Significance
The structural properties of a membrane-associating protein at the water–membrane interfaces are intimately linked to its biological function, but they are difficult to characterize using existing biophysical tools. We identify the existence of a distinct intrinsic gradient of water diffusion across the lipid bilayer, encompassing a thick surface hydration layer above the lipid membrane surface, and debut an approach to exploit this gradient as a highly sensitive ruler to determine the topology, immersion depth, and location of a membrane associating protein, including the segments residing well above the membrane surface, that are otherwise difficult to resolve. This study demonstrates a potential of a broadly applicable approach for the structure–dynamics–function study of membrane proteins, membrane systems, and beyond.
Keywords: electron paramagnetic resonance, site-directed spin labeling, protein-lipid interaction, liposomes, Parkinson disease
Abstract
Knowing the topology and location of protein segments at water–membrane interfaces is critical for rationalizing their functions, but their characterization is challenging under physiological conditions. Here, we debut a unique spectroscopic approach by using the hydration dynamics gradient found across the phospholipid bilayer as an intrinsic ruler for determining the topology, immersion depth, and orientation of protein segments in lipid membranes, particularly at water–membrane interfaces. This is achieved through the site-specific quantification of translational diffusion of hydration water using an emerging tool, 1H Overhauser dynamic nuclear polarization (ODNP)-enhanced NMR relaxometry. ODNP confirms that the membrane-bound region of α-synuclein (αS), an amyloid protein known to insert an amphipathic α-helix into negatively charged phospholipid membranes, forms an extended α-helix parallel to the membrane surface. We extend the current knowledge by showing that residues 90–96 of bound αS, which is a transition segment that links the α-helix and the C terminus, adopt a larger loop than an idealized α-helix. The unstructured C terminus gradually threads through the surface hydration layers of lipid membranes, with the beginning portion residing within 5–15 Å above the phosphate level, and only the very end of C terminus surveying bulk water. Remarkably, the intrinsic hydration dynamics gradient along the bilayer normal extends to 20–30 Å above the phosphate level, as demonstrated with a peripheral membrane protein, annexin B12. ODNP offers the opportunity to reveal previously unresolvable structure and location of protein segments well above the lipid phosphate, whose structure and dynamics critically contribute to the understanding of functional versatility of membrane proteins.
Membrane proteins (MPs) constitute more than 25% of all human proteins. They not only represent the largest class of drug targets, but they also perform essential functions ranging from sensing, signaling, to transporting ions and solutes (1). MP function is critically associated with its structure, not only the conformations within the hydrophobic core of the bilayer but also of the protein segments located at or near the hydrophilic lipid headgroup region (1–4). This water–membrane interface comprises a large portion (∼20%) of the total MP in the lipid bilayer. Many functional domains of MPs are found to extend outward from the outer leaflet of the lipid bilayer into the water–membrane interface, exposing a heterogeneous environment capable of mediating a wide range of noncovalent interactions in the extracellular matrix. For instance, the membrane-associating domain of peripheral MP is essential for several key cellular functions by regulating the dynamics and morphology of cellular membranes (5). These functional domains of MPs are commonly highly disordered, display heterogeneous conformations, are often associated with parts of the cell matrix, or transiently bind to a cell surface in crowded environments (4). Thus, it is difficult to study the structure and dynamics of these protein segments at water–membrane interfaces by using traditional analytical tools. For instance, although X-ray crystallography and solution-state NMR spectroscopy offer atomic-level information of MPs, they have limitations associated with the size, dynamics, and complexity of MPs in membrane-mimetic systems (6).
Electron paramagnetic resonance (EPR) spectroscopy combined with site-directed spin labeling (SDSL) is an alternative approach for accessing local structural and dynamic information of MPs by means of probing dynamics and coupling of stable nitroxide radicals, termed spin labels, attached to biomolecular sites of interest. Its unique benefit is that the measurement is not intrinsically limited by the size or complexity of protein and its environments (7). Continuous wave (CW) EPR line shape analysis can track conformational changes of protein segments and their interactions with other biomolecular assemblies (8). Pulsed dipolar EPR methods provide intra- or interprotein distances between 2- and 7-nm length scales in various biological environments (9). Moreover, the local protein structure with respect to the lipid bilayer surface can be determined from the collision frequency (or solvent accessibility) of a single nitroxide radical labeled on consecutive protein residues to paramagnetic reagents, such as nickel-ethylenediaminediacetic acid (Ni-EDDA) (water-soluble) and O2 (lipid-soluble), via power saturation EPR measurements (10). Although this collision gradient approach has been well established to determine the immersion depth and topology of various MP structural elements in lipid bilayers (10–18), it has lower or, in some cases, no resolution when the protein segment is located more than 5 Å away from the lipid phosphate toward the solvent (13, 14). This limitation is because the collision frequency of nitroxide radicals with paramagnetic reagents measured by this method relies on their concentration gradient along the bilayer, which vanishes in the region of a distance >5 Å above the phosphate level. Thus, the collision frequency becomes indistinguishable from that in bulk water. As MPs’ domains close to water–membrane interfaces are highly tuned to their functional role, alternative and/or more sensitive methods for characterizing their structural features and locations are needed.
A synthetic lipid bilayer comprises well-defined hydrophobic dimensions, with a substantially steep gradient of water density connecting the surface hydration layers of the membrane and a nearly dehydrated core of hydrocarbon bilayer (19). A less obvious, but equally important, characteristic is the differential hydration dynamics across the lipid bilayer that is determined by the interfacial energy between hydration water and the lipid molecules, as well as the spatial confinement of hydration water within the bilayers. Given that these properties are thought to play a key role in tuning the structure and function of MPs in the lipid membrane (20), it motivates us to closely investigate the diffusion characteristics of hydration water across the lipid bilayer. Although it is now well accepted that the water diffusivity within the interior and near the surface of the lipid bilayer is significantly (5- to 11-fold) retarded (21–23) compared with that of bulk water (i.e., 2.3 × 10−9 m2/s at 25 °C), the mechanism by which water passively diffuses across the bilayer is not well established. Although water diffusivity is assumed to be homogeneous across the bilayer interior in some studies (24, 25), recent computational works have suggested local heterogeneities in water diffusivity across the bilayer, possibly mediated by transient pores (26). Diverging viewpoints are further fueled by the sparsity of direct experimental measurements. Among others, an experimental observation of differential hydration dynamics across the lipid bilayer has been reported on large unilamellar vesicles (LUVs, 200 nm in diameter) made of 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP), countering the homogeneous diffusion model (23). These measurements were enabled by Overhauser dynamic nuclear polarization (ODNP)-enhanced NMR relaxometry that probes the local translational diffusion of hydration water around the specific sites of biological samples under physiological conditions (27–33).
