Significance
Our data demonstrate that differences in host genotype that affect the carbohydrate landscape of the distal gut interact with diet to alter the composition and function of resident microbes in a diet-dependent manner.
Keywords: host–microbial mutualism, intestinal microbiota, metabolomics
Abstract
We investigate how host mucus glycan composition interacts with dietary carbohydrate content to influence the composition and expressed functions of a human gut community. The humanized gnotobiotic mice mimic humans with a nonsecretor phenotype due to knockout of their α1–2 fucosyltransferase (Fut2) gene. The fecal microbiota of Fut2− mice that lack fucosylated host glycans show decreased alpha diversity relative to Fut2+ mice and exhibit significant differences in community composition. A glucose-rich plant polysaccharide-deficient (PD) diet exerted a strong effect on the microbiota membership but eliminated the effect of Fut2 genotype. Additionally fecal metabolites predicted host genotype in mice on a polysaccharide-rich standard diet but not on a PD diet. A more detailed mechanistic analysis of these interactions involved colonization of gnotobiotic Fut2+ and Fut2− mice with Bacteroides thetaiotaomicron, a prominent member of the human gut microbiota known to adaptively forage host mucosal glycans when dietary polysaccharides are absent. Within Fut2− mice, the B. thetaiotaomicron fucose catabolic pathway was markedly down-regulated, whereas BT4241–4247, an operon responsive to terminal β-galactose, the precursor that accumulates in the Fut2− mice, was significantly up-regulated. These changes in B. thetaiotaomicron gene expression were only evident in mice fed a PD diet, wherein B. thetaiotaomicron relies on host mucus consumption. Furthermore, up-regulation of the BT4241–4247 operon was also seen in humanized Fut2− mice. Together, these data demonstrate that differences in host genotype that affect the carbohydrate landscape of the distal gut interact with diet to alter the composition and function of resident microbes in a diet-dependent manner.
The human colon contains tens of trillions of microbes that participate in a dynamic set of mutualistic interactions with their host (1). The colon provides its microbial inhabitants with an environment rich in nutrients, including complex dietary plant polysaccharides and host-derived glycans secreted in the mucus layer and on the surface of the epithelium. Intestinal mucins are a rich source of carbohydrates, comprised of nearly 50–80% oligosaccharides by weight (2). Consumption of mucus glycans by many gut species has been well documented (3–5), yet few studies have detailed the global impact of changes in the host glycan landscape upon microbiota composition and function.
Fucose residues are common in the mucin glycans of individuals that possess a functional copy of the α1–2 fucosyltransferase (FUT2) gene. The terminal position of fucose on host glycans places this sugar at the interface of microbiota–mucus interactions. The FUT2 gene, also known as the secretor gene, encodes a galactoside 2-α-l-fucosyltransferase 2 (EC 2.4.1.69) responsible for adding a l-fucose residue in α1–2 linkage to the terminal β-d-galactose residue of gut mucus glycans (6). Approximately 20% of humans lack functional copies of this gene (known as “nonsecretors”) and, as a result, lack terminal fucose residues in their distal gut mucin (7). The importance of this residue in host–pathogen interaction is suggested by the observation that susceptibility to infection with Norwalk (8) and respiratory viruses (9), Vibrio cholerae (10), Campylobacter jejuni (11), and Helicobacter pylori (12) is affected by secretor status. Recently, genome-wide association studies have found nonsecretors have increased susceptibility to chronic inflammatory conditions linked to the gut microbiota, such as Crohn's disease (13) and primary sclerosing cholangitis (14).
The importance of mucosal fucose in mediating interactions between gut microbes and their host has been explored (15–17). Signals from the microbiota are required for Fut2 expression and mucus fucosylation in adult mice (6). The ability of the prominent gut symbiont, Bacteroides thetaiotaomicron, to induce fucosylation in intestinal mucin when introduced into germ-free (GF) mice depends on B. thetaiotaomicron’s ability to catabolize fucose (18). Bacteroides fragilis possesses a fucose salvage pathway that enables host-derived fucose to be recycled to its capsular polysaccharide, a pathway that enhances the bacterium’s fitness in vivo (19). FUT2 has been shown to interact with host disease status to influence alpha diversity (differences within a community) and beta diversity (differences between communities) in fecal microbial communities from individuals with and without Crohn's disease (20). Many of the bacteria known to interact with host fucose are also adept at using dietary carbohydrates, and diet is known to be one of the key factors influencing microbiota composition and function (21–23).
The simultaneous exposure of our microbiota to both host and environmental factors raises an important question: How do host genotype and diet interact to shape gut microbial communities? To address this question, we have studied the effect of fucosylation of gut glycans on human-derived gut microbial community composition and metabolism as a function of dietary glycan composition in a gnotobiotic mouse model. We used Fut2−/− animals (24), a model of human nonsecretors, which were rederived as GF to enable colonization with an intact uncultured human microbiota or with a single prominent human gut symbiont that is able to adaptively forage fucosylated mucosal glycans, depending on the availability of dietary polysaccharides. We show how mice harboring a transplanted human gut microbiota can be used to study the interplay of host fucosylated glycans and dietary glycan availability on the composition and function of the microbiota under well defined and controlled environmental conditions (25). Our results reveal that the impact of host fucosylation on gut microbial composition is diet dependent and that changes in gut microbial composition are accompanied by changes in the fecal and urinary metabolomes that discriminate Fut2-deficient animals from those that produce fucosylated glycans. Furthermore, whole-genome transcriptional profiling of B. thetaiotaomicron in monoassociated gnotobiotic mice provided direct insight into how the availability of host fucosylated glycans directly affects the functions that it expresses in the distal gut. Together, these findings illustrate an approach for identifying and testing hypotheses about the relative contributions of human mucosal glycan phenotype and dietary carbohydrate composition to the dynamic operations of the human gut microbiota in health or disease (26).
Results
Lack of Fucosylation in the Distal Gut Mucus Significantly Alters Microbial Diversity and Composition.
