Abstract
Protein isoforms/splice variants can play important roles in various biological processes and can potentially be used as biomarkers or therapeutic targets/mediators. Thus, there is a need for efficient and, importantly, accurate methods to distinguish and quantify specific protein isoforms. Since protein isoforms can share a high percentage of amino acid sequence homology and dramatically differ in their cellular concentration, the task for accuracy and efficiency in methodology and instrumentation is challenging. The analysis of intact proteins has been perceived to provide a more accurate and complete result for isoform identification/quantification in comparison to analysis of the corresponding peptides that arise from protein enzymatic digestion. Recently, novel approaches have been explored and developed which can possess the accuracy and reliability important for protein isoform differentiation and isoform-specific peptide targeting. In this review, we discuss the recent development in methodology and instrumentation for enhanced detection of protein isoforms as well as the examples of their biological importance.
Keywords: protein isoforms/splice variants, accurate and complete identification/quantification, intact protein analysis, novel proteomic approaches
1 Introduction
The analysis of a complex proteome with a broad dynamic range of protein concentrations is especially challenging. In addition, different forms of the same protein may be present due to alternative splicing, polymorphism and post-translation modifications (PTMs) which adds to the proteome complexity. Although protein isoforms can originate from separate genes, e.g. actin isoforms [1], single gene can code for multiple proteins due to the process called alternative (differential) mRNA splicing which results in multiple protein forms that comprise different peptide sequences. Each isoform can have various biological roles [2-4]. An example of a protein and its spliced variant is demonstrated in Figure 1. It shows the alignment of the amino acid sequences of two protein isoforms, transcription factor 2 (A1EC66_RAT, 558 amino acid sequence length) and transcription factor 2 splice variant (A1EC67_RAT, 532 amino acid sequence length) which are encoded by the same gene Hnf1b and differ due to alternative splicing.
Figure 1.
The alignment of amino acid sequences of two protein isoforms encoded by the same gene Hnf1b which differ due to alternative splicing shown for transcription factor 2 (A1EC66; A1EC66_RAT, 558 amino acid sequence length) and transcription factor 2, splice variant (A1EC67; A1EC67_RAT, 532 amino acid sequence length). The alternatively spliced mRNA results in protein A1EC67 in which amino acid sequence NQTVQSSGNMTDKSSQDQLLFLFPEF is missing. The initiator methionine (M) is removed in the mature protein.
As the methods for the quantification of splice variants expression developed over years [5-7], the estimated percentage of human gene products that went through alternative splicing was identified as 35 % [8], 60 % [3] as to high as 95 % [9] but it was not straightforward how many of the predicted splice variants are actually functional [10-11]. The isoforms/splice variants are involved in many biological processes and are connected to various stages in a number of pathologies [12-13]. As such, they have been used as biomarkers [14-16] and therapy targets [17-18]. For example, specific protein isoforms have been shown to impact thyroid pathological conditions [19], they are involved in various types of cancer [20-22], multiple sclerosis [23], heart hypertrophy [24], autoimmune diseases [25] and in diabetes [26] as well as in the regulation of embryonic stem cell pluripotency and reprogramming [27]. Yet, the exact roles of most splice variants/isoforms are still poorly understood. This is, in part, because of the lack of techniques to differentiate the isoforms and accurately identify/quantify them.
Two basic proteomic platforms are commonly used in protein analysis. In the first platform, proteins are enzymatically digested into peptide fragments prior to mass spectrometry (MS) identification (bottom-up). In the second approach, MS is performed on intact proteins (top-down) [28]. This classification is a MS-centric point of view, i.e. whether intact proteins or the resulting peptides after protein enzymatic cleavage are introduced into MS instrument. However, pre-fractionation of intact proteins followed by enzymatic digestion of proteins with peptides introduced to MS can be considered as combination of both top-down and bottom-up approaches. Although different analytical separation methods or their various combinations are often applied in the pre-fractionation step in order to simplify the protein mixture and increase the proteome coverage by MS, direct analysis on the liquid chromatography - tandem MS (LC-MS/MS) instrument is another possibility. While the bottom-up MS platform is conventionally used for protein identification and in high throughput manner, the top-down MS approach was not so frequently applied due to many factors but basically because of the lack of methodology available and compatible with MS and due to requirements for efficient MS instruments [29-33]. However, it is the analysis of intact proteins which can provide the accurate and more complete information, e.g. on the size of the proteins and on the quantification and precise characterization of splice forms and PTMs [32-39]. For example, the bottom-up approach detects only a certain number of peptides of the original protein due to various constraints during separation and MS analysis (isoelectric point, molecular mass and hydrophobicity range limitation) and thus, the important peptides which are unique to isoform can be undetected and missed. This is not the case with the top-down approach in which the whole protein amino acid sequence is analyzed.
The top-down approach has been used in targeted proteomics (single protein or simple protein mixture where the protein amino acid sequences are known) and as a complimentary/integrated method to bottom-up [40-42]. The possibility of using a top-down approach in large-scale discovery platform increased in the last years and has been reported [43-44].
Protein isoforms are often reported in the MS-based protein datasets obtained from a discovery proteomic study. However, the question is: Are they truly unique isoforms of the same proteins? Considering the false-positive rates for the protein identifications in bottom-up approach, protein/peptide redundancy in the databases and the manner in which isoform names are assigned by various database search engines, one must be careful in reporting distinct protein isoforms. Only if a peptide amino acid sequence that is unique to a specific isoform is observed, the protein isoform can be unambiguously assigned. This leads to the other questions: What is required to increase the probability of observing peptides that are specific to isoforms and quantify them? Are there certain methods that can improve the chance to identify protein isoforms? Can intact protein separation enhance the number and accuracy of isoform identifications?
Each step of proteomic pipeline will be important to improve the current status of protein isoform identification process with the separation methods, MS and efficient MS data processing to be the cornerstones. Recently, improvements to existing technologies as well as new methods have been developed for efficient and accurate peptide/protein identification and quantification in targeted proteomics. These targeted approaches are complementary to the established discovery proteomics. Targeted MS-based methods can be used as antibody-free validation tools or can stand alone as potential tools to selectively analyze specific protein isoforms. In this review, we will concentrate on novel technical improvements published in recent years that are related to (intact) protein/protein isoform analysis in gel-free format as well as on the examples of proteins whose isoforms have been identified to have important biological implications.
2 Separation techniques with emphasis on novel approaches
To be able to improve protein analysis and the detection of protein isoforms, new separation methodology platforms are needed in addition to the methods already used. Moreover, the separation techniques for intact proteins have to keep the proteins in soluble forms and have to be compatible with subsequent direct MS analysis [45]. As this special issue is dedicated to gel-free separation methods, we will not discuss gel-based methodologies (e.g. 1-D and 2-D gel electrophoresis and two-dimensional difference gel electrophoresis (2-D DIGE)) even though these methods are often applied in protein/isoform analysis. In addition to the improvements discussed in the following sections, there have been a number of reviews published that provide complementary information [33,38,45-47].
2.1 The improvement in LC
The common LC technique used for protein separation is reversed phase LC (RPLC) which separates proteins based on hydrophobicity. For intact proteins (high molecular weights, tertiary structures), C4 or C5 alkyl chain stationary phases with pore size particles of 300 Å are recommended [48], as they have less retention than longer carbon chain phases such as C18. This does not preclude the use of these long carbon chain matrices for protein separation. It depends on the composition and characteristics of the proteins comprising a particular sample, including e.g. their hydrophobicities, that dictates which chromatographic column yields better result in terms of separation efficiency, resolution and irreversible protein adsorption onto column. For example, butyl (C4) instead of octyl (C8) and octadecyl (C18) RP stationary phases was selected for its excellent separation ability, based on the comparison of column separation efficiency, adsorption of intact proteins and sample analysis. [49]. The less retentive C8 RP packing was used for intact protein analysis in 2-D separation of RPLC coupling with capillary isoelectric focusing [50]. On the contrary, better separation efficiency and resolution was accomplished with C18 column compared to C4 column for standard intact protein mixture [51], although the risk of protein loss due to stronger C18 column retention is always involved and it needs to be taken into consideration when deciding on what column matrix to use.
