Abstract
Depletion of mitochondrial DNA (mtDNA) or treatment with mitochondrial poison CCCP initiates mitochondrial stress signaling, which operates through altered Ca2+ homeostasis. In C2C12 rhabdomyoblasts and A549 human lung carcinoma cells mitochondrial stress signaling activates calcineurin and a number of Ca2+ responsive factors including ATF, NFAT, CEBP/δ and CREB. Additionally, PKC and MAP kinase are also activated. A number of nuclear gene targets including those involved in Ca2+ storage/release (RyR1, calreticulin, calsequestrin), glucose metabolism (hexokinase, pyruvate kinase, Glut4), oncogenesis (TGFβ1, cathepsin L, IGFR1, melanoma antigen) and apoptosis (Bcl-2, Bid, Bad, p53) are upregulated. Mitochondrial stress in both C2C12 myoblasts and A549 cells induced morphological changes and invasive phenotypes. These cells also showed markedly increased resistance to etoposide-induced apoptosis that is a hallmark of highly invasive tumors. Our results describe a new mechanism of altered nuclear gene expression and phenotypic changes triggered by mitochondrial dysfunction and mtDNA damage.
Keywords: Mitochondrial transmembrane potential, Mitochondrial stress signaling, Cytosolic free Ca2+, Calcineurin, Transcription regulation
1. Introduction
Mitochondria-to-nucleus retrograde signaling analogous to the RTG signaling pathway described in Yeast (Parikh et al., 1987; Liao and Butow, 1991; Butow and Avadhani, 2004) has also been described in mammalian cells (Biswas et al., 1999; Amuthan et al., 2002). The retrograde signaling in mammalian cells, also known as mitochondrial stress signaling, was described initially in C2C12 skeletal myoblasts (rhabdomyoblasts) and later confirmed in human lung carcinoma A549 cells (Biswas et al., 1999; Amuthan et al., 2002). Mitochondrial stress was defined as altered mitochondrial membrane potential (ΔΨm), induced either by treating cells with ethidium bromide to partially deplete the mtDNA content, or with the mitochondria-specific ionophore CCCP (carbonyl cyanide m-chlorophenyl hydrazone). These treatments resulted in elevated cytosolic free Ca2+ ([Ca2+]c) and activation of Ca2+/calmodulin responsive calcineurin and their related factors. A similar retrograde signaling involving increased [Ca2+]c was also observed in rat pheochromocytoma cells treated with the mitochondrial uncoupler carbonyl cyanide p-(trifluoromethoxy) phenylhydrazone (FCCP) (Luo et al., 1997). Using ρ° human fibrosarcoma 143B cells and a MERRF (myoclonic epilepsy with ragged red fibers) cybrid cell line carrying the mutated mitochondrial tRNALys (A8344G), Arnould et al. (2002) showed that respiratory deficiency and an associated increase in cytosolic free Ca2+ induced the activation of CaMK IV, which in turn activated CREB by protein phosphorylation. In all of these cell systems, removal of free Ca2+ by specific chelators abrogated the activation of various Ca2+ responsive factors as also the stress signal induced transcription activation of nuclear target genes, confirming the role of Ca2+ in retrograde signaling.
It is known that induced accumulation of misfolded proteins causes a stress response pathway in the endoplasmic reticulum (Mori et al., 1993; Travers et al., 2000). Recent studies suggest the occurrence of an analogous stress response pathway in the mitochondrial compartment as well. The accumulation of a mutant form of misfolded ornithine transcarbamylase (OTC) in the mitochondrial matrix induced a stress response that activated CEBP homology protein, CHOP, and was associated with increased expression of nuclear gene-encoded stress response proteins, Cpn60, Cpn10, mtDNAJ, and ClpP (Zhao et al., 2002). The transcriptional activity of CHOP is modulated by its association with CEBP/β and ATF2 (Averous et al., 2004). Notably, ATF2 and some members of the CEBP family such as CEBP/δ are also activated by retrograde signaling initiated by disruption of ΔΨm (Biswas et al., 1999), suggesting some degree of similarity between the two mitochondrial stress signaling pathways.
In the present study, we have further elucidated the mechanism of mitochondrial stress signaling and determined the nuclear target genes responding to mitochondrial stress signaling in C2C12 myocytes. Additionally, we have studied the nature of factors responsible for the transcription upregulation of catepsin-l, a key target gene of mitochondrial retrograde (alternatively called as mitochondrial stress) signaling. Our results point to an unusual cooperativity between NFκB and CEBP factors in the mitochondrial stress-mediated activation of nuclear target genes.
2. Materials and methods
2.1. Generation of mtDNA-depleted cells and culture conditions
Murine C2C12 skeletal myoblasts (ATCC CRL1772) were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and 0.01% gentamicin. Depletion of mtDNA by ethidium bromide treatment and isolation of C2C12 clone 2 with 20% of control cell mtDNA content was as described before (Biswas et al., 1999). Reverted cells represent clone 2 grown for 17 division cycles in the absence of ethidium bromide and contain 70% of control cell mtDNA. To ensure steady mtDNA levels, aliquots of the same cell isolate were used in all experiments and the mtDNA contents were routinely assayed in each experiment.
