unique zinc-ribbon insertion is shown in magenta, and the zinc ion in yellow. (C) Structural overlay of cytosolic nucleic acid sensors, human cGAS (blue) and human OAS (pink). (D) Electrostatic surface potential of cGAS (left); a conserved positively charged nucleic-acid binding cleft equivalent to the OAS dsRNA-binding site as observed in the structure of an OAS–dsRNA complex (right, PDB code 4IG8). See also Figures S2 and S3.
The structure of cGAS reveals an evolutionary link with the human dsRNA sensor OAS. Upon recognition of cytosolic dsRNA, OAS produces the second messenger 2′–5′ oligo-adenylate (Hovanessian et al., 1977; Kerr and Brown, 1978), which triggers innate immunity by binding and activating RNase L and translation arrest (Baglioni et al., 1978; Hovanessian et al., 1979). In line with their roles as cytoplasmic sensors that signal the presence of foreign RNA and DNA through the production of second-messenger nucleic acids, OAS and cGAS contain an NTase core domain that is structurally conserved (Fig. 1C). In contrast to the catalytic domain, the more divergent C-domain is rotated in cGAS with respect to its orientation in the OAS structure, consistent with altered geometry enabling cGAS to accommodate dsDNA.
Adjacent to the conserved enzymatic scaffold of cGAS and OAS is a positively charged cleft at the interface between the N-terminal extension and C-domain alpha-helical lobe (Fig. 1C, D). When compared to the crystal structure of dsRNA-bound OAS (Donovan et al., 2013), the location of the positively charged cleft in cGAS suggests that OAS and cGAS likely use a similar binding surface to engage double-stranded nucleic-acid ligands (Fig. 1D). Insertion of the H(X5)CC(X6)C zinc-ribbon binding motif between residues 389 and 405 induces structural rearrangement of the cGAS C-domain, relative to OAS. The zinc coordination site buttresses a charged loop that alters the geometry of the positive binding cleft, consistent with the different nucleic acid binding specificity of cGAS and OAS enzymes, as discussed below.
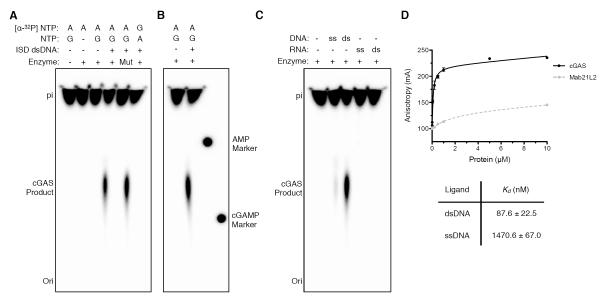
Previous studies, which relied on detection of cGAMP by mass spectrometry or indirect immune-stimulation assays requiring cellular extracts (Sun et al., 2013), did not analyze cGAS product species and activating conditions directly. Using purified components, we reconstituted DNA-dependent cyclic dinucleotide production by cGAS and analyzed the products using thin layer chromatography. Minimal cGAS activity requires GTP, ATP and an activating dsDNA ligand (Fig. 2A). cGAS dinucleotide synthesis activity is abolished by E225A and D227A mutations to the catalytic active site (Fig. 2A, “Mut”), confirming the specificity of our in vitro reconstitution system. Surprisingly, the cGAS GMP–AMP dinucleotide product migrates differently from chemically synthesized 3′–5′ linked cGAMP (Fig. 2B and fig. S4A). Concurrent experiments revealed that the cGAS product is a hybrid cyclic nucleotide containing a non-canonical 2′–5′ glycosidic linkage (Diner et al., 2013).
Fig. 2. In Vitro Reconstitution of cGAS Dinucleotide Signaling.
(A,B) Thin layer chromatography analysis of cGAS cyclic dinucleotide synthesis. Purified full-length cGAS was incubated with substrate nucleotides and immune-stimulatory DNA (ISD DNA) as indicated. An E225A/D227A mutation to the cGAS active site (Mut) ablates cyclic dinucleotide production. Dotted radioactive spots corresponding to UV-shadowed AMP and 3′–5′ linked cGAMP markers demonstrate the product of cGAS activity is a noncanonical dinucleotide product (see also fig. S4). (C) cGAS activity is strictly dependent on dsDNA activation. (D) Fluorescence anisotropy analysis of cGAS and related Mab21L2 NTase binding to dsDNA. See also Figure S4.