In another study [52], the compatibility of superficially porous (SP) resin (C18; a thin layer of porous silica on a solid core of silica) with online capillary LC/MS for label-free intact protein analysis was demonstrated. It showed improved chromatographic resolution, speed, sensitivity and reproducibility with sub-ultrahigh pressure LC (sub-UPLC) limits. The SPLC/MS platform was used for analysis of intact proteins from mouse heart homogenate and from HeLa S3 acid soluble extracts at low cell count (5 × 104 cells per analysis). The analysis of protein sample from mouse heart homogenate utilizing SP stationary phase followed by LTQ-Orbitrap FT-MS was fast (30 min), sensitive with only 500 ng of total protein loaded and yielded in 106 protein forms detected in a single high-resolution scan. For HeLa acid extracts, two replicated analyses yielded in total 343 protein forms identified in 30 min time per each analysis.
Contrary to the particle-packed columns, monolithic columns are made of a single piece of porous cross-linked polymer or porous silica. Due to their unique structure, monolithic columns enable fast separation with low backpressure and high resolution. As well, the macroporous structure and excellent separation efficiency make them well suited for analysis of proteins [53-54]. They can be used as stationary phase columns for various LC modes, e.g. reversed phase [55], ion exchange [56] or electrochromatography [57]. A new type of monolithic trapping column with high mechanical strength was tested in application to trap the intact proteins in on-line capillary LC [58]. The monolithic trapping column fabricated by entrapping of C8 RP particles through a sol-gel network based on methyltriethosilane chemistry showed a long-term stability for four protein mixtures with protein recoveries on average of 99.3%. The method was used for analysis of mouse liver intact proteins as well. In another study [56], high resolution separation of intact glycoprotein isoforms was performed by using weak anion exchange (WAX) monolithic capillary. The comparison with the capillary zone electrophoresis (CZE) showed that although decrease in resolution for several glycoisoforms was observed, the WAX column provided better selectivity (more glycoisoforms was observed).
2.2 HIC
Hydrophobic interaction chromatography (HIC) is an LC-based method which is especially suitable for the separation of large biomolecules, e.g. proteins, and it separates proteins based on their hydrophobicity [59-61]. Contrary to RPLC, HIC uses an aqueous solution during the separation. This helps to keep the proteins in intact forms without denaturation by organic solvents. Similarly to RPLC of intact proteins, medium hydrophobic C4 stationary phase is preferred, and the retention of proteins to the stationary phase is realized primarily by adding of high concentration of salt in water (e.g. up to 2 M ammonium sulfate; salt out effect). By decreasing the salt concentration during the gradient, the proteins are eluted from the column in order of increasing hydrophobicity. As shown previously [62], the selectivity of particular protein for HIC and RPLC columns depends on the protein structure during HIC and RPLC process (hydrophilic exterior of native protein in HIC (low retention) vs the exposure of more hydrophobic interior of the same protein due to organic solvent denaturation in RPLC (high retention)). The examination of protein function recovery has shown that 10-200 μg of lactic dehydrogenase and alpha-chymotrypsin were recovered from a HIC column (20 minutes linear gradient from 1.0 M sodium sulfate in 10 mM potassium phosphate buffer (pH 7.0) to 10 mM potassium phosphate buffer (pH 7.0)) with 90% and 86% of enzymatic activity, respectively. The enzymatic activity recovery from RPLC column (20-min linear gradient from 0.1% trifluoroacetic acid (TFA) in water to 0.1% TFA in 2-propanol-water (60:40) at flow-rate 1 mL/min) was 55-91 % for alpha-chymotrypsin and no activity was recovered for beta-glucosidase. Thus, depending on the downstream analysis needs and whether denatured or intact (functioning) protein is required will dictate use of HIC or RPLC.
Recently, a new approach was reported on separation of native proteins by using on-line combination of weak-cation exchange chromatography (WCX) and HIC in a single column with a single phase (2D-LC; 150 mm × 4.6 mm, silica-based; particle size 5 μm, pore size 22 nm) [63]. In this approach, an on-line buffer exchange (from WCX to HIC mode) was performed and the fractions from the 1st dimension were collected and subsequently re-injected into the same column and re-separated in a second retention mode. For native proteins, the separation selectivity of this method was better and the resolution was comparable to when the analysis was carried out using the two individual conventional chromatographic columns. In another study, the performance of HIC was compared to an immune-affinity column that was used for the depletion of highly abundant proteins of medium hydrophobicity from plasma [64]. Both methods showed similar reproducibility. For this instance, HIC can be considered as complementary to immune affinity method, since depletion of alpha-1-antitrypsin and albumin was incomplete by HIC but HIC method yielded twice amount of spots compared to immune affinity method as observed by subsequent 2-D gel electrophoresis.
Similarly to affinity chromatography and ion exchange chromatography, HIC has the ability to concentrate proteins. It has been used to concentrate and purify green fluorescent protein from bacterial lysate [65] and recombinant streptokinase from the Escherichia coli cell lysate [66] as well as for the detection and analysis of the serum prostate-specific antigen (PSA). In human prostate cancer, free PSA in serum contains a significant fraction of precursor form (pPSA) [67].
2.3 The other separation methods
The capillary electrophoresis (CE) coupled to ESI-MS employing microfabricated and monolithically integrated CE-ESI microchip has been used for intact protein analysis [68]. The material of the chip was a commercially available inorganic-organic hybrid polymer. Contrary to silica-based chips, it did not require any chemical or physical surface modification in order to reduce non-specific interactions with proteins. In another study [69], chip-type asymmetric (cross)-flow field-flow fractionation (AF4) channel was directly coupled with MS for top-down protein analysis. In this method, the size-dependent separation occurs in a channel with external field force applied perpendicularly to the carrier-flow that contains the sample. With this arrangement, desalting and purification of proteins was achieved by using of the exiting cross-flow during the AF4 operation and by MS-compatible buffer.
2.4 Recent multidimensional separation formats
Recently, a novel separation technology was reported [70] in which multiplexed gel-eluted liquid fraction entrapment electrophoresis (mGELFrEE; size-based separation) was performed simultaneously using 8 parallel glass gel columns. After SDS removal by chloroform/methanol/water precipitation, this method can be followed by LC-MS/MS for analysis of intact proteins. The major advantage of mGELFrEE is the separation of intact proteins in solution phase. The mGELFrEE device distributes the sample among multiple gel columns and thus, it allows the increase in the sample load while maintaining the electrophoretic resolution with well-resolved liquid fractions and high protein solubility during separation and sample collection. It can be a part of the multiplexed high-throughput 2-dimensional liquid electrophoretic (2D LE) platform with solution IEF (sIEF) coupled to mGELFrEE device. It is the analog to 2D PAGE but with an avoidance of low protein recovery and laborious steps prior to analysis by MS. This platform was used for analysis of S. cerevisiae proteome in the pI range from 3.8 to 7.8 and the Mw range from 10 kDa to 150 kDa with separation time of 3.25 h (1 mg of proteins from S. cerevisiae). Although followed by LC-MS/MS analysis of peptides after enzymatic digestion of the proteins in the 2D LE fractions in this particular study, recently, the method was extended and enhanced for using in top-down approach [37]. In this study [37], the 2D LE separation comprising sIEF and GELFrEE is combined with nanocapillary LC and MS for large-scale whole protein analysis of nuclear and cytosolic extracts of HeLa S3 cells. After separation, the detergent was removed by precipitation and the mGELFrEE fractions were resuspended in 5% acetonitrile/0.2% formic acid and injected into nano-RPLC coupled to linear ion trap FT MS (12T). For a protein sample of 0.5-1 mg, the peak capacity of the system (including MS) was 20-fold higher than that achieved using high-resolution 2-D gels (compare 100000 to 5000 peak capacity for the proteins below 25 kDa) with identification of proteins up to 105 kDa. More than 3000 protein forms (due to PTMs or mRNA splicing) were identified, most of which had not been previously detected.