2.2. Isolation of mitochondria and sub cellular fractions
Cells were homogenized in a medium containing 300 mM sucrose, 10 mM Tris–HCl (pH 7.5), 10 mM NaCl, 5 mM MgCl2, 1 mM NaVO4, 100 µM molybdic acid, 10 mm NaF, 100 mM EDTA, 1 mM phenylmethylsulfonyl fluoride and protease inhibitor mix (1 protease inhibitor cocktail tablet per 50 ml, Roche Molecular Biochemicals, Indianapolis, IN, USA) in a Dounce homogenizer and fractionated into nuclear, post-mitochondrial and mitochondrial fractions as described before (Biswas et al., 1999). Protein concentration was determined by using the method of Lowry et al. (1951).
2.3. Differential RNA display and Northern hybridization
Total RNA was isolated from the cultured cells using the “TRIZOL” reagent (Invitrogen). Differential display was carried out as described (Liang and Pardee, 1992) using total RNA. RNA (400 µg each) from control and mtDNA-depleted cells was reverse transcribed and the primer extension products were amplified by PCR using primer sets supplied as part of a kit from Vista Inc., CA, USA. The primer extension products showing different intensities between the control and mtDNA-depleted cells were cloned and sequenced. The DNA fragments were also used as probes for Northern blot analysis. Northern hybridization with 32P-labeled DNA probes was carried out under standard conditions (Schleicher & Schuell Laboratory Manual) using 30 µg of total RNA. Gel-purified, double-stranded DNA probes were labeled with 32P-dCTP (6000 Ci/mmol−1, Dupont, NEN) by random primer extension using the klenow polymerase. RNA loading was normalized by hybridizing the stripped blots with a 32P-labeled 18S DNA probe. The Northern blots were imaged and quantified using Bio-Rad GS-525 molecular imager.
2.4. TUNEL assay for measuring apoptosis
Both normal and mtDNA-depleted cells were grown on lysine-coated glass cover slips in 6-well plates. The cells were treated with 25 µM etoposide for 4 h to induce apoptosis. The extent of nuclear DNA breaks characteristic of cells undergoing apoptosis was measured by TUNNEL assay where cells were labeled with anti-Digoxigenin peroxidase using ApopTag Peroxidase, an in situ apoptosis detection kit from Intergen Company, as per manufacturer’s instructions. The cover slips were mounted on slides and examined under bright field microscopy using Olympus BX61 microscope.
2.5. Western blot analysis
Proteins (30 µg each) from each subcellular fraction were resolved on polyacrylamide gels and subjected to immunoblot analysis using respective antibodies (Biswas et al., 2003). Antibodies against BAD, Bax, Bid, Bcl-2, Bcl-XL, Caspases 3, 8 and 9 were purchased from Santa Cruz Biotechnology. The immunoblots were developed using the Pierce Super signal West Femto maximum sensitivity substrate kit, imaged and quantified in a Bio-Rad Fluor-S imaging system.
2.6. Cell transfection and assay of transcription activity
Cathepsin L promoter DNA (sequence −221 to +47) was amplified from the mouse genomic DNA based on the published sequence (Troen et al., 1991) and cloned in pGL3 vector (Promega, Madison, WI). Transfections were carried out in control C2C12 or mtDNA-depleted cells using Fugene 6 reagent (Roche Molecular Biochemicals, Indianapolis, IN) using the manufacturer’s suggested protocol. Promoter DNA construct (1 µg) and 0.5 µg of renila luciferase construct as an internal control were used in each transfection. The luciferase activity was assayed using Dual-Luciferase reporter assay system from Promega. Co-transfection with various cDNA constructs was carried out using 0.2 µg of expression construct.
