Significance
Penicillin binding protein 2a imparts to the human pathogen Staphylococcus aureus resistance to β-lactam antibiotics. Our structural characterization of the allosteric basis governing its resistance mechanism identifies a basis for the design of new antibacterials that can both activate and inhibit this key resistance enzyme.
Keywords: allosteric mechanism, antibiotic resistance, X-ray crystallography
Abstract
The expression of penicillin binding protein 2a (PBP2a) is the basis for the broad clinical resistance to the β-lactam antibiotics by methicillin-resistant Staphylococcus aureus (MRSA). The high-molecular mass penicillin binding proteins of bacteria catalyze in separate domains the transglycosylase and transpeptidase activities required for the biosynthesis of the peptidoglycan polymer that comprises the bacterial cell wall. In bacteria susceptible to β-lactam antibiotics, the transpeptidase activity of their penicillin binding proteins (PBPs) is lost as a result of irreversible acylation of an active site serine by the β-lactam antibiotics. In contrast, the PBP2a of MRSA is resistant to β-lactam acylation and successfully catalyzes the dd-transpeptidation reaction necessary to complete the cell wall. The inability to contain MRSA infection with β-lactam antibiotics is a continuing public health concern. We report herein the identification of an allosteric binding domain—a remarkable 60 Å distant from the dd-transpeptidase active site—discovered by crystallographic analysis of a soluble construct of PBP2a. When this allosteric site is occupied, a multiresidue conformational change culminates in the opening of the active site to permit substrate entry. This same crystallographic analysis also reveals the identity of three allosteric ligands: muramic acid (a saccharide component of the peptidoglycan), the cell wall peptidoglycan, and ceftaroline, a recently approved anti-MRSA β-lactam antibiotic. The ability of an anti-MRSA β-lactam antibiotic to stimulate allosteric opening of the active site, thus predisposing PBP2a to inactivation by a second β-lactam molecule, opens an unprecedented realm for β-lactam antibiotic structure-based design.
The inexorable spread of bacterial resistance mechanisms against β-lactam antibiotics is a critical clinical concern. The resistance mechanism used by methicillin-resistant Staphylococcus aureus (MRSA) is acquisition of a set of genes that are induced on β-lactam exposure (1, 2). The key resistance enzyme is a unique, monofunctional dd-transpeptidase designated as penicillin binding protein 2a (PBP2a) that is refractory to inhibition by virtually all β-lactam antibiotics. In the MRSA bacterium, PBP2a catalyzes, in concert with the transglycosylase activities of other penicillin binding proteins (PBPs), the biosynthesis of the bacterial cell wall (3–5). The structure of this cell wall is a peptidoglycan polymer, comprised of glycan strands consisting of a repeating disaccharide motif [N-acetylglucosamine-N-acetylmuramylpentapeptide (NAG-NAM pentapeptide)], wherein adjacent glycan strands are cross-linked by PBP2a using peptide stems found on each NAM saccharide. The cell wall encases the entire bacterium as a single molecule, and its integrity is indispensible to the organism’s survival (6). The PBPs are the lethal targets of the β-lactam antibiotics as a result of irreversible acylation of the active site serine.
The earlier structure determination for PBP2a (7) showed a closed active site conformation. Because the dd-transpeptidase site must accommodate two strands of peptidoglycan simultaneously—requiring an active site volume in excess of 1,000 Å3 (8, 9)—there must also exist an open conformation for PBP2a, which was preceded by structural studies with other PBPs (10–14). The mechanistic paradox in the case of PBP2a is not simply separate open and closed states but a mechanism that biases the open state to the peptidoglycan. Nature often resolves these paradoxes by allostery (15). Indeed, we have shown that synthetic samples of the peptidoglycan bind to PBP2a in a saturable manner and effect a conformational change that correlates to faster rates for PBP2a inactivation by β-lactams with enhanced PBP2a affinity (16). These observations suggested a structural model, wherein the binding of nascent peptidoglycan to a remote allosteric site opens the active site to both substrates and β-lactam inactivators. We disclose X-ray structures of PBP2a that confirm the presence of this allosteric site, reveal its location as 60 Å removed from the active site, and identify its ligands. Moreover, binding of these ligands to the allosteric site imparts conformational opening of the active site. Lastly, we document that ceftaroline, a new β-lactam antibiotic (Fig. 1A) that recently has received Food and Drug Administration approval for use in the treatment of MRSA infections, has the ability to trigger this conformational change and thus, enables access to the active site by a second ceftaroline molecule. These observations explain the mechanism for the manifestation of the physiological role of PBP2a, explain the advantageous anti-MRSA activity of ceftaroline, identify the basis for understanding emerging mutations in the gene for PBP2a that confer resistance to ceftaroline, and provide the context for future structure-based design of anti-MRSA β-lactams that will evade these mutations.