3 Mass spectrometry and data processing
3.1 General approach
To analyze and identify intact proteins/protein isoforms in top-down MS approach, there are special requirements for the MS instrumentation (e.g. high mass resolving power to resolve the overlapping fragment ions and mass accuracy for high confidence) and for data processing compared to the analysis of peptides obtained following protein digestion. Mostly, high-resolution Fourier transform ion cyclotron resonance (FTICR) is applied for high mass accuracy measurement of intact protein ion mass (mass errors < 2 ppm) [29,32-33] with a specialized software to perform the data analysis [71-72]. For protein mixtures, the isolation of a single molecular ion for MS/MS analysis can provide protein identification of far higher reliability and confidence and can directly characterize amino acid sequence errors and variations [73]. Thus, it can point out the protein isoforms and PTMs. Nevertheless, top-down MS has been performed not only with FTICR MS but other approaches have been used including various MS instruments and different types of dissociation of intact proteins into fragments, such as MALDI TOF MS [74], LTQ Orbitrap MS [75], Orbitrap Elite MS [37,76], Triple TOF 5600 System [77] with either collision-induced dissociation (CID) [78], electron capture dissociation (ECD) [79] or electron transfer dissociation (ETD) [80].
During data analysis, the extensive MS spectra datasets are generated which are compared and matched to existing protein databases (e.g. UniProt/SwissProt, UniProt/TrEMBL, NCBInr and Ensembl) using search engines (e.g. Sequest, Mascot, X!tandem, OMSSA and various custom-made softwares) with pre-selected stringent rules and limitation criteria (e.g. monoisotopic/average masses, semi-/full-enzyme search, number of missed cleavages, variable/fixed modifications and post-search analysis). To increase the proteome coverage, multiple search engines can be used since they use different search algorithms. The major problems with identification are protein name and peptide redundancy. In different databases (and even in the same database), a protein with identical amino acid sequence can have various protein names and accession numbers. Although the efforts have occurred to remove these redundancies from the databases, the investigators still need to run additional amino acid sequence homology and clustering algorithms to avoid an incorrect and redundant reporting. Another problem is peptide redundancy. Since many proteins share amino acid sequence homology, especially the protein isoforms originated from the same gene (splice variants), the peptide sequence identified can match several different proteins making the assignment of the peptide to protein/protein isoform ambiguous. Unambiguous protein isoform identification can be made only if peptide sequence that is unique to the specific isoform is detected in the MS. This is a problem in bottom-up approach since only a part of the proteome is identified due to e.g. limitation in pI and/or mass range of instrumentation or peptide insufficient ionization. And thus, the isoform specific peptide sequence/sequences can be lost during pre-separation, enzyme digestion and mass spectrometry analysis. It can be helpful to digest the sample with multiple enzymes since they will generate different peptides, increasing the probability of detecting peptides comprising isoform specific sequences. Intact protein analysis is advantageous, since one can observe the complete amino acid sequence. Moreover, the use of both methods (top-down and bottom-up) as a complimentary/integrated approach for protein/protein isoform identification could be a good option as well.
As mentioned previously, the lack of appropriate methodology and efficient MS instruments in the past made the analysis of intact proteins less exploited. Over years, the studies have been reported analyzing the intact proteins from low-mass proteome (5-25 kDa) to protein sizes of 80 kDa [81-85] and up to 229 kDa in protein size if combination of electrospray additives, heated vaporization and bond dissociation was used [86], and the possibility of using a top-down approach in large-scale discovery platform increased in the last years and has been reported [43-44]. The more detailed description and summary on top-down MS can be found in the recent reviews [32-34,36,40,73,80].
3.2 MRM and SWATH (do we really need to analyze isoforms in intact forms to get accurate isoform identification/quantification?)
The ability to carry out target peptide analysis using high accuracy MS instrumentation in which peptides represent unique protein isoforms evokes the question whether this method or intact proteins analysis is more effective in the identification of protein isoforms. The former approach is advantageous if many isoform-specific peptides could be accurately detected in a single MS run. An additional advantage would be if accurate quantification of these isoform specific peptides was also possible. Multiple reaction monitoring (MRM) or selected reaction monitoring (SRM) is a sensitive and accurate targeted MS-based method that has recently been used in proteomics for quantification of selected (known/targeted) peptides (representing unique proteins) in complex mixtures [87-89]. MRM requires a priori information regarding proteins and their potential isoforms in order to select the appropriate isoform specific peptides. Based on this information, one can select the appropriate pairs of parent-product ions (transitions) for the various peptides of interest. Compared to classical quantitative methods (ELISA, Western blot), the advantages of MRM technique are: i) the ability to carry out both relative and absolute quantification, ii) simultaneously quantified multiple proteins/peptides, iii) it can be antibody-free (although enrichment methods can be used to enhance sensitivity) iv) differentiation between protein isoforms (unique gene/splice variants), v) detection of multiple PTMs vi) it can be used to analyze any body fluid and tissue/cells and vii) small sample quantities are needed (unless enrichment is required). Although the development of the assay for each target protein is still time consuming, it needs to be performed only once, and once established, it can be universally used for a given instrument. The MRM technique is usually performed on triple quadrupole MS instruments which score high quantitative capabilities. However, the sample can be further analyzed on other MS instruments (e.g. LTQ Orbitrap, Orbitrap Elite, TripleTOF 5600 System) if qualitative analysis is the goal. For example, two protein isoforms, serum amyloid A1 and 2 (SSA1 and SSA2), were selected from crude serum samples and differentiated by MRM [90]. As acute SSA exists in two isoforms with 92 % homology between their amino acid sequences and only SAA1-specific antibody is available, SSA2 cannot be detected by classical antibody-based methods. In the study mentioned, MRM assay was developed for differential measurement of SSA1 and SSA2 in clinical crude serum samples from 99 healthy controls and 100 lung adenocarcinoma patients. While most of the healthy control samples had small/no MS/MS peaks of the MRM targeted peptides, 10- to 34-fold increases in lung cancer samples were observed.
Among the major features of LC-MS/MS instruments belong e.g. resolving power, peak capacity, mass accuracy, sensitivity and rates of MS/MS spectra acquiring. In terms of these features, the performances of MS instruments differ as they excel in only some of these characteristics. For example, hybrid Q-TOF MS instruments have excellent spectral acquisition rates while hybrid FT MS instruments (Q-FTICR, LTQ FT) exhibit high resolution power and mass accuracy [91]. But even high resolution (RP ∼ 30000 FWHM) and high mass accuracy (∼ 2 ppm) MS instruments available to date are able to identify only limited fractions of proteins in complex protein samples, and one of the reasons is a relatively slow MS/MS acquisition. Very recently, novel acquisition strategy called “sequential window acquisition of all theoretical fragment-ion spectra” (SWATH MS) [92-93] was developed. Unlike a discovery MS approach, in which MS instruments use data dependent acquisitions (a fixed number of precursor ions selected from a survey MS scan are subjected to subsequent MS/MS analysis and resulting fragment ion spectra are searched against database), SWATH MS strategy uses a data independent acquisition (DIA) and acquires fragment ion spectra of all analytes in a single sample injection by cycling through whole LC range and recording the consecutive survey scans and fragment ion spectra for all precursors in the series of isolated windows (swaths) [92]. This acquisition method produces fragment ion spectra of all precursor ions within a pre-determined precursor retention time and m/z space and compiles them into complex fragment ion maps with the dimensions of retention time, fragment ion m/z and intensity. The analysis of the fragment ion maps requires a targeted data extraction strategy that exploits a priori informations that have been previously obtained from MS spectral libraries of various cells and organisms. In order to uniquely identify and quantify the peptides from the fragment ion maps, many parameters (fragment ion signals, relative intensities, chromatographic profiles) are matched to the known coordinates and parameters of each specific peptide of interest [92]. This approach allows quantification of the proteins in a number typically possible by MS/MS of tryptic peptides with the accuracy and reproducibility reached by MRM. Moreover, DIA used in SWATH MS strategy allows re-examination of the acquired fragment ion spectra in silico for any new protein without further data acquisition. It has been demonstrated that SWATH MS targeted data extraction strategy is also valuable tool for unambiguous assignment of PTMs [92]. This can also be used to target the peptides consisting of the isoform-specific amino acid sequences.
