Significance
Gene identification for complex behavioral traits, alcoholism in particular, has been largely unsuccessful, in part because of the rarity of many causative variants and the heterogeneity and small effect size of the causal loci. Artificially selected animals may be valuable in gene identification. We identified a causal role of metabotropic glutamate receptor 2 (mGluR2) in altered alcohol preference via genomic sequencing of selectively bred rats, linkage analysis in the F2 rats, and function validation in mGluR2 null mice. Our findings represent a valuable contribution to understanding the neurobiology of alcoholism and a strategy for gene identification for complex behavior traits.
Keywords: gene identification, selectively bred lines
Abstract
Identification of genes influencing complex traits is hampered by genetic heterogeneity, the modest effect size of many alleles, and the likely involvement of rare and uncommon alleles. Etiologic complexity can be simplified in model organisms. By genomic sequencing, linkage analysis, and functional validation, we identified that genetic variation of Grm2, which encodes metabotropic glutamate receptor 2 (mGluR2), alters alcohol preference in animal models. Selectively bred alcohol-preferring (P) rats are homozygous for a Grm2 stop codon (Grm2 *407) that leads to largely uncompensated loss of mGluR2. mGluR2 receptor expression was absent, synaptic glutamate transmission was impaired, and expression of genes involved in synaptic function was altered. Grm2 *407 was linked to increased alcohol consumption and preference in F2 rats generated by intercrossing inbred P and nonpreferring rats. Pharmacologic blockade of mGluR2 escalated alcohol self-administration in Wistar rats, the parental strain of P and nonpreferring rats. The causal role of mGluR2 in altered alcohol preference was further supported by elevated alcohol consumption in Grm2 −/− mice. Together, these data point to mGluR2 as an origin of alcohol preference and a potential therapeutic target.
Alcoholism is a moderately to highly heritable disease (1, 2). The search for genetic variation influencing this complex disorder has yielded limited success. In human populations, candidate gene analyses have established roles for polymorphisms of ADH1B and ALDH2, both enzymes involved in alcohol metabolism (3). Genome-wide association studies (4–7) have implicated several genomic regions. However, the genes and functional variants that account for the genetic association signals remain largely unknown. Complex behavioral disorders may be influenced by many different genes. Most functional alleles are rare or uncommon, also contributing to genetic heterogeneity that hampers locus identification in population samples. Individual variants influencing alcoholism are also likely to be probabilistic in their actions, with limited effect sizes on risk of the disease itself. All of these factors pose significant challenges for identification of genes and functional variants contributing to alcoholism by population-based analyses in people.
Model organisms, including selectively bred lines (8–10), offer a potentially powerful framework for genomic analyses to identify genes and their functional variants that contribute to complex disorders. The selective breeding process reduces genetic heterogeneity and enriches to high frequency variants that influence the targeted phenotype. Alcohol-preferring (P) and nonpreferring (NP) rats, a seminal rat model of alcoholism, were bred from Wistar rats by 30–70 generations of bidirectional selection for alcohol preference. These rats model human alcoholism in several ways. They voluntarily consume excessive amounts of alcohol with sustained high blood alcohol concentrations, consume alcohol for CNS effects, show signs of intoxication, and exhibit relapse-like behavior (11). Quantitative trait loci (QTLs) for alcohol preference and consumption, including a large chromosome 4 region, were identified (12, 13). In addition, neurobehavioral differences reminiscent of findings in some human alcoholics have been observed between the two lines, including higher anxiety in P rats (14–16). However, the identities of the functional variants altering alcohol consumption and preference in P and NP rats as well as the genes in which they reside have thus far remained largely elusive.
To identify genes and functional variants influencing alcohol preference, we performed genomic sequencing to uncover coding sequence variants that segregate between P and NP rats. The influence of the segregating variants on alcohol preference was identified by QTL analysis in the F2 rats generated from intercrossing inbred P (iP) and NP (iNP) rats. The causal effect on alcohol preference was further validated by pharmacological challenge and by phenotyping knockout mice. Our strategy led to the identification of Grm2 as a locus influencing alcohol preference. Loss of metabotropic glutamate receptor 2 (mGluR2) function contributes to elevated alcohol consumption.
Results
Genomic Sequencing Identifies Genetic Variants Segregating Between P and NP Rats.
We first identified sequence differences in gene-coding regions between P and NP rats by exome sequencing. We detected 129,170 SNPs in six individually sequenced P rats and six NP rats from independent litters. Among these SNPs, 25,715 showed a uniform homozygous difference between P and NP rats, indicating that they were either selected for alcohol preference or genetically fixed by inbreeding. We comparatively sequenced four Wistar rats to measure the degree of inbreeding and reduction of exonic genetic diversity in P and NP rats (SI Appendix, Fig. S1). A genomic map of the SNPs highlighting genetic differences between P and NP rats (SI Appendix, Fig. S2) revealed several large segregating blocks. These blocks harbored potentially functional loci linked to alcohol preference. Among 2,579 coding sequences (CDSs) segregating SNPs mapping to 1,068 genes (SI Appendix, Table S1), there were two stop codons mapping to separate genes (Grm2 *407 and Lcn2 *137). Thirty-one missense variants were predicted by Polyphen (17) and SIFT (18) to have damaging effects on protein function (SI Appendix, Table S2). To establish that segregating, potentially functional variants were not merely randomly fixed, we performed linkage analysis for the two stop codons and 22 potentially damaging missense variants located in independent genomic regions. We measured alcohol preference and consumption in 384 F2 rats generated by cross-breeding iP and iNP rats. These iP and iNP rats had been produced by 19 generations of inbreeding following the initial 30 generations of selection for alcohol preference (13). Four of the variants in three independent genomic regions (Grm2 C407*, Lcn2 Q137*, Sspo V1868M, and Fam190a P82L; SI Appendix, Table S2 and Fig. S3) were significantly linked to alcohol preference, and among these four variants we have clearly defined the functionality of the Grm2 stop codon and its effects on alcohol preference.
Grm2 *407 Results in Loss of mGluR2 in P Rats.