3. Results and discussion
3.1. Nature of mitochondrial stress signaling
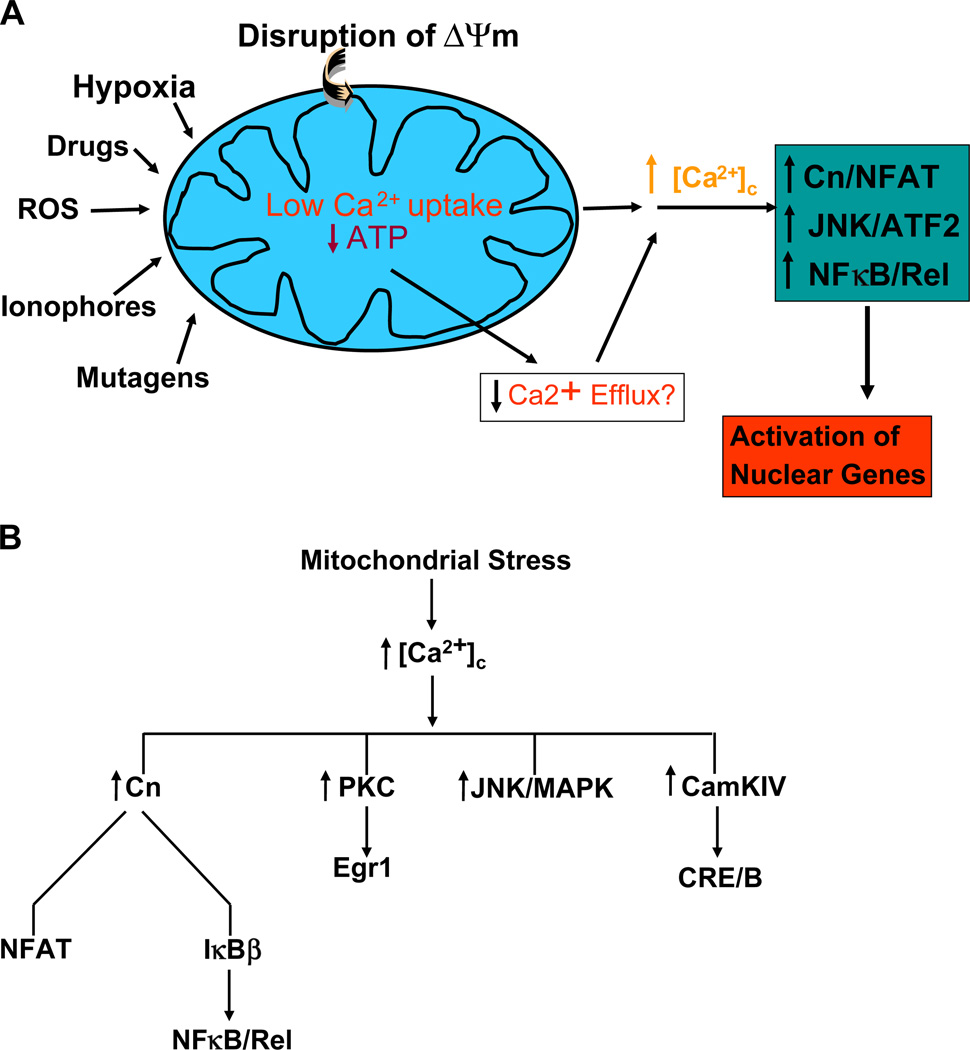
Mitochondria are important players in Ca2+ storage and homeostasis as they take up large amounts of the divalent cation transiently and release it to the cytoplasm as part of the intracellular Ca2+ traffic and signaling (Rizzuto et al., 1993; Babcock et al., 1997; Ichas et al., 1997). In previous studies, we showed that disruption of mitochondrial ΔΨm either by depletion of mtDNA or treatment with mitochondria-specific ionophore, CCCP (Biswas et al., 1999), in C2C12 myoblasts and A549 lung carcinoma cells (Amuthan et al., 2002) caused a steady and sustained elevation of cytosolic free Ca2+ ([Ca2+]c). We proposed that a combination of impaired mitochondrial uptake of Ca2+ and low rate of Ca2+ efflux because of limited ATP production are reasons for the observed increase in [Ca2+]c (see Fig. 1A).
Fig. 1.
Mitochondrial retrograde signaling in mammalian cells through altered Ca2+ levels and activation of Ca2+ responsive factors. (A) A proposed model for mitochondrial stress signaling caused by disruption of ΔΨm either by mtDNA depletion, or treatment with drugs/chemicals that affect mitochondrial function. An upward arrow indicates a net increase in concentration (such as free Ca2+), activation of factors or upregulation of transcription. A downward arrow indicates inactivation of factors or lower activity. (B) List of transcription factors and signaling proteins known to be activated by different forms of stress induced retrograde signaling.
A sustained three- to 8-fold increase of steady-state Ca2+ as part of retrograde signaling led to the activation of calcineurin, which in turn activated NFAT and NFκB. In addition, Ca2+-dependent PKC, JNK and MAPK are activated, which then activated ATF2, CEBP/δ, CREB, Egr-1 and CHOP in C2C12 myocytes and A549 cells. In ρ° fibrosarcoma cells, mitochondrial stress induced CamK IV and CREB, while mitochondrial stress induced by over-expression of misfolded protein induced MAPK and its responsive CHOP. These results are summarized in Fig. 1B.
3.2. Nuclear gene targets of mitochondrial stress signaling
The nuclear gene targets were identified by differential display of RNA from control and mtDNA-depleted C2C12 cells and A549 lung carcinoma cells. In some cases, differential expression of genes was also identified initially by Western blot analysis. In all cases, however, the differential expression was ascertained and quantified by Northern blot hybridization of RNA from control and mtDNA-depleted cells. In Table 1, results of northern blot hybridization have been expressed as fold mRNA increase or decrease in relation to control cells. It is seen that mitochondrial stress affects the expression of a wide spectrum of genes including those involved in Ca2+ storage and release (RyR1, RyR2, calreticulin, calsequestrin), glucose uptake and metabolism (Glut 4, IGF1R, hexokinase, phosphoenol pyruvate carboxykinase, IRS1), genes involved in oncogenesis (cathepsin L, mouse myeloma antigen, cJun, RelA, cRel, TGFβ1, p53, and cMyc), and apoptosis (Bcl-2, Survivin, BAD, Bax, Bid), and mitochondrial structure and function (COX Vb, COX IV, TOM40).
