Significance
The migration of cells during embryo development or in cancer often occurs collectively as a group. Underlying mechanisms have been studied in the zebrafish lateral line, in which Wnt signaling regulates the spatially restricted expression of two chemokine receptors required for directional migration of the cell cluster along the track of Sdf1a ligand. We show that a Hox transcription factor, hoxb8a, is essential for correct migration and acts downstream of Wnt signals to regulate the spatial expression of both chemokine receptors. These findings uncover a Wnt-Hox–chemokine receptor pathway that may act widely in the control of directional cell migration.
Keywords: directional cell migration, Wnt-Hox-chemokine receptor pathway
Abstract
The posterior lateral line primordium in zebrafish provides an amenable model to study mechanisms of collective cell migration. The directed migration of the cell cluster along the path of Sdf1a chemokine requires two receptors, Cxcr4b and Cxcr7b, which are expressed in the leading and trailing part of the primordium, respectively. The polarized expression of receptors is regulated by Wnt signaling, but downstream players mediating this control remain to be found. Here, we show that the Hox homeobox gene Hoxb8a is a critical component that acts downstream of the Wnt pathway to coordinate the expression of both chemokine receptors. We find that Hoxb8a is expressed in the leading part of the primordium and is required for the correct speed and extent of migration. Hoxb8a expression is dependent upon Wnt activity and needed both for cxcr4b expression and to repress and thus restrict cxcr7b expression to the trailing zone of the primordium. In the absence of Wnt activity, overexpressed Hoxb8a is able to repress cxcr7b but not up-regulate cxcr4b expression. Together with results from expressing dominant activator and repressor constructs, these findings suggest that Hoxb8a is induced by and cooperates with Wnt signaling to up-regulate cxcr4b, and acts through multiple mechanisms to repress cxcr7b expression.
During collective migration, cells remain in contact with each other and move as a coherent group with distinct front and rear polarity (1). The formation of the posterior lateral line in zebrafish is an amenable model for studying the molecular control of collective movement. The posterior lateral line arises from a group of cells, the primordium, that migrates along the anteroposterior axis of the embryo and distributes the future sensory organs (neuromasts) at regular intervals (2). This directional migration requires the Sdf1a/Cxcl12a chemokine, which is present in the horizontal myoseptum of somites along the path, and its receptor Cxcr4b, which is expressed in the primordium (3–5). Migration also requires Cxcr7b, which is expressed in the trailing part of the primordium and deposited cells, and has been proposed to provide directionality by acting as a sink for Sdf1a (6–8).
In the leading part of the primordium, activation of the canonical Wnt pathway regulates multiple aspects of posterior lateral line development, including proliferation, proneuromast formation, and primordium migration (9–13). By regulating the expression of specific Fibroblast Growth Factors (Fgfs) and the Fgf inhibitor Sef1, Wnt signaling induces and restricts Fgf signaling to the trailing zone. In turn, the Fgf pathway triggers epithelial morphogenesis of the neuromasts (14–17) and restricts Wnt activity to the leading part. This double-feedback interaction is essential for the asymmetric expression of chemokine receptors required for directional migration (13, 16). The regulation of chemokine receptor expression by Wnt signaling is in part mediated by Lef1, which acts in parallel with unidentified components (9). This raises the question as to whether a transcription factor(s) up-regulated by Wnt signaling mediates the regulation of chemokine receptor expression.
Hox genes encode transcription factors best known for their conserved role in axial patterning during development (18–20). They contain a conserved homeodomain that mediates binding to DNA (21) and share similar DNA binding preferences in vitro, with their in vivo specificity thought to result from interactions with cofactors, such as three amino acid loop extension (TALE) proteins of the PBX and MEIS families (22, 23). In addition, Hox genes have been implicated in the regulation of cell migration, for example of subpopulations of neurons along the anteroposterior axis of the hindbrain (24–26). In Caenorhabditis elegans, a Hox gene is required for the migration of specific neuroblasts under the control of the Wnt pathway (27). However, functions of Hox genes in cell migration remain largely unexplored, and it is therefore important to identify downstream target genes that mediate the guidance of migrating cells. In this study, we show that Hoxb8a is a critical component in a network required for directional migration of the lateral line primordium, in which it acts downstream of Wnt signaling to control the spatial expression of the chemokine receptors, cxcr4b and cxcr7b.
Results
Hox Genes Are Expressed in the Leading Zone of the Migrating Primordium.
Searches of the ZFIN database (http://zfin.org) identified several hox genes expressed in the posterior lateral line primordium: hoxb8a, hoxc8a, hoxb6a, hoxc6a, hoxc4a, and hoxd4a. We therefore analyzed their expression pattern in detail by whole mount in situ hybridization (ISH) during the formation and migration of the primordium, from 18 to 48 h postfertilization (hpf). Only hoxb8a and hoxb6a mRNAs were detected in the primordium before the onset of migration, which occurs at 22 hpf (28). At early stages, both genes were expressed in the leading two-thirds of the primordium, with an anterior limit of expression located in L1, the first forming proneuromast (2) (Fig. 1A and Fig. S1). During migration, from 24 to 48 hpf, hoxb8a expression was maintained in the leading zone and down-regulated in the trailing zone and deposited cells (Fig. 1 B and C and Fig. S1). The hoxb6a showed a similar expression pattern initially but was not expressed after 35 hpf (Fig. S1). The other hox genes were up-regulated at later stages during migration, with similar expression in the leading zone of the primordium (Fig. S1). These expression patterns prompted us to examine potential roles of hox genes in the migrating primordium.
Fig. 1.