SWATH MS acquisition technology can be used on a new hybrid quadrupole-TOF MS instrument (TripleTOF 5600 System, AB SCIEX) [91]. This MS instrument combines high score MS characteristics that have not been previously found simultaneously in a single MS device. It possesses high resolution and mass accuracy (like the LTQ Orbitrap) with high spectral acquisition rates and sensitivity (like the Q-TOF) and quantitative capabilities (like the triple quadrupoles). Thus, TripleTOF 5600 System appears to be able to perform accurate identification and quantification of the sample in a single platform. The TripleTOF 5600 System was applied to analyze the global proteome of Sacharomyces cerevisiae lysate [91] as well as in a study of small molecules such as bosentan and its metabolites in urine [93].
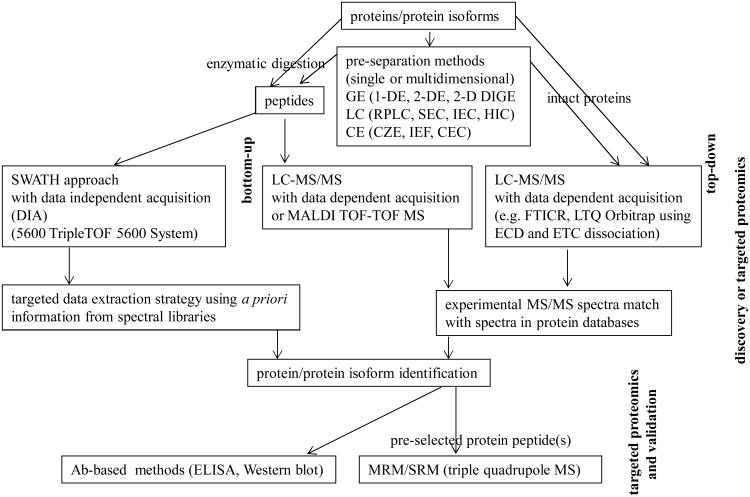
The top-down, bottom-up and MRM and SWATH methods are schematically shown in Figure 2 as the parts of proteomic strategy for protein/protein isoform identification.
Figure 2.
The top-down, bottom-up and MRM and SWATH methods schematically shown as the parts of proteomic strategy for proteins/protein isoform identification.
GE – gel electrophoresis, SEC – size exclusion chromatography, IEC – ion exchange chromatography, HIC – hydrophobic interaction chromatography, CE – capillary electrophoresis, CZE – capillary zone electrophoresis, IEF – isoelectric focusing, CEC – capillary electrochromatography.
4 Protein isoforms/splice variants and biological relevance
Although the biological functions and roles are known for many proteins, the knowledge about alternative spliced isoforms of a particular protein is often sparse. This includes whether the isoform i) is expressed, and if so, in what cell type and at what stage of development, ii) is altered with disease or iii) has distinct cell localization and function. In this section, we will provide several examples to illustrate that specific protein isoform/splice variant can alter biological functions and physiopathologies, highlighting the importance of their reliable and accurate identifications. Examples of protein isoforms/splice variants known to be relevant to various biological functions and diseases based on literature search are summarized in Table 1.
Table 1.
The examples of protein isoforms/splice variants known to be relevant to various biological functions and diseases (based on literature).
| Protein isoforms/splice variants | Function/disease | Ref. |
|---|---|---|
| Interferon receptor (IFNAR) isoforms | multiple sclerosis | [23] |
| Enigma homolog splice variants (ENH 1-4) | ENH1 promotes the expression of cardiac muscle hypertrophy markers, high levels of ENH4 prevent these changes | [24] |
| B-cell activating factor (BAFF) isoforms | autoimmune diseases | [25] |
| T-cell factor 7-like 2 (TCF7L2) splice variants | related to the development of type 2 diabetes | [26] |
| Forkhead family transcription factor (FOXP1) embryonic stem cell-specific isoform (FOXP1-ES) | embryonic stem cell pluripotency and reprogramming regulation | [27] |
| Vascular endothelial growth factor (VEGF) splice variants | anti-angiogenic properties in cancer therapy | [98-99] |
| Insulin-like growth factor-1 (IGF-I) splice variants (IGF-IEa, IGF-IEb and IGF-IEc) | high resistance exercise | [100] |
| Interleukin IL37 and its splice variants | anti-inflammatory cytokines | [101] |
| Fibronectin splice variants | participate in lymphatic valve formation; prevent tissue fibrosis; mediate fibroblast to myofibroblast differentiation; pro-thrombotic role | [102-103] |
| Thrombopoietin splice variants | pro-apoptotic causing death of newly generated neurons; risk of myocardial infarction | [105-106] |
| Neuregulin splice variants | breast cancer; prostate cancer | [22,107] |
| Osteopontin splice variants (OPNa, OPNb and OPNc) | OPNa is a major isoform in both healthy and non-small cell lung cancer (NSCLC) and it is elevated in NSCLC | [108] |
| p53 family (TP53, TP63 and TP73) splice variants | high expression levels contribute to tumorigenesis; affect tumor response to therapy | [109] |
The cytokines represent a broad group of proteins, many of which have a number of isoforms [94]. For example, the splice variants of fibroblast growth factor (FGF-1 and FGF-2), transforming growth factor (TGF-β1, TGF-β2 and TGF-β3), insulin-like growth factor (IGF-1 and IGF-2), macrophage colony-stimulating factor (CSF-1 and CSF-4), interleukin family (IL-1α and IL-1β) and platelet-derived, placenta and vascular endothelial growth factors (PDGF, PGF and VEGF). The cytokines and their splice variants are involved in a broad number of biological processes, in variety of diseases and they have been targeted as therapeutic agents [95-96]. A single gene gives a rise to several distinct isoforms of VEGF which differ in their expression patterns and properties [97]. The targeted decrease in VEGF level and/or the blocking of VEGF receptors resulting in the inhibition of downstream signaling pathways have been demonstrated to be therapeutic anti-angiogenesis approach for partial treatment of various cancer types. Although VEGF splice variants have mostly pro-angiogenic features, several novel splice variants of VEGF with anti-angiogenic properties were identified recently and were tested for more efficient anti-angiogenesis therapy [98-99]. Another study was dedicated to IGF-I and to better understanding the function mechanism of three splice variants (IGF-IEa, IGF-IEb and IGF-IEc) [100]. IGF-I produced in skeletal muscle is known to play the role in skeletal muscle development, growth and repair. IGF-IEa, IGF-IEb and IGF-IEc are up-regulated (in different time courses) in the muscles of elderly people and they cause high resistance exercise. Although some of the IL-1 family cytokines are known to be important parts of development and inflammation regulation (e.g. IL-1α and IL-1β), the function of IL-37 was not well explained yet. Recently, IL-37 was studied as a new anti-inflammatory cytokine of the IL-1 family together with its five different splice variants [101].
Fibronectin as a component of extracellular matrix is another example of protein that exists in many alternatively spliced isoforms [102] and it was only recently associated with a human disorder. An extensive review was published [103] discussing the roles of alternatively spliced fibronectin isoforms which includes participation in lymphatic valve formation, in prevention of tissue fibrosis, in fibroblast to myofibroblast differentiation and fibronectin isoform pro-thrombotic roles.