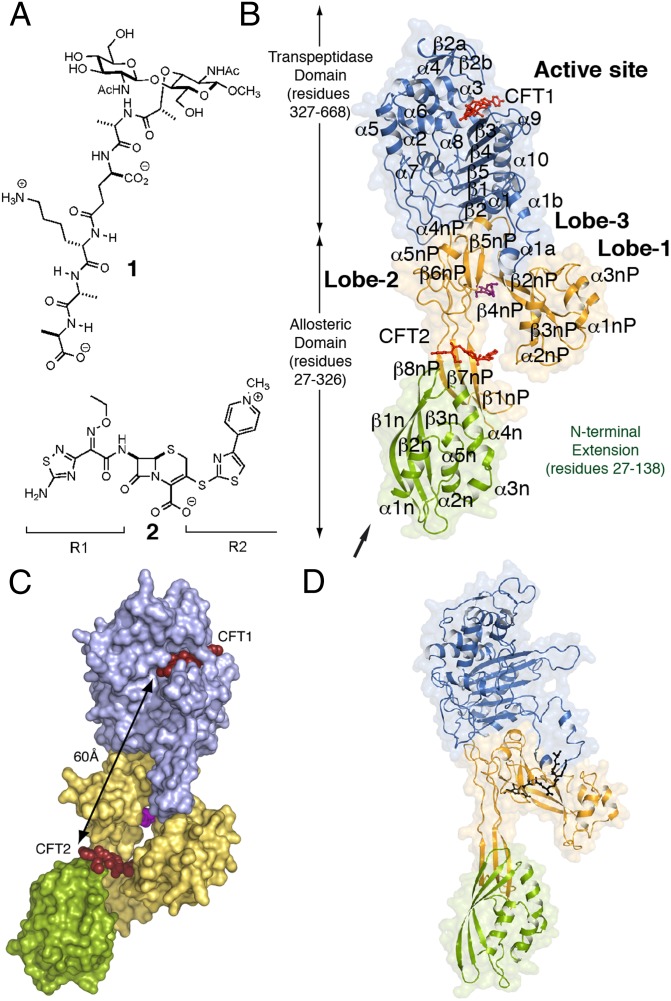
Fig. 1.
Domains of PBP2a and key ligands. (A) The chemical structures of a synthetic NAG-NAM(pentapeptide) (1) and ceftaroline (2). The R1 and R2 groups of 2 are labeled. (B) Ribbon representation of PBP2a acylated by ceftaroline. The N-terminal extension is colored in green, the remaining allosteric domain is colored in gold, and the transpeptidase (TP) domain is colored in blue. These domain colors are retained in all other figures. Two molecules of ceftaroline (capped sticks in red) are found in complex with protein: one covalently bound as an acyl-enzyme in the TP domain (CFT1) and one intact at the allosteric domain (CFT2). A muramic acid saccharide (capped sticks in magenta) is found at the center of the allosteric domain. The arrow indicates the point of attachment of the membrane anchor. (C) The solvent-accessible surface representation for PBP2a is shown. The distance between the two ceftaroline molecules is 60 Å. (D) Ribbon representation of PBP2a in complex with 1 (black sticks). This view is rotated ∼45° on the y axis compared with the view of C.
Results
The crystal structures of a soluble construct of PBP2a (lacking the N-terminal membrane anchor) in three different complexes were solved (Fig. 1 and Table S1). These complexes include the synthetic peptidoglycan 1 (Complex 1) (Fig. 1A) and PBP2a–ceftaroline complexes obtained by either soaking (Complex 2) or cocrystallization (Complex 3). The subsequently assigned allosteric site is located in the formerly termed nonpenicillin binding (7) domain—hereafter referred to as the allosteric domain—at the intersection of Lobe 1 (residues 166–240), Lobe 2 (residues 258–277), Lobe 3 (residues 364–390), and the top of the N-terminal extension domain (Fig. 1 B and C). The distance between the allosteric site and the transpeptidase active site is 60 Å (Fig. 1C). All three complexes show two (labeled as chains A and B) PBP2a protein molecules in the asymmetric unit. Unless noted, our discussion concentrates on the structure of molecule A.
Both soaking and cocrystallization of PBP2a with ceftaroline gave acylation of the active site serine by ceftaroline (soaking, only A; cocrystallization, both A and B) (Table 1). Moreover, noncovalently bound ceftaroline was seen in both crystals (Fig. 1 and Table 1). Additional electron density seen in a cleft of the allosteric domain was modeled as a muramic acid saccharide (Fig. 1B and Fig. S1). Because inclusion of a muramic saccharide was not used in our crystallization experiments, this molecule must have been carried through the protein purification. Importantly, soaking of the PBP2a crystals with the synthetic peptidoglycan fragment 1 displaced the muramic saccharide by the NAM pentapeptide segment of structure 1 (Complex 1) (Fig. 1D and Fig. S1).
Table 1.
Ligands bound to the different PBP2a complexes
| Complex 1 |
Complex 2 |
Complex 3 |
||||
| Chain | A | B | A | B | A | B |
| Active site | — | — | CFT1 | — | CFT1 | CFT1 |
| Allosteric site | 1 | 1 | MUR, CFT2 | MUR* | MUR, CFT2 | MUR, CFT2 |
Residual electron density was observed for CFT2 but not included because of its poor quality.
Ceftaroline Recognition at PBP2a Transpeptidase Active Site.
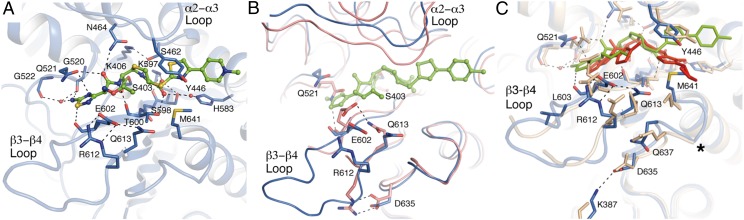
Acylation of the catalytic Ser403 by ceftaroline is observed in both Complexes 2 and 3 (Fig. 2A, Table 1, and Fig. S1). A comparison of the ceftaroline acyl-enzyme of Complex 2 with that of the structure of Complex 1 revealed a conformational change spanning the distance between the allosteric and active sites. Motion within the active site occurs for loop α2–α3 (∼2.5 Å movement up from the Cα atom of Tyr446), loop β3–β4 [the loop was not seen in the first PBP2a structure (7) because of its mobility and now protrudes ∼10 Å distance between Cα of Arg612 and Cα of Gln607], and the loop β5–α10. These motions create room for antibiotic/ligand binding (Fig. 2B). In particular, the active site conformational changes seen at strand β3 and the N terminus of helix α2 coincide with serine acylation. In the absence of ceftaroline, S403 is distant and thus, incapable of acting as a nucleophile (7). Furthermore, serine acylation by ceftaroline twists strand β3 (Fig. 2B), which was seen previously with PBP2a acyl-enzyme structures derived from other β-lactams (7) (Fig. S2) and other PBP acyl-enzyme structures (11, 17). The ceftaroline R1 segment (Figs. 1A and 2A) provokes a dramatic conformational change involving the interaction among Q521, E602, and R612 (Fig. 2B). As a consequence, the R612···D635 salt bridge is disrupted, with the R612 side chain moving to engage E602 to form a new salt bridge (Fig. 2B). This salt bridge swap is one component of an extended conformational change that intimately links occupancy of the allosteric site to opening of the active site.