We observed robust cGAS catalytic activity only in the presence of dsDNA (Fig. 2C). While single-stranded DNA substrates weakly stimulate catalysis, we detected no dinucleotide synthesis in the presence of ssRNA or dsRNA ligands or in the absence of nucleic acids (Fig. 2C). Strict DNA-stimulated activity was not observed for murine cGAS (Sun et al., 2013), suggesting that the human variant has evolved more stringent ligand-activation requirements. The construct used for structural studies, lacking the poorly conserved ~150 amino-acid N-terminus, retains enzymatic activity and DNA selectivity, indicating all domains required for dsDNA detection and immune signaling are present in our crystal structure (fig. S4B). Fluorescence anisotropy experiments confirmed that cGAS specifically engages dsDNA (Kd ~87.6 nM), whereas a related human NTase Mab21L2 lacking the zinc-ribbon domain insertion cannot interact as robustly with DNA substrates (Fig. 2D). cGAS had a dramatically reduced affinity for ssDNA (Kd ~1.5 μM), in agreement with the inability of single-stranded nucleic acids to stimulate enzymatic activity (Fig. 2D). The affinity of cGAS for dsDNA decreases for dsDNA ligands shorter than two helical turns (fig. S4C). This finding is consistent with previous results demonstrating that at least 20–30 base pairs of dsDNA are required for efficient stimulation of innate immunity (Ablasser et al., 2009; Karayel et al., 2009; Stetson and Medzhitov, 2006).
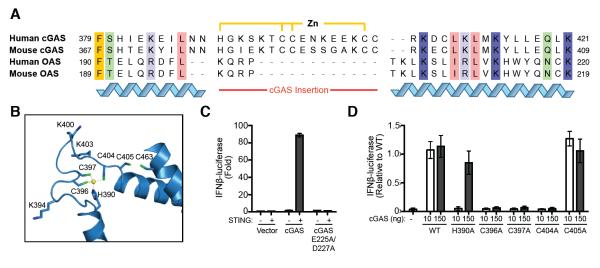
The zinc-ribbon structural domain is conserved among vertebrate cGAS members, but is not found in other OAS and related NTase family members (Fig. 3A). Zinc coordination in cGAS is by an atypical H(X5)CC(X6)C motif that resembles most closely HCCC-type zinc-ribbons found in TAZ domains (Laity et al., 2001). In the human cGAS structure, the first pair of adjacent cysteine residues (C396 and C397) each coordinate the zinc ion. In the second cysteine pair, C404 completes the coordination, and neighboring C405 flips out to form a paired cysteine interaction with downstream C463 from alpha helix 11 (Fig. 3A,B).
Fig. 3. cGAS Zinc-Ribbon Domain Is Essential for Interferon Signaling.
(A) Sequence alignment of human and murine cGAS and OAS cytosolic sensors. The unique cGAS zinc-ribbon insertion domain and coordinating residues are indicated. (B) Structural details of the zinc coordination site. Highly conserved amino acids are labeled. (C) Reconstitution of STING-dependent cGAS signaling in cells. Luciferase production under control of the interferon-β (IFNβ) promoter demonstrates that DNA-stimulated cGAS signaling only activates the interferon pathway in the presence of STING; cGAS E225A/D227A contains two point mutations in the enzymatic active site. (D) Mutational analysis of the cGAS zinc-ribbon motif. Substitutions to the zinc coordination motif (H390A, C396A, C397A, C404A) abolish cGAS activity when expressed at low levels (10 ng), and overexpression demonstrates only H390A retains weakened signaling potential (150 ng). Error bars represent the SD from the mean of at least three independent experiments.