In this work, we extend this finding by demonstrating that the gradient of water diffusion not only exists within the bilayer interior, but also spans about 20–30 Å above the phosphate level, eventually interfacing bulk water. This finding is consistent with recent studies identifying the existence of a thick hydration shell (>20 Å thickness) on biomolecular surfaces (34). Based on this observation, we debut a unique approach that employs this extended gradient of hydration dynamics as a sensitive and high-resolution molecular ruler for probing conformation and location of MPs within the bilayer and near the water–membrane interface, extending to the distance of 20–30 Å above the phosphate level. In this work, α-synuclein (αS), a 14-kDa amyloid protein implicated in Parkinson disease (PD), was used as an illustrative system to showcase the herein introduced approach. It has been known that its amphipathic N terminus (residues 1–60) can transform into an extended α-helix upon binding to a negatively charged small unilamellar vesicle (SUV) surface, whereas the C terminus (residues 96–140) remains unstructured and does not bind to the lipid membrane (11, 12, 35–38). The residues 61–95 of αS contain a highly hydrophobic self-aggregating sequence that is essential for αS fibrillation (39). It is generally thought that the interaction between different αS aggregated species and the cell membrane is directly implicated in the pathological and physiological roles of PD (39–41). However, little is known about the conformational states of αS in the disease mechanism of PD, fueled by challenges in characterizing the structural states of membrane-bound proteins in the aggregation processes. Although the C terminus does not seem to play a key role in the αS fibrillation, recent studies have suggested that it may be critical for preventing the formation of rapid αS filament assemblies (42–44). Our unique approach demonstrates that the local hydration dynamics at the αS protein–membrane interfaces can sensitively register the relative position of the protein residues that are embedded within the lipid bilayer, as well as located more than 5 Å above the lipid phosphate. Importantly, this result implies that the intrinsic hydration dynamics gradient along the bilayers must dominate over the intrinsic hydration dynamics of the protein surface. We verify this to be the case by studying a model peripheral MP, annexin B12 (17, 18, 45), whose surface hydration dynamics are found to scale with distance up to 30 Å from the phosphate level toward bulk water, suggesting the potential generality of this method. Thus, ODNP is uniquely applicable to characterizing the structural features of MPs’ regions that reside at the water–membrane interface. By integrating this powerful tool with other mature biophysical tools, such as power saturation EPR and CW EPR line shape analysis, previously inaccessible questions can now be addressed, such as whether a MP segment is bound to the surface or immersed in the bilayer, whether the structural motifs of a MP orient along the membrane surface, whether hydration water resides between the MP segment and the membrane surface, and what part of the unstructured MP segments resides close to the membrane surface or extends into bulk water.
Results and Discussion
Approach to Quantify Local Hydration Dynamics at Biomolecular Interfaces.
ODNP measurements were used to quantify the diffusion dynamics of hydration water around nitroxide spin labels tethered to the specific sites of protein or lipid membrane systems, while the local conformational dynamics at the same sites of protein or lipid membrane were concurrently evaluated by CW EPR line shape analysis. ODNP relies on extracting the efficiency of dipolar relaxation between the electron spin of a nitroxide radical and the nuclear spin of nearby water protons. The efficiency of the electron–nuclear cross-relaxation is critically modulated by the water diffusivity within 5–10 Å around the spin label (28–30). The distance-dependent dipolar coupling between the water protons and the electron spin of the radical functionalized at biomolecular interfaces permits the selective characterization of the diffusivity of hydration water within 5–10 Å (encompasses two to four hydration layers) around the tethered spin label, whose dynamic characteristics is defined by the nearby biomolecular surface. Given that the frequency for probing hydration dynamics by NMR relaxometry is extended from the traditional upper limit of hundreds of megahertz to 10 GHz, and given that the efficiency of electron–nuclear cross-relaxation yields up to 300-fold NMR signal amplification, ODNP is rendered much more sensitive than conventional NMR relaxometry methods.
In ODNP, the electron–proton dipolar cross-relaxation that is critically mediated through the translational diffusion of the hydration water is driven by the saturation of the EPR transition, giving rise to NMR signal enhancement (27–30). At extrapolated maximum saturation, the NMR signal enhancement can be expressed as (27–30)
where ξ is the coupling factor, f is the leakage factor accounting for paramagnetic-enhanced proton relaxation over all proton relaxation mechanisms, smax is the maximum saturation factor for the electron spin (i.e., smax = 1 for full saturation achieved for slow-tumbling molecular systems) (28), and |γe/γN| is the ratio of the gyromagnetic ratios of the electron and proton spins, giving 658. ξ is the key parameter describing the efficiency of electron–proton cross-relaxation and can be obtained directly from the measurement of Emax and the proton T1 relaxation rates in the presence and absence of the paramagnetic radicals (28–30). The translational correlation time, τ, of hydration water around the radical can then be extracted from ξ, when explicit spectral density functions are modeled (27), as described in SI Text.