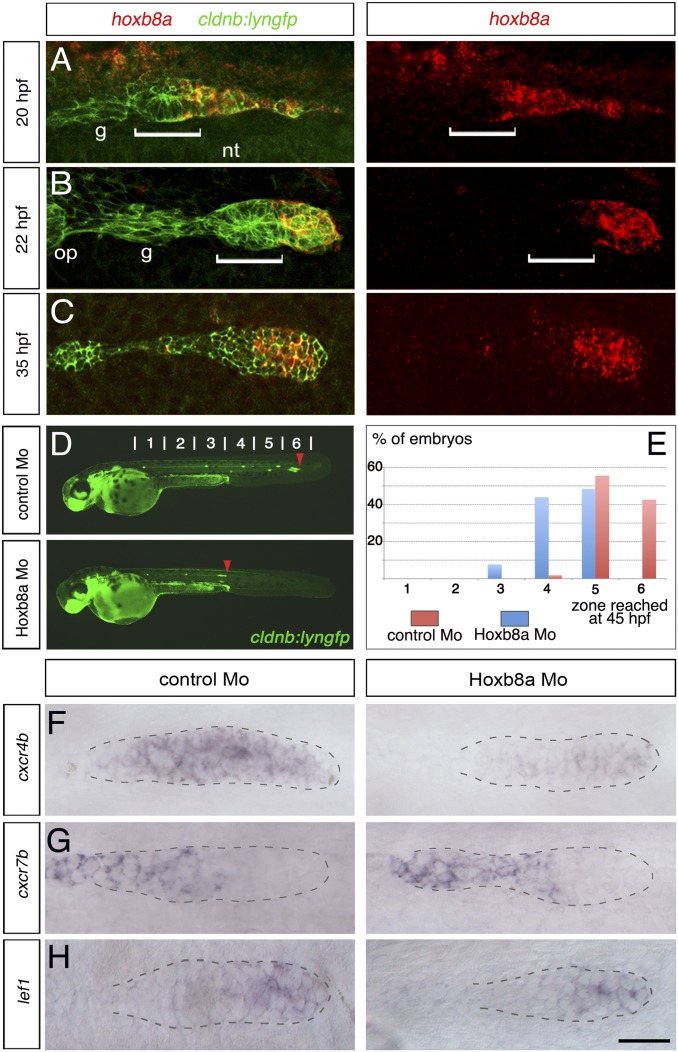
hoxb8a is required for proper migration of the posterior lateral line primordium and expression of the chemokine receptor cxcr4b. Anterior to the left. Detection of hoxb8a mRNA (red) in cldnb:lyngfp embryos (green) at (A) 20, (B) 22, and (C) 35 hpf. Dorsal view in A, lateral views in B and C. White brackets indicate the width of the first forming proneuromast (L1). (D) Fuorescence images of cldnb:lyngfp embryos at 45 hpf injected with control and hoxb8a Mo. Red arrowheads indicate the position of the primordium. (E) Quantification of the migration of the primordium at 45 hpf (control Mo, n = 54; hoxb8a Mo, n = 91). ISH to detect expression of (F) cxcr4b, (G) cxcr7b, and (H) lef1 at 30 hpf. The primordium is outlined with dotted line. g, Posterior lateral line sensory ganglion; op, otic placode; nt, neural tube. (Scale bar: 25 μm.)
Hoxb8a Knockdown Leads to Posterior Lateral Line Defects.
As several hox genes are coexpressed in the migrating primordium, they potentially have redundant functions. To investigate their role, we carried out antisense morpholino (Mo) knockdowns of hoxb8a and hoxb6a, the only hox genes expressed before the onset of migration. Splice-blocking morpholinos were designed and their efficiency ascertained by RT-PCR (Fig. S2). We found similar effects of single knockdown of these genes on primordium migration (Fig. S2), but double knockdown had major effects on embryo morphology, likely reflecting roles in axial patterning. We therefore focused our analysis on hoxb8a as it is expressed throughout the migration process (Fig. 1 A−C and Fig. S1). Following knockdown of hoxb8a, decreased migration of the primordium was observed at 45 hpf (Fig. 1 D and E). Time-lapse imaging from 24 hpf (Movie S1) and quantification of the distance traveled by the primordium at 26 hpf (Fig. S2) revealed a decreased speed during the early phase of migration. Later on, the delayed primordia either continued to migrate at reduced speed to the tip of the tail or stalled in the second half of the trunk region. These changes to migration of the primordium were not due to altered sdf1a expression, as this was unaffected by hoxb8a knockdown (Fig. S3). Similar lateral line phenotypes were observed in embryos injected with an ATG-blocking hoxb8a Mo, and as shown below, the specificity is corroborated by other loss of function approaches. In addition to changes in migration, we observed a decreased cell number in the primordium of Hoxb8a morphant embryos, and reduction of BrdU incorporation at 24 hpf (Fig. S4). Taken together, these results show that Hoxb8a is required for the normal speed and distance of migration, as well as for normal proliferation of primordium cells.
Hoxb8a Is Required for cxcr4b Expression in the Primordium.
To understand why migration is affected in hoxb8a morphants, we analyzed the expression of cxcr4b and cxcr7b that are required for directional migration. We found that expression of cxcr7b was unchanged in hoxb8a morphants (Fig. 1G), but a marked reduction in cxcr4b expression was detected at 30 hpf (60% of morphants, n = 84) compared with control Mo injected embryos (Fig. 1F and Fig. S2). A similar decrease in cxcr4b expression was observed in embryos injected with the ATG-blocking hoxb8a Mo. We conclude that Hoxb8a is required for normal expression of cxcr4b in the primordium. In view of the crucial role of cxcr4b in directional migration, the decrease in its expression could account for the migration delay in hoxb8a morphants.
One potential explanation of these findings is that Hoxb8a knockdown perturbs the Wnt/Fgf network that is required for correct patterning and migration of the primordium. To investigate this, we analyzed the expression of lef1 and pea3, which are readouts of Wnt and Fgf signaling, respectively (13, 16, 17). The expression of both genes appeared unchanged in hoxb8a morphants (Fig. 1H and Fig. S5). Moreover, injection of hoxb8a Mo did not affect the expression of fgf10, fgf3, and the Fgf inhibitor sef1 that are regulated by Wnt signaling (13), or of fgfr1, a target of the Fgf pathway (13) (Fig. S5). These data indicate that the decreased cxcr4b expression and primordium migration upon Hoxb8a knockdown are not due to an alteration in Wnt and Fgf signaling pathways. Consistent with this, the formation and morphology of neuromasts were normal in hoxb8a morphants (Fig. 1D, Fig. S2, and Movie S1).
Overexpression of Hoxb8a Throughout the Primordium Down-Regulates cxcr7b.