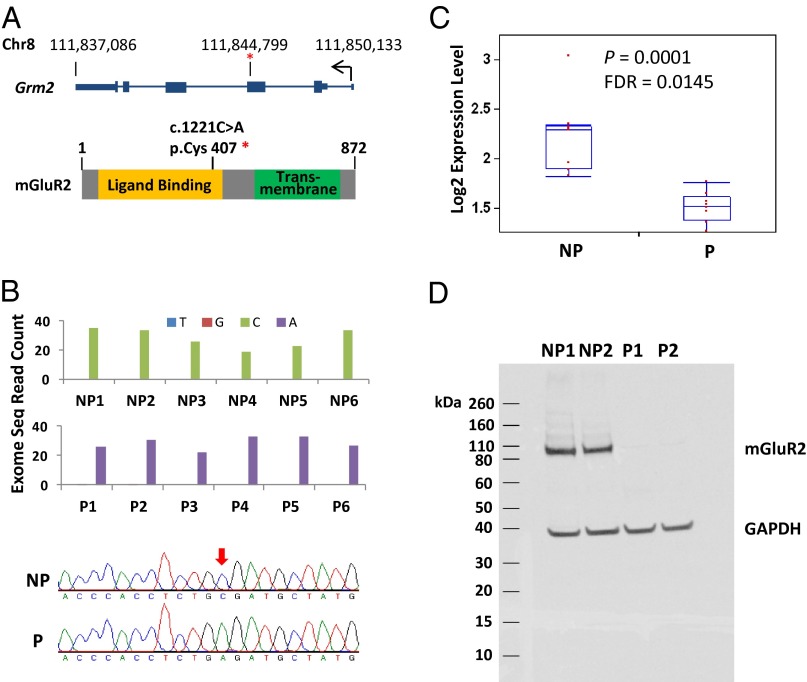
All P rats were homozygous for the Grm2 407 stop codon [c.1221C>A, p.Cys407*, located at nucleotide 111,844,799 of chromosome 8 (Baylor 3.4/rn4) on the minus strand encoding Grm2; Fig. 1A]. The stop codon was absent in NP rats. The *407 allele is predicted to produce a truncated protein containing a partial ligand-binding domain but none of the seven transmembrane domains of mGluR2. Homozygosity of P rats (*407/*407) and NP rats (C407/C407) was verified by Sanger sequencing of genomic DNA in these 12 rats (Fig. 1B) followed by genotyping 125 P rats and 59 NP rats from different litters. Therefore, at generation 70, Grm2 C407* alleles fully segregate between NP and P rats. mRNA was also sequenced and the allelic segregation was also confirmed in the transcripts of P and NP rats. Significantly lower levels of Grm2 transcript were found in brain of P rats (Fig. 1C), possibly reflecting nonsense-mediated decay. In contrast with NP rats in which mGluR2 protein expression was clearly detected in brain by Western blot using a monoclonal antibody directed against the mGluR2 N-terminal region (19), expression of mGluR2 protein was undetectable in P rats (Fig. 1D), confirming loss of this receptor.
Fig. 1.
Grm2 *407 results in the loss of mGluR2 protein expression in P rats. (A) Genomic location of the Grm2 *407 variant. (B) (Upper) Exome sequencing read counts for each nucleotide at nt 111,844,799 of Chr 8, in six NP and six P rats. (Lower) Sanger sequencing validation for Grm2 *407 in the six NP and six P rats. Red arrow indicates the nucleotide position. (C) Hippocampal mRNA levels (normalized counts) measured by RNA-Seq. (D) Western blot of hippocampal mGluR2 protein in NP and P rats. A mouse monoclonal anti-mGluR2 N-terminal antibody (ab15672; Abcam) was used. A GAPDH antibody (MAB374; Millipore) was used as control.
mGluR-Mediated Synaptic Depression Is Impaired in P Rats.
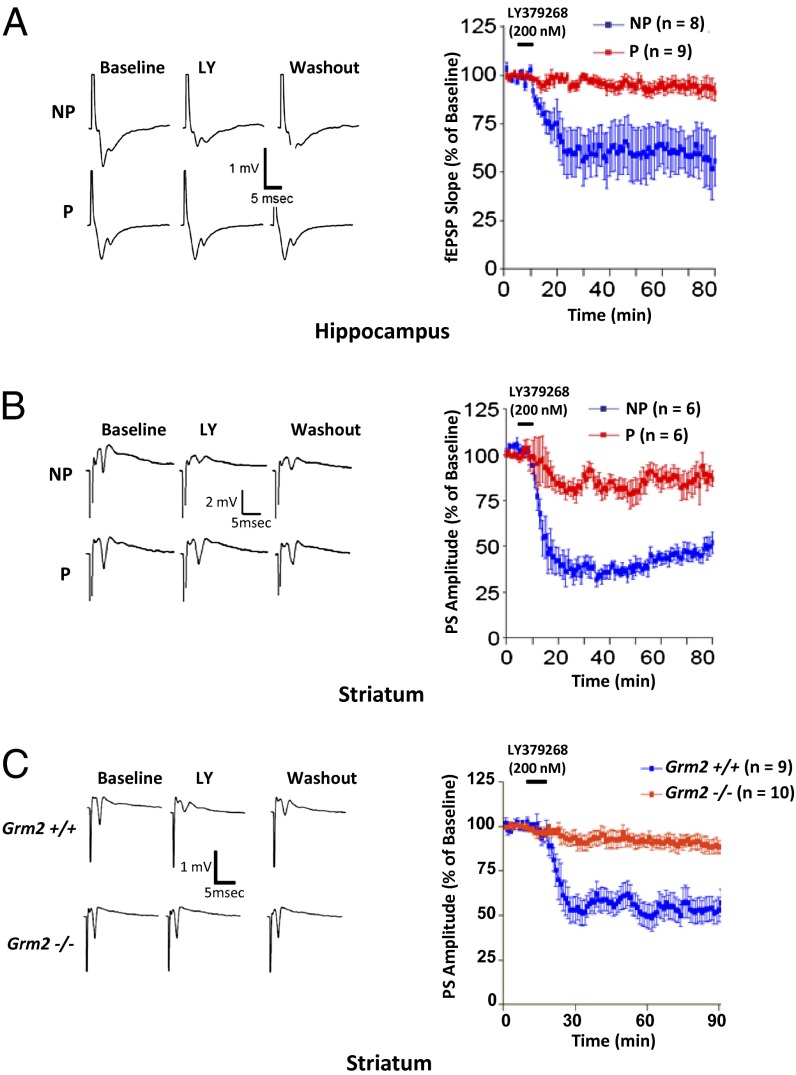
mGluR2 along with mGluR3 is a group II metabotropic glutamate receptor. These receptors are primarily located presynaptically and function as auto-receptors. mGluR2 is thought to be the receptor primarily responsible (20) for the group II mGluR-mediated presynaptic depression of glutamatergic neurons projecting onto striatum (21). Using field potential recordings, we examined effects of the group II mGluR agonist LY379268 on the synaptically evoked field excitatory postsynaptic potential (fEPSP) or population spike (PS) in dentate gyrus (DG)/hippocampal (Fig. 2A) and striatal slices (Fig. 2B) from adult (2- to 3-mo-old) P and NP rats. Whereas application of 200 nM LY379268 produced a 40–60% decrease below baseline levels in both regions in NP rats, consistent with the synaptic depression previously observed in DG and striatum of Sprague–Dawley rats (20, 21), we observed less than 10% depression in P rats, consistent with uncompensated loss of mGluR2 function.