Fig. 2.
Interaction of PBP2a with ceftaroline at the active site. (A) View of ceftaroline within the active site is shown in capped sticks, with carbons in green, oxygens in red, nitrogens in blue, and sulfur in yellow. (B) Structural contrast between Complex 1 (pink tubes) and the PBP2a ceftaroline acyl-enzyme (blue tubes). Relevant amino acids are represented in capped sticks, and the two important active site loops are labeled. (C) Structural comparison of the ceftaroline interaction at the PBP2a active site compared with ceftobiprole. Superposition of the active site of ceftaroline-acyl-PBP2a (CFT-PBP2a) with ceftobiprole-acyl-PBP2a (PDB ID code 4DKI). Ceftaroline is drawn as green sticks, and ceftobiprole is drawn as red sticks. Side chains of residues are shown in capped sticks for the CFT-PBP2a (blue) and the ceftobiprole-acyl-PBP2a (brown). Polar interactions in ceftaroline-acyl-PBP2a are shown as dashed lines. *The disordered region found in the ceftobiprole-acyl-PBP2a around M641. The crystal structure of PBP2a in complex with ceftobiprole (PDB ID code 4DKI) also shows residual electron density at the same position in the allosteric domain as occupied by ceftaroline, and it also exhibits some of the modifications observed in the ceftaroline complex.
A crystal structure of the PBP2a acyl-enzyme complex of ceftobiprole, also a broad spectrum cephalosporin, was published recently (18). Although both the ceftaroline and ceftobiprole PBP2a acyl-enzyme structures give similar structural changes at strand β3 and the N terminus of helix α2, additional changes were seen as a consequence of the recognition of the R1 and R2 segments in the ceftaroline-derived acyl-enzyme (Fig. 2C). The R1 interaction disrupted the critical Q521···E602 hydrogen bond interaction, with motion of E602 engaging R612 in a salt bridge interaction. This salt bridge is not observed in the ceftobiprole complex as a result of a different conformation for L603 (Fig. 2C). The orientations of the R2 groups in the respective ceftaroline– and ceftobiprole–PBP2a acyl-enzymes are very different. In the ceftaroline complex, Y446 on the α2–α3 loop (which seems to serve as a gatekeeper to the active site) interacts with M641 to close the active site and hold ceftaroline within a narrow cleft. In contrast, in the ceftobiprole complex, Y446 and M641 sandwich the R2 substituent. The ceftobiprole-derived acyl-enzyme presents disorder in the M641 region (18), and no model was built between M641and D638 (Fig. 2C). This disorder also affects other surrounding residues (Q613, Q637, and D635) and results in a D635 conformation that excludes the salt bridge interaction with K387 (from Lobe 3) that is observed in the ceftaroline complex. The β3–β4 loops of the two acyl-enzymes are quite different.
Allosteric Peptidoglycan Binding Site in PBP2a.
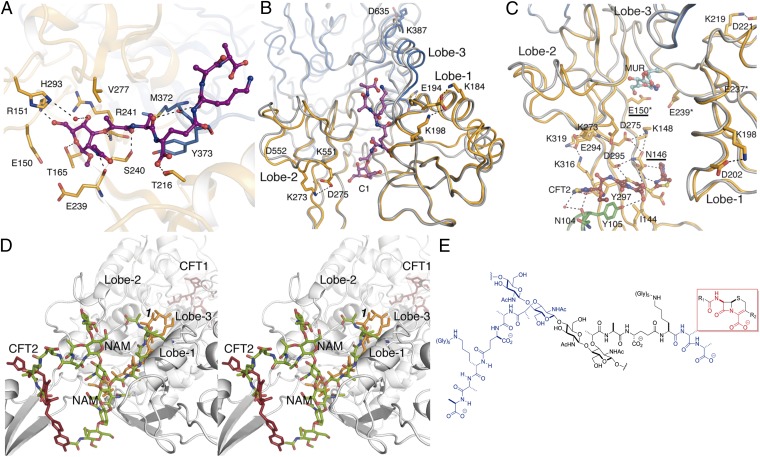
The locations of the peptidoglycan analog and the intact form of ceftaroline (Fig. 3 A–C) identify the allosteric domain, the existence of which was inferred from kinetic studies (16). The evidence supporting the identity of this site as allosteric includes the chemical nature of its ligands (peptidoglycan and ceftaroline), the saturable binding behavior displayed by both ligands (16, 19), its distance from the active site, and lastly, the substantial conformational change resulting in the opening of the active site. Our discussion of this conformational change begins with the structure of peptidoglycan mimetic 1 bound in a 25-Å-long groove formed by Lobes 1–3 (Complex 1) (Figs. 1D and 3A). The NAM pentapeptide moiety of 1 is stabilized by both polar and hydrophobic interactions mediated by residues contributed by all three lobes (Fig. 3A). A comparison (Fig. 3B) of this structure with the structure of apo PBP2a [Protein Data Bank (PDB) ID code 1VQQ] shows that the positioning of these lobes changes as a result of complex formation with 1: the distance between Lobes 1 and 2 is around 3 Å shorter in Complex 1 than the apo enzyme. Complex 1 shows eight unique salt bridge interactions (not present in apo PBP2a) inside Lobes 1 and 2 (Table S2), which interconnect these lobes with the transpeptidase domain. Despite the absence of substrate in the active site of Complex 1, the side chain conformation of Q521—a critical residue in ceftaroline stabilization (see above)—changed to allow different hydrogen bond contacts (Q521 interacts with G520 and S400 in apo-PBP2a but K604 and E602 in Complex 1).