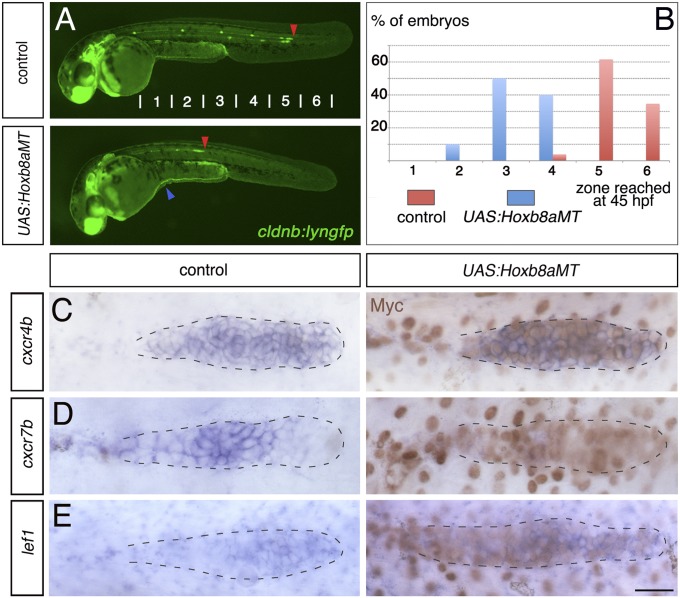
These findings raised the question of why hox gene expression is restricted to the leading region of the primordium and down-regulated in the most mature proneuromast before deposition. To address this, we overexpressed Hoxb8a throughout the primordium and deposited cells. A Myc-tagged full-length Hoxb8a protein (Hoxb8aMT) was used to generate a UAS:Hoxb8aMT line, which was crossed with a cldnb4.2kb:gal4 line in which Gal4 (29) is expressed under control of the cldnb proximal promoter. This Gal4 line drives continuous and mosaic expression of UAS-driven genes in the primordium and deposited cells from 18 to 20 hpf and throughout the migration (Fig. S6). A potential complicating factor is that transgene expression from the cldnb promoter also occurs in surface ectoderm (5), so we analyzed the possibility of a nonautonomous effect on sdf1a expression in the somites; however, we found no such change after expression of normal or mutant Hoxb8a. Embryos overexpressing Hoxb8aMT in the posterior lateral line showed a major delay in migration, with the primordium stalling in the middle of the trunk at around 30 hpf (Fig. 2 A and B). Live imaging revealed that these primordia have uncoordinated collective migration (Movie S2), a behavior previously observed when cxcr7b function is impaired (7, 13). To further investigate the cause of this migration defect, we analyzed cxcr4b and cxcr7b expression. Hoxb8aMT overexpression did not affect cxcr4b expression in the primordium nor activate its expression in the deposited cells, but was sufficient to down-regulate cxcr7b in the back of the primordium and deposited cells (Fig. 2 C and D and Fig. S7). A loss of cxcr7b expression was previously observed upon expansion of Wnt activity throughout the primordium (13). However, lef1 expression was still restricted to the leading zone, as in the control situation (Fig. 2E), indicating that cxcr7b down-regulation is not due to expanded Wnt activity. We conclude that Hoxb8a in the leading zone restricts cxcr7b expression to the trailing part of the primordium, and that down-regulation of hoxb8a in the trailing zone is therefore necessary for normal migration. However, Hoxb8a overexpression is not sufficient to up-regulate cxcr4b in the trailing zone.
Fig. 2.
Overexpression of Hoxb8a leads to down-regulation of cxcr7b. (A) Fluorescent images of cldnb:lyngfp in cldnb4.2:gal4 x UAS:Hoxb8aMT embryos and control siblings at 45 hpf. The blue arrowhead shows the yolk defect seen in all cldnb4.2:gal4 x UAS:Hoxb8aMT embryos. Red arrowheads indicate the position of the primordium. (B) Quantification of the migration at 45 hpf (control, n = 20; cldnb4.2:gal4 x UAS:Hoxb8aMT, n = 26). Expression of (C) cxcr4b, (D) cxcr7b, and (E) lef1 in cldnb4.2:gal4 x UAS:Hoxb8aMT and control siblings at 28 hpf. ISH was followed by Myc immunostaining (brown). (Scale bar: 25 μm.)
A Dominant Repressor Form of Hoxb8a Down-Regulates cxcr4b and cxcr7b Expression.
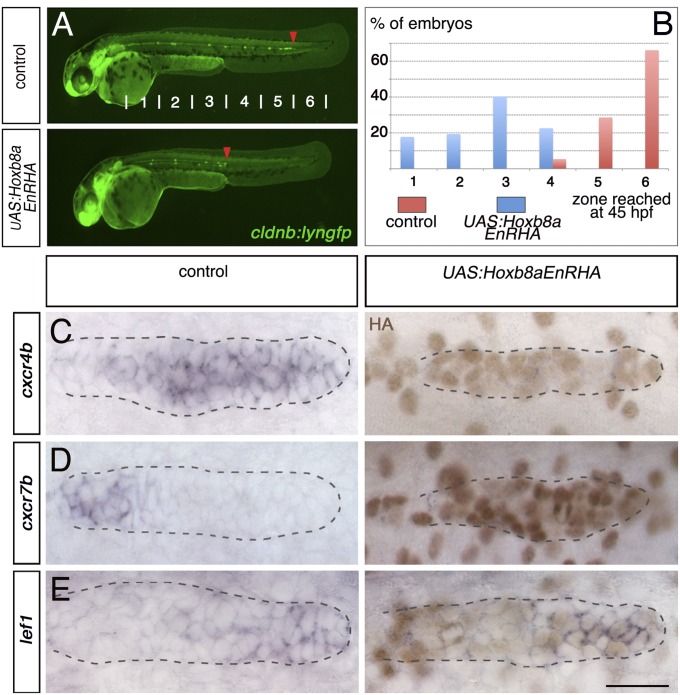
Hox proteins can act as transcriptional activators or repressors, depending on the context and nature of available cofactors (22, 23, 30). To further investigate Hoxb8a function, we generated a stable transgenic line expressing a dominant repressor form of Hoxb8a (fusion with the Engrailed repressor domain) (31) under the control of UAS sequences (UAS:Hoxb8aEnRHA). Overexpression of this repressor form in the primordium by use of the cldnb4.2kb:gal4 line was found to result in strong migration defects (Fig. 3 A and B). In live imaging experiments, the primordia showed major defects in their migration, with stretched morphology, reminiscent of phenotypes observed in cxcr4b mutant and cxcr7b morphant embryos (Movies S3 and S4) (5, 7). Consistent with these observations, the expression of both chemokine receptors was down-regulated by the dominant repressor form of Hoxb8a (Fig. 3 C and D and Fig. S8). In contrast, lef1 expression appeared to be normal (Fig. 3E), and thus the decreased expression of the chemokine receptors is not due to perturbation of Wnt signaling. These results further implicate Hoxb8a in regulation of cxcr4b and cxcr7b.