In addition to regulation of thrombopoiesis (the process of generating of blood platelets), thrombopoietin (THPO) and its splice variants (six in human and nine in mouse) were found to be expressed even in the tissues not directly related to thrombopoiesis (brain, liver and kidney) [104-105]. As well, THPO splice variants have been detected in cerebrospinal fluid (CSF) of developing central nervous system (CNS) at different levels compared to adult CNS [105], and THPO was shown to be strongly pro-apoptotic and causing the death of newly generated neurons [105]. THPO isoforms have also been associated with risk of myocardial infarction at a young age [106].
Neuregulins (NRG 1-4) and their corresponding splice variants play important roles in mammary gland development, they are expressed at high levels in breast cancer [22] and they are related to prostate cancer [107]. For example, NRG 4 has at least five splice variants (A1, A2, B1, B2 and B3), their expressions are cell specific and they differ in subcellular location. In prostate cancer, each variant has a different degree and pattern of expression and function e.g. the effects on cell architecture and motility [107].
Enigma homolog (ENH) is a protein with four splice variants which have been detected in the heart with different expression patterns in embryonic, neonatal and adult stages [24]. ENH 2, 3 and 4 are predominantly expressed in adult heart while ENH 1 is an embryonic isoform. In rat neonatal cardiomyocytes, high levels of ENH1 promote the expression of hypertrophy markers and they increase cell volume. On the contrary, high levels of ENH4 prevent these changes.
In pancreatic islet, four splice variants of T-cell factor 7-like 2 (TCF7L2; also known as TCF4) were detected in higher expression levels compared to other organ tissues. Although the molecular mechanism still remains to be clarified, the studies have shown that pancreatic islet is a major organ for TCF7L2-dependent risk of developing type 2 diabetes (T2D) [26].
Osteopontin splice variants (OPNa, b and c) are involved in cancer metastasis and progression but the isoform specific expression patterns and particular functions are not well understood due to, in part, the absence of reliable methods to distinguish among the isoforms. Recently, a mass spectrometric approach was developed to identify and quantify OPN isoforms in human plasma [108]. The approach consisted of immunocapture method applied prior to analysis of isoform specific tryptic peptides by MRM MS. It was demonstrated that OPNa is a major isoform in both healthy and non-small cell lung cancer (NSCLC) and it is elevated in NSCLC whilst there is no difference in expression levels of OPNb and OPNc isoforms in healthy individuals compared to NCCLC patients.
The p53 protein family consists of TP53, TP63 and TP73 genes, each of them produces various spliced variants. They are involved in many various cellular functions and it was shown that their high expression levels contribute to tumorigenesis and can affect tumor response to some therapies [109].
5 Concluding remarks
The subset of protein isoforms and splice variants have been shown to be biological distinct and involved in various biological processes. This strongly suggests that many other protein isoforms are of importance as well. The improvement in routinely applied proteomic methods and progress in development of novel approaches, techniques and instrumentation promise the possibility for better and more precise protein analysis as well as for specific protein isoform analysis. In proteomic discovery methods, the approaches of intact protein separation or direct MS analysis have been proposed to enhance detection of protein isoforms/splice variants. However, advancing commercial MS instrumentation and data processing software, such as triple quadrupoles for targeted protein isoform analysis by MRM/SRM or newly developed SWATH approach in TripleTOF 5600 System will allow the investigation and targeting of selected isoform-specific peptides (and perhaps proteins) in large numbers for enhanced detection. When coupled with quantitative standards (e.g. IS), it should allow for greatly improved analysis of protein isoforms to determine their expression profiles across the cell types, disease states and during development and aging. Linking the identification and quantification of a particular isoform to disease state is the first important step, followed by determination of the functional consequences. This will be necessary to completely understand the impact of biological diversity evoked by the expression of various protein isoforms.
Acknowledgments
Supported by the National Heart, Lung, and Blood Institute Proteomic Initiative - contract NHLBI-HV-10-05 (2) (JEV), by NHLBI PO1 HL10026 (Glycoconjungates and Cardiovascular Disease) (JEV), by the AHA grant in-aid (JEV), and by institutional support RVO: 68081715 (MS).
Footnotes
Conflict of interest: The authors have declared no conflict of interest.
References
- 1.Perrin BJ, Ervasti JM. The actin gene family: function follows isoforms. Cytoskeleton. 2010;67:630–634. doi: 10.1002/cm.20475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grabowski PJ, Black DL. Alternative RNA splicing in the nervous system. Prog Neurobiol. 2001;65:289–308. doi: 10.1016/s0301-0082(01)00007-7. [DOI] [PubMed] [Google Scholar]
- 3.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 4.Blencowe BJ. Alternative splicing: New insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 5.Bracco L, Throo E, Cochet O, Einstein R, et al. Methods and platforms for the quantification of splice variants' expression. Prog Mol Subcell Biol. 2006;44:1–25. doi: 10.1007/978-3-540-34449-0_1. [DOI] [PubMed] [Google Scholar]
- 6.Schindler S, Heiner M, Platzer M, Szafranski K. Comparison of methods for quantification of subtle splice variants. Electrophoresis. 2009;30:3674–3681. doi: 10.1002/elps.200900292. [DOI] [PubMed] [Google Scholar]
- 7.Howard BE, Heber S. Towards reliable isoform quantification using RNA-SEQ data. BMC Bioinformatics. 2010;11(3):S6. doi: 10.1186/1471-2105-11-S3-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mironov AA, Fickett JW, Gelfand MS. Frequent alternative splicing of human genes. Genome Res. 1999;9:1288–1293. doi: 10.1101/gr.9.12.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan Q, Shai O, Lee LJ, Frey BJ, et al. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 10.Sorek R, Shamir R, Ast G. How prevalent is functional alternative splicing in the human genome? Trends Genet. 2004;20:68–71. doi: 10.1016/j.tig.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 11.Lareau LF, Green RE, Bhatnagar RS, Brenner SE. The evolving roles of alternative splicing. Curr Opin Struct Biol. 2004;14:273–282. doi: 10.1016/j.sbi.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Baralle D, baralle M. Splicing in action: assessing disease causing sequence changes. J Med Genet. 2005;42:737–748. doi: 10.1136/jmg.2004.029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 14.Brinkman BM. Splice variants as cancer biomarkers. Clin Biochem. 2004;37:584–594. doi: 10.1016/j.clinbiochem.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 15.Yi Q, Tang L. Alternative spliced variants as biomarkers of colorectal cancer. Curr Drug Metab. 2011;12:966–974. doi: 10.2174/138920011798062355. [DOI] [PubMed] [Google Scholar]