Fig. 3.
The structure of the allosteric domain and its interactions with ligands. (A) Interactions of the peptidoglycan 1 (capped sticks in magenta) bound at the allosteric site. (B) View of compound 1 (in magenta) bound at the allosteric site. We superimpose the apo PBP2a structure (PDB ID code 1VQQ) in gray onto this structure. The unique salt bridge interactions formed on 1 binding are represented as dotted lines. (C) View of ceftaroline (sticks with carbon atoms in dark red) bound noncovalently at the allosteric site. This complex also shows the muramic acid (MUR) saccharide (in cyan) at 1 o’clock. We superimpose the apo PBP2a structure in gray onto this structure. The unique salt bridge interactions formed on ceftaroline binding are represented as dotted lines. Mutations in clinical isolates for ceftaroline-resistant (N146 and E150) and ceftobiprole- or l-695,256–resistant (E150*, E239*, and E237*) are labeled. (D) Stereoview of the allosteric site. The structures of 1 (orange) and ceftaroline (red) are from our crystal structures. The computational model of the extended peptidoglycan is shown in green. (E) Composite model shown in D is based on the crystal coordinates for compound 1 (at 9 o’clock; blue) and the backbone atoms of ceftaroline (the boxed structure in red) for the d-Ala-d-Ala moiety at 3 o’clock (blue) per the hypothesis by Tipper and Strominger (21). The intervening atoms depicted in black correspond to the second NAG-NAM unit.
Complexes 2 and 3 showed a noncovalently bound ceftaroline in the allosteric site at the interface between the N-terminal extension and Lobe 2 (Fig. 3C and SI Materials and Methods) and ∼17 Å distant from the site occupied by 1 (measured from the β-lactam ring of the ceftaroline to MurNAc ring of 1). The β-lactam moiety of ceftaroline contacts the side chains of Y297 and Y105, the R2 group of ceftaroline is ensconced in a pocket formed by I144 and Y105, and the R1 group makes polar interactions with N104 and K76 (Fig. 3C). The extensive salt bridge interactions observed previously in Complex 1 are mainly maintained in Complexes 2 and 3, whereas unique ones appear to interconnect these lobes to the transpeptidase domain (Table S2). Unique salt bridge interactions around the ceftaroline molecule in the allosteric site are shown in Fig. 3C.
Peptidoglycan strands adopt a right-handed helical conformation in solution (20). Computational placement into the PBP2a allosteric site of a hexasaccharide peptidoglycan strand having this conformation by superimposition on the experimental density for bound peptidoglycan 1 in Complex 1 (Fig. 3D) threads this longer peptidoglycan through the allosteric domain. Two well-defined and consecutive binding sites for its full-length pentapeptide stems are identified, just as would be expected to exist in nascent peptidoglycan. Tipper and Strominger (21) conceptualized the β-lactam antibiotics as mimics of the acyl-d-Ala-d-Ala terminus of the peptidoglycan to explain the ability of the β-lactam to inhibit the dd-transpeptidase activity by irreversible serine acylation (8). Because a ceftaroline molecule is found noncovalently bound to the allosteric site, we inquired whether its β-lactam substructure coincided with the location of the d-Ala-d-Ala terminus of the pentapeptide stem. The two structures superimpose (Fig. 3 D and E). The allosteric domain recognizes at once the peptide stems of nascent peptidoglycan and the β-lactam backbone of a cephalosporin with anti-MRSA activity as a mimetic of the d-Ala-d-Ala terminus. These observations are consistent (indeed, prerequisite) with requirements of binding of nascent peptidoglycan with the full-length stem peptide at the allosteric site as a trigger for the opening of the active site of the transpeptidase domain, which would, in turn, take as its exclusive substrate the nascent peptidoglycan with the full-length peptide stem. We propose that the function of the allosteric domain is to open the active site for catalysis of transpeptidation only when nascent peptidoglycan is present.
Ligand Binding and Allosteric Conformational Change.
Allosteric enzymes contain at least two distinct binding sites, an active site and an allosteric site, the latter of which influences the activity of the former. Binding of effector molecules (ligands) to the allosteric site propagates a conformational change to enable catalysis. Allostery requires flexibility in the protein. This characteristic gives rise to populations of conformers that interconvert on various timescales, exhibiting amino acid networks that communicate between distant sites (22). Our PBP2a structures provide a structural context for allosteric regulation of its activity. Our evidence rests on the self-consistency of kinetic study (16, 19) with the structural consequences of the occupancy of two separate ligands (peptidoglycan 1 and ceftaroline) to a domain remote from the active site. The smaller conformational change occurred for 1 (Complex 1) (Figs. S3 and S4), and the larger conformational change occurred for ceftaroline (Complexes 2 and 3) (Figs. S3 and S4). The interaction of 1 at the allosteric site keeps the active site closed but initiates conformational change in Lobes 1–3 by increasing the number of salt bridge interactions among them and to the transpeptidase domain. The segment of the β3 strand that is proximal to the active site (and flush against Lobe 3 of the catalytic domain) (Fig. 1) twists on ceftaroline occupancy of the allosteric site. As β3 moves, its β-sheet interacts with Lobe 3 and the mobile β3–β4 loop. The β3–β4 loop was not seen in the first structure of PBP2a (7) and is also absent in our Complex 1. This loop is seen in the allosteric complexes of PBP2a with ceftaroline. The mobility of the β3–β4 loop correlates to the catalytic reactivity of the PBP enzymes (7). Stabilization of the loop in the open conformations lengthens the active site by more than 10 Å (to a total length of 23 Å). Hence, the allosterically induced conformational change doubles the surface area of the active site.