Fig. 2.
Impairment of group II mGluR-mediated synaptic depression in hippocampus and striatum of P rats (A and B) and Grm2−/− mice (C). (Left) Representative voltage traces of field potentials evoked in dentate gyrus/ hippocampus (A) and striatum (B) of P and NP rats and striatum of Grm2−/− mice (C) before and after application of group II mGluR agonist LY, and after prolonged artificial cerebrospinal fluid wash. Calibrations in the upper left-hand trace apply to all traces. (Right) Normalized fEPSP slope or PS amplitude (mean ± SD) during the application (black line) of LY to hippocampal (A) or striatal (B) slices of P and NP rats and striatal slices of Grm2−/− mice (C).
Loss of mGluR2 Increases Alcohol Consumption and Preference in iP/iNP F2 Rats.
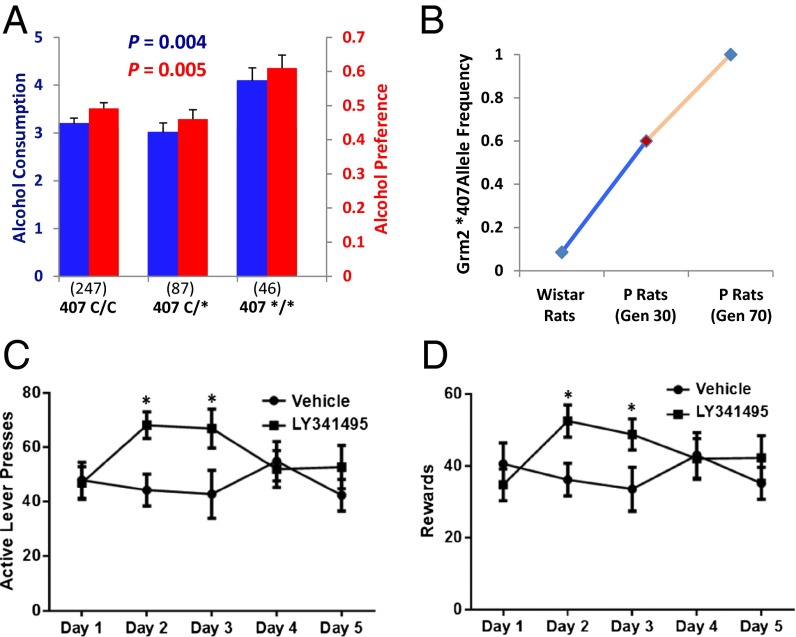
In F2 rats in which the P and NP genetic backgrounds of Grm2 genotypes have been scrambled by meiotic recombination, homozygosity for the Grm2 stop codon led to a 32% increase in alcohol consumption and a 28% increase in preference (Fig. 3A). There was no difference in alcohol consumption and preference between heterozygotes and noncarriers of the stop codon, indicating that the effect of the stop codon on alcohol preference may be recessive. The overall frequency of *407 in the F2 rats was 0.236, indicating that at the 30th generation of selection not all P rats were *407 carriers (although the allele frequency may already have been as high as 0.5). To evaluate the *407 frequency in the parents of these F2s, we genotyped five inbred P and four inbred NP rats that were the grandparents of these particular F2s, an experiment that was limited to the samples retrospectively available. In these animals, three of the five P grandparents were homozygous for the stop codon. None of the other rats, including the P rats, carried *407. DNA from the original Wistar stock used to make P and NP rats was unavailable; however, we were able to investigate the frequency of the *407 allele in this commonly used rat strain by genotyping 64 outbred Wistar rats from Charles River Laboratories, a widely used commercial source, all from independent litters. In these outbred Wistar rats, there were 11 C407/*407 heterozygotes, and no homozygotes, for an allele frequency of 0.086. Therefore, approximately one in six Charles River Wistar rats carried the Grm2 stop codon. The reciprocal change in frequency of Grm2 *407 (Fig. 3B) in P and NP rats, the loss of mGluR2 protein in P rats, linkage of *407 to alcohol preference and consumption, and location of Grm2 in a large genomic block segregating between P and NP rats all point to selection of Grm2 *407 for alcohol preference.
Fig. 3.