- 16.Omenn GS, Yocum AK, Menon R. Alternative splice variants, a new class of protein cancer biomarker candidates: findings in pancreatic cancer and breast cancer with systems biology implications. Dis Markers. 2010;28:241–251. doi: 10.3233/DMA-2010-0702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hilmi C, Guyot M, Pages G. VEGF splice variants: possible role of anti-angiogenesis therapy. J Nucleic Acids. 2012;2012:162692. doi: 10.1155/2012/162692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blair CA, Zi X. Potential molecular targeting of splice variants for cancer treatment. Indian J Exp Biol. 2011;49:836–839. [PMC free article] [PubMed] [Google Scholar]
- 19.Miranda ER, De Marco L, Soares MM. Splicing variants impact in thyroid normal physiology and pathological conditions. Arq Bras Endocrinol Metabol. 2009;53:709–715. doi: 10.1590/s0004-27302009000600003. [DOI] [PubMed] [Google Scholar]
- 20.Srebrow A, Kornblihtt AR. The connection between splicing and cancer. J Cell Sci. 2006;119:2635–2641. doi: 10.1242/jcs.03053. [DOI] [PubMed] [Google Scholar]
- 21.Pio R, Montuenga LM. Alternative splicing in lung cancer. J Thorac Oncol. 2009;4:674–678. doi: 10.1097/JTO.0b013e3181a520dc. [DOI] [PubMed] [Google Scholar]
- 22.Hayes NV, Gullick WJ. The neuregulin family of genes and their multiple splice variants in breast cancer. J Mammary Gland Biol Neoplasia. 2008;13:205–214. doi: 10.1007/s10911-008-9078-4. [DOI] [PubMed] [Google Scholar]
- 23.Gilli F. Role of differential expression of interferon receptor isoforms on the response of multiple sclerosis patients to therapy with interferon beta. J Interferon Cytokine Res. 2010;30:733–741. doi: 10.1089/jir.2010.0098. [DOI] [PubMed] [Google Scholar]
- 24.Yamazaki T, Walchli S, Fujita T, Ryser S, et al. Splice variants of enigma homolog, differentially expressed during heart development, promote or prevent hypertrophy. Cardiovasc Res. 2010;86:374–382. doi: 10.1093/cvr/cvq023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Youinou P, Pers JO. The late news on baff in autoimmune diseases. Autoimmun Rev. 2010;9:804–806. doi: 10.1016/j.autrev.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 26.Hansson O, Zhou Y, Renstrom E, Osmark P. Molecular function of TCF7L2: Consequences of TCF7L2 splicing for molecular function and risk for type 2 diabetes. Curr Diab Rep. 2010;10:444–451. doi: 10.1007/s11892-010-0149-8. [DOI] [PubMed] [Google Scholar]
- 27.Gabut M, Samavarchi-Tehrani P, Wang X, Slobodeniuc V, et al. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell. 2011;147:132–146. doi: 10.1016/j.cell.2011.08.023. [DOI] [PubMed] [Google Scholar]
- 28.Kelleher NL. Top-down proteomics. Anal Chem. 2004;76:197A–203A. [PubMed] [Google Scholar]
- 29.Bogdanov B, Smith RD. Proteomics by FTICR mass spectrometry: top down and bottom up. Mass Spectrom Rev. 2005;24:168–200. doi: 10.1002/mas.20015. [DOI] [PubMed] [Google Scholar]
- 30.Hung CW, Tholey A. Tandem mass tag protein labeling for top-down identification and quantification. Anal Chem. 2012;84:161–170. doi: 10.1021/ac202243r. [DOI] [PubMed] [Google Scholar]
- 31.Lee JL, Chen H, Liu T, Berkman CE, et al. High resolution time-of-flight mass analysis of the entire range of intact singly-charged proteins. Anal Chem. 2011;83:9406–9412. doi: 10.1021/ac202001z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tipton JD, Tran JC, Catherman AD, Ahlf DR, et al. Analysis of intact protein isoforms by mass spectrometry. J Biol Chem. 2011;286:25451–25458. doi: 10.1074/jbc.R111.239442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia BA. What does the future hold for Top Down mass spectrometry? J Am Soc Mass Spectrom. 2010;21:193–202. doi: 10.1016/j.jasms.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 34.Siuti N, Kelleher NL. Decoding protein modifications using top-down mass spectrometry. Nat Methods. 2007;4:817–821. doi: 10.1038/nmeth1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pesavento JJ, Mizzen CA, Kelleher NL. Quantitative analysis of modified proteins and their positional isomers by tandem mass spectrometry: human histone H4. Anal Chem. 2006;78:4271–4280. doi: 10.1021/ac0600050. [DOI] [PubMed] [Google Scholar]
- 36.Roth MJ, Forbes AJ, Boyne MT, 2nd, Kim YB, et al. Precise and parallel characterization of coding polymorphisms, alternative splicing, and modifications in human proteins by mass spectrometry. Mol Cell Proteomics. 2005;4:1002–1008. doi: 10.1074/mcp.M500064-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tran JC, Zamdborg L, Ahlf DR, Lee JE, et al. Mapping intact protein isoforms in discovery mode using top-down proteomics. Nature. 2011;480:254–258. doi: 10.1038/nature10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H, Hanash S. Intact-protein based sample preparation strategies for proteome analysis in combination with mass spectrometry. Mass Spectrom Rev. 2005;24:413–426. doi: 10.1002/mas.20018. [DOI] [PubMed] [Google Scholar]
- 39.Stastna M, Zhang P, Murphy A, Van Eyk JE. Muscle: Fundamental Biology and Mechanisms of Disease. In: Hill JA, Olson EN, editors. Cardiovascular proteomics: Assessment of protein post-translational modifications. Chapter 19. Academic Press; London: 2012. pp. 261–271. [Google Scholar]
- 40.McLafferty FW, Breuker K, Jin M, Han X, et al. Top-down MS, a powerful complement to the high capabilities of proteolysis proteomics. FEBS Journal. 2007;274:6256–6268. doi: 10.1111/j.1742-4658.2007.06147.x. [DOI] [PubMed] [Google Scholar]
- 41.Chalmers MJ, Mackay CL, Hendrickson CL, Wittke S, et al. Combined top-down and bottom-up mass spectrometric approach to characterization of biomarkers for renal disease. Anal Chem. 2005;77:7163–7171. doi: 10.1021/ac050983o. [DOI] [PubMed] [Google Scholar]
- 42.Millea KM, Krull IS, Cohen SA, Gebler JC, et al. Integration of multidimensional chromatographic protein separations with a combined “top-down” and “bottom-up” proteomic strategy. J Proteome Res. 2006;5:135–146. doi: 10.1021/pr050278w. [DOI] [PubMed] [Google Scholar]
- 43.Durbin KR, Tran JC, Zamdborg L, Sweet SM, et al. Intact mass detection, interpretation, and visualization to automate top-down proteomics on a large scale. Proteomics. 2010;10:3589–3597. doi: 10.1002/pmic.201000177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang H, Hanash S. Intact-protein analysis system for discovery of serum-based disease biomarkers. Methods Mol Biol. 2011;728:69–85. doi: 10.1007/978-1-61779-068-3_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doucette AA, Tran JC, Wall MJ, Fitzsimmons S. Intact proteome fractionation strategies compatible with mass spectrometry. Expert Rev Proteomics. 2011;8:787–800. doi: 10.1586/epr.11.67. [DOI] [PubMed] [Google Scholar]
- 46.Capriotti AL, Cavaliere C, Foglia P, Samperi R, et al. Intact protein separation by chromatographic and/or electrophoretic techniques for top-down proteomics. J Chromatogr A. 2011;1218:8760–8776. doi: 10.1016/j.chroma.2011.05.094. [DOI] [PubMed] [Google Scholar]
- 47.Staub A, Guillarme D, Schappler J, Veuthey JL, et al. Intact protein analysis in the biopharmaceutical field. J PharmBiomed Anal. 2011;55:810–822. doi: 10.1016/j.jpba.2011.01.031. [DOI] [PubMed] [Google Scholar]
- 48.Staub A, Zurlino D, Rudaz S, Veuthey JL, et al. Analysis of peptides and proteins using sub-2 μm fully porous and sub-3 μm shell particles. J Chromatogr A. 2011;1218:8903–8914. doi: 10.1016/j.chroma.2011.07.051. [DOI] [PubMed] [Google Scholar]