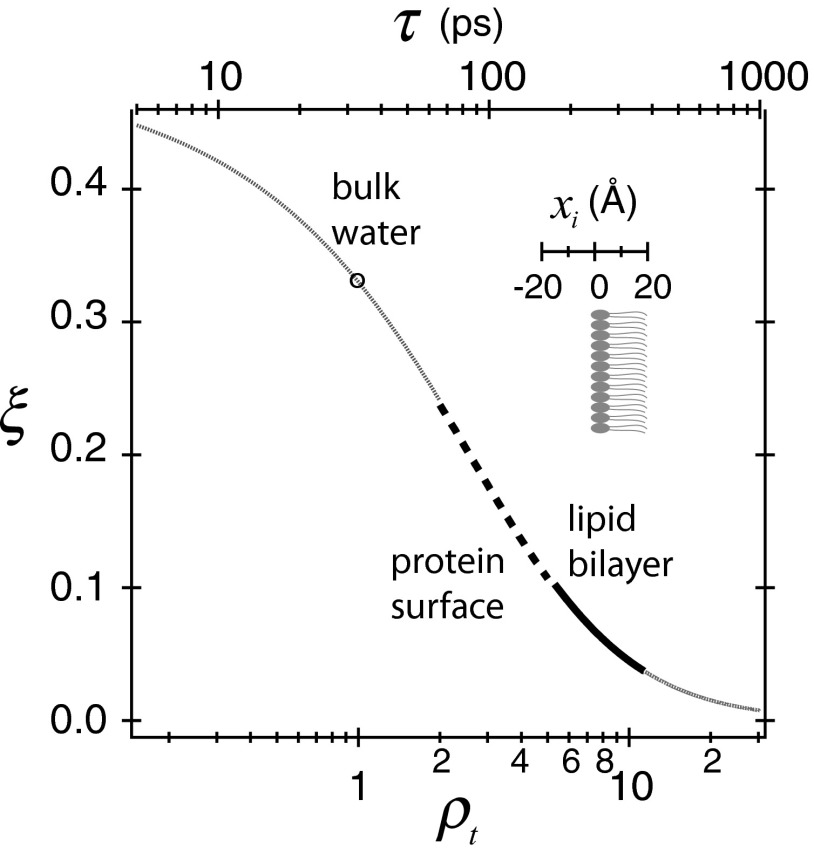
In this study, ODNP is performed using a modified X-band CW EPR spectrometer at a magnetic field of 0.35 T and at 25 °C, irradiating with moderate microwave powers (up to ∼4 W) at the electron Larmor frequency of ωe = 9.8 GHz. The choice of ωe of 9.8 GHz is critical, as the electron–nuclear cross-relaxation near this frequency is precisely modulated by the fast translational diffusion of loosely bound hydration water within the distance of closest approach (d) between the electron and 1H at biomolecular interfaces with translational correlation time, τ, on the order of tens to hundreds of picosecond, given the translational diffusivity of water, D, following D ∼ d2/τ (27–30). The range of τ accessible by ODNP at ωe = 9.8 GHz covers the dynamic range for hydration water surrounding solvent-exposed sites, as well as to some extent buried sites of proteins and other biomacromolecules (33). To qualitatively compare the hydration dynamics in different biological environments, we define the retardation factor of hydration dynamics, ρt = 〈τ〉/τbulk, which is the average translational correlation time of the local hydration water around a tethered spin label divided by that of bulk water (i.e., τbulk = 33 ps, measured by ODNP at 0.35 T using a small nitroxide radical dissolved in water) (30, 46). As illustrated in Fig. 1, the coupling between the electron spin and water proton is shown to be sensitively modulated within the dynamic range of hydration water at solvent-exposed surfaces of proteins (ρt = 2–5) (32, 47–49), as well as on the surfaces or within cores of lipid bilayers (ρt = 5–11) (21–23), affording the use of ODNP to probe conformational changes or interactions between proteins and lipid membranes by monitoring the changes in the local hydration dynamics at molecular interfaces. Crucially, biological samples at dilute concentrations (approximately tens of micromolars), of minute volumes (approximately a few microliters), and in an environment of excess water, lipids and other biological constituents at physiological temperature are experimentally accessible. Similar to CW EPR spectroscopy, ODNP is not fundamentally limited by the molecular weight, size, or the complexity of the sample system, permitting the study of site-specific hydration dynamics of MPs in lipid membrane environments under physiological conditions.
Fig. 1.
Coupling factor (ξ) is presented as a function of translational correlation time (τ) of hydration water at 0.35 T and at 25 °C. Retardation factor (ρt) of hydration water is referenced to the bulk water diffusivity. The distance (xi) with respect to the phosphate level and its relation with the retardation factor are shown in the Inset.
Probing Hydration Dynamics as a Function of Distance Along the Bilayer Normal.
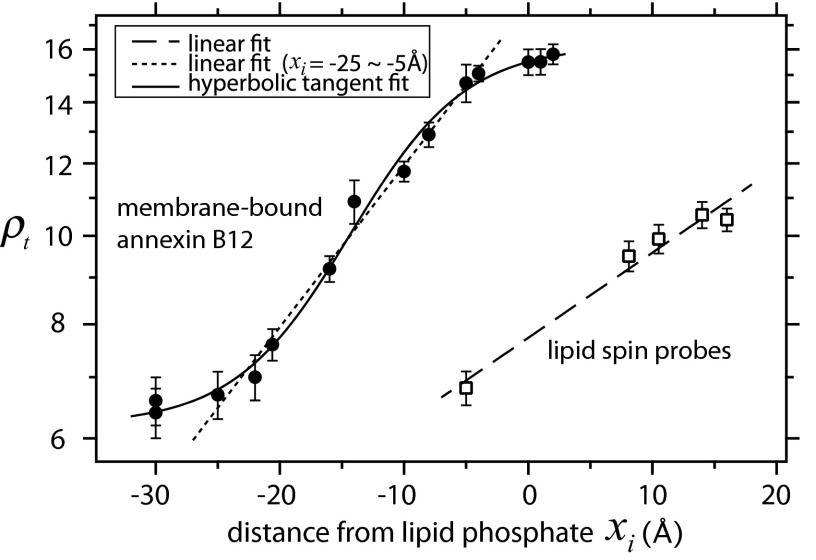
To obtain the diffusion profile of water penetration into the fully hydrated lipid bilayer composed of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine (POPS) in SUVs, we measured the ρt value of hydration water at distances above the lipid phosphate and at different bilayer depths using lipid spin probes: 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho(tempo)choline (TEMPO-PC) with a nitroxide radical attached to the choline moiety of PC and doxyl stearic acids (DSAs) with a nitroxide radical attached at various positions along the alkyl chains. The distance from the nitrogen of the nitroxide to the phosphate in TEMPO-PC was estimated to be ∼5 Å based on molecular models (50), whereas the immersion depths of the lipid spin probes were estimated from X-ray diffraction (51). The correlation between the distance from the lipid phosphate to the nitroxide of the lipid spin probes, xi, and the herein measured retardation factors, ρt, at the given spin-labeled sites of bilayers is presented in Fig. 2, where a positive xi value signifies the position below the phosphate group toward the bilayer and a negative xi value represents the position above the phosphate group toward bulk water. The experimental ODNP data in Fig. 2 are presented in Tables S1 and S2. We empirically found that the natural logarithm of the ρt value of the POPC/POPS bilayer increases approximately linearly with the immersion depth of the nitroxide spin probe within the bilayers. Most significantly, the surface hydration dynamics probed by TEMPO-PC at around 5 Å above the phosphate group of the lipid bilayer (50) is still 6.8-fold retarded compared with bulk water diffusivity, thereby providing a significant dynamic range for resolving the location of protein segments within the relatively thick hydration layers on the lipid membrane surfaces. In contrast, the solvent accessibility measured by power saturation EPR loses contrast when the spin label is >5 Å away from the phosphate level (13, 14).