Fig. 3.
Dominant repressor Hoxb8a down-regulates the expression of cxcr4b and cxcr7b. (A) Fluorescent images of cldnb:lyngfp in cldnb4.2:gal4 x UAS:Hoxb8aEnRHA embryos and control siblings at 45 hpf. Red arrowheads indicate the position of the primordium. (B) Quantification of the migration at 45 hpf (control, n = 56; cldnb4.2:gal4 x UAS:Hoxb8aEnRHA, n = 62). Expression of (C) cxcr4b, (D) cxcr7b, and (E) lef1 in cldnb4.2:gal4 x UAS:Hoxb8aEnRHA and control siblings at 30 hpf. ISH was followed by HA immunostaining (brown). (Scale bar: 25 μm.)
We noticed a reduction in cell number within the primordium upon overexpression of the dominant repressor form of Hoxb8a (Fig. S9). By analyzing embryos in which transgene expression is mosaic, we found that Hoxb8aEnRHA cell-autonomously reduces BrdU incorporation in the primordium at 24 hpf (Fig. S9), further supporting that Hoxb8a is also involved in the control of cell proliferation.
A Dominant Activator Form of Hoxb8a Activates cxcr4b Expression and Inhibits cxcr7b Expression.
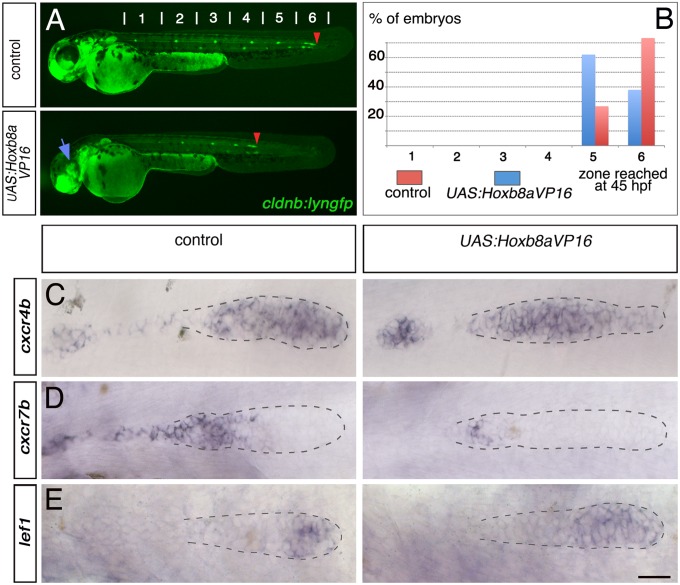
To further investigate how Hoxb8a functions, we generated a stable transgenic line expressing a dominant activator form (fusion with the VP16 activator domain) (31) under the control of UAS sequences (UAS:Hoxb8aVP16). Targeted overexpression of Hoxb8aVP16 mediated by the cldnb4.2:gal4 line resulted in a delay in primordium migration (Fig. 4 A and B and Movie S5). Strikingly, Hoxb8aVP16 overexpression led to an expansion of cxcr4b expression into the trailing zone and deposited cells at 28 hpf (Fig. 4C). Because full-length Hoxb8a was not sufficient to up-regulate ectopic cxcr4b expression (Fig. 2C and Fig. S7), the dominant activator form may override a need for a cofactor that is absent in the trailing zone. In contrast, the level of cxcr7b transcripts was reduced, although not completely abolished (Fig. 4D). The altered expression of chemokine receptors is likely to account for the delayed migration of the primordium (see Discussion). Lef1 expression was normal (Fig. 4E), confirming the uncoupling between cxcr4b/cxcr7b regulation by Hoxb8a and overall Wnt activity.
Fig. 4.
Dominant activator Hoxb8a activates cxcr4b expression and down-regulates cxcr7b. (A) Fluorescent images of cldnb:lyngfp in cldnb4.2:gal4 x UAS:Hoxb8aVP16 embryos and control siblings at 45 hpf. Blue arrow shows the morphological eye defect observed in all Hoxb8aVP16 embryos. Red arrowheads indicate the position of the primordium. (B) Quantification of the migration at 45 hpf (control, n = 50; cldnb4.2:gal4 x UAS:Hoxb8aVP16, n = 56). ISH showing expression of (C) cxcr4b, (D) cxcr7b, and (E) lef1 in cldnb4.2:gal4 x UAS:Hoxb8aVP16 and control siblings at 28 hpf. (Scale bar: 25 μm.)
Hoxb8a Down-Regulates cxcr7b Expression Downstream of Wnt Signaling.
Because ectopic activation of the Wnt pathway results in expansion of cxcr4b expression and loss of cxcr7b in the trailing primordium (13, 16), localized Wnt activity is thought to underlie the polarized expression of cxcr4b and cxcr7b. Our findings suggest that Hoxb8a does not act upstream of Wnt pathway activation, as lef1 expression appears unaffected following loss or gain of Hoxb8a function. To investigate whether Hoxb8a is a target of the Wnt pathway, we inhibited Wnt signaling using the hsp70:dkk1gfp line (32), as verified by the down-regulation of lef1 (Fig. 5 A and B). Overexpression of Dkk1 from 24 hpf led to a complete down-regulation of hoxb8a and hoxb6a expression at 30 hpf (Fig. 5 C−F), indicating that both hox genes are transcriptional targets of Wnt signaling in the leading part of the primordium.