Table 1.
Altered expression of nuclear genes in mtDNA depleted C2C12 and A459 cells
| Genes identified | mRNA levels (fold of control cells) | |
|---|---|---|
| Depleted | Reverted | |
| RyR1 | 18.0 | 3.0 |
| Calsequestrin | 4.1 | 1.1 |
| Calreticulin | 2.5 | 1.2 |
| Cathepsin | 5.0 | 2.0 |
| TGFβ1 | 5.0 | 1.1 |
| Epiregulin | 6.5 | 1.0 |
| Mouse melanoma antigen | 16.0 | 2.5 |
| P53 | 9.3 | 2.5 |
| cMyc | 4.2 | 1.8 |
| PEP carboxykinase | 3.0 | 1.5 |
| Hexokinase | 4.0 | 1.0 |
| 2 novel genes with no homology to published sequence | 4.0–5.3 | 2.0–3.0 |
| Tim44 | 8.0 | – – |
| Tom40 | 5.2 | – – |
| VDAC | 6.0 | – – |
| Bcl2 | 4.3 | 1.4 |
| Bid | 3.0 | – – |
| Bax | 2.5 | – – |
| Insulin Receptor Substrate 1 | 0.5 | 0.7 |
| Glut4 | 5.3 | 1.6 |
| 6 MtDNA transcript | 0.2 | 0.7–0.9 |
Primer extension products showing increased or decreased RNA levels in mtDNA-depleted cells were amplified by PCR, cloned and sequenced to identify the genes. The DNA fragments were subsequently used as probes in Northern blot hybridization with RNA from control, mtDNA-depleted and reverted cells. The relative RNA levels have been expressed as fold of control cell level that was considered as 1.
3.3. Resistance to etoposide-mediated apoptosis in mtDNA-depleted cells
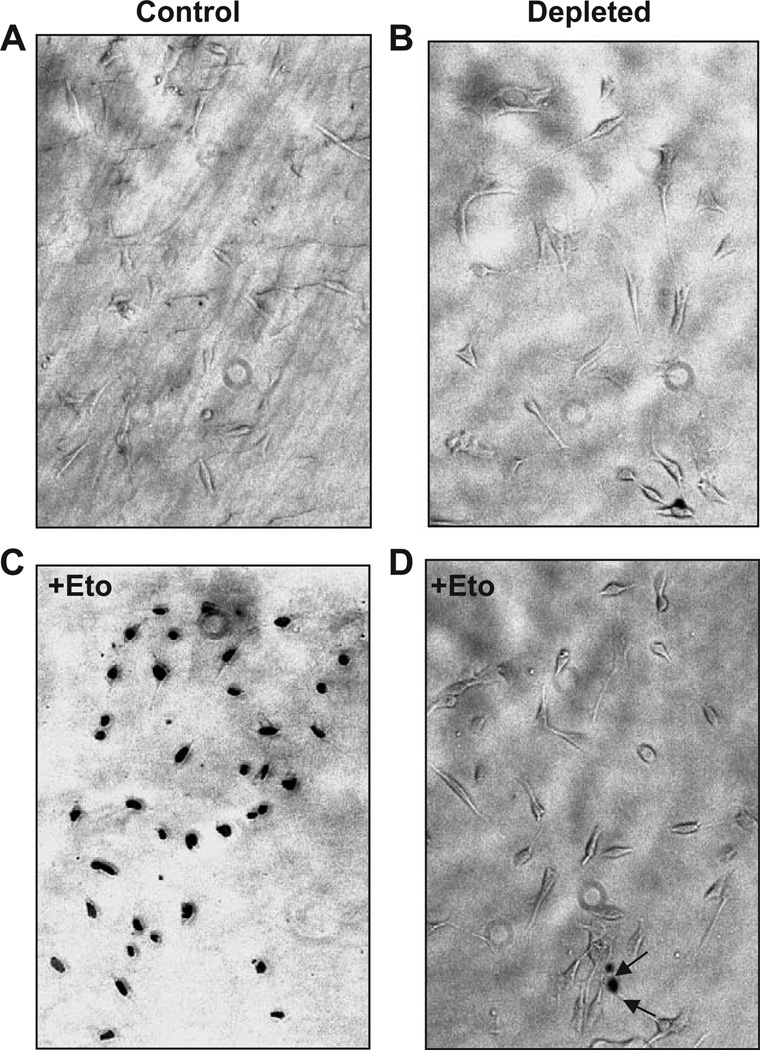
Consistent with the overexpression of a number of genes implicated in oncogenesis (Table 1), studies from our laboratory showed that both C2C12 and A549 cells subjected to mitochondrial stress showed vastly increased invasive behavior in both in vitro Matrigel and in vivo xenotransplantation systems. To understand the basis for altered cell morphology and increased invasive behavior of mtDNA-depleted cells, we investigated the sensitivity or resistance of these cells to etoposide-mediated apoptosis. It is seen from Fig. 2 that nearly 95% of control C2C12 cells underwent apoptosis in response to 25 µM etoposide. Apoptotic cells exhibit nuclear blebbing and intense nuclear staining characteristic of fragmented DNA (Chen et al., 1995). In the case of mtDNA-depleted cells, however, only 5–10% of cells were positive for TUNEL assay. These results show that mtDNA-depleted cells are significantly more resistant to etoposide-mediated apoptosis compared to normal cells. Although not shown, mtDNA-depleted A549 cells also showed relative resistance to apoptosis.