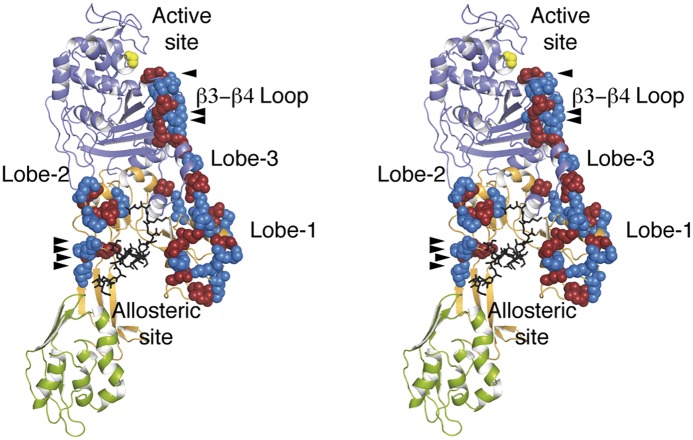
Binding of either peptidoglycan 1 or ceftaroline to the allosteric domain produces unique salt bridge interactions spanning the entire distance from the allosteric site to the active site. Molecular dynamics simulations indicated that binding of larger peptidoglycan chains at allosteric sites increases displacements of lobes of the allosteric domain (Fig. S3) and the number of salt bridge interactions both inside each lobe and also linking lobes among them and with the transpeptidase domain (Table S2). The conformational state resulting from salt bridge swapping across the 60-Å edge connecting the allosteric and active sites is shown in Fig. 4. The salt bridge transitions propagate in a manner akin to falling dominos.
Fig. 4.
Stereoview of the allosteric signal propagation in PBP2a. Binding of the peptidoglycan (black) at the allosteric site propagates a unique network of salt bridge interactions extending between the allosteric and catalytic domains. The seven salt bridge interactions seen by crystallography are identified with arrowheads. An additional 17 salt bridge interactions were predicted by molecular dynamics (Table S2). The catalytic serine (yellow) and the acidic (red) and basic (blue) residues of the salt bridge interactions are shown as spheres. Peptidoglycan binding at the allosteric site stimulates this domino effect commencing from the allosteric site (Lobes 1 and 2) to Lobe 3 onto the β3–β4 loop. The changes in the β3–β4 loop of the active site as a result of formation of the unique salt bridge network are detailed in Fig. 2.
Two conformational states for the residues interconnecting the allosteric and active sites are implicated by our X-ray structures and our molecular dynamics simulations. Unfortunately, kinetic assays to characterize allosteric opening and closing of the active site do not exist. To provide additional evidence for our falling dominos proposal, we set out to disrupt the process by mutating residues along the path of its propagation. Mutational change within the path is anticipated to disrupt the allosteric communication. The consequence of each mutation was measured by examination of the rate constant for acylation of the active site serine by nitrocefin, a chromogenic cephalosporin.
We mutated the gene, expressed the proteins, and purified to homogeneity 12 mutant variants of PBP2a (Table S3). These variants incorporated single, double, and triple mutations dispersed in all three regions along the path of conformational change: the network connecting Lobe 3 with the β3–β4 loop of active site, the network between Lobes 2 and 3, and the network between Lobe 2 and allosteric site. The ability of ceftaroline to trigger the allosteric opening of the active site was assessed for each variant. Two of these variants (K387A-D635A and D343A-E389A-D635A) showed a lack of active site acylation, notwithstanding the fact that their overall protein fold was the same as the WT, which was judged by their far UV circular dichroism spectra. However, four (E389A-K634A, K188A-D367A, K148A-D295A, and E267A-E268A) showed rate constants similar to those of the rate constants of the WT enzyme (k2/Ks of 2,490 ± 1,490 M−1s−1). All remaining variants (Table S3) exhibited diminished ability to undergo active site acylation by ceftaroline, indicative of impaired allosteric triggering (k2/Ks in the range of 600–1,560 M−1s−1). We also explored compound 1 as the allosteric trigger with WT and four of the mutant variants (K387A-D635A, D343A-E389A-D635A, K188A-K219A, and E294A) (Table S4). The WT protein experienced enhancement of the active site acylation in the presence of ligand 1. The K387A-D635A double mutant failed to enable access to the active site, and acylation of the D343A-E389A-D635A triple mutant was severely impaired in the presence of 1. In the other two cases, the presence of 1 did not affect the intrinsic rate of active site acylation. Hence, the ligand could not enhance conformational change, indicative of a lack of allosteric trigger.
These results suggest a larger effect for mutations closer to the active site. Several routes for the propagation of the allosteric trigger might exist (Fig. 4), with some redundancy to accommodate the convergence of the orchestrated motion to lead to the opening of the active site. The area near the active site might be the bottleneck for the process, which is reflected by the fact that mutants K387A-D635A and D343A-E389A-D635A are unable to experience acylation at the active site.
Discussion
Antibiotic resistance has rendered many classes of antibiotics clinically obsolete. MRSA strains exhibiting broad resistance to β-lactam antibiotics were identified in the early 1960s and persist to this day as the cause of serious infection. Ceftaroline (23) exemplifies the exhaustive efforts to the discovery of new β-lactam antibiotics efficacious against MRSA. We suggest that the β-lactams that have emerged from empirical optimization owe their success, in part, to an ability to bind at the allosteric domain of PBP2a to predispose its transpeptidase active site to facilitate β-lactam acylation (19). Using peptidoglycan 1 as a substrate mimetic and ceftaroline as an MRSA-effective β-lactam, we see structural transitions within PBP2a that are fully consistent with this hypothesis. These transitions call attention to the central domain of this unusual monofunctional PBP as a regulatory component. Because ceftaroline was introduced to the clinic in 2010, one might expect already the emergence of a resistant mutant. This event has happened. Two ceftaroline-resistant MRSA clinical isolates show point mutations remote from the active site (24): one isolate has two PBP2a point mutations (N146K and E150K), whereas the second strain retains these two mutations and adds a third mutation (H351N). Whereas the ability of distal mutations to impart resistance phenotypes is well-recognized, our studies provide a structural interpretation for these mutations, because they directly alter the allosteric site (Fig. 3C and Fig. S5). Interestingly, in vitro selection of MRSA strains resistant to ceftobiprole (25) or l-695,256 (an investigational β-lactam) (26) reveals additional amino acid mutations (such as E150K, E237K, or E239K) that also locate to the core of the PBP2a allosteric site (Fig. 3C). More importantly, we have been able to disrupt this communication between the two sites by additional mutations, which support the invocation of the allosteric effect for the function of PBP2a.