Fig. 2.
Distance dependence of retardation factor (ρt) at specific sites of small unilamellar vesicles composed of POPC:POPS (7:3) (□) and annexin B12 bound on the surface of large unilamellar vesicles composed of POPC:POPS (1:2) in the presence of 1 mM Ca2+ (●) at 25 °C. Error bars represent SDs. The fit parameters are summarized in supporting information.
To test the generality and the validity of this intrinsic hydration dynamics gradient-based ruler along the bilayer normal at distances of >5 Å above the lipid phosphate, we examined another membrane-associating protein—a Ca2+-dependent peripheral MP, annexin B12. The membrane-bound form of annexin B12 has a convex shape on phosphatidylserine (PS)-rich membrane surfaces with a known tertiary structure (45), whose membrane binding is mediated by Ca2+ ions (17, 18). The crystallographic data suggest that the most distant portion of the membrane-bound annexin B12 is around xi = −30 Å (above the phosphate), whereas its deepest residue is xi ∼2 Å (below the phosphate) (45), as illustrated in Fig. S1. Fig. 2 presents a plot of the retardation factors at selected water-exposed residues of annexin B12 bound on the LUV surface composed of POPC and POPS with varying distance, xi, with respect to the phosphate level. Interestingly, we revealed that the natural logarithm of the retardation factor again monotonically decreases with the distance away from the bilayer surface, whereas a simple linear regression of the measured data is not suitable, especially at distant water-exposed sites of the membrane-bound annexin B12. Rather, the distance dependence of the lnρt value across the protein surface along bilayer normal follows a hyperbolic tangent function edged by a significant retardation factor up to xi = −30 Å (solid curve in Fig. 2), whereas an approximately linear relation was only found between xi = −5 and −25 Å (dotted line in Fig. 2). To understand whether Ca2+ influences the absolute values of hydration dynamics on the POPC/POPS bilayer surface studied herein, we compare the ODNP data in the presence and absence of Ca2+. We found that upon addition of 1 mM CaCl2 to the POPC/POPS LUV solution, the hydration dynamics measured at the bare membrane surface substantially slows by 54% (τ increases from 257 to 474 ps), likely because Ca2+ rigidifies the PS and neighboring PC headgroups (52). All experimental data yielding ρt values are presented in Table S2. This finding suggests that, although a qualitatively similar hydration dynamics gradient was found in different lipid bilayer systems, the absolute diffusivities of the hydration water alter significantly, likely depending on the lipid composition, lipid membrane curvature, charge density of the phospholipid headgroups, buffer condition, and the presence of ions or osmolytes. For instance, a similar linear correlation between the retardation factor and immersion depth of lipid bilayers was observed in different bilayer systems, regardless of their surface charges or vesicle sizes, whereas their absolute water diffusivities are quite different (Fig. S2). Detailed investigation into this dependence merits future studies, in particular considering the potential effect of membrane curvature on tuning the hydration dynamics on the membrane surface (53) and the interactions of curvature-sensitive MPs with lipid membranes.
Topology and Immersion Depth of α-Synuclein at the Water–Membrane Interface.
The approximately linear relation found between the lnρt value and corresponding distance with respect to the phosphate group, xi, can be used as a calibration curve to determine the relative distance of specific protein residues in the lipid bilayer. Here, we use αS as an example to examine whether the ODNP-derived hydration dynamics gradient can indeed provide structural information of a membrane-associating protein with higher sensitivity and spatial resolution. Previous studies have revealed that when αS binds to intact negatively charged SUVs, its amphipathic repeats comprising ∼90 N-terminal residues form an extended α-helix, located parallel to the membrane surface, whereas the disordered C terminus has not been observed to directly interact with the lipid membrane (11, 12, 35–38).
We first measured hydration dynamics at selected sites of monomeric αS between residues 76 and 108 in the absence of lipid membranes, as shown in Fig. 3. The results show that the ρt value exhibits a small dispersion between 2.4 and 4.0, suggesting that hydration water is relatively freely, and roughly evenly, diffusing near the protein surface in the unbound state. This result displays typical characteristics of hydration water around solvent-exposed protein surfaces with ρt values found between 2 and 5 (32, 47–49), confirming that αS in solution is in an overall unstructured state, where each residue is significantly exposed to bulk water.
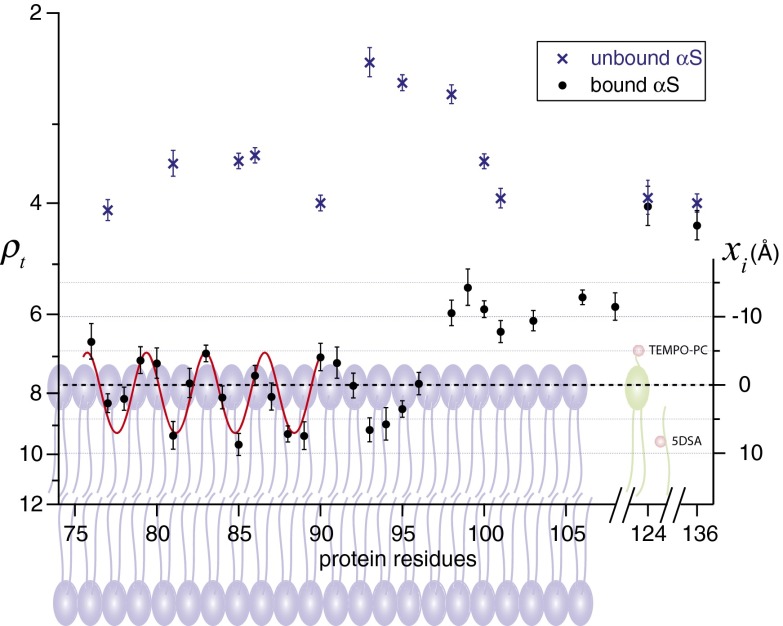
Fig. 3.
Retardation factor (ρt) at various residues of membrane-bound and free α-synuclein measured by ODNP. The distance (xi) of nitroxide radicals with respect to the phosphate group of the POPC/POPS bilayers is presented. Error bars represent SDs. The red curves are least-squares fits of data to a cosine function. The representative phospholipids and lipid spin probes are shown in the background. Nitroxide radicals of lipid spin probes are represented in red balls.