Impairment of mGluR2 function increases alcohol consumption and preference in iP × iNP F2 rats and Wistar rats. (A) Alcohol consumption (grams of ethanol per kilogram body weight per day) and preference (ethanol/total fluid) were compared across the Grm2 C407* genotype groups (numbers of rats in parentheses) in F2 rats by ANOVA (consumption: df = 2, F = 5.582, P = 0.004; preference: df = 2, F = 5.309, P = 0.005). (B) Grm2 *407 frequencies in the Wistar rats and P rats. Wistar rats (n = 64) and P rats at generation 70 (n = 139) were directly genotyped. Frequency in P rats at generation 30 was inferred from the F2 (n = 380) genotypes and the genotypes of five inbred grandparental P rats of the F2s. The drinking phenotypes were selected every generation before and at generation 30 and every three generations thereafter. (C and D) mGluR2/3 antagonism escalates alcohol self-administration in Wistar rats. After 14 d of training, vehicle or LY341495 (3 mg/kg) (n = 16 per group) were injected (i.p.) 30 min before sessions for five consecutive days. (C) Total numbers of lever presses (mean ± SD) in the 30-min session for each group are shown [two-way ANOVA (treatment and day): F4, 56 = 6.58, P = 0.0002]. (D) numbers of lever presses for 20% alcohol during the intervals when reward was available [F4, 56 = 3.42, P = 0.014]. *Significant by Newman–Keuls test.
Blockade of mGluR2 in Wistar Rats Escalates Alcohol Self-Administration.
We tested the effect of mGluR2 blockade on alcohol intake of Wistar rats with an mGluR2/3 antagonist, LY341495. We used Wistar rats because P rats do not express functional mGluR2, and NP rats are strongly averse to alcohol. The alcohol aversion of NP rats is likely influenced by unidentified loci selected for the trait and may obscure effects of mGluR2 antagonism. Injection of LY341495 (3 mg/kg i.p. daily for 5 d) into Wistar rats trained in an operant self-reinforcement paradigm resulted in significant, although possibly short-lived, escalation of active lever pressing (Fig. 3C) and self-administration of 20% alcohol (Fig. 3D). These results indicate that mGluR2 influences alcohol self-reinforcement, although it is necessary to exercise caution in interpreting the results because in the purely pharmacobehavioral analysis the lack of specific mGluR2 antagonist makes it difficult to distinguish the roles of mGluR2 and mGluR3 and the transient effects may imply compensation by the glutamate system, possibilities warranting further investigation. Overall, these results are also convergent with prior pharmacological studies using two structurally related nonselective mGluR2/3 agonists, LY379268 and LY404039, that suppressed relapse to alcohol seeking in rats (22, 23).
Alcohol Consumption and Preference Are Elevated in mGluR2 Null Mice.
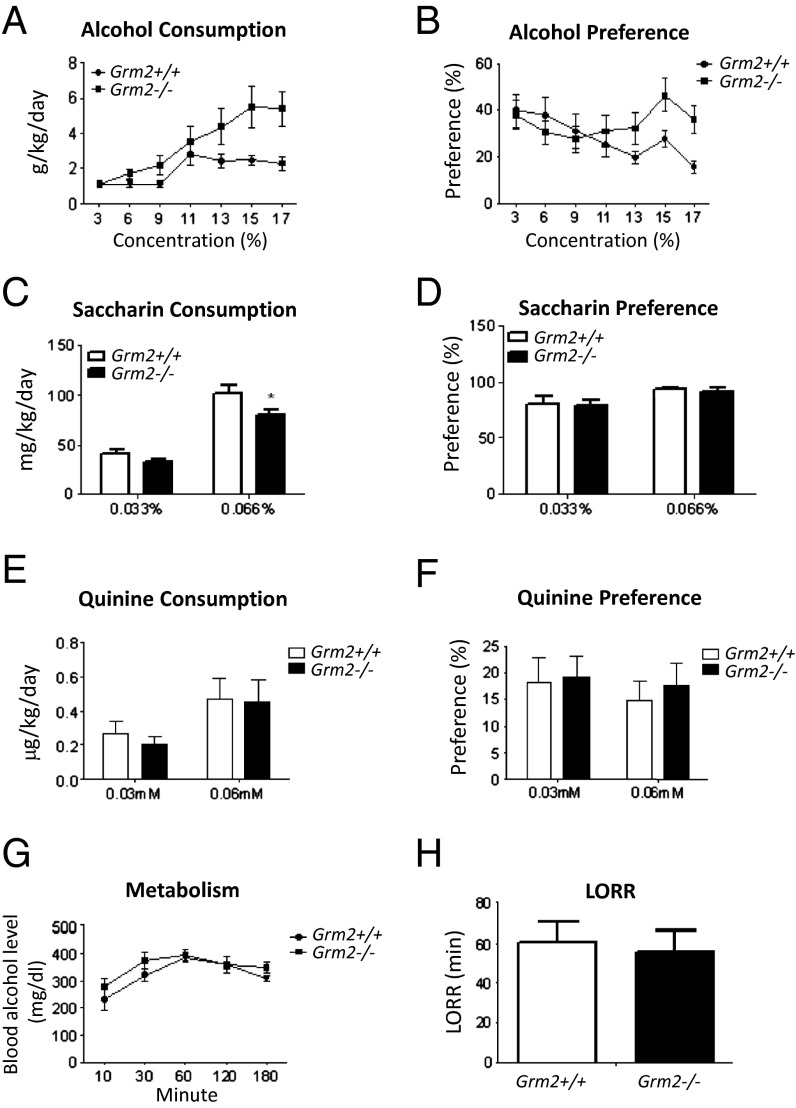
To further validate whether loss of mGluR2 causally contributes to elevated alcohol consumption, we examined alcohol drinking in Grm2−/− mice (24). Unlike P and NP rats that differ at many other genetic loci in addition to Grm2, which might be compromising factors for validation via pharmacological or genetic manipulations of P rats, Grm2 −/− and +/+ mice provided a controlled model for comparison. We confirmed the loss of the mGluR2 protein in Grm2−/− mice (SI Appendix, Fig. S4) using the monoclonal antibody that we also used to show loss of the receptor protein in P rats. The loss of mGluR2 in Grm2−/− mice also led to an uncompensated impairment of mGluR agonist-induced depression of PS amplitude (Fig. 2C), in parallel with the effect of the stop codon in P rats. We then used a two-bottle free-choice model and an escalation procedure in which alcohol concentration was gradually increased from 3 to 17% over 80 d. Grm2−/− mice escalated their alcohol consumption to a much greater degree than controls (Fig. 4A). Separation between the genotypes was most pronounced at the two highest alcohol concentrations at which consumption motivated by pharmacological alcohol effects is known to dominate over intake for taste, calories, or other nonpharmacological effects (25). Preference for alcohol was also differentially affected (Fig. 4B). Whereas controls decreased their preference at high alcohol concentrations in a manner typically seen owing to the aversive taste, a sustained preference was observed in Grm2−/− mice, indicating that differences in consumption were not simply driven by altered total fluid intake. Furthermore, preference for saccharin did not differ between genotypes, and in contrast to their increased alcohol consumption saccharin consumption was decreased in the Grm2 −/− mice at the higher concentration. Neither consumption nor preference for quinine differed between genotypes (Fig. 4 C–F). These results indicate that the observed differences in alcohol consumption were not a result of taste differences. Finally, deletion of mGluR2 did not alter alcohol metabolism and elimination, or sedative effects of alcohol as measured by loss of righting reflex (Fig. 4 G and H). Together with our findings identifying a Grm2 stop codon as a genetic factor altering alcohol preference in the selectively bred P rats and that blockade of mGluR2 escalates alcohol intake in Wistar rats, the Grm2 null mice data support a causal role of mGluR2 in consumption of alcohol for its pharmacological properties.