To examine the role of zinc ion coordination and to extend our biochemical studies to cellular interferon signaling in the context of the endoplasmic reticulum-adaptor protein STING (Burdette and Vance, 2013; Ishikawa and Barber, 2008; Sun et al., 2009; Zhong et al., 2008), we tested the function of site-specific cGAS protein mutations in a cell-based assay. Using an interferon-β stimulated promoter cassette upstream of firefly luciferase, we first confirmed that intracellular cGAS signaling requires the presence of STING to confer second-messenger detection (Fig. 3C). As expected, a double mutation to the cGAS active site (E225A/D227A) that prevents cyclic dinucleotide synthesis abolished interferon signaling (Fig. 3C). Mutations at each position in the zinc coordination site near the DNA binding groove also ablated or severely impaired detectable interferon response activation (Fig. 3D), confirming that this motif is essential for innate immune signaling, but C405A mutation did not inhibit interferon signaling, indicating that the paired cysteine interaction with alpha helix 11 is not critical for cytosolic DNA recognition (Figure 3D).
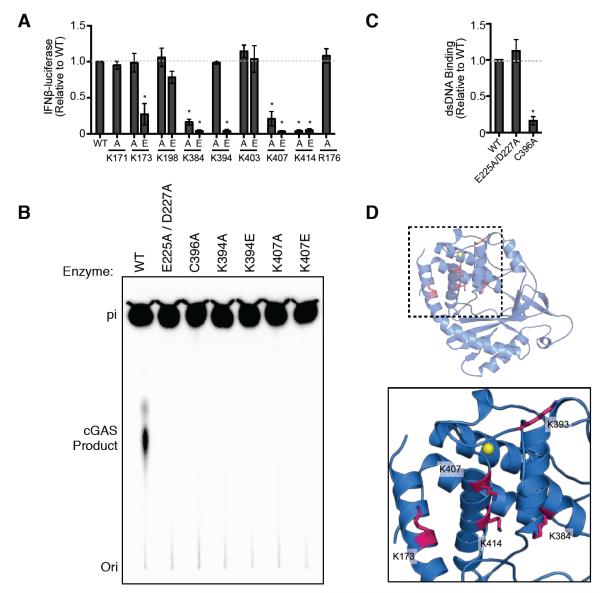
We also examined conserved positively charged positions along the potential DNA-binding cleft (fig. S3). Single alanine or glutamate substitutions along the N-terminal alpha helical extension and within the conserved zinc-ribbon loop dramatically reduced the ability of cGAS to detect cytosolic DNA, demonstrating the importance of conserved residues in this region of the protein (Figure 4A). Biochemical analysis with purified human cGAS confirmed mutations to the catalytic active site, zinc-coordination motif and conserved charged cleft ablates DNA-stimulated dinucleotide synthesis activity (Figure 4B). While catalytic active site mutants retained affinity for dsDNA, a C396A mutation disrupting the zinc-coordination motif prevented dsDNA interactions (Figure 4C). These data show that dsDNA engagement is critical for activation of the enzymatic potential of cGAS, and that conserved, positively charged aminoacids along with the unique zinc-ribbon insertion are essential for DNA recognition (Figure 4D). We note that the presence of the unstructured N-terminal tail greatly enhanced the stability of cGAS protein during purification (data not shown), hinting that the N-terminus may play a further role in stabilization or auto-inhibition as observed with other innate immune cellular receptors (Sun et al., 2013).
Fig. 4. cGAS Zinc-Ribbon and Positive DNA-binding Cleft Are Essential for DNA Recognition and Catalytic Activity.
(A) Reconstitution of STING-dependent cGAS signaling in cells as described in Figure 3. Single alanine and glutamine mutations to conserved positively charged amino acids within the DNA-binding cleft demonstrate K173, K384, K407 and K414 are required for efficient cytosolic DNA detection. (B) In vitro reconstitution of cGAS dinucleotide synthesis using purified components as described in Figure 2 (*P < 0.001). Mutations to the catalytic active site (E225A/D227A), zinc-coordination motif (C396A) and conserved DNA-binding cleft (K394A, K394E, K407A, K407E) all abolish DNA-stimulated enzymatic activity. (C) The ability of mutant cGAS enzymes to engage a 45 bp ISD dsDNA substrate was measured by fluorescence polarization using 2 μM of purified protein as described in Figure 2D (*P < 0.001). Mutations to the catalytic active (E225A/D227A site do no disrupt DNA interactions while disruption of the zinc-coordination motif drastically inhibits the ability of cGAS to interact with dsDNA. Error bars represent the SD from the mean of at least three independent experiments. See also Figure S1C.