Additionally, the retardation factor of membrane-bound αS at consecutive spin-labeled sites from residue 76–108, as well as two additional residues 124 and 136 of the C-terminal end, were examined (Fig. 3). The first observation is that all sites, except residues 124 and 136, display measurably higher retardation factors (ρt = 5.4–9.7) in the membrane-bound states compared with their unbound states, implying the existence of the interactions between membrane surfaces and the C-terminal segment of αS that was previously assumed to be simply water exposed (11, 12). The ODNP data of unbound and bound αS, together with representative plots of 1H NMR signal enhancements vs. microwave power, are summarized in Tables S3 and S4 and Fig. S3.
By examining the periodicity of the hydration dynamics along the protein sequence in quantitative detail, we found the retardation factor between residues 76 and 90 to exhibit periodic oscillation every 3.48 ± 0.08 residues, as extracted from the least-squares fits to a cosine function, constituting close to an ideal α-helix structure (with 3.67 residues per helical turn or 11 residues per three turns) (11, 12). We then carefully evaluated the periodicity by using the α-helix periodicity index, αPI, based on a harmonic analysis with discrete Fourier transformation (Fig. S4) (54, 55). This method has been previously used to facilitate the secondary structure assignment of MPs by power saturation EPR (15, 16). The protein segment with αPI ≥ 2 is considered to be an α-helical structure with statistical significance (54, 55). We found an αPI value of 3.38 for the retardation factors of membrane-bound αS between residues 76 and 90, confirming a significant α-helical structure. Interestingly, the retardation factors of membrane-bound αS between residues 90 and 96, representing the transition region between the α-helix and the disordered C terminus, yield an αPI value of 0.13, indicating these residues are not part of an α-helix. It is likely that the hydrophobic residues (Phe-94 and Val-95) of this domain facilitate the penetration below the phosphate level to form a more extended loop than an α-helix.
The positions of membrane-bound αS residues with respect to the lipid phosphate can be determined by referencing their retardation factors against the calibration curve obtained from lipid spin probes, as shown in Fig. 2. We observe that the immersion depth of the helical bottom lies in the range of 8–10 Å, with the center of the helix located 1–3 Å below the phosphate level (Fig. 4). This result is in excellent agreement with the first observation of this shallow insertion into bilayers made by power saturation EPR (11, 12), where the immersion depth of the bottom portion of the α-helix was estimated at 11 Å and the center of the helix was at 1–4 Å below the lipid phosphate (11, 12). Our findings are also consistent with recent fluorescence and neutron reflectometry results, reporting an immersion depth of the α-helical bottom of around 9–14 Å (56).
Fig. 4.
Structural representation of topology and immersion depth of membrane-bound α-synuclein. The red ribbon was the topology obtained from ODNP measurements, whereas the gray ribbon was observed from the literature (11, 12). Black solid line: phosphate level; black dashed line: center of the helix; black dotted line: bottom of the helix.
Interestingly, we found that the hydration dynamics between residues 98 and 108, which are thought to be part of an entirely bulk water-exposed C terminus, exhibit still significantly retarded hydration dynamics. Their retardation factors are distinctly different from those of the same αS residues free in solution. This observation implies that this segment still resides closely within the surface hydration layers spanning 5–15 Å above the lipid phosphate group and experiences a more stable network of hydration water through which water diffusion is significantly retarded compared with in bulk water. Interestingly, 9 of the 11 amino acids between residues 98 and 108 are either polar or negatively charged, making this segment highly favorable for binding to positively charged trimethyl ammonium groups of the choline moieties that protrude to the solvent-exposed headgroup region of the bilayer. Noteworthily, the ρt values at residues 124 and 136 of membrane-bound αS, which are much closer to the end of the C terminus, display unaltered hydration dynamics within errors to those of the unbound state. This finding solidifies the idea that the C-terminal region, following the extended α-helical segment, gradually winds out and dangles outward to the solvent, where only the very end of the C terminus is fully solvated in bulk water (Fig. 4). This finding implies that the transition region still resides within the surface hydration layers of the outer leaflet of the lipid membrane. Moreover, ODNP reveals that the C terminus of membrane-bound αS displays a distinct landscape of hydration dynamics that may have functional implications. Its disordered and flexible structural configuration could regulate the interaction with constituents in solution, such as ligands, proteins, or metals, implicated in modulating the biological functions of αS (57, 58). Furthermore, a recent study showed that the highly charged C terminus of free αS can efficiently inhibit its fibril formation driven by electrostatic repulsions (44). Our result demonstrates that the beginning portion of the C terminus is in close proximity to the membrane surface—this possibly tucks away the αS and slows its aggregation by preventing free inter-αS interactions. ODNP could be used in future studies to track the conformational changes of αS or other amyloid proteins on membrane surfaces during their aggregation process.
Conclusion
In this work, we introduce a unique tool to determine and refine the topology and the immersion depth of membrane-associating proteins using the intrinsic diffusion gradient of hydration water across a lipid bilayer as a sensitive ruler. We characterize the dependence of the local water diffusivity on the distance across the bilayer normal in bare lipid membranes and on the surface of membrane-bound annexin B12. We find the intrinsic hydration dynamics gradient to extend around 20–30 Å above the phosphate level and to dominate over that of the protein surface, both of which are prerequisites for the general applicability of this intrinsic ruler. However, the absolute diffusivities of hydration water is expected to vary with membrane surface properties (e.g., membrane curvature, roughness, or charge density), buffer conditions, and to some extent, the structural nature of protein embedded, requiring a proper experimental calibration. We demonstrate the application of this intrinsic ruler method on the example of membrane-bound αS that we found to form an extended α-helix between residues 76 and 90, with its long axis of the helical center slightly embedded 1–3 Å below the lipid phosphates, in good agreement with earlier findings (11, 12). We further observed that the beginning portion of the C terminus is located within the distinctly characteristic hydration layers off the membrane surface, whereas the very end of the C terminus is entirely exposed to bulk water. In general, by exploiting the predetermined relationship between the retardation of hydration dynamics and its corresponding distance with respect to the lipid phosphate, ODNP provides a critical supplement to structural biology tools to characterize the topology, immersion depth, and orientation of membrane-active biological constituents, including transmembrane proteins, amyloid fibrils, and membrane-active peptides. A key strength of this approach is that the measurements are carried out in solution under physiological conditions, while the constituents can be associated with lipid membranes with virtually any size or complexity. Overall, ODNP provides a unique capability with enhanced resolution, site specificity, and sensitivity for identifying the structure and location of MPs, especially with finer resolution in the interfacial volume at distances up to 20–30 Å above the phosphate level. We believe ODNP will offer new opportunities for deepening the understanding of MP structure, organization, and function.