Fig. 4.
Grm2 knockout increases alcohol consumption and preference in mice. (A) With increasing alcohol concentrations, Grm2−/− mice escalated their alcohol consumption to a greater degree than wild-type controls (repeated measures ANOVA; genotype, F1,84 = 4.6, P < 0.05; concentration (conc), F6,84 = 14.8, P < 0.001; conc × genotype interaction, F6,84 = 3.9, P < 0.002; Newman–Keuls post hoc: P = 0.02 for 15% and P = 0.02 for 17% alcohol, n = 8 per group). (B) Alcohol preference remained relatively unaltered in Grm2−/− mice while decreasing in wild-type mice (genotype, F1,84 = 0.89, not significant; conc, F6,84 = 3.66, P < 0.003; conc × genotype ANOVA interaction, F6,84 = 4.0, P < 0.01; n = 8 per group). (C–F) Taste preferences for saccharin (D) and quinine (F), sweet and bitter, two taste components for alcohol, were not altered in Grm2−/− mice (n = 11 per group, C–F). Grm2−/− mice consumed less saccharin (C) at a 0.066% concentration (genotype, F1, 20 = 3.21, not significant; conc F1, 20 = 135, P < 0.001; conc × genotype ANOVA interaction, F1, 20 = 4.49, P < 0.05, Newman–Keuls post hoc P = 0.001). No differences in quinine consumption were observed between Grm2−/− mice and wild-type mice (E). (G) Blood alcohol concentrations did not differ between genotypes when measured for 3 h following injection of a large bolus dose of alcohol, 4 g/kg (n = 5). (H) There were no differences in duration of loss righting reflex (LORR) induced by alcohol (4 g/kg) between Grm2−/− and wild-type mice (n = 7–8 per group).
Differential Gene Expression Converges with Genetic Difference Between P and NP.
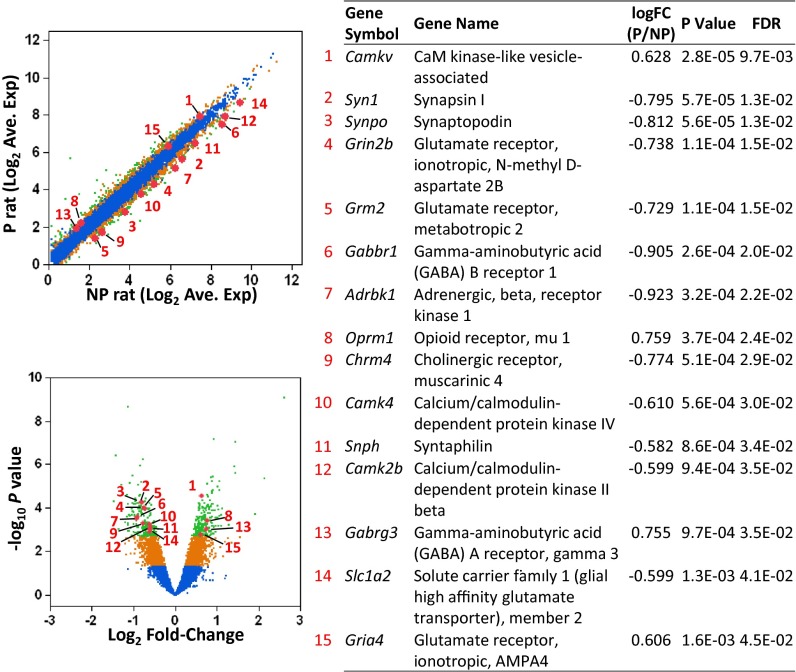
Hippocampus, as well as several other brain regions including prefrontal cortex, is known to be critically involved in drug/alcohol addictions. We performed hippocampal transcriptome analysis of P and NP rats. The results point to a global alteration in synaptic function. Among 11,406 annotated genes profiled by RNA sequencing (RNA-Seq), 485 were differentially expressed at false discovery rate <0.05 following correction for genome-wide testing (Fig. 5 and SI Appendix, Table S3). Quantitative RT-PCR was performed, validating the differential expression observed by RNA-Seq (SI Appendix, Fig. S5). Differentially expressed genes were significantly enriched among genes containing segregating SNPs located in CDS and UTRs (SI Appendix, Table S4), indicating the potential involvement of cis-regulatory elements in these genes. We included the 485 differentially expressed genes in functional annotation analysis. Using twofold enrichment as a cutoff (adjusted P < 0.05, Benjamini), several functional domains were identified, including calmodulin binding, synapse, and neuronal projection (SI Appendix, Tables S5 and S6). Of particular interest was overrepresentation of genes involved in glutamate, GABA, opioid, cholinergic, and adrenergic transmission (Fig. 5), including Grm2, Grin2b, Gria4, Slc1a3, Slc1a2, Gabbr1, Gabrg3, Oprm1, Chrm4, and Adrbk1. We also examined in striatum a group of genes involved in synaptic gluamatergic functions defined by KEGG pathway using quantitative RT-PCR (qRT-PCR) and observed significant differential expression between P and NP rats for the genes in this pathway (combined P value = 2.54 × 10−6; SI Appendix, Fig. S6). Thus, differences in gene expression between P and NP rats converge broadly with their genetic differences.