We argue that ceftaroline binds noncovalently to the allosteric domain in the identical site otherwise occupied by the acyl-d-Ala-d-Ala terminus of the full-length peptide stem of the nascent peptidoglycan. The complex of Streptococcus pneumoniae PBP2x with cefuroxime is another possible example for such mimicry (27). One molecule of cefuroxime binds to the active site of PBP2x, whereas another binds noncovalently to a so-called PBP and ser/thr kinase-associated (PASTA) domain, distal to the active site. Among the many resistance mutations that occur in PBP2x, some occur not at the active site but at or near the PASTA domain, affecting binding of the β-lactam to the PASTA site (27, 28). Although neither the function of the PASTA domain nor the structural basis for its recognition of β-lactam antibiotics is fully certain, the structural mobility of the PASTA domain (29) may likewise be governed by the presence of nascent peptidoglycan or β-lactam antibiotics (28, 30).
None of the solved X-ray structures for other PBPs of S. aureus (PBP2, PBP3, and PBP4) (31, 32) or the close homolog of S. aureus PBP1, the PBP2x of S. pneumoniae, have even a remnant of the allosteric domain (Fig. S6), and hence, these PBPs cannot exhibit the allostery that we find for PBP2a. However, PBP5fm from Enterococcus faecium (33) may be a close kin of PBP2a. Its structure shows equivalents for Lobes 2 and 3 and the presence of a Lobe 1 (not seen; as a result of structural disorder). PBP5fm may be a second example of a PBP subject to the allosteric control, which we have invoked here for PBP2a. How PBP2a and likely also PBP5fm acquired allostery is not known. In light of the fact that the mecA gene for PBP2a was acquired from a non-S. aureus source, this allostery might have been a random event that was selected as beneficial to the organism. This event has taken place multiple times (34).
The implications of this proposed allosteric regulation must eventually embrace the intimate cooperation between PBP2a and the biosynthetic transglycosylases (4). Allosteric communication may be mediated by both changes in local conformation and global changes in the protein dynamics. Both factors may operate in PBP2a. Although X-ray diffraction cannot give a precise description on protein dynamics, crystallographic B factors account for thermal motion of the atoms. In this sense, the high B factors observed in the active site loops and the external regions of lobes of all of the PBP2a structures up to now reported (7, 18) and the equivalent regions of PBP5fm from E. faecium (33) are noteworthy.
The persistence of MRSA as a pathogen, the continuing proliferation of its antibiotic-resistance mechanisms, and the extraordinary difficulty of empirical structural optimization collectively demand new strategies for antibiotic discovery, which are exemplified by the discoveries of non–β-lactams targeting PBP2a (35, 36) and unique targets synergistically lethal with existing β-lactams (37–45). Binding of an allosteric effector can influence protein function, and thus, allosteric binding sites can be targets for new drugs. Our PBP2a structural studies conceptually unify these two strategies: by allowing the identification of unique structures—β-lactam or otherwise—with binding to the allosteric site that predisposes PBP2a to inactivation and by the ability to use design rather than empiricism for their optimization.
Materials and Methods
Protein Crystallization.
Crystals were obtained at 4 °C in a precipitant solution consisting of 20% (vol/vol) PEG 550 monomethyl ether, 880 mM NaCl, 100 mM Hepes (pH 7.0 buffer), and 16 mM CdCl2. Details are in SI Materials and Methods.
Data Collection, Phasing, and Model Refinement.
Diffraction datasets were collected at synchrotron radiation facilities. Structures were solved by molecular replacement and then refined (statistics shown in Table S5). The allosteric domain presents high B factors, which were also observed in previous PBP2a structures. These B factors are similar to B factors observed for the ligands at the allosteric site. Details are in SI Materials and Methods.
Computational Methods.
A hexasaccharide peptidoglycan strand, solved previously by NMR, was docked on Complex 1. The first NAM pentapeptide stem was built into the hexasaccharide based on the structure of the compound 1 ligand. The resulting complex was solvated and subjected to a molecular dynamics simulation, and energy was minimized. Details are in SI Materials and Methods.
Cloning, Expression Purification of PBP2a Mutants, and Determination of Kinetic Parameters.
Twelve mutant variants of PBP2a (single, double, and triple mutations) along the path of the conformational change between the allosteric and active sites were produced. The consequence of each mutation was measured by examination of the rate constant for acylation of the active site serine by nitrocefin. Details are in SI Materials and Methods.
Supplementary Material
Acknowledgments
Ceftaroline fosamil was a gift from Robert Bonomo. We thank Pavel Afonine for help with feature-enhanced maps. L.I.L. is a Pew Latin American Fellow in the Biomedical Sciences and supported by The Pew Charitable Trust. Work in the United States was supported by National Institutes of Health Grants AI090818 and AI104987, and work in Spain was supported by Grants BFU2011-25326 (from the Spanish Ministry of Economy and Competitiveness) and S2010/BMD-2457 (from the Autonomous Government of Madrid).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 3ZG5 (Complex 1), 3ZFZ (Complex 2), and 3ZG0 (Complex 3)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300118110/-/DCSupplemental.