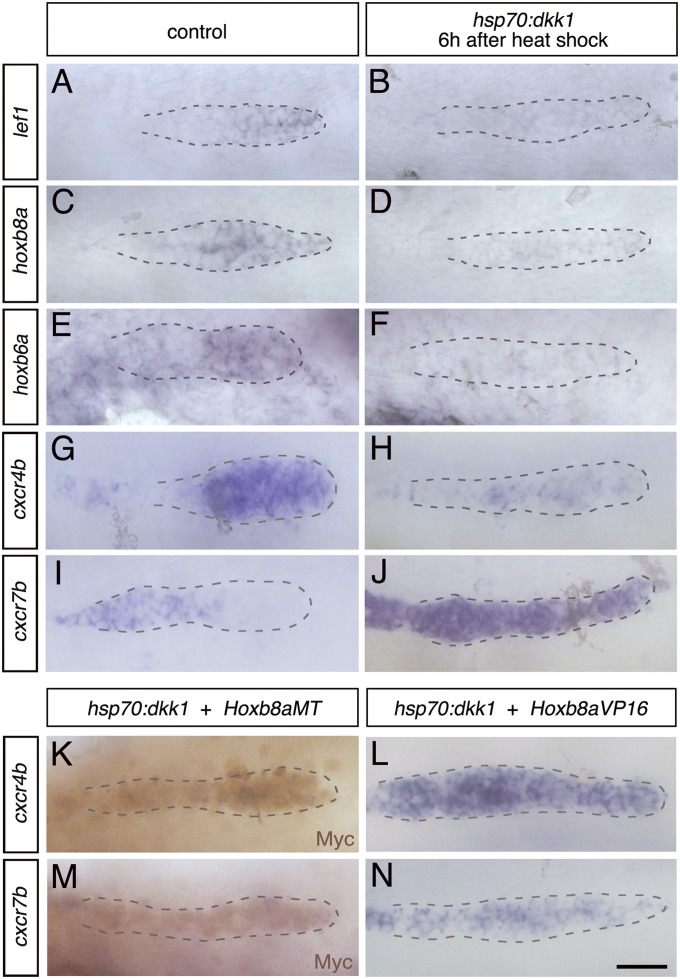
Fig. 5.
Hoxb8a is a target of Wnt signaling and is sufficient to down-regulate cxcr7b upon inhibition of Wnt pathway. ISH showing expression of (A and B) lef1, (C and D) hoxb8a, (E and F) hoxb6a, (G and H) cxcr4b, and (I and J) cxcr7b in hsp70:dkk1gfp embryos and control heat-shocked siblings. Embryos were heat-shocked at 24 hpf and fixed at 30 hpf. ISH to detect expression of (K) cxcr4b and (M) cxcr7b followed by Myc immunostaining (brown) in hsp70:dkk1gfp/cldnb4.2:gal4 x UAS:Hoxb8aMT embryos. ISH to detect expression of (L) cxcr4b and (N) cxcr7b in hsp70:dkk1gfp/cldnb4.2:gal4 x UAS:Hoxb8aVP16 embryos. The number of embryos indicated are pooled from three independent experiments and they are as follows: A, 57; B, 60; C, 62; D, 58; E, 41; F, 63; G, 66; H, 40; I, 67; J, 67; K, 23; L, 55; M, 38; N, 46. (Scale bar: 25 μm.)
In previous studies, inactivation of the Wnt pathway with the hsp70:dkk1gfp line led to an expansion of cxcr7b expression into the leading zone and no apparent change in levels of cxcr4b expression (10, 13), suggesting that cxcr4b expression is not regulated by the Wnt pathway. We found that 3 h after heat shock induction of dkk1, there was a major down-regulation in lef1, hoxb6a, and hoxb8a expression, with a mild decrease in cxcr4b and spreading of cxcr7b expression (Fig. S10), which became robust by 6 h (Fig. 5 G and H). Our results thus reveal that, in addition to confining cxcr7b expression to the trailing zone, Wnt activity is also required for long-term maintenance of normal cxcr4b expression levels in the primordium.
Our findings show that the Wnt pathway regulates Hoxb8a expression, which in turn regulates cxcr4b and cxcr7b. This raises the question of whether Hoxb8a is sufficient to rescue the effect of blocking Wnt activity on chemokine receptor expression. To investigate this, we crossed hsp70:dkk1gfp/cldnb4.2:gal4 double transgenic fish with UAS:Hoxb8aMT or UAS:Hoxb8aVP16 fish. In triple transgenic embryos, Wnt signaling is inhibited upon heat shock, and Hoxb8aMT or Hoxb8aVP16 are present in the primordium due to forced expression. Embryos were heat-shocked at 24 hpf, and Dkk1gfp positive and negative embryos analyzed at 30 hpf. We found that overexpression of Hoxb8aVP16, but not Hoxb8aMT, restores normal levels of cxcr4b expression when Wnt signaling is inhibited (compare Fig. 5 K and L to H). This suggests that Hoxb8a needs a Wnt-dependent cofactor for cxcr4b activation, whose requirement is bypassed in the dominant activator form. Strikingly, in hsp70:dkk1 embryos, cxcr7b expression was abolished by Hoxb8aMT (compare Fig. 5 M and J), and decreased by Hoxb8aVP16 (compare Fig. 5 N and J). Hoxb8a or its dominant activator form are therefore sufficient to down-regulate cxcr7b in the absence of Wnt signaling.
Discussion
Our findings have unraveled a regulatory network between Wnt signaling, a Hox transcription factor, and chemokine receptors, which is required for migration of the posterior lateral line primordium. Wnt signaling activates Hoxb8a expression in the leading zone of the primordium, which in turn maintains normal levels of cxcr4b expression and restricts cxcr7b expression to the trailing zone. Hoxb8a is thus a critical component in the network that provides directionality for collective cell migration.
Hoxb8a Mediates Differential Expression of cxcr4b and cxcr7b.