- 49.Hong G, Gao M, Yan G, Guan X, et al. Optimization of two-dimensional high performance liquid chromatographic columns for highly efficient separation of intact proteins. Se Pu. 2010;28:158–162. doi: 10.3724/sp.j.1123.2012.00158. [DOI] [PubMed] [Google Scholar]
- 50.Yu W, Li Y, Deng C, Zhang X. Comprehensive two-dimensional separation in coupling of reversed-phase chromatography with capillary isoelectric focusing followed by MALDI-MS identification using on-target digestion for intact protein analysis. Electrophoresis. 2006;27:2100–2110. doi: 10.1002/elps.200500820. [DOI] [PubMed] [Google Scholar]
- 51.Wang Y, Balgley BM, Rudnick PA, Lee CS. Effects of chromatography conditions on intact protein separations for top-down proteomics. J Chromatogr A. 2005;1073:35–41. doi: 10.1016/j.chroma.2004.08.140. [DOI] [PubMed] [Google Scholar]
- 52.Roth MJ, Plymire DA, Chang AN, Kim J, et al. Sensitive and reproducible intact mass analysis of complex protein mixture with superficially porous capillary reversed-phase liquid chromatography mass spectrometry. Anal Chem. 2011;83:9586–9592. doi: 10.1021/ac202339x. [DOI] [PubMed] [Google Scholar]
- 53.Strancar A, Podgornik A, Barut M, Necina R. Short monolithic columns as stationary phases for biochromatography. Adv Biochem Eng Biotechnol. 2002;76:49–85. doi: 10.1007/3-540-45345-8_2. [DOI] [PubMed] [Google Scholar]
- 54.Xie S, Allington RW, Frechet JM, Svec F. Porous polymer monoliths: an alternative to classical beads. Adv Biochem Eng Biotechnol. 2002;76:87–125. doi: 10.1007/3-540-45345-8_3. [DOI] [PubMed] [Google Scholar]
- 55.Iwasaki M, Sugiyama N, Tanaka N, Ishihama Y. Human proteome analysis by using reversed phase monolithic silica capillary columns with enhanced sensitivity. J Chromatogr A. 2012;1228:292–297. doi: 10.1016/j.chroma.2011.10.059. [DOI] [PubMed] [Google Scholar]
- 56.Liu J, Ren L, Liu Y, Li H, et al. Weak anion exchange chromatographic profiling of glycoprotein isoforms on a polymer monolithic capillary. J Chromatogr A. 2012;1218:276–282. doi: 10.1016/j.chroma.2011.08.079. [DOI] [PubMed] [Google Scholar]
- 57.Eeltink S, Svec F. Recent advances in the control of morphology and surface chemistry of porous polymer-based monolithic stationary phases and their application in CEC. Electrophoresis. 2007;28:137–147. doi: 10.1002/elps.200600573. [DOI] [PubMed] [Google Scholar]
- 58.Guan X, Yan G, Gao M, Hong G. Intact-protein trapping columns for proteomic analysis in capillary high-performance liquid chromatography. J Chromatogr A. 2010;1217:6875–6881. doi: 10.1016/j.chroma.2010.08.051. [DOI] [PubMed] [Google Scholar]
- 59.Fausnaugh JL, Pfankoch E, Gupta S, Regnier FE. High-performance hydrophobic interaction chromatography of proteins. Anal Biochem. 1984;137:464–472. doi: 10.1016/0003-2697(84)90114-3. [DOI] [PubMed] [Google Scholar]
- 60.Goheen SC, Engelhorn SC. Hydrophobic interaction high-performance liquid chromatography of proteins. J Chromatogr. 1984;317:55–65. [Google Scholar]
- 61.Cummins PM, O'Connor BF. Hydrophobic interaction chromatography. Methods Mol Biol. 2011;681:431–437. doi: 10.1007/978-1-60761-913-0_24. [DOI] [PubMed] [Google Scholar]
- 62.Fausnaugh JL, Kennedy LA, Regnier FE. Comparison of hydrophobic-interaction and reversed-phase chromatography of proteins. J Chromatogr. 1984;317:141–155. doi: 10.1016/s0021-9673(01)91654-1. [DOI] [PubMed] [Google Scholar]
- 63.Geng X, Ke C, Chen G, Liu P, et al. On-line separation of native proteins by two-dimensional liquid chromatography using a single column. J Chromatogr A. 2009;1216:3553–3562. doi: 10.1016/j.chroma.2009.01.085. [DOI] [PubMed] [Google Scholar]
- 64.Mahn A, Reyes A, Zamorano M, Cifuentes W, et al. Depletion of highly abundant proteins in blood plasma by hydrophobic interaction chromatography for proteomic analysis. J chromatogr B Analyt Technol Biomed Life Sci. 2010;878:1038–1044. doi: 10.1016/j.jchromb.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 65.Murphy PJ, Stone OJ, Anderson ME. Automated hydrophobic interaction chromatography column selection for use in protein purification. J Vis Exp. 2011 doi: 10.3791/3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Palani S, Gueorguieva L, Rinas U, Seidel-Morgenstern A, et al. Recombinant protein purification using gradient-assisted simulated moving bed hydrophobic interaction chromatography. Part I: selection of chromatographic system and estimation of adsorption isotherms. J Chromatogr A. 2011;1218:6396–6401. doi: 10.1016/j.chroma.2011.06.111. [DOI] [PubMed] [Google Scholar]
- 67.Mikolajczyk SD, Grauer LS, Millar LS, Hill TM, et al. A precursor form of PSA (pPSA) is a component of the free PSA in prostate cancer serum. Urology. 1997;50:710–714. doi: 10.1016/S0090-4295(97)00449-4. [DOI] [PubMed] [Google Scholar]
- 68.Sikanen T, Aura S, Franssila S, Kotiaho T. Microchip capillary electrophoresis-electrospray ionization-mass spectrometry of intact proteins using uncoated Ormocomp microchips. Anal Chim Acta. 2012;711:69–76. doi: 10.1016/j.aca.2011.10.059. [DOI] [PubMed] [Google Scholar]
- 69.Kim KH, Moon MH. Chip-type asymmetrical flow field-flow fractionation channel coupled with mass spectrometry for top-down protein identification. Anal Chem. 2011;83:8652–8658. doi: 10.1021/ac202098b. [DOI] [PubMed] [Google Scholar]
- 70.Tran JC, Doucette AA. Multiplexed size separation of intact proteins in solution phase for mass spectrometry. Anal Chem. 2009;81:6201–6209. doi: 10.1021/ac900729r. [DOI] [PubMed] [Google Scholar]
- 71.Taylor GK, Kim YB, Forbes AJ, Meng F, et al. Web and database software for identification of intact proteins using “top down” mass spectrometry. Anal Chem. 2003;75:4081–4086. doi: 10.1021/ac0341721. [DOI] [PubMed] [Google Scholar]
- 72.Durbin KR, Tran JC, Zamdborg L, Sweet SM, et al. Intact mass detection, interpretation, and visualization to automate Top-Down proteomics on a large scale. Proteomics. 2010;10:3589–3597. doi: 10.1002/pmic.201000177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Breuker K, Jin M, Han X, Jiang H, et al. Top-down identification and characterization of biomolecules by mass spectrometry. J Am Soc Mass Spectrom. 2008;19:1045–1053. doi: 10.1016/j.jasms.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fagerquist CK, Sultan O. Induction and identification of disulfide-intact and disulfide reduced β-subunit of Shiga toxin 2 from Escherichia coli O157:H7 using MALDI-TOF-TOF-MS/MS and top-down proteomics. Analyst. 2011;136:1739–1746. doi: 10.1039/c0an00909a. [DOI] [PubMed] [Google Scholar]
- 75.Poulsen K, Bahl JM, Tanassi JT, Simonsen AH, et al. Characterization and stability of transhyretin isoforms in cerebrospinal fluid examined by immunoprecipitation and high-resolution mass spectrometry of intact protein. Methods. 2012;56:284–292. doi: 10.1016/j.ymeth.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 76.Michalski A, Damoc E, lange O, Denisov E, et al. Ultra high resolution linear ion trap Orbitrap mass spectrometer (Orbitrap Elite) facilitates top down LC MS/MS and versatile peptide fragmentation modes. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.O111.013698. O111.013698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Simmons D. A practical introduction to analysis of intact proteins on the TripleTOF™ 5600 System. 2011 AB SCIEX. publication number: 3240911-01, www.absciex.com.