References
- 1.Llarrull LI, Fisher JF, Mobashery S. Molecular basis and phenotype of methicillin resistance in Staphylococcus aureus and insights into new β-lactams that meet the challenge. Antimicrob Agents Chemother. 2009;53(10):4051–4063. doi: 10.1128/AAC.00084-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oliveira DC, de Lencastre H. Methicillin-resistance in Staphylococcus aureus is not affected by the overexpression in trans of the mecA gene repressor: A surprising observation. PLoS One. 2011;6(8):e23287. doi: 10.1371/journal.pone.0023287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pinho MG, Filipe SR, de Lencastre H, Tomasz A. Complementation of the essential peptidoglycan transpeptidase function of penicillin-binding protein 2 (PBP2) by the drug resistance protein PBP2A in Staphylococcus aureus. J Bacteriol. 2001;183(22):6525–6531. doi: 10.1128/JB.183.22.6525-6531.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pinho MG, de Lencastre H, Tomasz A. An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc Natl Acad Sci USA. 2001;98(19):10886–10891. doi: 10.1073/pnas.191260798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim C, et al. Properties of a novel PBP2A protein homolog from Staphylococcus aureus strain LGA251 and its contribution to the β-lactam-resistant phenotype. J Biol Chem. 2012;287(44):36854–36863. doi: 10.1074/jbc.M112.395962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao Z, Kahne D, Kishony R. Distinct single-cell morphological dynamics under β-lactam antibiotics. Mol Cell. 2012;48(5):705–712. doi: 10.1016/j.molcel.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim D, Strynadka NC. Structural basis for the β lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus. Nat Struct Biol. 2002;9(11):870–876. doi: 10.1038/nsb858. [DOI] [PubMed] [Google Scholar]
- 8.Lee W, et al. A 1.2-Å snapshot of the final step of bacterial cell wall biosynthesis. Proc Natl Acad Sci USA. 2001;98(4):1427–1431. doi: 10.1073/pnas.98.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi Q, Meroueh SO, Fisher JF, Mobashery S. A computational evaluation of the mechanism of penicillin-binding protein-catalyzed cross-linking of the bacterial cell wall. J Am Chem Soc. 2011;133(14):5274–5283. doi: 10.1021/ja1074739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dessen A, Mouz N, Gordon E, Hopkins J, Dideberg O. Crystal structure of PBP2x from a highly penicillin-resistant Streptococcus pneumoniae clinical isolate: A mosaic framework containing 83 mutations. J Biol Chem. 2001;276(48):45106–45112. doi: 10.1074/jbc.M107608200. [DOI] [PubMed] [Google Scholar]
- 11.Pernot L, et al. A PBP2x from a clinical isolate of Streptococcus pneumoniae exhibits an alternative mechanism for reduction of susceptibility to β-lactam antibiotics. J Biol Chem. 2004;279(16):16463–16470. doi: 10.1074/jbc.M313492200. [DOI] [PubMed] [Google Scholar]
- 12.Macheboeuf P, et al. Active site restructuring regulates ligand recognition in class A penicillin-binding proteins. Proc Natl Acad Sci USA. 2005;102(3):577–582. doi: 10.1073/pnas.0407186102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lovering AL, De Castro L, Lim D, Strynadka NC. Structural analysis of an “open” form of PBP1B from Streptococcus pneumoniae. Protein Sci. 2006;15(7):1701–1709. doi: 10.1110/ps.062112106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fedarovich A, Nicholas RA, Davies C. Unusual conformation of the SxN motif in the crystal structure of penicillin-binding protein A from Mycobacterium tuberculosis. J Mol Biol. 2010;398(1):54–65. doi: 10.1016/j.jmb.2010.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Changeux JP. Allostery and the Monod-Wyman-Changeux model after 50 years. Annu Rev Biophys. 2012;41:103–133. doi: 10.1146/annurev-biophys-050511-102222. [DOI] [PubMed] [Google Scholar]
- 16.Fuda C, et al. Activation for catalysis of penicillin-binding protein 2a from methicillin-resistant Staphylococcus aureus by bacterial cell wall. J Am Chem Soc. 2005;127(7):2056–2057. doi: 10.1021/ja0434376. [DOI] [PubMed] [Google Scholar]
- 17.Macheboeuf P, Contreras-Martel C, Job V, Dideberg O, Dessen A. Penicillin binding proteins: Key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol Rev. 2006;30(5):673–691. doi: 10.1111/j.1574-6976.2006.00024.x. [DOI] [PubMed] [Google Scholar]
- 18.Lovering AL, et al. Structural insights into the anti-methicillin-resistant Staphylococcus aureus (MRSA) activity of ceftobiprole. J Biol Chem. 2012;287(38):32096–32102. doi: 10.1074/jbc.M112.355644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Villegas-Estrada A, Lee M, Hesek D, Vakulenko SB, Mobashery S. Co-opting the cell wall in fighting methicillin-resistant Staphylococcus aureus: Potent inhibition of PBP 2a by two anti-MRSA β-lactam antibiotics. J Am Chem Soc. 2008;130(29):9212–9213. doi: 10.1021/ja8029448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meroueh SO, et al. Three-dimensional structure of the bacterial cell wall peptidoglycan. Proc Natl Acad Sci USA. 2006;103(12):4404–4409. doi: 10.1073/pnas.0510182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tipper DJ, Strominger JL. Mechanism of action of penicillins: A proposal based on their structural similarity to acyl-D-alanyl-D-alanine. Proc Natl Acad Sci USA. 1965;54(4):1133–1141. doi: 10.1073/pnas.54.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodey NM, Benkovic SJ. Allosteric regulation and catalysis emerge via a common route. Nat Chem Biol. 2008;4(8):474–482. doi: 10.1038/nchembio.98. [DOI] [PubMed] [Google Scholar]
- 23.Saravolatz LD, Stein GE, Johnson LB. Ceftaroline: A novel cephalosporin with activity against methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 2011;52(9):1156–1163. doi: 10.1093/cid/cir147. [DOI] [PubMed] [Google Scholar]