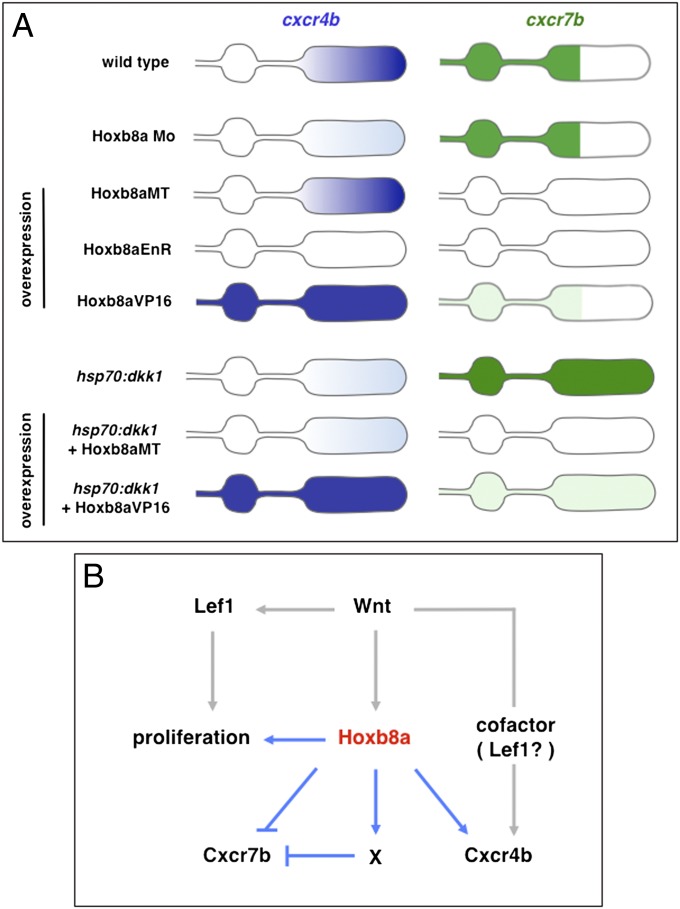
Based on finding that several hox genes are expressed in the developing posterior lateral line in a dynamic and restricted fashion, we investigated the function of hoxb8a with gain and loss of function approaches. Our findings, summarized in Fig. 6A, reveal that Hoxb8a is involved in the differential regulation of the chemokine receptors and in cell proliferation in the primordium. Hoxb8a is required to maintain normal levels of cxcr4b expression in the primordium, and this involves transcriptional activation, because dominant repressor and activator forms of Hoxb8a abolish and ectopically activate cxcr4b expression, respectively. The cxcr4b expression is not completely abolished in Hoxb8a morphants, and this could be due to incomplete knockdown of Hoxb8a (Fig. S2) and/or functional overlap with coexpressed hox genes or Lef1 (9). These factors may also contribute to functional overlap in regulation of cxcr7b expression, which does not expand toward the leading zone in hoxb8a morphants, whereas it is repressed by Hoxb8a overexpression.
Fig. 6.
Summary and working model for the regulation of cxcr4b and cxcr7b by Hoxb8a. (A) Summary of changes in the expression of cxcr4b and cxcr7b following experimental manipulations. (B) Proposed working model. In the leading primordium, Wnt signaling activates the expression of Hoxb8a, which in turn regulates the chemokine receptors cxcr4b and cxcr7b. Hoxb8a is likely to activate cxcr4b with a Wnt-dependent cofactor that could be Lef1. Hoxb8a down-regulates cxcr7b through two possible mechanisms: activation of a repressor X and repression. Hoxb8a also regulates cell proliferation within the primordium and another branch downstream of Wnt signaling controls proliferation via Lef1.
The effects of Hoxb8a dominant activator and repressor overexpression suggest a complex relationship between Hoxb8a and cxcr7b expression. We find that Hoxb8a dominant repressor down-regulates cxcr7b expression, which seems to be a specific effect, as lef1 (Fig. 3E), eya1 (Fig. S8), and cldnb:lyngfp transgene (Fig. 3A) expression are not affected. This finding argues that cxcr7b down-regulation involves Hoxb8a-mediated transcriptional repression. Intriguingly, overexpression of a dominant active form of Hoxb8a also led to down-regulation of cxcr7b expression. The simplest explanation is that Hoxb8a acts through two parallel mechanisms to restrict cxcr7b expression to the trailing zone, one involving Hoxb8a-mediated repression and the other the activation of an unknown repressor (Fig. 6B).
Inhibition of Fgf signaling and/or activation of the Wnt pathway lead to concurrent cxcr7b down-regulation and expansion of cxcr4b expression into the trailing zone, both of which may contribute to the strong migration defects seen in these situations (13, 16). It is informative to compare these results with the effects of manipulating Hoxb8a function. In Hoxb8aMT-overexpressing embryos, cxcr7b expression is abolished but cxcr4b expression is not expanded, and migration of the primordium is strongly affected. Migration defects are weaker in Hoxb8aVP16-overexpressing embryos, in which cxcr7b expression is only partly decreased and cxcr4b expression is expanded. Thus, the severity of the migration delay correlates with the decrease in cxcr7b expression levels rather than with expansion of cxcr4b expression. These findings are consistent with the major role of cxcr7b in directional migration, in which it is proposed to create anisotropy in Cxcr4b activation by acting as a sink for Sdf1a (8).
Hoxb8a Acts Downstream of Wnt Signaling.
Because the polarized distribution of cxcr4b and cxcr7b expression is altered upon modulation of Wnt signaling, Hoxb8a may act downstream of Wnt activity to regulate their expression. Indeed, we found that hoxb8a (and hoxb6a) expression is abolished upon Wnt inhibition, showing that hoxb8a is a direct or indirect transcriptional target of the Wnt pathway in the leading zone (Fig. 6B). Whereas there was a major decrease in lef1 and hoxb8a expression 3 h after Dkk1 induction, cxcr4b expression had only a mild decrease at this stage, but was strongly down-regulated by 6 h. This slower change in cxcr4b expression may reflect perdurance of Hoxb8a protein and/or of cxcr4b mRNA after Dkk1 induction. Although previous studies (10, 13) had not observed decreased cxcr4b expression after blocking Wnt activity, this is likely explained by these studies having analyzed expression at earlier stages after Dkk1 induction.
Hoxb8a overexpression is sufficient to down-regulate cxcr7b in the presence or absence of Wnt signaling. In contrast, in the context of Wnt pathway inhibition, dominant activator but not full-length Hoxb8a is able to rescue normal cxcr4b levels. Together with the observation that dominant activator but not full-length Hoxb8a is sufficient to activate cxcr4b in the trailing zone and deposited cells, these findings suggest a model in which Wnt signaling is required for leading zone expression of both Hoxb8a and a cofactor required for up-regulation of cxcr4b (Fig. 6B). This cofactor could be Lef1, as this component of the Wnt pathway contributes to activation of a minimal cxcr4b promoter (9). Because lef1 knockdown does not affect cxcr4b expression, it acts in parallel with other factors that compensate for its function (9). Taken together, this suggests a feed-forward mechanism in which Wnt signaling acts by inducing and cooperating with Hoxb8a in the regulation of cxcr4b (Fig. 6B). These findings set the stage for investigating the relationship between Hoxb8a and other factors that regulate cxcr4b and cxcr7b expression (9, 33).
Experimental Procedures
Fish Strains and Transgenesis.