Concurrent with our structural analysis of the human cGAS enzyme, Patel and colleagues determined the structure of murine cGAS bound to dsDNA and product dinucleotide (Gao et al., 2013). The overall sequence identity of human and murine cGAS is ~55% (fig. S3), and structures of both enzymes now reveal that rapid mammalian evolution has occurred in patches along the surface of the enzyme, indicating of positive selection and host-pathogen conflict (Daugherty and Malik, 2012). The critical role of cGAS in innate immunity and cytosolic DNA detection (Sun et al., 2013), suggests that the mechanisms by which intracellular dsDNA pathogens subvert cGAS-dependent DNA recognition may help detect regulation of cGAS enzymatic activity and cytosolic signaling.
Recent discovery of cGAS as a cytosolic DNA sensor is an important advance in the field of innate immunity (Sun et al., 2013), and the structure described here presents essential molecular details of cGAS biochemistry. The structure of human cGAS, revealing the similar folds of cGAS and OAS, implicates a common evolutionary ancestor as the origin of a family of structurally related but functionally distinct cytosolic nucleic acid sensors. Although multiple duplications of the OAS genes had been considered to be an outlier grouping of restriction factors, it is now clear that the OAS/cGAS NTase scaffold has evolved as part of a second messenger system to rapidly generate and amplify di- and oligo-nucleotide signals upon pathogen recognition. cGAS and OAS constitute a new family of catalytic OAS-like second-messenger receptors (OLRs), which together with Toll-like receptors (TLRs) and RIG-I-like receptors (RLRs) comprise the front line of immune defense against foreign pathogens.
Experimental Procedures
Protein Purification
Full-length human cGAS and cGAS truncations were PCR-amplified from a previously described interferon-stimulated gene cDNA library (kind gift from J. Schoggins and C. Rice, Rockefeller Univ.; (Schoggins et al., 2011) and cloned into a custom pET vector optimized for E. coli expression of an N-terminal 6xHis-MBP-TEV fusion protein (Kranzusch and Whelan, 2011). Proteins were over-expressed at 16°C in BL21-RIL DE3 E. coli (along with pRARE2 human tRNA plasmid) (Agilent) grown in 2xYT media for 20 h following induction with 0.5 M IPTG. Recombinant protein was purified by successive Ni-NTA affinity, Heparin ion-exchange, and Superdex 75 chromatography steps. Cells were lysed by sonication in 20 mM HEPES pH 7.5, 400 mM NaCl, 10% glycerol, 30 mM imidazole, 1 mM PMSF (supplemented with Complete Protease Inhibitor, Roche), 1 mM TCEP. Clarified lysate was bound to Ni-NTA agarose (Qiagen), and resin was washed with lysis buffer supplemented to 1 M NaCl prior to elution of bound protein using lysis buffer supplemented to 300 mM imidazole. MBP-tagged proteins were concentrated to ~30–40 mg mL−1 and digested with Tobacco Etch Virus protease for ~16 h at 4°C. cGAS was separated from MBP on a 5 mL Heparin HiTrap column (GE Life Sciences) using a linear gradient of 250–1000 mM NaCl. Proteins were further purified by size exclusion chromatography on a Superdex 75 16/60 column in 20 mM HEPES pH 7.5, 150 mM KCl and 1 mM TCEP. Eluted protein was concentrated to ~10–20 mg mL−1 and used immediately in crystallography experiments or flash-frozen in the presence of 10% glycerol in liquid nitrogen and stored at −80°C for biochemical experiments.
Mutant cGAS variants were purified as described for the wildtype human enzyme except, instead of TEV-digestion, MBP-tagged proteins were dialyzed overnight at 4°C against buffer containing 20 mM HEPES pH 7.5, 150 mM KCl, 10% glycerol and 1 mM TCEP. Wiltdype and mutant MBP-tagged cGAS enzymes were concentrated to ~10–12 mg mL−1, flash-frozen in liquid nitrogen and stored at −80°C for biochemical experiments.