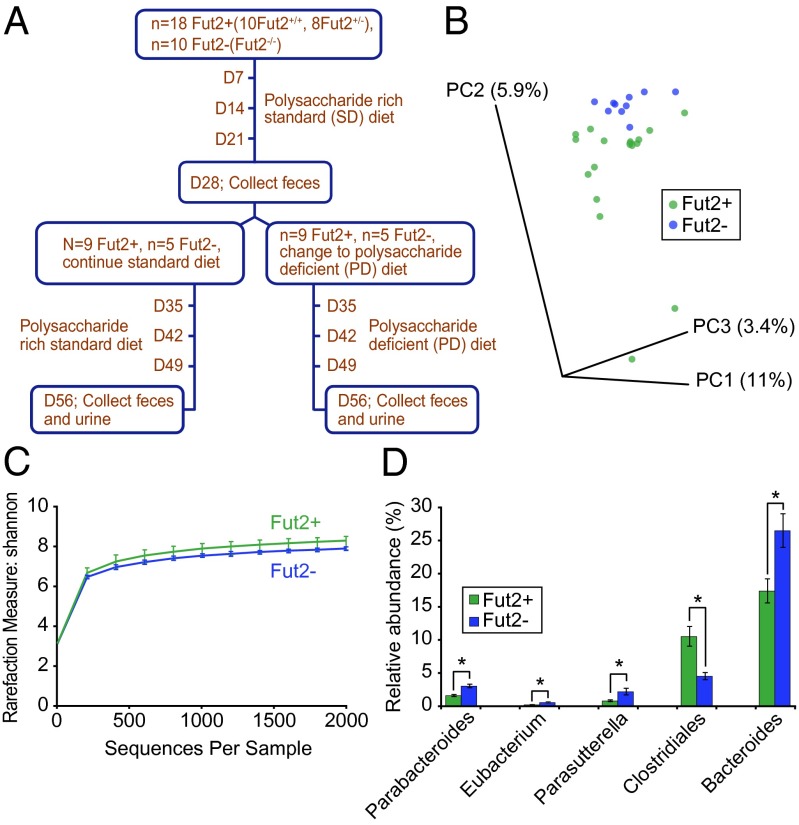
To test the hypothesis that the presence or absence of terminal fucose residues on host mucus and epithelial glycans would impact the microbiota, C57BL/6J Fut2−/− mice (27) were rederived as GF. GF Fut2-deficient Fut2−/−, wild-type Fut2+/+ (WT), and heterozygous Fut2+/− (HT) mice were colonized with feces obtained from a healthy human donor (secretor) and were maintained on a standard (SD) diet. Fecal samples were collected 4 wk (D28) after humanization from Fut2−/− (n = 10), WT (n = 10), and HT mice (n = 8) and subjected to 454 pyrosequencing of amplicons generated from the V3-V5 variable region of the bacterial 16S rRNA gene (Fig. 1A). Data analysis and normalization were performed by using QIIME 1.4.0 as described in Methods. Fecal communities were compared by using UniFrac, a phylogeny-based distance metric that measures the degree to which any two microbiota share branch length on a Bacterial tree of life.
Fig. 1.
Effect of gut mucus fucosylation on the diversity and composition of a human microbiota transplanted into gnotobiotic mice fed a plant polysaccharide-rich standard diet. (A) Experimental design. (B) Unweighted UniFrac-based PCoA plot of fecal microbial communities from humanized Fut2+ (n = 18) and Fut2− (n = 10) mice. (C) Rarefaction curves using Shannon measure of alpha diversity in fecal samples obtained from humanized Fut2+ (n = 18) and Fut2− (n = 10) animals. (D) Significant genus level differences (P < 0.05; ANOVA, FDR < 5%) in gut microbial communities of humanized gnotobiotic Fut2+ and Fut2− mice. Data are shown as averages ± SEM. *P < 0.05.
The 16S rRNA-based microbiota community composition analysis of fecal samples obtained from mice of all three genotypes, Fut2−/−, Fut2+/−, Fut2+/+, maintained on SD diet revealed that engraftment of the human gut microbiota following humanization of GF mice was successful: All bacterial phyla, classes, and 90% (35/39) of genus-level taxa in the donor sample were detected among the recipient mice. There was no significant difference in the interindividual variation versus the intraindividual variation in humanized WT and HT mice (as measured by unweighted UniFrac distance metric) on the SD diet at 4 wk (SI Appendix, Fig. S1A). Moreover, there was no significant difference in either alpha or beta diversity between humanized WT and HT mice (SI Appendix, Fig. S1 B and C), consistent with both genotypes expressing fucose on their intestinal mucin. Bacterial 16S rRNA datasets generated from the fecal microbiota of WT and HT humanized mice were therefore combined (referred to as Fut2+) and compared with 16S rRNA data generated from the fecal microbiota of humanized Fut2−/− mice (referred to as Fut2−). Principal coordinate analysis (PCoA) of unweighted UniFrac distances revealed clustering of the transplanted human microbial communities based on the Fut2 status of recipient mice [Fig. 1B; P < 0.05, permutational multivariate analysis of variance (PERMANOVA)]. A significant decrease in alpha diversity by using Shannon index was observed in the fecal microbiota of Fut2− compared with Fut2+ mice (Fig. 1C; P < 0.05, two-sample t test). However, a significant difference in alpha diversity was not seen by using Chao1 index (SI Appendix, Fig. S2), suggesting that there is a difference in evenness (relative abundance of species) and not richness (total number of species).
Followup analysis disclosed significant differences in the relative abundance of bacterial taxa at multiple taxonomic levels [SIAppendix, Table S1; as judged by ANOVA with false discovery rate (FDR) < 5%]. Members of the genera Parabacteroides, Eubacterium, Parasutterella, Bacteroides, and family Lachnospiraceae were significantly more abundant in Fut2− mice, whereas an unclassified genus within the order Clostridiales was more abundant in Fut2+ mice (Fig. 1D). Collectively, these data show that the availability of fucosylated glycans affects the overall diversity and composition of a human gut microbiota. Our findings are consistent with the decreased alpha diversity and an increase in an operational taxonomic unit (OTU) within Lachnospiraceae in nonsecretors reported in a recent survey of humans (20).