Materials and Methods
Preparation of Spin-Labeled α-Synuclein.
Single cysteine mutants of α-synuclein were expressed in BL21(DE3)pLysS Escherichia coli cells, and the cell pellet was resuspended in lysis buffer (100 mM Tris⋅HCl, 300 mM NaCl, 1 mM EDTA, pH 8) (11, 12). After boiling the cell lysate and precipitating with HCl, the supernatant was dialyzed against a buffer (20 mM Tris⋅HCl, 1 mM EDTA, and 1 mM DTT, pH 8). Two rounds of anion exchange chromatography were performed, and proteins were eluted with a salt gradient of 0–1 M NaCl. DTT was added to the samples to a final concentration of 1 mM for 30 min. DTT was then removed by size exclusion PD-10 columns (GE Healthcare) in buffer A (10 mM Hepes and 100 mM NaCl, pH 7.4). A fivefold molar excess of the 2,2,5,5-tetramethylpyrroline-3-yl-methanethiosulfonate (MTSL) spin label (Toronto Research Chemicals) was incubated with the protein samples for 1 h at 25 °C. Excess spin label was removed by buffer exchange using PD-10 columns. Spin-labeled proteins were concentrated using Amicon Ultra centrifugal filters (Millipore). Protein concentration was quantified using Micro BCA protein assay kit (Thermo Scientific). The SUVs composed of POPC and POPS (7:3, wt:wt) were prepared in buffer A using a sonication method. The average diameter of SUV was about 25 nm, as measured by dynamic light scattering (Zetasizer Nano ZS; Malvern Instruments). The following lipid spin probes were used for ODNP measurements: TEMPO-PC and n-doxyl stearic acid (n = 5, 7, 10, 12); 62.5 mM lipid and 500 μM spin probe were used. The chemical structure of the lipids and lipid spin probes used in this work are summarized in Fig. S5. Proteins were incubated with SUVs at a 1:250 protein to lipid molar ratio in buffer A at 25 °C for 1 h, followed by several rounds of washes using a 100-kDa cutoff centrifugal filter unit (Millipore) to remove the unbound proteins.
Preparation of Spin-Labeled Annexin B12.
The expression and purification of the annexin B12 used in this study were previously described (17, 18). The preparation of MTSL spin-labeled annexin B12 cysteine mutants is similar to that of spin-labeled αS, except equilibrated in buffer B (20 mM Hepes, pH 7.4, and 100 mM NaCl). LUVs composed of POPS and POPC in a 2:1 molar ratio were prepared in buffer B through an extrusion method using 1,000-nm filters. Spin-labeled annexin B12 (150 μg) was incubated with 1.5 mg of LUVs and 1 mM CaCl2 and allowed to react for at least 15 min at 25 °C. Lipids were pelleted at 16,000 × g for 20 min, the supernatant was removed, and the pellet was resolubilized in 6 μL of buffer B and subjected to ODNP measurements. The distances between the lipid phosphates and the selected residues of membrane-bound annexin B12 were approximated based on a molecular model of annexin B12’s crystal structure on a membrane surface (45) using Discovery Studio Client 3.0 (Accelrys) and known depths of its membrane-binding surface (17, 18).
ODNP Experiments.
1H ODNP experiments were performed at a 0.35-T electromagnet, operating at a 14.8-MHz 1H Larmor frequency and at 9.8-GHz electron Larmor frequency. A 3.5-μL sample was loaded in a 0.6-mm-inner-diameter quartz capillary tube (Fiber Optic Center), and both ends were sealed with bee wax. The capillary was mounted on a homebuilt NMR probe with a U-shaped RF coil. The EPR signal was then acquired by Bruker X-band EMX CW EPR spectrometer with a dielectric microwave resonator (Bruker ER-4123D) at 25 °C. During the ODNP experiments, the center field of nitroxide hyperfine transition lines was irradiated continuously at various microwave powers while the 1H NMR signal was recorded. Dry air continuously flowed through the EPR cavity at a flow rate of 8 L/min to prevent the sample heating. 1H T1 relaxation measurements were performed by an inversion-recovery pulse sequence operated by a Bruker Avance spectrometer at a 0.35-T electromagnet. The theory and instrumentation of ODNP are summarized in the SI Text.
Supplementary Material
Acknowledgments
S.H. acknowledges support by the 2011 NIH Directors New Innovator Award. C.-Y.C. and S.H. made use of the Materials Research Laboratory (MRL) Central Facilities supported by the National Science Foundation (NSF) through the Materials Research Science and Engineering Centers under Grant DMR 1121053. The MRL is a member of the NSF–funded Materials Research Facilities Network (www.mrfn.org). This work was also supported by National Institutes of Health Grants GM063915 and AG027936 (to R.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1307678110/-/DCSupplemental.
