Significance
Adhesion G protein-coupled receptors (GPCRs) are expressed in many developing organs, immune cells, and cancer cells, suggesting that they might play an important role in physiological and pathological functions. Compared with their potential importance, their function and signaling mechanisms are poorly understood. Disruption of the G protein-coupled receptor 126 (Gpr126) gene in mice leads to lack of myelination in the peripheral nervous system (PNS) and heart abnormalities. Similarly, the zebrafish mutant line gpr126st49 exhibits PNS abnormalities but, in contrast, no heart phenotype. Here we provide an explanation for these discrepancies. The presented data suggest that in the heart, the N-terminal fragment of Gpr126 can act independently as a ligand or coreceptor. Taken together, our data provide evidence of tissue- and domain-specific adhesion GPCR function.
Abstract
Despite their abundance and multiple functions in a variety of organ systems, the function and signaling mechanisms of adhesion G protein-coupled receptors (GPCRs) are poorly understood. Adhesion GPCRs possess large N termini containing various functional domains. In addition, many of them are autoproteolytically cleaved at their GPS sites into an N-terminal fragment (NTF) and C-terminal fragment. Here we demonstrate that Gpr126 is expressed in the endocardium during early mouse heart development. Gpr126 knockout in mice and knockdown in zebrafish caused hypotrabeculation and affected mitochondrial function. Ectopic expression of Gpr126-NTF that lacks the GPS motif (NTFΔGPS) in zebrafish rescued the trabeculation but not the previously described myelination phenotype in the peripheral nervous system. These data support a model in which the NTF of Gpr126, in contrast to the C-terminal fragment, plays an important role in heart development. Collectively, our analysis provides a unique example of the versatile function and signaling properties of adhesion GPCRs in vertebrates.
Members of the adhesion G protein-coupled receptor (GPCR) family are expressed in many developing organs (e.g., nervous system and reproductive organs), immune cells, and cancer cells, suggesting that they might play an important role in physiological and also in pathological functions (1). Compared with their potential importance, the function and signaling mechanisms of adhesion GPCRs are poorly understood. Gpr126 (DREG) is expressed in mice heart and somites during embryogenesis and in the adult lung (2). It has been shown that both human and mouse Gpr126 can be cleaved at the GPS site by an endogenous proteolytic process resulting in a membrane-bound 35-kDa protein containing a seven-transmembrane domain [C-terminal fragment (CTF)] and an extracellular soluble protein [N-terminal fragment (NTF)], which might be further cleaved resulting in a 70-kDa soluble protein containing a complement, Uegf, Bmp1 (CUB) and a pentraxin (PTX) domain (2). These data indicated that the membrane-bound CTF can act as an independent receptor and the soluble NTF can act as a ligand or coreceptor for unknown receptors. This hypothesis is supported by the recent finding that the GAIN/GPS structure of latrophilin, another adhesion GPCR, operates independently of the CTF in Caenorhabditis elegans fertility (3). It has also been reported that the NTF of the brain angiogenesis inhibitor 1 (BAI1) inhibits endothelial cell proliferation and angiogenesis in mice (4, 5) and that the NTFs of EMR2 and GPR56 can cross-react with the CTF of latrophilin (6).
Studies on Gpr126 knockout mice have shown that disruption of the Gpr126 gene leads to fully penetrant embryonic lethality with cardiac abnormality (7). In another Gpr126 knockout mouse line from Taconic (T-Gpr126−/−) (8), most mutants die in utero, although a few mice survive to postnatal stages. T-Gpr126−/− mice are characterized both by a lack of myelination in the peripheral nervous system (PNS) and by multiple defects in peripheral nerves. A myelination phenotype has also been observed in the zebrafish mutant line gpr126st49 (9). Several experiments including rescue experiments with forskolin indicate that cAMP and PKA are involved in the GPR126-mediated pathway initiating myelination (9, 10). However, although mouse and zebrafish Gpr126 are true orthologs (7), no heart phenotype has been described in the zebrafish mutant line. The gpr126st49 mutation introduces a premature stop codon before the GPS motif. This raises the possibility that gpr126st49 mutant fish still express a functional fragment of NTF (amino acid 1–783, hereafter referred to as NTFΔGPS) but no CTF. Therefore, we hypothesize that the NTF of Gpr126 functions independently of its CTF during heart development in contrast to the role of Gpr126 in the PNS.