Diet Overrides the Effect of Gut Mucus Fucosylation on Microbial Composition.
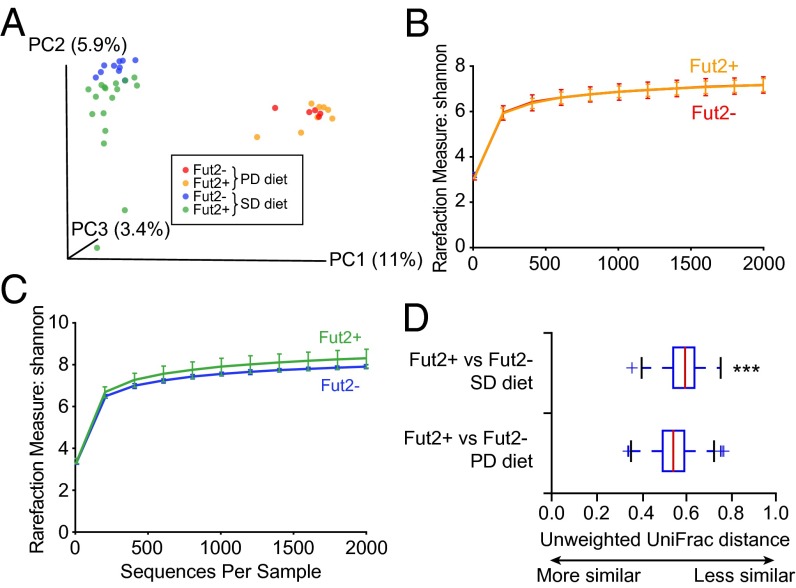
We hypothesized that a diet lacking complex polysaccharides, which increases microbiota reliance upon host mucus as an alternate source of glycans (5), would exacerbate Fut2 genotype-associated differences in microbial diversity and composition in our humanized gnotobiotic mice. Therefore, after 4 wk on the SD diet, half of the humanized Fut2− (n = 5) and half of the humanized Fut2+ mice (n = 9; 4 Fut2+/+; 5 Fut2+/−) were switched to a polysaccharide-deficient (PD) diet rich in glucose (68% wt/vol) for an additional 4 wk, whereas the remaining mice continued on the SD diet rich in complex polysaccharides. At the end of this 4-wk period (day 56 after gavage with the human microbiota sample), fecal microbial communities from all mice were characterized (Fig. 1A). PCoA of unweighted UniFrac distances showed primary clustering by diet 4 wk after switching to the PD diet, but failed to show the discrete clustering of Fut2+ and Fut2− animals (Fig. 2A; P > 0.05, PERMANOVA), as was observed on the SD diet. No difference in alpha diversity was apparent as determined by the Shannon index between humanized gnotobiotic Fut2− and Fut2+ mice on the PD diet (D56, Fig. 2B; P > 0.05, two-sample t test) and no statistically significant differences in the relative abundance of bacterial taxa (at any level) could be detected. In contrast, Fut2− mice that continued on SD diet continued to show decreased alpha diversity compared with their Fut2+ counterparts (D56, Fig. 2C; P < 0.05, two-sample t test), similar to D28.
Fig. 2.
Dietary influence on the effect of gut mucus fucosylation on microbial diversity. (A) Unweighted UniFrac-based PCoA plot of gut microbial communities from humanized Fut2+ and Fut2− mice fed the SD diet for 28 d (sampling on D28; n = 28 animals) followed by a PD diet for an additional 28 d (sampling on D56; n = 14 animals). (B and C) Rarefaction curves using Shannon measure of alpha diversity from fecal samples obtained from humanized Fut2+ and Fut2− mice fed a PD diet (B; n = 14) or SD diet (C; n = 14) collected on D56. (D) Unweighted UniFrac distance plot of fecal samples collected weekly for 4 wk after the diet switch on day 29 from mice that remained on the SD diet or that were switched to the PD diet. “+” represents values outside the interquartile range. ***P < 0.0001.
We extended our analysis by comparing fecal samples collected weekly for 4 wk after the day 29 diet change from animals that remained on SD diet, to those that were switched to PD diet. Unweighted UniFrac revealed that during this time period, the transplanted human donor fecal microbiota had a significant difference in its overall phylogenetic configuration in Fut2+ versus Fut2− mice as they were consuming the SD diet (Fig. 2D; P < 0.01, PERMANOVA). In contrast, no significant differences were observed between Fut2+ and Fut2− animals while they were fed the PD diet (Fig. 2D, P > 0.05, PERMANOVA). Collectively, these data demonstrate that elimination of dietary polysaccharides has an overall greater impact on the structural configuration of the human gut microbiota than host genotype. In addition, PD diet appears to minimize the effect of genotype on microbiota composition, contrary to our initial hypothesis. We reasoned that this lack of compositional change may be due to the restrictive nature of the diet, enriching for efficient host mucus glycan users that are capable of functional accommodation to the difference in host genotype.
Urinary and Fecal Metabolites Show Genotype-Dependent Changes on Standard Diet.