Crystallization and Structure Determination
Full-length cGAS protein was digested with increasing amounts of trypsin at 25°C for 30 min to identify stable constructs for crystallography trials. Trypsin reactions were terminated by addition of 1 mM PMSF for SDS-PAGE analysis or an equal volume of 6 M guanidine hydrochloride for mass-spectrometry. A human cGAS 157–522 amino acid construct was designed based on mass spectrometry results and phylogenetic alignment, and the cGAS truncation was purified as described above. Initial crystals of cGAS 157–522 were obtained at 18°C in 1:1 hanging drops set with 10 mg mL−1 protein and 50 mM KCl, 10 mM MgCl2, 15% PEG-6000 well solution after 36 h of growth, and subsequently optimized in 15-well hanging drop trays (Qiagen) using 1.5:0.5 drops with 8 mg mL−1 protein and 44 mM KCl, 10 mM MgCl2, 25 mM Tris pH 7.0, 15 mM Tris pH 9.0, 6.9% PEG-6000. Crystals were harvested with nylon loops and cryo-protected by incubation in well solution supplemented to 25% ethylene glycol for 30–60 s prior to flash-freezing in liquid nitrogen. Initial native X-ray data were measured under cryo-conditions at the Lawrence Berkeley National Laboratory Advanced Light Source (Beamline 8.3.1) and zinc anomalous data were measured at the Stanford Synchrotron Radiation Lightsource (Beamlines 11.1 and 12.2). Selenium-substituted cGAS 157–522 was purified using identical conditions, and crystals of this sample grew in 44 mM KCl, 10 mM MgCl2, 25 mM Tris pH 7.0, 15 mM Tris pH 9.0, 9.7% PEG-6000. These crystals were optimized using micro- and streak-seeding of crushed native crystals using a Kozak whisker. X-ray diffraction data from selenium-containing crystals were measured at the Stanford Synchrotron Radiation Lightsource (Beamline 12.2).
X-ray diffraction data were processed using XDS and SCALA (Kabsch, 2010). Indexed crystals belonged to the orthorhombic spacegroup P21212 with 1 copy of cGAS in the asymmetric unit. The zinc site was identified using HySS / PHENIX (Adams et al., 2010) and SOLVE / RESOLVE was used to calculate an initial map (Terwilliger, 1999). Following initial model building in Coot (Emsley and Cowtan, 2004), iterative rounds of model building and refinement using PHENIX were conducted until all interpretable electron density was modeled. Using selenium anomalous data, the five selenium sites were located by molecular-replacement phasing and used to verify the register and position of the cGAS model.
In vitro Reconstitution of cGAS Cyclic Dinucleotide Synthesis
DNA-dependent human cGAS cyclic dinucleotide synthesis was reconstituted using recombinant full-length cGAS and a 45 bp double-stranded immune-stimulatory (ISD) DNA (Integrated DNA Technologies) (Stetson and Medzhitov, 2006). cGAS (final concentration ~2 μM) or equal volumes of gel-filtration buffer were incubated with ISD dsDNA (final concentration ~2 μM) in the presence of 25 μM ATP and GTP and [α-32P]-labeled ATP or GTP (~10 μCi) as indicated. All reactions included 50 mM KCl, 5 mM Mg(OAc)2, 50 mM Tris pH 7.0, 1 mM TCEP, 0.1 mg mL−1 BSA (NEB), and reactions were incubated at 37°C for 1.5 h. Reactions were terminated with the addition of 5 units of alkaline phosphatase (New England Biolabs) and incubation at 37°C for 30 min. 1 μl of each reaction was spotted onto a PEI-cellulose F thin-layer chromatography plate (EMD-Biosciences), and reaction products were separated using 1.5 M KH2PO4 (pH 3.8) as solvent. Plates were dried at 80°C for 30 min, and radiolabeled products were detected with a phosphor-screen and Storm phosphorimager (GE Life Sciences). Where indicated, controls consisting of chemically synthesized AMP (Jena Biosciences) or 3′–5′ linked cGAMP (kind gift from S. Wilson and M. Hammond, Univ. of California, Berkeley) were imaged using a ~254 nm light for UV-shadowing and marked by spotting a dot of radiolabeled ATP prior to phosphor-screen exposure. Alternatively, reactions were carried out in the presence of 45 bp single-stranded ISD DNA (sequence: 5′ TACAGATCTACTAGTGATCTATGACTGATCTGTACATGATCTACA 3′) (Stetson and Medzhitov, 2006), or single-stranded and double-stranded RNA formed by annealing two chemically synthesized RNA oligomers (sequence: 5′ CGGUAGAGCUCACAUGAUGG 3′) (Integrated DNA Technologies).