Fig. 5.
Differential gene expression in hippocampus of P and NP rats measured by RNA-Seq. Average expression levels (normalized log2 read counts, Upper Left) of 11,406 detected genes are compared between P and NP rats. The fold change (P vs. NP) and nominal P values (−log10, t test) for each gene are also plotted (Lower Left). Significance of difference between P and NP for each gene is color-coded: blue, not significant; orange, P < 0.05; green, false discovery rate <0.05. Fifteen differentially expressed genes among those in the overrepresented functional domains are highlighted and details of their expression differences are listed in the table (Right).
Discussion
Using genomic sequencing in P and NP rats, we identified a Grm2 stop codon variant that fully segregates between the two artificially selected rat lines. P rats are homozygous for the Grm2 *407, which results in the loss of the mGluR2 and largely uncompensated impairment of group II mGluR-mediated synaptic depression. Transcriptome analysis revealed perturbation of expression of genes involved in synaptic transmission. The role of mGluR2 in altered alcohol preference was supported by multiple lines of evidence. Loss of mGluR2 in the F2 rats derived from crossing P and NP rats increased alcohol consumption and preference. Blockade of mGluR2 by an mGluR2/3 antagonist escalated self-administration of alcohol in Wistar rats. The causal link was further validated in Grm2−/− mice, which exhibited significantly elevated alcohol consumption and preference.
mGluR2 is primarily located presynaptically and plays critical roles in glutamatergic functions. Dysfunction of mGluR2 can cause perturbation of glutamate transmission and related networks of neurocircuitries that may be more readily measured at molecular and physiological levels, with stronger effect sizes, as was illustrated by a functional NPY locus whose effects were diluted from the molecular level of RNA and protein to the behavioral level of anxiety (26). The consequences of molecular perturbation for complex behavior may be broader and less specific (pleiotropic), and thus more difficult to detect. Alcohol dependence is a complex behavior frequently associated with other behavioral disorders. Disruption of mGluR2-related signaling was shown in prior studies to have broad effects on addictive behaviors (27, 28). However, as shown here, whereas the small effect size of a locus in a complex behavioral trait can pose a challenge for gene discovery, model organisms represent a sensitive framework for identification and also for multilevel functional validation.
Pharmacologic blockade of mGluR2 was performed in Wistar rats, the parental strain, because P and NP rats are not suitable for this type of testing. P rats do not express functional mGluR2, thus preventing meaningful studies that use drugs targeting this receptor. In NP rats, the test would be whether an antagonist increases alcohol consumption, because alcohol preference of NP rats is so low that the effect of the agonist would be limited by a floor effect. However, the NP line is known to be strongly averse to alcohol. This is likely to reflect prominent effects of other presently unidentified loci selected for this trait. This would likely obscure effects of mGluR2 antagonism, because the effect size contributed by disruption of this locus is relatively modest. P and NP rats were selected from Wistar rats that express functional mGluR2 (unlike P rats) and are not averse to alcohol (unlike NP rats). Our results in Wistar rats provide convergent evidence that loss of functional mGluR2 increases alcohol preference, although caution has to be exercised in interpreting the results because of lack of the selectivity of the available antagonists for mGluR2 and mGluR3, and the observed effect was relatively brief. However, this concern is addressed by our findings in mGluR2 null mice, which provide clear functional validation (Grm2 −/− vs. Grm2 +/+) in a constant background, including intact mGluR3 function.
We profiled hippocampal gene expression in P and NP rats by transcriptome sequencing because functions of this region including short- and long-term memory and synaptic plasticity are important for drug/alcohol-associated learning and memory. The hippocampus directly projects excitatory efferents to the ventral striatum and can also activate dopaminergic neurons of the ventral tegmental area, explaining its involvement in cue-conditioned dopamine release. We also used qRT-PCR to measure the expression of a group of genes involved in synaptic glutamatergic function in striatum, following up the altered expression of this gene group in hippocampus. There is an overall pattern of altered expression of genes involved in neural development and synaptic functions, especially the ones that function in glutamate, GABA, opioid, cholinergic, and adrenergic transmission. This pattern is consistent with the lack of mGluR2 receptor in P rats but more securely points to neuronal differences between P and NP rats that influence alcohol-drinking behaviors. The gene expression differences between P and NP rats are thus consistent and convergent with their genetic and phenotypic differences and are likely to be influenced by their overall genetic differences or the interaction of Grm2 C407* with other loci. Further studies on Grm2 −/− mice or rats engineered for the knockout of Grm2 could clarify this point.
Genetic variations of GRM2 are also likely to influence alcohol-related phenotypes in people. However, variants as functionally damaging as the rat Grm2 stop codon are probably rare or uncommon and therefore may not be readily detectable by association analysis. Rare and uncommon variants altering function that are rare worldwide may be abundant in families and founder populations and linkable to behavior, such as the HTR2B (29) and MAOA (30) stop codons. Our sequencing of GRM2 exonic regions in two human founder populations (Finns and Plains Indians, n = 216) and genotyping of several missense variants in GRM2 reported in the 1000 Genomes Project and dbSNP in the Finnish (n = 735) and Plains Indian (n = 466) samples did not identify any relatively abundant variant that is associated with alcohol dependence. These results illustrate the challenges of gene and functional variant discovery in humans. They also point to the value of selectively bred model organisms in gene identification for complex traits.