Embryos were obtained by natural spawning and raised at 28.5 °C (34). The primordium was visualized using the cldnb:lyngfp line (5), and the hsp70:dkk1gfp line (32) used to inhibit Wnt signaling. The cldnb4.2:gal4 line (nim11) was generated by placing the Gal4 sequence (29) downstream of 4.2 kb of the proximal promoter of the cldnb gene. For overexpression, full-length Hoxb8a was tagged with 6xMyc at the C terminus (Hoxb8aMT). Hoxb8a dominant repressor and activator forms were generated by fusing Engrailed repressor (HA tagged, Hoxb8aEnRHA) or VP16 activator (Hoxb8aVP16) domains (31), respectively, C-terminal to full-length Hoxb8a sequence. Hoxb8aMT, Hoxb8aEnRHA, and Hoxb8aVP16 were placed under the control of UAS regulatory sequences and used to establish stable transgenic lines (nim12, -13, and -14, respectively) using Tol2 mediated integration (35).
In Situ Hybridization and Immunostaining.
Whole mount ISH with digoxigenin-labeled RNA probes was performed as described (36). Detection with fluorescent substrate used the Tyramide TSA-Plus Palette System (Perkin-Elmer). The size of the chemokine receptor expression domains has been shown to change during the deposition cycle of the primordium (12). For cxcr4b and cxcr7b ISH, care was taken to compare primordia with similar deposition states. Immunostaining used the following primary antibodies: anti-GFP (rabbit, Torrey Pines Biolabs), anti-Myc (mouse, clone 9E10, Santa Cruz), anti-HA (rat, Roche), anti-VP16 (mouse, Santa Cruz), and anti-BrdU (mouse, Sigma). For some experiments, Myc or HA immunostainings were performed after ISH; this did not work for VP16 immunostaining. To identify Hoxb8aMT-, Hoxb8aEnRHA-, or Hoxb8aVP16-expressing embryos among their siblings, we used the yolk morphology defect (Fig. 2A), the overall reduction in embryo size and eye morphology defect (Figs. 3A and 4A), or ectopic expression of cxcr4b in the olfactory placode (Fig. S11), respectively.
Morpholino Knockdown and Heat-Shock Inductions.
Antisense Mo were obtained from Gene Tools LLC. Four to ten nanograms of Mo were coinjected with 8 ng p53 Mo (37, 38). With doses exceeding 7 ng, injections of hoxb8a Mo led to some embryos with morphological alterations combined with somitic defects and disruption of Sdf1a expression, likely due to early functions of Hoxb8a. We therefore used a dose of 5 ng, leading to a partial knockdown of the gene (Fig. S2), such that embryo morphology (Fig. 1D) was not affected. Morpholino sequences used are as follows:
Control Mo: 5′ CCTCTTACCTCAGTTACAATTTATA 3′
Hoxb8a ATG Mo: 5′AGCTCATCTTTTACTGCTGTTGGTG 3′
Hoxb8a splice blocker (SB) Mo: 5′ TTTCTGTCTCACCTTGAGGTCGCAT 3′
Hoxb6a SB Mo: 5′ TTACCGAAGGTCCCTGTCCATGAGA 3′
p53 Mo: 5′ GCGCCATTGCTTTGCAAGAATTG 3′
To inhibit Wnt signaling, hsp70:dkk1gfp (32) embryos were heat-shocked at 24 hpf for 30 min at 37 °C and fixed at 30 hpf for analysis.
Confocal and Time Lapse Imaging.
Cldnb:lyngfp embryos were dechorionated manually, immobilized in 0.005% Tricaine (Sigma), and mounted in 3% (wt/vol) Methylcellulose in 0.5X Danieau’s solution. Movies were recorded at 24–25 °C on a Leica DMIRE2 SP2 upright confocal microscope.
BrdU Incorporation.
Embryos were incubated at 28.5 °C in 10 mM BrdU or 10% (vol/vol) DMSO at 24 hfp for 1 h, washed and fixed, and then processed for BrdU immunostaining.
Statistical Analysis.
Graphs show means ± sem (SD of the mean). P values correspond to two-tailed Student t test analysis. For quantification of the migration at 45 hpf, the trunk of the embryo was subdivided into six zones of equal size (Fig. 1D). For each zone, the proportion of embryos in which the primordium had reached the zone was calculated and plotted.
Supplementary Material
Acknowledgments
We thank Dr. Elke Ober for critically reading the manuscript and the staff of the fish facility of the National Institute for Medical Research for their work. This work was supported by the Medical Research Council (U117532048). M.A.B. was supported by fellowships from the Fondation Fyssen (France) and the European Molecular Biology Organisation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1306282110/-/DCSupplemental.
References
- 1.Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10(7):445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- 2.Ghysen A, Dambly-Chaudière C. Development of the zebrafish lateral line. Curr Opin Neurobiol. 2004;14(1):67–73. doi: 10.1016/j.conb.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 3.David NB, et al. Molecular basis of cell migration in the fish lateral line: Role of the chemokine receptor CXCR4 and of its ligand, SDF1. Proc Natl Acad Sci USA. 2002;99(25):16297–16302. doi: 10.1073/pnas.252339399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Q, Shirabe K, Kuwada JY. Chemokine signaling regulates sensory cell migration in zebrafish. Dev Biol. 2004;269(1):123–136. doi: 10.1016/j.ydbio.2004.01.020. [DOI] [PubMed] [Google Scholar]
- 5.Haas P, Gilmour D. Chemokine signaling mediates self-organizing tissue migration in the zebrafish lateral line. Dev Cell. 2006;10(5):673–680. doi: 10.1016/j.devcel.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 6.Dambly-Chaudière C, Cubedo N, Ghysen A. Control of cell migration in the development of the posterior lateral line: Antagonistic interactions between the chemokine receptors CXCR4 and CXCR7/RDC1. BMC Dev Biol. 2007;7:23. doi: 10.1186/1471-213X-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valentin G, Haas P, Gilmour D. The chemokine SDF1a coordinates tissue migration through the spatially restricted activation of Cxcr7 and Cxcr4b. Curr Biol. 2007;17(12):1026–1031. doi: 10.1016/j.cub.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 8.Boldajipour B, et al. Control of chemokine-guided cell migration by ligand sequestration. Cell. 2008;132(3):463–473. doi: 10.1016/j.cell.2007.12.034. [DOI] [PubMed] [Google Scholar]
- 9.Gamba L, Cubedo N, Lutfalla G, Ghysen A, Dambly-Chaudiere C. Lef1 controls patterning and proliferation in the posterior lateral line system of zebrafish. Dev Dyn. 2010;239(12):3163–3171. doi: 10.1002/dvdy.22469. [DOI] [PubMed] [Google Scholar]