Fluorescence anisotropy DNA interaction studies were carried out using 5′ fluorescein derived DNA oligomers with the ISD DNA sequence and indicated sizes (Integrated DNA Technologies). Using the same buffer conditions as during cyclic dinucleotide synthesis reactions, cGAS was incubated with DNA for 30 min at 25°C prior to fluorescence polarization measurements using a fluorimeter (Perkin Elmer). Polarization data were converted to anisotropy, and data from independent experiments were combined and analyzed with GraphPad Prism software to determine binding constants. As indicated, statistical significance was calculated using an unpaired, two-tailed t test.
Cell-based IFN β-Luciferase Assay
293T cells were plated into tissue-culture treated 96-well plates for transfection. Cells were transfected as indicated, along with IFNβ-firefly luciferase (a kind gift from J.U. Jung, University of Southern California, Los Angeles) and TK promoter-Renilla luciferase reporter plasmids. At 24 h post transfection, cells were lysed in passive lysis buffer (Promega) for 15 min. Luminescence was measured on a Veritas Microplate Luminometer (Turner Biosystems) using the Dual-Luciferase Reporter Assay System according to manufacturer’s instructions (Promega). The relative IFN-β expression was calculated by normalizing firefly luciferase to Renilla luciferase activity. Mutations in cGAS were generated by site-directed mutagenesis using the QuikChange methodology (Stratagene). As indicated, statistical significance was calculated using an unpaired, two-tailed t test.
Supplementary Material
Highlights.
· Crystal structure of the human cytosolic DNA sensor
· cGAS and OAS comprise a new family of innate immune receptors
· Unique zinc-ribbon motif insertion within cGAS confers DNA specificity
Acknowledgements
All structural and biochemical work was performed by P.J.K. with supervision by J.M.B. and J.A.D. In cell signaling assays were performed by A.S.Y.L. The manuscript was written by P.J.K. and J.A.D., and all authors contributed to editing the manuscript and support the conclusions. The authors are grateful to the staff at beamlines 11.1 and 12.2 of the Stanford Synchrotron Radiation Lightsource (SSRL) and beamline 8.3.1 of the Lawrence Berkeley National Lab Advanced Light Source for technical assistance; T. Doukov (Stanford, SSRL) for assistance with data collection and analysis; M. Jinek (Univ. of Zurich), A. Lyubimov, and J. Cate (Univ. of California, Berkeley) for advice on phase determination and model building; D. King (HHMI, Univ. of California, Berkeley) for mass spectrometry; J. Schoggins and C. Rice (Rockefeller Univ.) for reagents; M. Solovykh for technical assistance; and Y. Bai, S. Floor and members of the Doudna and Berger labs for helpful comments and discussion. This work was funded by HHMI (J.A.D.), G. Harold and Leila Y. Mathers Foundation (J.M.B.) and the NIGMS Center for RNA Systems Biology (A.S.Y.L.). J.A.D. is an HHMI Investigator.