We next explored whether we could detect differences in functionality of the microbiotas of Fut2− and Fut2+ mice on the SD and PD diet. To do so, we used nontargeted ultra performance liquid chromatography (UPLC)-MS to profile metabolites in the urine and feces collected from three humanized Fut2+ and Fut2− mice each on PD diet (D28–D56) and SD diet (D0–D56). In the urine, we identified a larger number of significantly different features between Fut2+ and Fut2− mice fed the SD diet compared with mice fed the PD diet (SI Appendix, Fig. S3 A and B): 117 features of the 1,443 total detected (8%) were defined as significantly different (>twofold change, P < 0.05; two-sample t test; 70 features more abundant in Fut2+; 47 more abundant in Fut2−) (SI Appendix, Fig. S3A and Table S2). In contrast, only 12 of 942 detected features (1.2%) were significantly different between Fut2+ and Fut2− mice fed the PD diet (SI Appendix, Fig. S3B and Table S2). Similarly, in fecal samples, 13 features of the 563 total detected (2.3%) were significantly different between the Fut2+ and Fut2− mice on the SD diet (SI Appendix, Fig. S3C and Table S4), whereas only 3 of the 404 features (0.7%) were significantly different between the two genotypes on the PD diet (SI Appendix, Fig. S3D and Table S5).
Procrustes analysis (comparing two distance matrices developed using different mechanisms) revealed that the goodness of fit was significant (P < 0.05 with 1,000 permutations) when comparing UniFrac distances (for 16S rRNA based compositional data) to Hellinger distances (for fecal metabolomic data), with both data types showing significant agreement in detecting the diet-induced changes to the microbial communities and fecal metabolomes (SI Appendix, Fig. S4).
One drawback of Procrustes analysis is the reduction of large datasets to distance matrices. Therefore, we next used a supplementary robust statistical method, sparse canonical correlation analysis, to identify correlations among metabolites, OTUs, and fecal samples. The analysis revealed several metabolites and OTUs to be strongly correlated and the striking separation between individual samples on either PD diet or SD diet (SI Appendix, Fig. S5).
Machine learning (useful for identifying patterns in complex datasets) using the commonly adopted classifier for microarray analysis, the Random Forests classifier (Breimen: www.springerlink.com/content/u0p06167n6173512), suggested that fecal metabolites can classify samples as being from Fut2+ or Fut2− mice on the SD diet with 84% accuracy. The most discriminatory metabolites contributing to 0.3% accuracy or more each had the following m/z values: 309.1670, 252.0847, 270.1779, and 332.2429. (See SI Appendix, Table S6 for a full listing of metabolites that contribute to genotypic discrimination.) However, in mice fed the PD diet, the distinction between Fut2+ and Fut2− by fecal metabolites was no better than chance, suggesting that in this dietary context, similarly composed microbial communities on PD diet are also metabolically similar. We also applied Random Forests analysis to the urine metabolite datasets. We found that the metabolite that most discriminates Fut2+ from Fut2− animals on the SD diet had m/z value 324.1563 (0.4% contribution), whereas the one that discriminates them most on the PD diet had m/z value 340.1752 (0.3% contribution).
Together, these data demonstrate that fecal metabolites show strong agreement with 16S rRNA-based microbial community data in diet-induced alteration of microbial communities. If fed the PD diet, microbial communities from both humanized Fut2+ and Fut2− mice have similar community composition and metabolomic profile.
Imputed Community Functional Capacity Changes Reflect Dependence on Host Mucin Glycans.
Because of the abundance of genome sequencing data for microbiota-derived strains, it is possible to infer metagenomes from 16S rRNA profiles, an approach that has proven informative in previous studies (28). In an effort to better understand the shift in functional capacity of the microbiota that can result from compositional changes, we imputed the metagenomes of the communities by using our 16S rRNA sequencing data. Normalizing for 16S rRNA copy numbers and summing the genes that mapped to Kyoto Encyclopedia of Genes and Genomes (KEGG) orthologs across pathways, the KEGG pathways relating to carbohydrate metabolism and membrane transport occupied the largest share of genes across all pathways (SI Appendix, Table S7). Investigating those genes involving carbohydrate metabolism revealed that 75% (FDR < 0.05) of the 53 glycoside hydrolase families (GHs) identified in all metagenomes combined were significantly enriched in mice on the PD diet compared with those on the SD diet (SI Appendix, Table S8), including the β-galactosidases (GH35) that are capable of hydrolyzing terminal host mucin glycans.
We next hypothesized that the microbiota would optimize for mucin-associated GHs between the SD and PD diets and potentially reveal an effect of genotype. Plotting comparisons by individual (SI Appendix, Fig. S6A) and average percent enrichment (SI Appendix, Fig. S6B) in significant mucin-associated GHs reveals the shift in functional capacities that correspond to specific community compositional changes (SI Appendix, Fig. S6C). These findings suggest that the extent of dependence on host mucin glycans can be influenced by both diet and genotype (SI Appendix, Fig. S6 A and B). Differences in GHs in humanized Fut2+ and Fut2− mice on SD and PD diet are outlined in SI Appendix, Tables S9–S12. The differential change in host mucus glycan related GHs in the two genotypes reflects change in functionality of the community based around glycan harvesting that would be difficult to detect by using our UPLC-MS–based metabolomics approach.
The prominence of carbohydrate metabolism and several glycoside hydrolases in the inferred metagenomic response, combined with the known biochemical change in the mouse intestinal mucus, led us to wonder whether functional changes in individual microbes induced by host genotypic differences might be identified using a more targeted approach in a simplified community. Specifically we wondered whether the presence or absence of host fucose in mucus could result in gene expression changes of carbohydrate utilization genes in mucus using species.
A Case Study of a Prominent Member of the Human Gut Microbiota Known to Adaptively Forage on Host Fucosylated Glycans.
Alterations in microbiota glycan foraging that may occur as a result of changes in host distal gut glycan landscape may not be apparent in metabolomic signatures. Therefore, we investigated how functional niches (“professions”) occupied by a prominent saccharolytic human gut symbiont, B. thetaiotaomicron, are impacted by host fucosylation status. B. thetaiotaomicron has the capacity to use a broad array of dietary and host mucus carbohydrates including fucose (3, 21, 29). Moreover, in the absence of dietary polysaccharides, B. thetaiotaomicron relies primarily on host mucus glycans because a majority of simple sugars like glucose are absorbed in the small intestine. We hypothesized that a host genetic mutation that alters distal gut fucosylation patterns would impact the host carbohydrates accessed by B. thetaiotaomicron in vivo—a change in functionality that should be evident in altered gene expression of the symbiont.