Fig. 2.
Resistance to etoposide-mediated apoptosis in mtDNA-depleted C2C12 cells. Control (A and C), and mtDNA-depleted (B and D) cells were subjected to apoptosis assay before (A and B) or after treatment with 25 µM etoposide for 4 h to induce apoptosis. The extent of nuclear DNA breaks was measured by TUNEL assay with anti Digoxigenin peroxidase ApopTag Peroxidase system as described in the Materials and methods section.
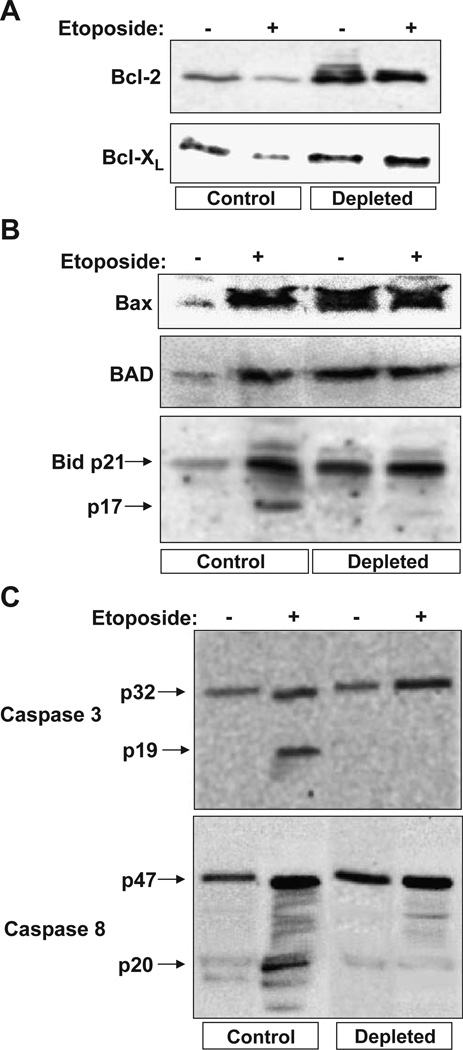
The mechanism of mitochondrial stress-mediated resistance to apoptosis was investigated by analyzing the relative levels of pro- and anti-apoptotic proteins in the mitochondrial and cytosolic fractions by western blot analysis. Fig. 3A shows that the levels of anti-apoptotic Bcl-2 was increased 3- to 4-fold in mtDNA-depleted cells, while the level of Bcl-XL was increased only marginally. It is also seen that etoposide reduced the levels of both anti-apoptotic proteins in control but was without effect in depleted cells. It is known that outer mitochondrial membrane associated Bcl-2 sequesters pro-apoptotic proteins BAD, Bax and Bid by heterodimerization thus preventing apoptosis (Tsujimoto, 1998; Hattori et al., 2000). In contrast to known mechanisms of resistance to apoptosis under various physiological and pathological conditions, mitochondria from cells subjected to mitochondrial stress also contain elevated levels of Bax, BAD and Bid, although these cells resist cytochrome c release in response to added etoposide (Fig. 2). As seen from Fig. 3B the mitochondrial contents of BAD, Bax and Bid were increased 2- to 3-fold in mtDNA-depleted cells. It is noteworthy that in mtDNA-depleted cells, most of the antibody cross-reactive bid was detected as unprocessed 21-kDa (p21) species as opposed to the presence of significant processed p17 (17 kDa), active tBid in etoposide-treated control cells (Fig. 3B). These results suggested the possibility that the cytoplasmic caspase 8, which is one of the early caspases that is involved in the processing of p21 Bid into active p17 tBid was absent or defective in mtDNA-depleted cells. It is known that caspase 8, which is one of the early caspases involved in the activation of downstream caspases, is affected in mtDNA-depleted cells. Consistent with this possibility, immunoblot in Fig. 3C shows that both caspases 3 and 8, whose combined action is required for the execution of the apoptotic signal, are detected in inactive (unprocessed) forms in mtDNA-depleted cells even after treatment with etoposide. Although not shown caspase 9 also remained in the unprocessed form in mtDNA-depleted cells. These results therefore suggest that despite an increased steady state level of pro-caspase 8, its inability to undergo processing in mtDNA-depleted cells may be the primary cause of etoposide-mediated resistance to apoptosis in mtDNA-depleted cells.