- 78.Lourette N, Smallwood H, Wu S, Robinson EW, et al. A top-down LC-FTICR MS-based strategy for characterizing oxidized calmodulin in activated macrophages. J Am Soc Mass Spectrom. 2010;21:930–939. doi: 10.1016/j.jasms.2010.02.027. [DOI] [PubMed] [Google Scholar]
- 79.Zhang H, Cui W, Wen J, Blankenship RE, et al. Native electrospray and electron-capture dissociation in FTIR mass spectrometry provide top-down sequencing of a protein component in an intact protein assembly. J Am Soc Mass Spectrom. 2010;21:1966–1968. doi: 10.1016/j.jasms.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim M, Pandey A. Electron transfer dissociation mass spectrometry in proteomics. Proteomics. 2012;12:530–542. doi: 10.1002/pmic.201100517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee JE, Kellie JF, Tran JC, Tipton JD, et al. A robust two-dimensional separation for top-down tandem mass spectrometry of the low mass proteome. J Am Soc Mass Spectrom. 2009;20:2183–2191. doi: 10.1016/j.jasms.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ge Y, Lawhorn BG, ElNaggar M, Strauss E, et al. Top down characterization of larger proteins (45 kDa) by electron capture dissociation mass spectrometry. J Am Chem Soc. 2002;124:672–678. doi: 10.1021/ja011335z. [DOI] [PubMed] [Google Scholar]
- 83.Kellie JF, Catherman AD, Durbin KR, Tran JC, et al. Robust analysis of the yeast proteome under 50 kDa by molecular-mass-based fractionation and top-down mass spectrometry. Anal Chem. 2012;84:209–215. doi: 10.1021/ac202384v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tipton JD, Tran JC, Catherman AD, Ahlf DR, et al. Nano-LC FTICR tandem mass spectrometry for top-down proteomics: routine baseline unit mass resolution of whole cell lysate proteins up to 72 kDa. Anal Chem. 2012;84:2111–2117. doi: 10.1021/ac202651v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vellaichamy A, Tran JC, Catherman AD, Lee JE, et al. Size-sorting combined with improved nanocapillary liquid chromatography-mass spectrometry for identification of intact proteins up to 80 kDa. Anal Chem. 2010;82:1234–1244. doi: 10.1021/ac9021083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Han X, Jin M, Breuker K, McLafferty FW. Extending top-down mass spectrometry to proteins with masses greater than 200 kilodaltons. Science. 2006;314:109–112. doi: 10.1126/science.1128868. [DOI] [PubMed] [Google Scholar]
- 87.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kitteringham NR, Jenkins RE, Lane CS, Elliott VL, et al. Multiple reaction monitoring for quantitative biomarker analysis in proteomics and metabolomics. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:1229–1239. doi: 10.1016/j.jchromb.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 89.Malmstrom L, Malstrom J, Selevsek N, Rosenberger G. Automated workflow for large-scale selected reaction monitoring experiments. J Proteome Res. 2012;11:1644–1653. doi: 10.1021/pr200844d. [DOI] [PubMed] [Google Scholar]
- 90.Sung HJ, Jeon SA, Ahn JM, Seul KJ, et al. Large-scale isotype-specific quantification of serum amyloid A 1/2 by multiple reaction monitoring in crude sera. J Proteomics. 2012;75:2170–2180. doi: 10.1016/j.jprot.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 91.Andrews GL, Simons BL, Young JB, Hawkridge AM. Performance characteristics of a new hybrid quadrupole time-of-flight tandem mass spectrometer (Triple TOF 5600) Anal Chem. 2011;83:5442–5446. doi: 10.1021/ac200812d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gillet LC, Navarro P, Tate S, Roest H. Targeted data extraction of the MS/MS spectra generated by data independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics. 2012 doi: 10.1074/mcp.O111.016717. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hopfgartner G, Tonoli D, Varesio E. High-resolution mass spectrometry for integrated qualitative and quantitative analysis of pharmaceuticals in biological matrices. Anal Bioanal Chem. 2012;402:2587–2596. doi: 10.1007/s00216-011-5641-8. [DOI] [PubMed] [Google Scholar]
- 94.Atamas SP. Alternative splice variants of cytokines: making a list. Life Sci. 1997;61:1105–1112. doi: 10.1016/s0024-3205(97)00243-9. [DOI] [PubMed] [Google Scholar]
- 95.Steinman L. Nuanced roles of cytokines in three major human brain disorders. J Clin Invest. 2008;118:3557–3563. doi: 10.1172/JCI36532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Blair CA, Zi X. Potential molecular targeting of splice variants for cancer treatment. Indian J Exp Biol. 2011;49:836–839. [PMC free article] [PubMed] [Google Scholar]
- 97.Robinson CJ, Stringer SE. The splice variants of vascular endothelial growth factor (VEGF) and their receptors. J Cell Sci. 2001;114:853–865. doi: 10.1242/jcs.114.5.853. [DOI] [PubMed] [Google Scholar]
- 98.Bates DO, Harper SJ. Therapeutic potential of inhibitory VEGF splice variants. Future Oncol. 2005;1:467–473. doi: 10.2217/14796694.1.4.467. [DOI] [PubMed] [Google Scholar]
- 99.Hilmi C, Guyot M, Pages G. VEGF spliced variants: possible role of anti-angiogenesis therapy. J Nucleic Acids. 2012;2012:162692. doi: 10.1155/2012/162692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Velloso CP, Harridge SD. Insulin-like growth factor-I E peptides: implications for aging skeletal muscle. Scand J Med Sci Sports. 2010;20:20–27. doi: 10.1111/j.1600-0838.2009.00997.x. [DOI] [PubMed] [Google Scholar]
- 101.Boraschi D, Lucchesi D, Hainzl S, Leitner M, et al. IL-37: a new anti-inflammatory cytokine of the IL-1 family. Eur Cytokine Netw. 2011;22:127–147. doi: 10.1684/ecn.2011.0288. [DOI] [PubMed] [Google Scholar]
- 102.White ES, Baralle FE, Muro AF. New insights into form and function of fibronectin splice variants. J Pathol. 2008;216:1–14. doi: 10.1002/path.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.White ES, Muro AF. Fibronectin splice variants: understanding their multiple roles in health and disease using engineered mouse models. IUBMB Life. 2011;63:538–546. doi: 10.1002/iub.493. [DOI] [PubMed] [Google Scholar]
- 104.Marcucci R, Romano M. Thrombopoietin and its splicing variants: structure and functions in thrombopoiesis and beyond. Biochim Biophys Acta. 2008;1782:427–432. doi: 10.1016/j.bbadis.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 105.Dame C, Wolber EM, Freitag P, Hofmann D, et al. Trombopoietin gene expression in the developing human central nervous system. Brain Res Dev Brain Res. 2003;143:217–223. doi: 10.1016/s0165-3806(03)00134-2. [DOI] [PubMed] [Google Scholar]
- 106.Webb KE, Martin JF, Hamsten A, Eriksson P, et al. Polymorphisms in the thrombopoietin gene are associated with risk of myocardial infarction at a young age. Atherosclerosis. 2001;154:703–711. doi: 10.1016/s0021-9150(00)00633-x. [DOI] [PubMed] [Google Scholar]
- 107.Hayes NV, Blackburn E, Boyle MM, Russell GA, et al. Expression of neuregulin 4 splice variants in normal human tissues and prostate cancer and their effects on cell motility. Endocr Relat Cancer. 2010;18:39–49. doi: 10.1677/ERC-10-0112. [DOI] [PubMed] [Google Scholar]
- 108.Wu J, Pungaliya P, Kraynov E, Bates B. Identification and quantification of osteopontin splice variants in the plasma of lung cancer patients using immunoaffinity capture and targeted mass spectrometry. Biomarkers. 2012;17:125–133. doi: 10.3109/1354750X.2011.643485. [DOI] [PubMed] [Google Scholar]
- 109.Wei J, Zaika E, Zaika A. p53 family: Role of protein isoforms in human cancer. J Nucleic Acids. 2012;2012:687359. doi: 10.1155/2012/687359. [DOI] [PMC free article] [PubMed] [Google Scholar]