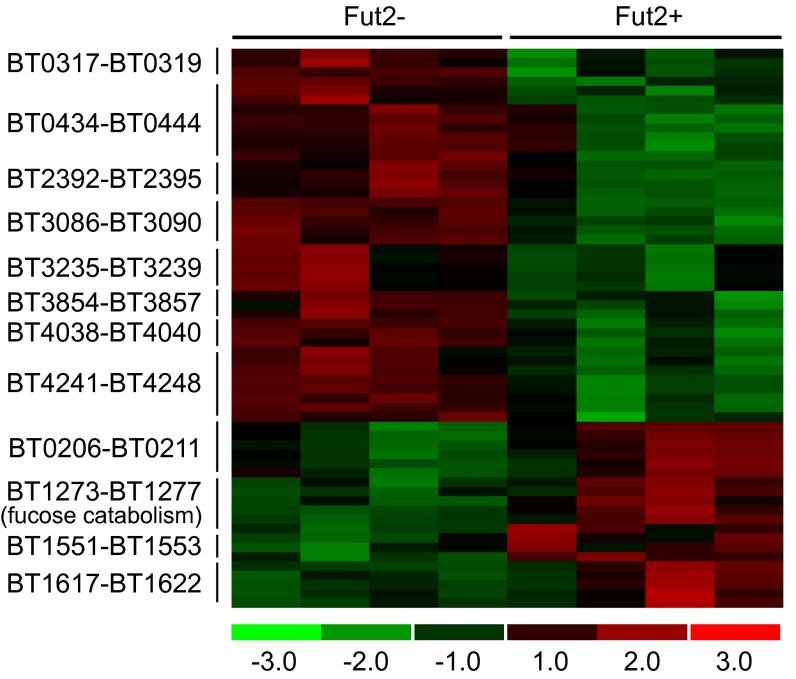
Fut2+ (n = 4) and Fut2− (n = 4) adult GF male mice that were fed a PD diet were colonized with B. thetaiotaomicron and killed after 10 d. In all mice, cecal contents including the mucus layer were removed, and RNA was extracted for transcriptional profiling of B. thetaiotaomicron by using custom GeneChips containing probe pairs derived from 4,719 of the 4,779 predicted B. thetaiotaomicron genes. A total of 636 B. thetaiotaomicron genes were identified as differentially regulated on the PD diet between the Fut2− and Fut2+ mice (FDR 7.5%, fold change > 1.2; q value <5%, see Methods for detailed description of gene filtering). A total of 421 B. thetaiotaomicron genes were up-regulated and 215 genes were down-regulated in the Fut2+ mice relative to Fut2− mice on the PD diet (SI Appendix, Table S13). The genes involved in fucose catabolism (BT1273–77 encoding l-fucose isomerase, l-fuculose-1-phosphate aldolase, l-fuculose kinase, fucose mutarotase, and l-fucose permease) were up-regulated in Fut2+ mice (2.2–3.7-fold), consistent with the presence of fucosylated glycans in the host mucosa (Fig. 3).
Fig. 3.
Fucosylation determines transcriptional responses of B. thetaiotaomicron in the intestines of monocolonized Fut2− and Fut2+ mice fed a PD diet. Heat map showing significantly regulated genes within 11 PULs (associated with SusC/SusD-like gene pairs) and the fucose catabolic operon (n = 4 mice per genotype). All genes shown exhibit significant differences between the two host genotypes with the exception of BT0438 and BT3856. Colors indicate the deviation of a gene’s signal above (red) and below (green) its mean expression value across all eight samples.
B. thetaiotaomicron possesses 88 polysaccharide utilization loci (PULs) in its genome (3) that are characterized by genes encoding homologs of SusC and SusD, outer membrane proteins that bind and import glycans. These highly regulated PULs specialize in acquisition and metabolism of different types of carbohydrates. Therefore, the relative expression levels of these 88 loci provide a representation of what B. thetaiotaomicron is “eating” within a given condition. Eleven PULs exhibited significant differences in their levels of expression in Fut2+ versus Fut2− mice consuming the PD diet, consistent with marked changes in the nutrients being accessed by B. thetaiotaomicron in vivo (Fig. 3). Genes (BT3854–57, BT2392–95, BT0434–44, BT4038–40, BT0317–19, and BT4241–48) within six of these PULs have been shown to be more highly expressed by B. thetaiotaomicron in vivo under PD diet conditions where the reliance on adaptive foraging of host mucus increases (see SI Appendix, Table S14 for gene annotations) (3). Alternatively, whole genome transcriptional profiling of B. thetaiotaomicron failed to show any significant differences in expression of any PULs reported above when Fut2+ (n = 5) and Fut2− (n = 5) B. thetaiotaomicron-monoassociated mice were fed a SD diet rather than PD diet; only two genes involved in fucose utilization, BT1276 (fucose mutarotase) and BT1277 (l-fucose permease), showed detectable changes in expression (SI Appendix, Table S14). We concluded that in the absence of pressure to use host glycans (SD diet), host fucosylation status has minimal impact on B. thetaiotaomicron. However, dietary polysaccharide deficiency interacts with host fucosylation status to significantly impact B. thetaiotaomicron’s glycan foraging in vivo.