Alcohol preference in rats is also influenced by additional genetic factors. We also observed other potentially functional genetic variants that segregated between P and NP rats and that were linked to alcohol preference in F2 rats. These loci include Lcn2 Q137* (a stop codon), Sspo V1868M (a missense variant scored as damaging), and Fam190a P82L (a missense variant scored as damaging) (SI Appendix, Fig. S3), all in previously identified QTL regions (12, 13). Sspo and Fam190a are in high linkage disequilibrium within a chromosome 4 QTL region where they are 14 Mb apart, and they are likely to represent the linkage signal from the same functional locus that remains to be identified. Lcn2 *137 is itself a strong functional candidate allele for alcohol preference. However, loss of lipocalin2 protein (encoded by Lcn2) in P rats needs to be confirmed and its causal role for alcohol preference needs to be validated before we can exclude the possibility that this stop codon is a surrogate marker for a different functional allele related to alcohol preference.
Using genomic sequencing, combining with genetic linkage and functional validation, we detected a functional nonsense variant of Grm2 in P rats and uncovered a causal role of Grm2 in alcohol preference. Our results demonstrate that selectively bred animal models are a potentially invaluable resource for the identification of genes influencing complex behavioral traits. Gene identification for complex disorders in population-based studies, particularly alcohol dependence and abuse, has been largely unsuccessful, in part because of the rarity of many causative variants, and the heterogeneity and usually small effect size of the alleles. In model organisms causal alleles that are uncommon can be brought to high frequency by artificial selection. Identification of roles of these rare and uncommon alleles can thereby implicate genes, and pathways. Detection of functional variants in genetically selected strains such as the P rat offers an attractive discovery strategy and can also help identify candidate drug targets for pharmacotherapies of excessive alcohol use.
Materials and Methods
Animals.
P and NP rats at generation 70 were provided by the Indiana University Alcohol Research Center. iP × iNP F2 rats were described previously (13). Wistar rats were obtained from Charles River Laboratories. mGluR2+/+ and mGluR2 −/− mice were obtained from Lilly Research Laboratories and previously described (24). All research protocols were approved by the institutional review board of National Institute on Alcohol Abuse and Alcoholism.
Exome Sequencing and RNA-Seq Data Analysis.
Exome sequencing libraries were constructed using Agilent SureSelect protocols. Strand-specific cDNA libraries were constructed using an Illumina protocol with modifications. Genomic sequencing was carried out on a GAIIx sequencer and analyzed with GApipeline (Illumina). RNA-Seq data analysis has been previously described (31). SI Appendix, SI Materials and Methods gives details.
Variant Validation, Western Blots, Genotyping, qRT-PCR, and QTL Analysis.
Nonsense and missense variants were validated by Sanger sequencing. A mouse monoclonal anti-mGluR2 N-terminal antibody (ab15672, Abcam) was used to analyze mGluR2 expression from hippocampal proteins of P and NP rats and Grm2−/− and Grm2+/+ mice. Genotyping was performed by TaqMan assays. More details of these methods as well as qRT-PCR and F2 rats QTL analysis are described in SI Appendix, SI Materials and Methods.
Field Potential Recording in Brain Slices.
Brain slices of P and NP rats (2–3 mo old) and Grm2+/+ and −/− mice (10 wk old) containing the dorsal hippocampus or the striatum posterior to the head of the caudate nucleus were used in field potential recording. The fEPSP slope in DG recordings or the PS amplitude in striatum was measured. The group II mGluR agonist LY379268 (200 nM) was applied to the slice for 5 min via whole-bath superfusion, once recording was initiated and a stable field potential obtained. In some experiments the group II mGluR antagonist LY341495 (1 μM) was applied throughout the recording to validate the involvement of group II mGluR in the agonist action. SI Appendix, SI Materials and Methods gives details.
Alcohol Intake in Wistar Rats and in Grm2−/− Mice.
Effect of LY341495 on alcohol self-administration in male Wistar rats (300–400 g) was carried out in operant conditioning chambers. Alcohol consumption and preference in Grm2+/+ and Grm2 −/− mice (10 mo old) were measured using a two-bottle choice procedure. The details of these experiments are described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. William J. McBride and Dr. Lawrence Lumeng for their contributions to the derivation of the P/NP rats. We thank Dr. Lucinda Carr for her contribution to the F2 rat drinking data. We thank Dr. Tatiana Foroud for assistance with statistical genetics. We thank Cheryl Marietta and Longina Akhtar for assistance with data generation, Basel Baghal for assistance with data analysis, and Elisa Moore for assistance with bioinformatics. This work was supported by Indiana Alcohol Research Center Grant P60-AA007611.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1309839110/-/DCSupplemental.