References
- 1.Rosenbaum DM, Rasmussen SG, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459(7245):356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.White SH, von Heijne G. How translocons select transmembrane helices. Annu Rev Biophys. 2008;37:23–42. doi: 10.1146/annurev.biophys.37.032807.125904. [DOI] [PubMed] [Google Scholar]
- 3.von Heijne G. Membrane-protein topology. Nat Rev Mol Cell Biol. 2006;7(12):909–918. doi: 10.1038/nrm2063. [DOI] [PubMed] [Google Scholar]
- 4.Cho WH, Stahelin RV. Membrane-protein interactions in cell signaling and membrane trafficking. Annu Rev Biophys Biomol Struct. 2005;34:119–151. doi: 10.1146/annurev.biophys.33.110502.133337. [DOI] [PubMed] [Google Scholar]
- 5.Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9(2):99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 6.Raschle T, Hiller S, Etzkorn M, Wagner G. Nonmicellar systems for solution NMR spectroscopy of membrane proteins. Curr Opin Struct Biol. 2010;20(4):471–479. doi: 10.1016/j.sbi.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altenbach C, Oh KJ, Trabanino RJ, Hideg K, Hubbell WL. Estimation of inter-residue distances in spin labeled proteins at physiological temperatures: Experimental strategies and practical limitations. Biochemistry. 2001;40(51):15471–15482. doi: 10.1021/bi011544w. [DOI] [PubMed] [Google Scholar]
- 8.Hubbell WL, Cafiso DS, Altenbach C. Identifying conformational changes with site-directed spin labeling. Nat Struct Biol. 2000;7(9):735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 9.Jeschke G. DEER distance measurements on proteins. Annu Rev Phys Chem. 2012;63:419–446. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- 10.Altenbach C, Greenhalgh DA, Khorana HG, Hubbell WL. A collision gradient method to determine the immersion depth of nitroxides in lipid bilayers: Application to spin-labeled mutants of bacteriorhodopsin. Proc Natl Acad Sci USA. 1994;91(5):1667–1671. doi: 10.1073/pnas.91.5.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jao CC, Der-Sarkissian A, Chen J, Langen R. Structure of membrane-bound alpha-synuclein studied by site-directed spin labeling. Proc Natl Acad Sci USA. 2004;101(22):8331–8336. doi: 10.1073/pnas.0400553101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jao CC, Hegde BG, Chen J, Haworth IS, Langen R. Structure of membrane-bound alpha-synuclein from site-directed spin labeling and computational refinement. Proc Natl Acad Sci USA. 2008;105(50):19666–19671. doi: 10.1073/pnas.0807826105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frazier AA, et al. Membrane orientation and position of the C2 domain from cPLA2 by site-directed spin labeling. Biochemistry. 2002;41(20):6282–6292. doi: 10.1021/bi0160821. [DOI] [PubMed] [Google Scholar]
- 14.Frazier AA, Roller CR, Havelka JJ, Hinderliter A, Cafiso DS. Membrane-bound orientation and position of the synaptotagmin I C2A domain by site-directed spin labeling. Biochemistry. 2003;42(1):96–105. doi: 10.1021/bi0268145. [DOI] [PubMed] [Google Scholar]
- 15.Chakrapani S, Cuello LG, Cortes DM, Perozo E. Structural dynamics of an isolated voltage-sensor domain in a lipid bilayer. Structure. 2008;16(3):398–409. doi: 10.1016/j.str.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perozo E, Cortes DM, Cuello LG. Three-dimensional architecture and gating mechanism of a K+ channel studied by EPR spectroscopy. Nat Struct Biol. 1998;5(6):459–469. doi: 10.1038/nsb0698-459. [DOI] [PubMed] [Google Scholar]
- 17.Isas JM, et al. Calcium- and membrane-induced changes in the structure and dynamics of three helical hairpins in annexin B12. Biochemistry. 2005;44(50):16435–16444. doi: 10.1021/bi051751m. [DOI] [PubMed] [Google Scholar]
- 18.Isas JM, Langen R, Hubbell WL, Haigler HT. Structure and dynamics of a helical hairpin that mediates calcium-dependent membrane binding of annexin B12. J Biol Chem. 2004;279(31):32492–32498. doi: 10.1074/jbc.M402568200. [DOI] [PubMed] [Google Scholar]
- 19.Wimley WC, White H. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat Struct Biol. 1996;3(10):842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- 20.MacCallum JL, Tieleman DP. Hydrophobicity scales: A thermodynamic looking glass into lipid-protein interactions. Trends Biochem Sci. 2011;36(12):653–662. doi: 10.1016/j.tibs.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 21.Tobias DJ, Tu KC, Klein ML. Atomic-scale molecular dynamics simulations of lipid membranes. Curr Opin Colloid Interface Sci. 1997;2(1):15–26. [Google Scholar]
- 22.Hodges MW, Cafiso DS, Polnaszek CF, Lester CC, Bryant RG. Water translational motion at the bilayer interface: An NMR relaxation dispersion measurement. Biophys J. 1997;73(5):2575–2579. doi: 10.1016/S0006-3495(97)78286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kausik R, Han S. Dynamics and state of lipid bilayer-internal water unraveled with solution state 1H dynamic nuclear polarization. Phys Chem Chem Phys. 2011;13(17):7732–7746. doi: 10.1039/c0cp02512g. [DOI] [PubMed] [Google Scholar]
- 24.Reeves JP, Dowben RM. Water permeability of phospholipid vesicles. J Membr Biol. 1970;3(2):123–141. doi: 10.1007/BF01868011. [DOI] [PubMed] [Google Scholar]
- 25.Finkelstein A. Water and nonelectrolyte permeability of lipid bilayer membranes. J Gen Physiol. 1976;68(2):127–135. doi: 10.1085/jgp.68.2.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bemporad D, Essex JW, Luttmann C. Permeation of small molecules through a lipid bilayer: A computer simulation study. J Phys Chem B. 2004;108(15):4875–4884. [Google Scholar]
- 27.Hausser KH, Stehlik D. Dynamic nuclear polarization in liquids. Adv Magn Reson. 1968;3:79–139. [Google Scholar]
- 28.Armstrong BD, Han S. A new model for Overhauser enhanced nuclear magnetic resonance using nitroxide radicals. J Chem Phys. 2007;127(10):104508. doi: 10.1063/1.2770465. [DOI] [PubMed] [Google Scholar]
- 29.Armstrong BD, Han S. Overhauser dynamic nuclear polarization to study local water dynamics. J Am Chem Soc. 2009;131(13):4641–4647. doi: 10.1021/ja809259q. [DOI] [PubMed] [Google Scholar]
- 30.Franck JM, Pavlova A, Scott JA, Han S. Quantitative cw Overhauser effect dynamic nuclear polarization for the analysis of local water dynamics. Prog Nucl Magn Reson. 2013 doi: 10.1016/j.pnmrs.2013.06.001. 10.1016/j.pnmrs.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hussain S, Franck JM, Han S. Transmembrane protein activation refined by site-specific hydration dynamics. Angew Chem Int Ed Engl. 2013;52(7):1953–1958. doi: 10.1002/anie.201206147. [DOI] [PubMed] [Google Scholar]