- 10.McGraw HF, et al. Lef1 is required for progenitor cell identity in the zebrafish lateral line primordium. Development. 2011;138(18):3921–3930. doi: 10.1242/dev.062554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valdivia LE, et al. Lef1-dependent Wnt/β-catenin signalling drives the proliferative engine that maintains tissue homeostasis during lateral line development. Development. 2011;138(18):3931–3941. doi: 10.1242/dev.062695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aman A, Nguyen M, Piotrowski T. Wnt/β-catenin dependent cell proliferation underlies segmented lateral line morphogenesis. Dev Biol. 2011;349(2):470–482. doi: 10.1016/j.ydbio.2010.10.022. [DOI] [PubMed] [Google Scholar]
- 13.Aman A, Piotrowski T. Wnt/beta-catenin and Fgf signaling control collective cell migration by restricting chemokine receptor expression. Dev Cell. 2008;15(5):749–761. doi: 10.1016/j.devcel.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 14.Harding MJ, Nechiporuk AV. Fgfr-Ras-MAPK signaling is required for apical constriction via apical positioning of Rho-associated kinase during mechanosensory organ formation. Development. 2012;139(17):3130–3135. doi: 10.1242/dev.082271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ernst S, et al. Shroom3 is required downstream of FGF signalling to mediate proneuromast assembly in zebrafish. Development. 2012;139(24):4571–4581. doi: 10.1242/dev.083253. [DOI] [PubMed] [Google Scholar]
- 16.Nechiporuk A, Raible DW. FGF-dependent mechanosensory organ patterning in zebrafish. Science. 2008;320(5884):1774–1777. doi: 10.1126/science.1156547. [DOI] [PubMed] [Google Scholar]
- 17.Lecaudey V, Cakan-Akdogan G, Norton WH, Gilmour D. Dynamic Fgf signaling couples morphogenesis and migration in the zebrafish lateral line primordium. Development. 2008;135(16):2695–2705. doi: 10.1242/dev.025981. [DOI] [PubMed] [Google Scholar]
- 18.Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78(2):191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 19.Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nat Rev Genet. 2005;6(12):893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- 20.Narita Y, Rijli FM. Hox genes in neural patterning and circuit formation in the mouse hindbrain. Curr Top Dev Biol. 2009;88:139–167. doi: 10.1016/S0070-2153(09)88005-8. [DOI] [PubMed] [Google Scholar]
- 21.Gehring WJ, et al. Homeodomain-DNA recognition. Cell. 1994;78(2):211–223. doi: 10.1016/0092-8674(94)90292-5. [DOI] [PubMed] [Google Scholar]
- 22.Moens CB, Selleri L. Hox cofactors in vertebrate development. Dev Biol. 2006;291(2):193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 23.Slattery M, et al. Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell. 2011;147(6):1270–1282. doi: 10.1016/j.cell.2011.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Studer M, Lumsden A, Ariza-McNaughton L, Bradley A, Krumlauf R. Altered segmental identity and abnormal migration of motor neurons in mice lacking Hoxb-1. Nature. 1996;384(6610):630–634. doi: 10.1038/384630a0. [DOI] [PubMed] [Google Scholar]
- 25.Cooper KL, Leisenring WM, Moens CB. Autonomous and nonautonomous functions for Hox/Pbx in branchiomotor neuron development. Dev Biol. 2003;253(2):200–213. doi: 10.1016/s0012-1606(02)00018-0. [DOI] [PubMed] [Google Scholar]
- 26.Geisen MJ, et al. Hox paralog group 2 genes control the migration of mouse pontine neurons through slit-robo signaling. PLoS Biol. 2008;6(6):e142. doi: 10.1371/journal.pbio.0060142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maloof JN, Whangbo J, Harris JM, Jongeward GD, Kenyon C. A Wnt signaling pathway controls hox gene expression and neuroblast migration in C. elegans. Development. 1999;126(1):37–49. doi: 10.1242/dev.126.1.37. [DOI] [PubMed] [Google Scholar]
- 28.Breau MA, Wilson D, Wilkinson DG, Xu Q. Chemokine and Fgf signalling act as opposing guidance cues in formation of the lateral line primordium. Development. 2012;139(12):2246–2253. doi: 10.1242/dev.080275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Distel M, Wullimann MF, Köster RW. Optimized Gal4 genetics for permanent gene expression mapping in zebrafish. Proc Natl Acad Sci USA. 2009;106(32):13365–13370. doi: 10.1073/pnas.0903060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bami M, Episkopou V, Gavalas A, Gouti M. Directed neural differentiation of mouse embryonic stem cells is a sensitive system for the identification of novel Hox gene effectors. PLoS ONE. 2011;6(5):e20197. doi: 10.1371/journal.pone.0020197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muhr J, Andersson E, Persson M, Jessell TM, Ericson J. Groucho-mediated transcriptional repression establishes progenitor cell pattern and neuronal fate in the ventral neural tube. Cell. 2001;104(6):861–873. doi: 10.1016/s0092-8674(01)00283-5. [DOI] [PubMed] [Google Scholar]
- 32.Stoick-Cooper CL, et al. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development. 2007;134(3):479–489. doi: 10.1242/dev.001123. [DOI] [PubMed] [Google Scholar]
- 33.Kwan KM, et al. The Tol2kit: A multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236(11):3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- 34.Xu Q, Wilkinson DG. In situ hybridisation of mRNA with hapten labelled probes. In: Wilkinson DG, editor. Situ Hybridisation: A Practical Approach. 2nd Ed. Oxford: Oxford Univ Press; 1998. pp. 87–106. [Google Scholar]
- 35.Robu ME, et al. p53 activation by knockdown technologies. PLoS Genet. 2007;3(5):e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerety SS, Wilkinson DG. Morpholino artifacts provide pitfalls and reveal a novel role for pro-apoptotic genes in hindbrain boundary development. Dev Biol. 2011;350(2):279–289. doi: 10.1016/j.ydbio.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gamba L, Cubedo N, Ghysen A, Lutfalla G, Dambly-Chaudière C. Estrogen receptor ESR1 controls cell migration by repressing chemokine receptor CXCR4 in the zebrafish posterior lateral line system. Proc Natl Acad Sci USA. 2010;107(14):6358–6363. doi: 10.1073/pnas.0909998107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Westerfield M. The Zebrafish Book. Eugene: Univ of Oregon Press; 1993. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.