References
- 1.Goldman D, Oroszi G, Ducci F. The genetics of addictions: Uncovering the genes. Nat Rev Genet. 2005;6(7):521–532. doi: 10.1038/nrg1635. [DOI] [PubMed] [Google Scholar]
- 2.Prescott CA, Kendler KS. Genetic and environmental contributions to alcohol abuse and dependence in a population-based sample of male twins. Am J Psychiatry. 1999;156(1):34–40. doi: 10.1176/ajp.156.1.34. [DOI] [PubMed] [Google Scholar]
- 3.Thomasson HR, Crabb DW, Edenberg HJ, Li TK. Alcohol and aldehyde dehydrogenase polymorphisms and alcoholism. Behav Genet. 1993;23(2):131–136. doi: 10.1007/BF01067417. [DOI] [PubMed] [Google Scholar]
- 4.Bierut LJ, et al. Gene, Environment Association Studies Consortium A genome-wide association study of alcohol dependence. Proc Natl Acad Sci USA. 2010;107(11):5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edenberg HJ, et al. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol Clin Exp Res. 2010;34(5):840–852. doi: 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schumann G, et al. Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc Natl Acad Sci USA. 2011;108(17):7119–7124. doi: 10.1073/pnas.1017288108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Treutlein J, et al. Genome-wide association study of alcohol dependence. Arch Gen Psychiatry. 2009;66(7):773–784. doi: 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crabbe JC. Genetic animal models in the study of alcoholism. Alcohol Clin Exp Res. 1989;13(1):120–127. doi: 10.1111/j.1530-0277.1989.tb00296.x. [DOI] [PubMed] [Google Scholar]
- 9.Kaun KR, Azanchi R, Maung Z, Hirsh J, Heberlein U. A Drosophila model for alcohol reward. Nat Neurosci. 2011;14(5):612–619. doi: 10.1038/nn.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stead JD, et al. Selective breeding for divergence in novelty-seeking traits: heritability and enrichment in spontaneous anxiety-related behaviors. Behav Genet. 2006;36(5):697–712. doi: 10.1007/s10519-006-9058-7. [DOI] [PubMed] [Google Scholar]
- 11.Li TK, Lumeng L, Doolittle DP. Selective breeding for alcohol preference and associated responses. Behav Genet. 1993;23(2):163–170. doi: 10.1007/BF01067421. [DOI] [PubMed] [Google Scholar]
- 12.Bice P, et al. Genomic screen for QTLs underlying alcohol consumption in the P and NP rat lines. Mamm Genome. 1998;9(12):949–955. doi: 10.1007/s003359900905. [DOI] [PubMed] [Google Scholar]
- 13.Carr LG, et al. A quantitative trait locus for alcohol consumption in selectively bred rat lines. Alcohol Clin Exp Res. 1998;22(4):884–887. [PubMed] [Google Scholar]
- 14.Edenberg HJ, et al. Gene expression in the hippocampus of inbred alcohol-preferring and -nonpreferring rats. Genes Brain Behav. 2005;4(1):20–30. doi: 10.1111/j.1601-183X.2004.00091.x. [DOI] [PubMed] [Google Scholar]
- 15.Hwang BH, Stewart R, Zhang JK, Lumeng L, Li TK. Corticotropin-releasing factor gene expression is down-regulated in the central nucleus of the amygdala of alcohol-preferring rats which exhibit high anxiety: A comparison between rat lines selectively bred for high and low alcohol preference. Brain Res. 2004;1026(1):143–150. doi: 10.1016/j.brainres.2004.08.042. [DOI] [PubMed] [Google Scholar]
- 16.Liang T, et al. Candidate genes for alcohol preference identified by expression profiling in alcohol-preferring and -nonpreferring reciprocal congenic rats. Genome Biol. 2010;11(2):R11. doi: 10.1186/gb-2010-11-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 19.Neki A, et al. Pre- and postsynaptic localization of a metabotropic glutamate receptor, mGluR2, in the rat brain: an immunohistochemical study with a monoclonal antibody. Neurosci Lett. 1996;202(3):197–200. doi: 10.1016/0304-3940(95)12248-6. [DOI] [PubMed] [Google Scholar]
- 20.Johnson MP, et al. Metabotropic glutamate 2 receptor potentiators: receptor modulation, frequency-dependent synaptic activity, and efficacy in preclinical anxiety and psychosis model(s) Psychopharmacology (Berl) 2005;179(1):271–283. doi: 10.1007/s00213-004-2099-9. [DOI] [PubMed] [Google Scholar]
- 21.Lovinger DM, McCool BA. Metabotropic glutamate receptor-mediated presynaptic depression at corticostriatal synapses involves mGLuR2 or 3. J Neurophysiol. 1995;73(3):1076–1083. doi: 10.1152/jn.1995.73.3.1076. [DOI] [PubMed] [Google Scholar]
- 22.Rodd ZA, et al. The metabotropic glutamate 2/3 receptor agonist LY404039 reduces alcohol-seeking but not alcohol self-administration in alcohol-preferring (P) rats. Behav Brain Res. 2006;171(2):207–215. doi: 10.1016/j.bbr.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 23.Zhao Y, et al. Activation of group II metabotropic glutamate receptors attenuates both stress and cue-induced ethanol-seeking and modulates c-fos expression in the hippocampus and amygdala. J Neurosci. 2006;26(39):9967–9974. doi: 10.1523/JNEUROSCI.2384-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linden AM, et al. Anxiolytic-like activity of the mGLU2/3 receptor agonist LY354740 in the elevated plus maze test is disrupted in metabotropic glutamate receptor 2 and 3 knock-out mice. Psychopharmacology (Berl) 2005;179(1):284–291. doi: 10.1007/s00213-004-2098-x. [DOI] [PubMed] [Google Scholar]
- 25.Crabbe JC, Phillips TJ. Pharmacogenetic studies of alcohol self-administration and withdrawal. Psychopharmacology (Berl) 2004;174(4):539–560. doi: 10.1007/s00213-003-1608-6. [DOI] [PubMed] [Google Scholar]
- 26.Zhou Z, et al. Genetic variation in human NPY expression affects stress response and emotion. Nature. 2008;452(7190):997–1001. doi: 10.1038/nature06858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin X, et al. The mGluR2 positive allosteric modulator BINA decreases cocaine self-administration and cue-induced cocaine-seeking and counteracts cocaine-induced enhancement of brain reward function in rats. Neuropsychopharmacology. 2010;35(10):2021–2036. doi: 10.1038/npp.2010.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katner SN, et al. Alcohol-preferring (P) rats are more sensitive than Wistar rats to the reinforcing effects of cocaine self-administered directly into the nucleus accumbens shell. Pharmacol Biochem Behav. 2011;99(4):688–695. doi: 10.1016/j.pbb.2011.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bevilacqua L, et al. A population-specific HTR2B stop codon predisposes to severe impulsivity. Nature. 2010;468(7327):1061–1066. doi: 10.1038/nature09629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brunner HG, Nelen M, Breakefield XO, Ropers HH, van Oost BA. Abnormal behavior associated with a point mutation in the structural gene for monoamine oxidase A. Science. 1993;262(5133):578–580. doi: 10.1126/science.8211186. [DOI] [PubMed] [Google Scholar]
- 31.Zhou Z, Yuan Q, Mash DC, Goldman D. Substance-specific and shared transcription and epigenetic changes in the human hippocampus chronically exposed to cocaine and alcohol. Proc Natl Acad Sci USA. 2011;108(16):6626–6631. doi: 10.1073/pnas.1018514108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.