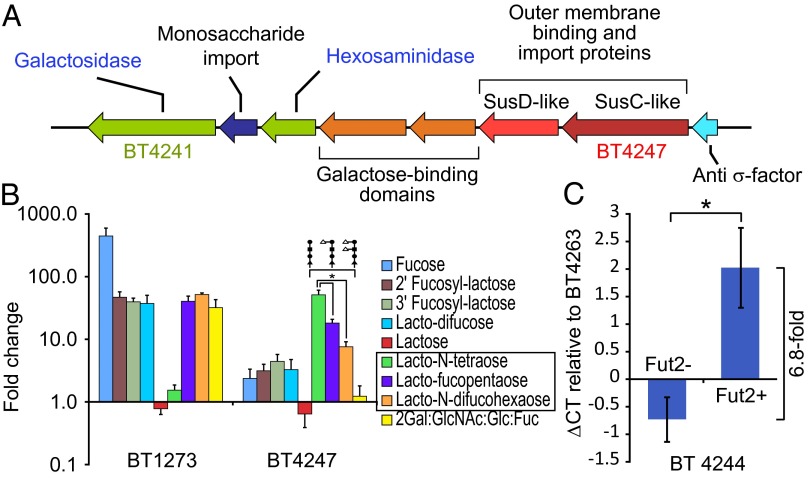
Of the PULs affected by host secretor status, BT4241–4248 was particularly notable because it includes genes that putatively encode a β-galactosidase (BT4241), a hexosaminidase (BT4243), and two proteins with galactose-binding domains (BT4244–5) in addition to paralogs of SusC and SusD (BT4246–7) (Fig. 4A). These genes appear to provide the requisite machinery to use host glycans that terminate with β-galactose. Furthermore, expression of this PUL is known to be induced in vitro in the presence of core-1 glycan structures terminating in β-galactose (3). The lack of fucosylated glycans in the stomach (antral region), cecum, and colon of Fut2− mice is accompanied by an accumulation of precursor structures that contain terminal β-galactose (27). The increased expression of the PUL containing BT4241–4248 in Fut2− mice suggested that the locus is responsive to host glycans in vivo that terminate in β-galactose and that masking of this galactose with fucosylation in the Fut2+ mice reduces expression of its component genes.
Fig. 4.
B. thetaiotaomicron responds to β-galactose–terminated glycans in the absence of fucose. (A) Gene content of the PUL encompassing BT4241–48. (B) Expression levels of BT1273 encoding fucose isomerase within the fucose catabolic operon and BT4243 (hexosaminidase) within the BT4241–4248 PUL as determined by qRT-PCR expressed as fold difference relative to growth in minimal medium with glucose. Averages ± SEM of three biological replicates are indicated. (C) Relative expression levels of BT4244 (putative galactose binding domain) in cecal contents of humanized Fut2+ (n = 4) and Fut2− (n = 5) mice fed a PD diet; data are normalized to the constitutively expressed glycolytic enzyme encoded by BT4263 (glyceraldehyde-3-phosphate dehydrogenase), as determined by qRT-PCR. *P < 0.05.
To test how the expression of this PUL is affected by glycan fucosylation, B. thetaiotaomicron was grown in minimal medium supplemented with single purified carbohydrates that vary in fucose composition (fucose, 2′ fucosyl-lactose, 3′ fucosyl-lactose, lacto-difucose, lactose, lacto-N-tetraose, lacto-fucopentaose, lacto-N-difucohexaose, or a mixture of unlinked component monosaccharides in the ratio 2Gal:GlcNAc:Glc:Fuc). Triplicate cultures of B. thetaiotaomicron cells were harvested midway through the logarithmic phase of growth in each condition; glucose-containing minimal medium was used as a control. Expression of BT4247 (SusC paralog within the putative β-galactose utilization operon) was monitored by quantitative RT-PCR (qRT-PCR). Expression of BT1273 was used as a control: It encodes a fucose isomerase gene located within the fucose utilization operon known to be transcriptionally responsive to the presence of fucose. BT1273 was significantly up-regulated compared with glucose as a baseline in the presence of all carbohydrate sources with one or two terminal fucose residues (2′ fucosyl-lactose, 46.9 ± 10.4-fold; 3′ fucosyl-lactose, 39.7 ± 5.7-fold; lacto-difucose, 37.3 ± 13.3-fold; lacto-fucopentaose, 40.3 ± 8.5-fold; lacto-N-difucohexaose, 51.7 ± 3.3-fold; free fucose 445.2 ± 149.6-fold; mean ± SEM, n = 3 biological replicates), and a monosaccharide mixture that represents the composition of host fucosylated glycans but contains no glycosidic linkages (2Gal:GlcNAc:Glc:Fuc, 31.9 ± 11-fold). However, expression of BT1273 was not induced in the presence of lactose or lacto-N-tetraose, two carbohydrates that lack fucose but possess the core glycan structures present in the other fucosylated glycans (Fig. 4B). Conversely, BT4247 was most prominently up-regulated compared with expression levels in glucose as a baseline, in the presence of lacto-N-tetraose, a tetrasaccharide that terminates in β-galactose (51.2 ± 9.6-fold). This up-regulation was attenuated upon addition of one fucose residue to lacto-N-tetrose (lacto-fucopentaose, 18.2 ± 2.7-fold; P < 0.05, two-sample t test compared with glucose as a baseline) or two fucose residues (lacto-N-difucohexaose, 7.6 ± 1.6-fold; P < 0.05 two-sample t test compared with glucose as a baseline) (Fig. 4B).
Together, these data are consistent with up-regulation of the PUL BT4241–8 in Fut2− compared with Fut2+ hosts and show that resident gut symbionts can exhibit remarkable versatility in glycan foraging with sensitivity to genetically dictated changes in the host mucosal glycan landscape. These data also provide an example of how host diet and genotype can intersect to alter the functionality within the gut microbiota.
Finally, we addressed whether the changes observed in expression of B. thetaiotaomicron genes in the Fut2− mice fed a PD diet were evident in humanized gnotobiotic mice. Cecal contents from humanized Fut2+ (n = 5) and Fut2− (n = 4) animals fed the PD diet described above (D56; Fig. 1A) were harvested, and cDNA was prepared and subjected to qRT-PCR. Expression of a marker gene within BT4241–8 (BT4244; putative galactose binding domain) was measured in all samples and normalized to expression of the constitutively expressed BT4263 (glyceraldehyde-3-phosphate dehydrogenase; B. thetaiotaomicron-specific primers), a part of the glycolytic pathway. The primers for BT4244 and BT4263 were designed to perfectly conserved regions of these genes within all three sequenced strains of B. thetaiotaomicron. BT4244 expression was significantly elevated (6.8-fold) in Fut2− mice relative to Fut2+ mice on this diet (Fig. 4C; ∆Ct, P < 0.05, two-sample t test). These data further underscore how studying the transcriptional responses of members of a defined gut community (even a community composed of one member) to defined environmental manipulations (diet, host genotype) can help predict transcriptional responses in more complex community contexts (30). They also illustrate how changes in microbial community function can be seen as a result of interactions between diet and host genotype, even in the absence of significant differences in community composition.