Fig. 3.
Steady-state levels of pro- and anti-apoptotic proteins and caspases in etoposide-treated control and mtDNA-depleted C2C12 cells. Cells treated with (25 µM for 4 h) or without etoposide were used for isolating the mitochondrial and cytosolic fractions as described in the Materials and methods section. 30 µg each of mitochondrial (A and B) and cytosolic (C) proteins was subjected to Western blot analysis using indicated antibodies. The immunoblot was developed using the Pierce Super signal West Femto substrate kit as described in the Materials and methods.
3.4. Transcriptional activation of nuclear target genes
Studies from our laboratory showed that a number of Ca2+ responsive factors including NFAT, ATF, CEBP/δ and NFκB are activated by mitochondrial stress (retrograde) signaling. We showed that NFκB activation by retrograde signaling involves a novel mechanism, distinct from the known cytokines, chemokines and other stress-mediated activation (Biswas et al., 2003). Under both genetic (mtDNA depletion) and chemical stress (CCCP treatment) conditions, cytosolic IκBβ levels are reduced by about 60–70% of control values, and nuclear cRel and p50 levels increase 2- to 3-fold, suggesting activation of the NFκB pathway (Biswas et al., 1999). Thus calcineurin plays a direct role in the activation of both NFAT and NFκB. There are also reports suggesting that CEBP/δ is alternatively activated through phosphorylation by a Ca2+-sensitive PKC (Mahoney et al., 1992). We therefore investigated the roles of these various factors on the level of expression of cathepsin L as a prototype nuclear target gene.
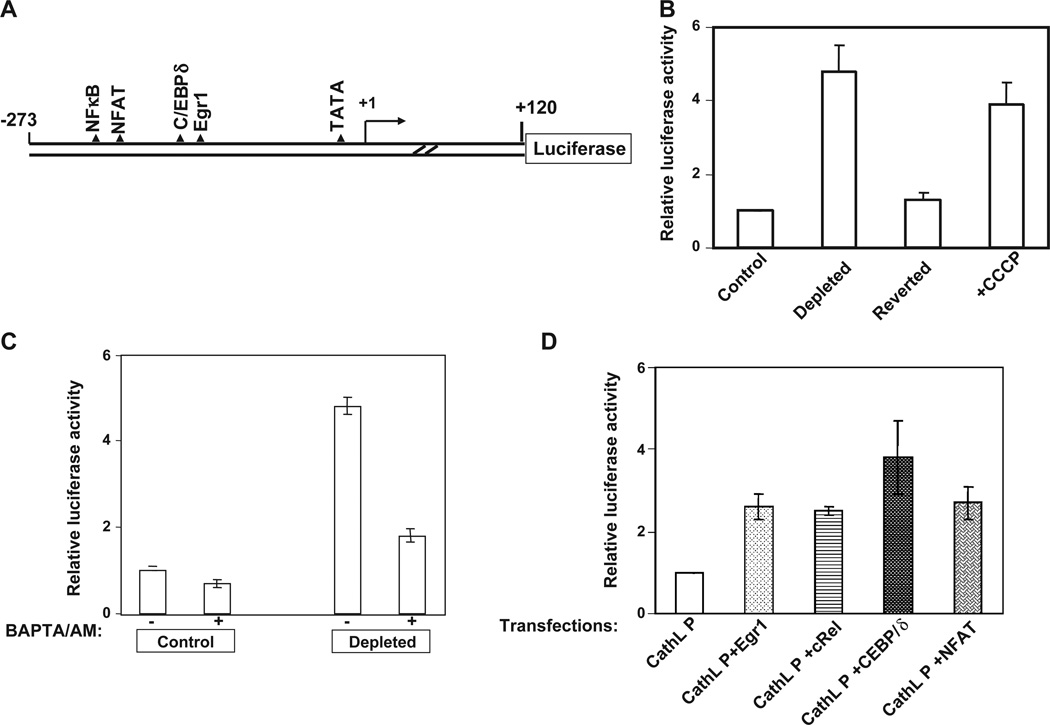
Fig. 4A shows a cartoon of murine cathepsin L promoter showing canonical sequence motifs for binding to NFκB, C/EBP, Egr-1, and NFAT. The promoter DNAwas amplified from genomic DNA and cloned into PGL-3 reporter plasmid vector (Promega Biotech Co. Madison, WI, USA) as described before (Amuthan et al., 2002). As shown in Fig. 4B, the 240-bp promoter DNA sequence was fully responsive to mitochondrial stress signaling as the promoter activity was induced 4- to 5-fold in mtDNA-depleted cells or cells treated with CCCP. Notably, the promoter activity in reverted cells with about 80% of control cell mtDNA content was only marginally higher than in control cells, suggesting the direct role of mtDNA depletion and mitochondrial stress signaling in induced transcription activity. It is also seen that addition of BAPTA/AM, a Ca2+ chelator abrogated mitochondrial stress-dependent increase of promoter activity in mtDNA-depleted C2C12 cells (Fig. 4C). The Ca2+ chelator inhibited promoter activity in control C2C12 cells only marginally. These results provide confirmatory evidence on elevated Ca2+ in mitochondrial stress signaling. Fig. 4C shows that the promoter activity was induced 2.3- to 4.5-fold by co-transfection with Egr-1, C/EBPδ, rel family factors cRel/p50, and NFAT suggesting their transcriptional activator role on the promoter activity. Although not shown, co-transfection with a combination of NFAT, C/EBPδ and c-Rel induced transcription by over 8-fold suggesting some degree of synergism between these factors. Although not shown, other potential target gene promoters, such as RyR1, Glut4 and hexokinase promoters, also respond to these factors in a synergistic manner. Although Rel family proteins are known to interact functionally and physically with various transcription factors (Bassuk et al., 1997), currently, there are no reported cases of functional synergy with NFκB, C/EBP and NFAT factors. We therefore believe that transcription activation by mitochondrial retrograde signaling may involve activation and recruitment of a distinct co-activator to the basal transcription complex.