- 32.Armstrong BD, et al. Site-specific hydration dynamics in the nonpolar core of a molten globule by dynamic nuclear polarization of water. J Am Chem Soc. 2011;133(15):5987–5995. doi: 10.1021/ja111515s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ortony JH, et al. Probing the hydration water diffusion of macromolecular surfaces and interfaces. New J Phys. 2011;13(1):015006. [Google Scholar]
- 34.Ebbinghaus S, et al. An extended dynamical hydration shell around proteins. Proc Natl Acad Sci USA. 2007;104(52):20749–20752. doi: 10.1073/pnas.0709207104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferreon ACM, Gambin Y, Lemke EA, Deniz AA. Interplay of alpha-synuclein binding and conformational switching probed by single-molecule fluorescence. Proc Natl Acad Sci USA. 2009;106(14):5645–5650. doi: 10.1073/pnas.0809232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lokappa SB, Ulmer TS. Alpha-synuclein populates both elongated and broken helix states on small unilamellar vesicles. J Biol Chem. 2011;286(24):21450–21457. doi: 10.1074/jbc.M111.224055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robotta M, et al. Direct evidence of coexisting horseshoe and extended helix conformations of membrane-bound alpha-synuclein. ChemPhysChem. 2011;12(2):267–269. doi: 10.1002/cphc.201000815. [DOI] [PubMed] [Google Scholar]
- 38.Bodner CR, Dobson CM, Bax A. Multiple tight phospholipid-binding modes of alpha-synuclein revealed by solution NMR spectroscopy. J Mol Biol. 2009;390(4):775–790. doi: 10.1016/j.jmb.2009.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fink AL. The aggregation and fibrillation of alpha-synuclein. Acc Chem Res. 2006;39(9):628–634. doi: 10.1021/ar050073t. [DOI] [PubMed] [Google Scholar]
- 40.Varkey J, et al. Membrane curvature induction and tubulation are common features of synucleins and apolipoproteins. J Biol Chem. 2010;285(42):32486–32493. doi: 10.1074/jbc.M110.139576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Outeiro TF, Lindquist S. Yeast cells provide insight into alpha-synuclein biology and pathobiology. Science. 2003;302(5651):1772–1775. doi: 10.1126/science.1090439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bertoncini CW, et al. Release of long-range tertiary interactions potentiates aggregation of natively unstructured alpha-synuclein. Proc Natl Acad Sci USA. 2005;102(5):1430–1435. doi: 10.1073/pnas.0407146102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dedmon MM, Lindorff-Larsen K, Christodoulou J, Vendruscolo M, Dobson CM. Mapping long-range interactions in alpha-synuclein using spin-label NMR and ensemble molecular dynamics simulations. J Am Chem Soc. 2005;127(2):476–477. doi: 10.1021/ja044834j. [DOI] [PubMed] [Google Scholar]
- 44.Levitan K, et al. Conserved C-terminal charge exerts a profound influence on the aggregation rate of α-synuclein. J Mol Biol. 2011;411(2):329–333. doi: 10.1016/j.jmb.2011.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cartailler JP, Haigler HT, Luecke H. Annexin XII E105K crystal structure: Identification of a pH-dependent switch for mutant hexamerization. Biochemistry. 2000;39(10):2475–2483. doi: 10.1021/bi992278d. [DOI] [PubMed] [Google Scholar]
- 46.Bennati M, Luchinat C, Parigi G, Türke MT. Water 1H relaxation dispersion analysis on a nitroxide radical provides information on the maximal signal enhancement in Overhauser dynamic nuclear polarization experiments. Phys Chem Chem Phys. 2010;12(22):5902–5910. doi: 10.1039/c002304n. [DOI] [PubMed] [Google Scholar]
- 47.Polnaszek CF, Bryant RG. Nitroxide radical induced solvent proton relaxation: Measurement of localized translational diffusion. J Chem Phys. 1984;81(9):4038–4045. [Google Scholar]
- 48.Jansson H, Kargl F, Fernandez-Alonso F, Swenson J. Dynamics of a protein and its surrounding environment: A quasielastic neutron scattering study of myoglobin in water and glycerol mixtures. J Chem Phys. 2009;130(20):205101. doi: 10.1063/1.3138765. [DOI] [PubMed] [Google Scholar]
- 49.Russo D, Hura G, Head-Gordon T. Hydration dynamics near a model protein surface. Biophys J. 2004;86(3):1852–1862. doi: 10.1016/S0006-3495(04)74252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Farahbakhsh ZT, Altenbach C, Hubbell WL. Spin labeled cysteines as sensors for protein-lipid interaction and conformation in rhodopsin. Photochem Photobiol. 1992;56(6):1019–1033. doi: 10.1111/j.1751-1097.1992.tb09725.x. [DOI] [PubMed] [Google Scholar]
- 51.Dalton LA, McIntyre JO, Fleischer S. Distance estimate of the active center of D-beta-hydroxybutyrate dehydrogenase from the membrane surface. Biochemistry. 1987;26(8):2117–2130. doi: 10.1021/bi00382a009. [DOI] [PubMed] [Google Scholar]
- 52.Boettcher JM, et al. Atomic view of calcium-induced clustering of phosphatidylserine in mixed lipid bilayers. Biochemistry. 2011;50(12):2264–2273. doi: 10.1021/bi1013694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kunding AH, et al. Intermembrane docking reactions are regulated by membrane curvature. Biophys J. 2011;101(11):2693–2703. doi: 10.1016/j.bpj.2011.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Donnelly D, Overington JP, Blundell TL. The prediction and orientation of alpha-helices from sequence alignments: The combined use of environment-dependent substitution tables, Fourier transform methods and helix capping rules. Protein Eng. 1994;7(5):645–653. doi: 10.1093/protein/7.5.645. [DOI] [PubMed] [Google Scholar]
- 55.Cornette JL, et al. Hydrophobicity scales and computational techniques for detecting amphipathic structures in proteins. J Mol Biol. 1987;195(3):659–685. doi: 10.1016/0022-2836(87)90189-6. [DOI] [PubMed] [Google Scholar]
- 56.Pfefferkorn CM, et al. Depth of α-synuclein in a bilayer determined by fluorescence, neutron reflectometry, and computation. Biophys J. 2012;102(3):613–621. doi: 10.1016/j.bpj.2011.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ulmer TS, Bax A, Cole NB, Nussbaum RL. Structure and dynamics of micelle-bound human alpha-synuclein. J Biol Chem. 2005;280(10):9595–9603. doi: 10.1074/jbc.M411805200. [DOI] [PubMed] [Google Scholar]
- 58.Eliezer D, Kutluay E, Bussell R, Jr, Browne G. Conformational properties of alpha-synuclein in its free and lipid-associated states. J Mol Biol. 2001;307(4):1061–1073. doi: 10.1006/jmbi.2001.4538. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.