Discussion
In this study, we investigate the effect of a null mutation in host fucosyltransferase-2 that affects the fucosylation status of the gut mucosa on gut microbiota composition and function and how this effect is altered by the carbohydrate content of the diet. Our results show that a genetically dictated change in gut carbohydrate landscape (the presence or absence of host fucose) significantly impacts gut microbial composition. The proportional representation of several genera within Bacteroidetes increase in abundance in Fut2-deficient (Fut2−/−) mice, including members of genus Bacteroides. Furthermore, we show that upon switching to PD diet, gut microbial compositional differences seen on SD diet are lost, illustrating that diet can exert a dominant effect over host genetics on the microbial composition, which corresponds to metabolomic features of these communities. Our results further highlight that microbial community composition alone is not reflective of the effect of diet on the gut microbiota. Our transcriptional data both in a simplified community (GF mice monocolonized with B. thetaiotaomicron) and in humanized mice clearly demonstrate a change in gut microbial community function in Fut2− mice when fed the PD diet despite a limited effect of genotype on community composition on this diet. The switch from using fucose to galactose by gut microbes represents an example of adaptive foraging as the carbohydrate landscape of the gut habitat changes.
The Fut2 gene encodes a galactoside 2-α-l-fucosyltransferase 2 (EC 2.4.1.69) enzyme that catalyzes the addition of terminal fucose residues, and the resulting H-type structure expressed on the surface of epithelial cells and in mucosal secretions is in a position to influence many types of host–microbe interactions (8–11). Nonsecretor individuals defined as having inactivating mutations in the human FUT2 gene have decreased susceptibility to H. pylori infection secondary to impairment of BabA-mediated adhesion of H. pylori to gastric mucosa as seen in Fut2−/− mice (31). Nonsecretors have decreased susceptibility to human noroviruses as noroviruses bind to α1–2 fucosylated glycans and as a result fail to show virus shedding or serum or salivary antibody responses (8). Secretor status also influences the composition of fucosylated glycans in human milk oligosaccharides. These oligosaccharides play important roles in the infant gut, providing protection from pathogens such as Campylobacter jejuni and norovirus by providing alternate binding sites and serving as nutrient substrates for Bifidobacterium spp. and Bacteroides spp (11, 32).
Recent genome-wide association studies have implicated FUT2 in the pathogenesis of Crohn's disease, primary sclerosing cholangitis, celiac disease, and type-1 diabetes (13, 14, 33, 34). Furthermore, nonsecretors at higher risk of Crohn's disease and primary sclerosing cholangitis have altered gut and biliary microbial communities compared with secretors: These nonsecretors exhibit changes in alpha diversity, increases in Firmicutes and decreases in Proteobacteria, as well as differences in Bacteroidetes, Actinobacteria, and Tenericutes in both the colonic and biliary microbiota (14, 20). The roles of altered host glycans in barrier integrity are likely numerous including physiological contributions to barrier strength, sensitivity of signaling pathways, and exclusion of pathogenic and invasive species either via direct interaction or by promoting microbiota adaptive foraging behavior that influences host responses (17).
Our findings reinforce the importance of assessing both host and dietary glycan function in preclinical models where both variables can be manipulated. Our findings further suggest that the effects of dietary interventions designed to impact microbial community function need to be interpreted in a holistic way with an understanding of what nutrient reservoirs exist for the community within the gut habitat.
Methods
All animal protocols were in accordance with Administrative Panel on Laboratory Animal Care, the Stanford Institutional Animal Care and Use Committee. Gene chip analyses were done by using custom Affymetrix B. thetaiotaomicron Genechips. Microbial community composition and imputed functionality analyses were based on datasets generated by using 16S rRNA amplicon pyrosequencing. Quantitative RT-PCR was performed by using gene-specific primers with SYBR Green (ABgene) in a MX3000P thermocycler (Stratagene). Metabolomic profiling of urine and feces was done by using UPLC-MS.
Please see detailed methods in SI Appendix.
Supplementary Material
Acknowledgments
We thank David O’Donnell and Maria Karlsson for rederivation of the Fut2-null mice as GF and Tim Vigers, Kathy Holt, Gail Ackermann, and Doug Wendel for help with data management. This work was supported by National Institutes of Health (NIH) Grants DK085025 (to J.L.S.), DK30292 (to J.I.G.), and GM86884 (to S.P.H.), NIH Pioneer Award DP1OD000964 (to D.A.R.), the Crohn’s and Colitis Foundation of America (J.I.G.), the Walter and Idun Berry Foundation (E.K.C.), and Thomas C. and Joan M. Merigan Endowment at Stanford Univeristy (to D.A.R.). S.A.S. is supported by a Smith Stanford Graduate Fellowship and National Science Foundation Fellowship Grant DGE-114747.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The GeneChip datasets have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE45641). The 16S sequence data have been deposited in the European Molecular Biology Laboratory (EMBL) nucleotide archive (accession no. ERP003630), and can also be found in the Quantitative Insights into Microbial Ecology (QIIME) database, www.microbio.me/qiime (study ID 1452).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1306070110/-/DCSupplemental.