Fig. 4.
Transcriptional activation of cathepsin L promoter (CathL P) by Ca2+ and calcineurin activated factors. (A) A cartoon of the mouse cathepsin L promoter cloned upstream of the luciferase reporter cDNA. The putative factor binding sites are shown. (B) Relative promoter activity in control, mtDNA-depleted cells containing only about 18% of mtDNA content, reverted cells containing about 80% of control cell mtDNA content and control cells treated with CCCP (25 µM for 4 h). Transfection of cells with 1 µg of promoter DNA and 0.5 µg of renila luciferase construct was carried out as described in the Materials and methods section. (C) The effect of Ca2+ chelator BAPTA/AM (30 µM added 2 h after transfection) on promoter activity in control and mtDNA-depleted cells. (D) Effects of overexpression of various Ca2+ responsive factors on transcription activation of cathepsin L promoter. Control C2C12 cells were co-transfected with 1 µg of promoter DNA, 0.2 µg each of cDNA constructs expressing indicated factors and 0.5 µg of renila luciferase DNA as described in the Materials and methods section.
3.5. Summary and conclusion
In extension to our previous studies (Biswas et al., 1999; Amuthan et al., 2001; Amuthan et al., 2002; Biswas et al., 2003), the present study shows that triggering of retrograde signaling by mitochondrial genetic and metabolic stress induce phenotypic changes, and altered profile of nuclear gene expression. A number of studies using ρ° tumor cells have yielded contrasting results showing that mtDNA depletion induced tumor formation with some tumor cell lines while it was inhibitory in others. In the case of both C2C12 and A549 cells, partial mtDNA depletion induced tumor formation and invasive behavior (Amuthan et al., 2001, 2002). Consistent with these observations, our present results show that in these cell lines partial mtDNA depletion induced resistance to etoposide-mediated apoptosis. Our results suggest that mtDNA-depleted C2C12 cells are unable to execute the apoptotic process due to a combination of increased Bcl-2 and Bcl-XL, and inability to process p21 Bid into active p17 tBid. The latter is probably due to inability to activate pro-caspase 8 and 3 into active caspases.
Results of this study also provide new information on the mechanism of stress-induced transcriptional modulation of nuclear target genes. First the observed upregulation of cathepsin L and also other nuclear target genes through mitochondrial stress signaling is dependent on elevated free Ca2+ since BAPTA/AM, a specific chelator of Ca2+ abrogated the stress-dependent transcriptional increase. Second, the transcription promoters that respond to mitochondrial stress such as cathepsin L contain binding sites for NFκB, NFAT, C/EBP that were previously shown to be activated as part of the mitochondrial retrograde signaling. Although not shown, promoters of other target genes are also transcriptionally activated by these same transcription factors suggesting a common feature of mitochondrial stress-mediated transcription activation. The mechanism of functional synergy and physical interaction between NFκB, NFAT, ATF and C/EBP factors is currently under investigation.
Mitochondrial mutations and associated mitochondrial dysfunction have been implicated in a large number of neuro-degenerative and retinal diseases, diabetes, aging and other age related diseases. It remains to be seen if altered ΔΨm and activation of retrograde signaling are contributing factors in the manifestations of these diseases.
Acknowledgement
This research was supported by NIH grant CA-22762.