Footnotes
Accession Numbers Coordinates of human cGAS have been deposited in the RCSB Protein Data Bank (PDB: 4KM5).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baglioni C, Minks MA, Maroney PA. Interferon action may be mediated by activation of a nuclease by pppA2′p5′A2′p5′A. Nature. 1978;273:684–687. doi: 10.1038/273684a0. [DOI] [PubMed] [Google Scholar]
- Burdette DL, Vance RE. STING and the innate immune response to nucleic acids in the cytosol. Nat Immunol. 2013;14:19–26. doi: 10.1038/ni.2491. [DOI] [PubMed] [Google Scholar]
- Daugherty MD, Malik HS. Rules of engagement: molecular insights from host-virus arms races. Annu Rev Genet. 2012;46:677–700. doi: 10.1146/annurev-genet-110711-155522. [DOI] [PubMed] [Google Scholar]
- Diner EJ, Burdette DL, Monroe KM, Wilson SC, Canlas C, Lemmens EE, Lauer P, Dubensky TW, Hammon MC, Vance RE. The innate immune DNA sensor cGAS produces a non-canonical cyclic-di-nucleotide that activates human STING. 2013. Submitted Manuscript. [DOI] [PMC free article] [PubMed]
- Donovan J, Dufner M, Korennykh A. Structural basis for cytosolic double-stranded RNA surveillance by human oligoadenylate synthetase 1. Proc Natl Acad Sci U S A. 2013;110:1652–1657. doi: 10.1073/pnas.1218528110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, Serganov AA, Liu Y, Jones RA, Hartmann G, et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the Metazoan Second Messenger Produced by DNA-Activated Cyclic GMP-AMP Synthase. Cell. 2013 doi: 10.1016/j.cell.2013.04.046. http://dx.doi.org/10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann R, Justesen J, Sarkar SN, Sen GC, Yee VC. Crystal structure of the 2′-specific and double-stranded RNA-activated interferon-induced antiviral protein 2′-5′-oligoadenylate synthetase. Mol Cell. 2003;12:1173–1185. doi: 10.1016/s1097-2765(03)00433-7. [DOI] [PubMed] [Google Scholar]
- Holm CK, Paludan SR, Fitzgerald KA. DNA recognition in immunity and disease. Curr Opin Immunol. 2013;25:13–18. doi: 10.1016/j.coi.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovanessian AG, Brown RE, Kerr IM. Synthesis of low molecular weight inhibitor of protein synthesis with enzyme from interferon-treated cells. Nature. 1977;268:537–540. doi: 10.1038/268537a0. [DOI] [PubMed] [Google Scholar]
- Hovanessian AG, Wood J, Meurs E, Montagnier L. Increased nuclease activity in cells treated with pppA2′p5′A2′p5′ A. Proc Natl Acad Sci U S A. 1979;76:3261–3265. doi: 10.1073/pnas.76.7.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan JC. Signaling organelles of the innate immune system. Cell. 2012;151:1168–1178. doi: 10.1016/j.cell.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karayel E, Burckstummer T, Bilban M, Durnberger G, Weitzer S, Martinez J, Superti-Furga G. The TLR-independent DNA recognition pathway in murine macrophages: Ligand features and molecular signature. Eur J Immunol. 2009;39:1929–1936. doi: 10.1002/eji.200939344. [DOI] [PubMed] [Google Scholar]
- Kerr IM, Brown RE. pppA2′p5′A2′p5′A: an inhibitor of protein synthesis synthesized with an enzyme fraction from interferon-treated cells. Proc Natl Acad Sci U S A. 1978;75:256–260. doi: 10.1073/pnas.75.1.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzusch PJ, Whelan SP. Arenavirus Z protein controls viral RNA synthesis by locking a polymerase-promoter complex. Proc Natl Acad Sci U S A. 2011;108:19743–19748. doi: 10.1073/pnas.1112742108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laity JH, Lee BM, Wright PE. Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol. 2001;11:39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
- Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Li Y, Chen L, Chen H, You F, Zhou X, Zhou Y, Zhai Z, Chen D, Jiang Z. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci U S A. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC. Reciprocal-space solvent flattening. Acta Crystallogr D Biol Crystallogr. 1999;55:1863–1871. doi: 10.1107/S0907444999010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Steitz TA. Mechanism of transfer RNA maturation by CCA-adding enzyme without using an oligonucleotide template. Nature. 2004;430:640–645. doi: 10.1038/nature02711. [DOI] [PubMed] [Google Scholar]
- Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.