- 24.Mendes RE, et al. Characterization of methicillin-resistant Staphylococcus aureus displaying increased MICs of ceftaroline. J Antimicrob Chemother. 2012;67(6):1321–1324. doi: 10.1093/jac/dks069. [DOI] [PubMed] [Google Scholar]
- 25.Banerjee R, Gretes M, Basuino L, Strynadka N, Chambers HF. In vitro selection and characterization of ceftobiprole-resistant methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2008;52(6):2089–2096. doi: 10.1128/AAC.01403-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katayama Y, Zhang HZ, Chambers HF. PBP 2a mutations producing very-high-level resistance to β-lactams. Antimicrob Agents Chemother. 2004;48(2):453–459. doi: 10.1128/AAC.48.2.453-459.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gordon E, Mouz N, Duée E, Dideberg O. The crystal structure of the penicillin-binding protein 2x from Streptococcus pneumoniae and its acyl-enzyme form: Implication in drug resistance. J Mol Biol. 2000;299(2):477–485. doi: 10.1006/jmbi.2000.3740. [DOI] [PubMed] [Google Scholar]
- 28.Maurer P, Todorova K, Sauerbier J, Hakenbeck R. Mutations in Streptococcus pneumoniae penicillin-binding protein 2x: Importance of the C-terminal penicillin-binding protein and serine/threonine kinase-associated domains for β-lactam binding. Microb Drug Resist. 2012;18(3):314–321. doi: 10.1089/mdr.2012.0022. [DOI] [PubMed] [Google Scholar]
- 29.Paracuellos P, et al. The extended conformation of the 2.9-Å crystal structure of the three-PASTA domain of a Ser/Thr kinase from the human pathogen Staphylococcus aureus. J Mol Biol. 2010;404(5):847–858. doi: 10.1016/j.jmb.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 30.Maestro B, et al. Recognition of peptidoglycan and β-lactam antibiotics by the extracellular domain of the Ser/Thr protein kinase StkP from Streptococcus pneumoniae. FEBS Lett. 2011;585(2):357–363. doi: 10.1016/j.febslet.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lovering AL, de Castro LH, Lim D, Strynadka NC. Structural insight into the transglycosylation step of bacterial cell-wall biosynthesis. Science. 2007;315(5817):1402–1405. doi: 10.1126/science.1136611. [DOI] [PubMed] [Google Scholar]
- 32.Yoshida H, et al. Crystal structures of penicillin-binding protein 3 (PBP3) from methicillin-resistant Staphylococcus aureus in the apo and cefotaxime-bound forms. J Mol Biol. 2012;423(3):351–364. doi: 10.1016/j.jmb.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 33.Sauvage E, et al. The 2.4-Å crystal structure of the penicillin-resistant penicillin-binding protein PBP5fm from Enterococcus faecium in complex with benzylpenicillin. Cell Mol Life Sci. 2002;59(7):1223–1232. doi: 10.1007/s00018-002-8500-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Enright MC, et al. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA) Proc Natl Acad Sci USA. 2002;99(11):7687–7692. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Contreras-Martel C, et al. Structure-guided design of cell wall biosynthesis inhibitors that overcome β-lactam resistance in Staphylococcus aureus (MRSA) ACS Chem Biol. 2011;6(9):943–951. doi: 10.1021/cb2001846. [DOI] [PubMed] [Google Scholar]
- 36.Sliwa A, et al. Unprecedented inhibition of resistant penicillin binding proteins by bis-2-oxoazetidinyl macrocyles. Med Chem Commun. 2012;3:344–351. [Google Scholar]
- 37.Huber J, et al. Chemical genetic identification of peptidoglycan inhibitors potentiating carbapenem activity against methicillin-resistant Staphylococcus aureus. Chem Biol. 2009;16(8):837–848. doi: 10.1016/j.chembiol.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 38.Lee SH, et al. Antagonism of chemical genetic interaction networks resensitize MRSA to β-lactam antibiotics. Chem Biol. 2011;18(11):1379–1389. doi: 10.1016/j.chembiol.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 39.Tan CM, et al. Restoring methicillin-resistant Staphylococcus aureus susceptibility to β-lactam antibiotics. Sci Transl Med. 2012;4:126ra35. doi: 10.1126/scitranslmed.3003592. [DOI] [PubMed] [Google Scholar]
- 40.Campbell J, et al. Synthetic lethal compound combinations reveal a fundamental connection between wall teichoic acid and peptidoglycan biosyntheses in Staphylococcus aureus. ACS Chem Biol. 2011;6(1):106–116. doi: 10.1021/cb100269f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Campbell J, et al. An antibiotic that inhibits a late step in wall teichoic acid biosynthesis induces the cell wall stress stimulon in Staphylococcus aureus. Antimicrob Agents Chemother. 2012;56(4):1810–1820. doi: 10.1128/AAC.05938-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown S, et al. Methicillin resistance in Staphylococcus aureus requires glycosylated wall teichoic acids. Proc Natl Acad Sci USA. 2012;109(46):18909–18914. doi: 10.1073/pnas.1209126109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Farha MA, et al. Inhibition of WTA synthesis blocks the cooperative action of PBPs and sensitizes MRSA to β-lactams. ACS Chem Biol. 2013;8(1):226–233. doi: 10.1021/cb300413m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harris TL, Worthington RJ, Melander C. Potent small-molecule suppression of oxacillin resistance in methicillin-resistant Staphylococcus aureus. Angew Chem Int Ed. 2012;51(45):11254–11257. doi: 10.1002/anie.201206911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Su Z, Yeagley AA, Su R, Peng L, Melander C. Structural studies on 4,5-disubstituted 2-aminoimidazole-based biofilm modulators that suppress bacterial resistance to β-lactams. ChemMedChem. 2012;7:2030–2039. doi: 10.1002/cmdc.201200350. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.