Abbreviations
- mtDNA
mitochondrial DNA
- CnA
calcineurin A
- ΔΨm
mitochondrial transmembrane potential
- CCCP
carbonyl cyanide m-chlorophenyl hydrazone
References
- Amuthan G, Biswas G, Zhang SY, Klein-Szanto A, Vijayasarathy C, Avadhani NG. Mitochondria-to-nucleus stress signaling induces phenotypic changes, tumor progression and cell invasion. EMBO J. 2001;20:1910–1920. doi: 10.1093/emboj/20.8.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amuthan G, Biswas G, Ananadatheerthavarada HK, Vijayasarathy C, Shephard HM, Avadhani NG. Mitochondrial stress-induced calcium signaling, phenotypic changes and invasive behavior in human lung carcinoma A549 cells. Oncogene. 2002;21:7839–7849. doi: 10.1038/sj.onc.1205983. [DOI] [PubMed] [Google Scholar]
- Arnould T, et al. CREB activation induced by mitochondrial dysfunction is a new signaling pathway that impairs cell proliferation. EMBO J. 2002;21:53–63. doi: 10.1093/emboj/21.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averous A, Bruhat C, Jousse V, Carraro G, Thiel P, Fafournoux Induction of CHOP expression by amino acid limitation requires both ATF4 expression and ATF2 phosphorylation. J. Biol. Chem. 2004;279:5288–5297. doi: 10.1074/jbc.M311862200. [DOI] [PubMed] [Google Scholar]
- Babcock DF, Herrington J, Goodwin PC, Park YB, Hille B. Mitochondrial participation in the intracellular Ca2+ network. J. Cell Biol. 1997;136:833–844. doi: 10.1083/jcb.136.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassuk AG, Anandappa RT, Leiden JM. Physical interactions between Ets and NF-kappaB/NFAT proteins play an important role in their cooperative activation of the human immunodeficiency virus enhancer in T cells. J. Virol. 1997;71:3563–3573. doi: 10.1128/jvi.71.5.3563-3573.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas G, et al. Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: a novel mode of inter-organelle crosstalk. EMBO J. 1999;18:522–533. doi: 10.1093/emboj/18.3.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas G, Anandatheerthavarada HK, Zaidi M, Avadhani NG. Mitochondria to nucleus stress signaling: a distinctive mechanism of NFkappaB/Rel activation through calcineurin-mediated inactivation of IkappaBbeta. J. Cell Biol. 2003;161:507–519. doi: 10.1083/jcb.200211104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butow RA, Avadhani NG. Mitochondrial signaling: the retrograde response. Mol. Cell. 2004;14:1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- Chen G, Branton PE, Shore GC. Induction of p53-independent apoptosis by hygromycin B: suppression by Bcl-2 and adenovirus E1B 19-kDa protein. Exp. Cell Res. 1995;221:55–59. doi: 10.1006/excr.1995.1351. [DOI] [PubMed] [Google Scholar]
- Hattori T, Ookawa N, Fujita R, Fukuchi K. Heterodimerization of Bcl-2 and Bcl-X(L) with Bax and Bad in colorectal cancer. Acta Oncol. 2000;39:495–500. doi: 10.1080/028418600750013410. [DOI] [PubMed] [Google Scholar]
- Ichas F, Jouaville LS, Mazat JP. Mitochondria are excitable organelles capable of generating and conveying electrical and calcium signals. Cell. 1997;89:1145–1153. doi: 10.1016/s0092-8674(00)80301-3. [DOI] [PubMed] [Google Scholar]
- Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- Liao X, Butow RA. RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell. 1991;72:61–71. doi: 10.1016/0092-8674(93)90050-z. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin Phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Luo Y, Bond JD, Ingram VM. Compromised mitochondrial function leads to increased cytosolic calcium and to activation of MAP kinases. Proc. Natl. Acad. Sci. U. S. A. 1997;94:9705–9710. doi: 10.1073/pnas.94.18.9705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney CW, Shuman J, McKnight SL, Chen HC, Huang KP. Phosphorylation of CCAAT-enhancer binding protein by protein kinase C attenuates site-selective DNA binding. J. Biol. Chem. 1992;267:19396–19403. [PubMed] [Google Scholar]
- Mori K, Ma W, Gething MJ, Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- Parikh VS, Morgan MM, Scott R, Clements LS, Butow RA. The mitochondrial genotype can influence nuclear gene expression in yeast. Science. 1987;235:576–580. doi: 10.1126/science.3027892. [DOI] [PubMed] [Google Scholar]
- Rizzuto R, Brini M, Murgia M, Pozzan T. Microdomains with high Ca2+ close to IP3-sensitive channels that are sensed by neighboring mitochondria. Science. 1993;262:744–747. doi: 10.1126/science.8235595. [DOI] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Troen BR, Chauhan SS, Ray D, Gottesman MM. Downstream sequences mediate induction of the mouse cathepsin L promoter by phorbol esters. Cell Growth Differ. 1991;2:23–31. [PubMed] [Google Scholar]
- Tsujimoto Y. Role of Bcl-2 family proteins in apoptosis: apoptosomes or mitochondria? Genes Cells. 1998;3:697–707. doi: 10.1046/j.1365-2443.1998.00223.x. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ. A mitochondrial specific stress response in mammalian cells. EMBO J. 2002;21:4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]