References
- 1.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336(6086):1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corfield AP, Carroll D, Myerscough N, Probert CS. Mucins in the gastrointestinal tract in health and disease. Front Biosci. 2001;6:D1321–D1357. doi: 10.2741/corfield. [DOI] [PubMed] [Google Scholar]
- 3.Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4(5):447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turroni F, et al. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc Natl Acad Sci USA. 2010;107(45):19514–19519. doi: 10.1073/pnas.1011100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sonnenburg JL, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307(5717):1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 6.Bry L, Falk PG, Midtvedt T, Gordon JI. A model of host-microbial interactions in an open mammalian ecosystem. Science. 1996;273(5280):1380–1383. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- 7.Kelly RJ, Rouquier S, Giorgi D, Lennon GG, Lowe JB. Sequence and expression of a candidate for the human Secretor blood group alpha(1,2)fucosyltransferase gene (FUT2). Homozygosity for an enzyme-inactivating nonsense mutation commonly correlates with the non-secretor phenotype. J Biol Chem. 1995;270(9):4640–4649. doi: 10.1074/jbc.270.9.4640. [DOI] [PubMed] [Google Scholar]
- 8.Lindesmith L, et al. Human susceptibility and resistance to Norwalk virus infection. Nat Med. 2003;9(5):548–553. doi: 10.1038/nm860. [DOI] [PubMed] [Google Scholar]
- 9.Raza MW, et al. Association between secretor status and respiratory viral illness. BMJ. 1991;303(6806):815–818. doi: 10.1136/bmj.303.6806.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaudhuri A, DasAdhikary CR. Possible role of blood-group secretory substances in the aetiology of cholera. Trans R Soc Trop Med Hyg. 1978;72(6):664–665. doi: 10.1016/0035-9203(78)90031-7. [DOI] [PubMed] [Google Scholar]
- 11.Ruiz-Palacios GM, Cervantes LE, Ramos P, Chavez-Munguia B, Newburg DS. Campylobacter jejuni binds intestinal H(O) antigen (Fuc alpha 1, 2Gal beta 1, 4GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. J Biol Chem. 2003;278(16):14112–14120. doi: 10.1074/jbc.M207744200. [DOI] [PubMed] [Google Scholar]
- 12.Ikehara Y, et al. Polymorphisms of two fucosyltransferase genes (Lewis and Secretor genes) involving type I Lewis antigens are associated with the presence of anti-Helicobacter pylori IgG antibody. Cancer Epidemiol Biomarkers Prev. 2001;10(9):971–977. [PubMed] [Google Scholar]
- 13.McGovern DP, et al. International IBD Genetics Consortium Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn’s disease. Hum Mol Genet. 2010;19(17):3468–3476. doi: 10.1093/hmg/ddq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Folseraas T, et al. Extended analysis of a genome-wide association study in primary sclerosing cholangitis detects multiple novel risk loci. J Hepatol. 2012;57(2):366–375. doi: 10.1016/j.jhep.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Terahara K, et al. Distinct fucosylation of M cells and epithelial cells by Fut1 and Fut2, respectively, in response to intestinal environmental stress. Biochem Biophys Res Commun. 2011;404(3):822–828. doi: 10.1016/j.bbrc.2010.12.067. [DOI] [PubMed] [Google Scholar]
- 16.Domino SE, et al. Cervical mucins carry alpha(1,2)fucosylated glycans that partly protect from experimental vaginal candidiasis. Glycoconj J. 2009;26(9):1125–1134. doi: 10.1007/s10719-009-9234-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guruge JL, et al. Epithelial attachment alters the outcome of Helicobacter pylori infection. Proc Natl Acad Sci USA. 1998;95(7):3925–3930. doi: 10.1073/pnas.95.7.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hooper LV, Xu J, Falk PG, Midtvedt T, Gordon JI. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc Natl Acad Sci USA. 1999;96(17):9833–9838. doi: 10.1073/pnas.96.17.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coyne MJ, Reinap B, Lee MM, Comstock LE. Human symbionts use a host-like pathway for surface fucosylation. Science. 2005;307(5716):1778–1781. doi: 10.1126/science.1106469. [DOI] [PubMed] [Google Scholar]
- 20.Rausch P, et al. Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proc Natl Acad Sci USA. 2011;108(47):19030–19035. doi: 10.1073/pnas.1106408108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sonnenburg ED, et al. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell. 2010;141(7):1241–1252. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu GD, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Domino SE, Zhang L, Gillespie PJ, Saunders TL, Lowe JB. Deficiency of reproductive tract alpha(1,2)fucosylated glycans and normal fertility in mice with targeted deletions of the FUT1 or FUT2 alpha(1,2)fucosyltransferase locus. Mol Cell Biol. 2001;21(24):8336–8345. doi: 10.1128/MCB.21.24.8336-8345.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turnbaugh PJ, et al. The effect of diet on the human gut microbiome: A metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1(6):6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448(7152):427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 27.Hurd EA, Holmén JM, Hansson GC, Domino SE. Gastrointestinal mucins of Fut2-null mice lack terminal fucosylation without affecting colonization by Candida albicans. Glycobiology. 2005;15(10):1002–1007. doi: 10.1093/glycob/cwi089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morgan XC, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13(9):R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martens EC, et al. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 2011;9(12):e1001221. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McNulty NP, et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med. 2011;3(106):106ra106. doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Magalhães A, et al. Fut2-null mice display an altered glycosylation profile and impaired BabA-mediated Helicobacter pylori adhesion to gastric mucosa. Glycobiology. 2009;19(12):1525–1536. doi: 10.1093/glycob/cwp131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marcobal A, Sonnenburg JL. Human milk oligosaccharide consumption by intestinal microbiota. Clin Microbiol Infect. 2012;18(Suppl 4):12–15. doi: 10.1111/j.1469-0691.2012.03863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smyth DJ, et al. FUT2 nonsecretor status links type 1 diabetes susceptibility and resistance to infection. Diabetes. 2011;60(11):3081–3084. doi: 10.2337/db11-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parmar AS, et al. Association study of FUT2 (rs601338) with celiac disease and inflammatory bowel disease in the Finnish population. Tissue Antigens. 2012;80(6):488–493. doi: 10.1111/tan.12016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.