Results
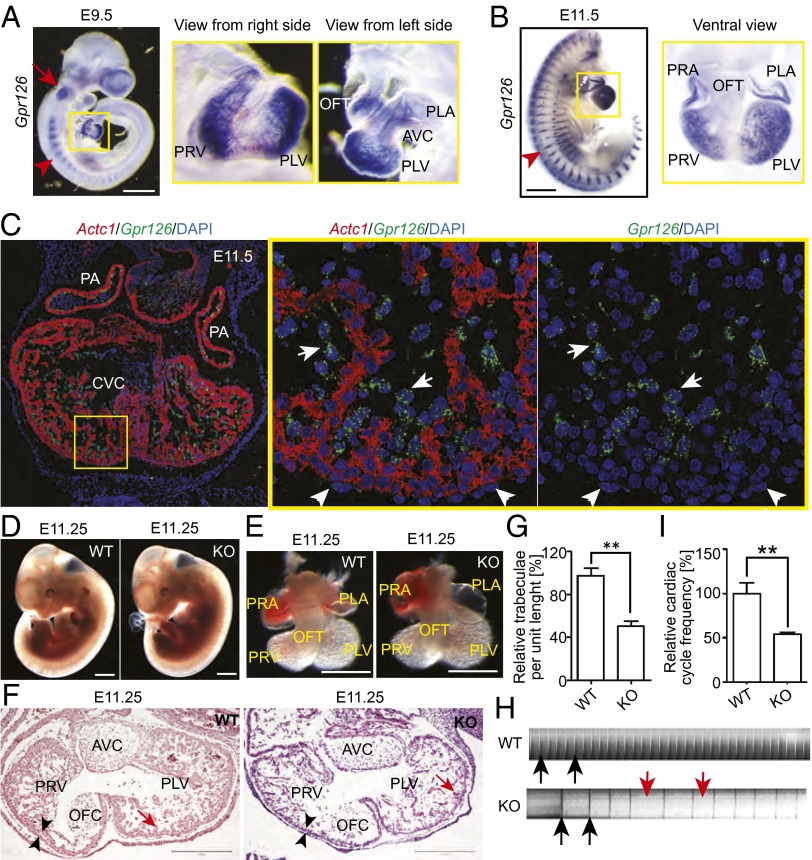
Large-scale RNA expression analysis identified Gpr126 as transiently expressed in the rat heart during embryogenesis (Fig. S1A). RT-PCR–based data showed a similar Gpr126 temporal expression pattern in mice and revealed the existence of two splice variants (Fig. S1 B and C). Based on in situ hybridization (ISH) analyses, Gpr126 is highly expressed in the heart, somites, and otic vesicle at E9.5 and E11.5 (Fig. 1 A and B). Cardiac Gpr126 expression is in the primitive ventricles, atrium, and outflow tract (OFT) at E9.5 and becomes restricted to the four chambers with no detectable expression in the interventricular region and OFT at E11.5 (Fig. 1 A and B). Multiplex ISH revealed that Gpr126 is prominently expressed in the endocardium covering trabeculae (Fig. 1C).
Fig. 1.
Expression pattern of Gpr126 and characterization of R-Gpr126−/− mice. (A and B) Whole-mount in situ hybridization (lateral view) showing Gpr126 expression in the otic vesicle (red arrow), somites (red arrowhead), and heart (yellow square) at E9.5 in primitive ventricles [primitive left ventricle (PLV) and primitive right ventricle (PRV)], atrium [primitive right atrium (PRA) and primitive left atrium (PLA)], and the outflow tract (OFT) and at E11.5 in all primitive chambers but not in the OFT and interventricular region. (Scale bar: 500 µm.) (C) Multiplex in situ hybridization against alpha cardiac actin (Actc1; red) and Gpr126 (green) combined with DAPI staining (nuclei; blue) on thin sections indicating Gpr126 expression in endocardial (white arrows) but not epicardial cells (white arrowheads) and cardiomyocytes (red). (D and E) Gross morphology of WT and KO embryos (D) and hearts (E) at E11.25 is not different. (Scale bar: 500 µm.) (F) Hematoxylin and eosin staining of heart sections indicates hypotrabeculation (red arrows) and thinner ventricular myocardium (arrowheads) in KO hearts. (Scale bar: 300 µm.) (G) Quantification of the trabeculation phenotype. WT value was set to 100%. (H) Kymograph analysis. KO hearts displayed cardiac arrhythmia and bradycardia. Black lines/peaks (black arrows) represent ventricular contraction. Red arrows indicate irregular time intervals. (I) Quantitative analysis of H. WT value was set to 100%. Data are mean ± SEM. AVC, atrioventricular cushion; KO, knockout; OFC, outflow tract cushion; WT, wild type. **P < 0.01.
Mating heterozygous Gpr126+/− mice [R-Gpr126+/−; The Institute of Physical and Chemical Research (Japan) (RIKEN)] yielded no homozygous knockout offspring at birth or E13.5, whereas at E11.25, offspring were produced in the expected Mendelian ratio (Fig. S1 D–F). R-Gpr126−/− mice at E11.25 exhibited mild pericardial edema, but gross morphology including the heart did not differ from wild-type (WT) littermates (Fig. 1 D and E). Histological analyses confirmed a previous report (7) that Gpr126−/− hearts are characterized by a thinner ventricular wall and fewer trabeculae (Fig. 1 F and G). To further support the importance of Gpr126 for heart development, we analyzed an additional Gpr126 knockout mouse line, T-Gpr126−/−, that had been used to demonstrate the importance of Gpr126 for PNS myelination (8). The authors reported that the majority of T-Gpr126−/− embryos did not survive until birth but did not analyze the cause of embryonic lethality or examine the heart. Histological analyses revealed that the hearts of this knockout mouse line at E11.25 are also affected, exhibiting hypotrabeculation and thinner trabeculae in comparison with their WT littermates (Fig. S2A).
Video and Kymograph analyses revealed that R-Gpr126−/− mice suffer from bradycardia along with cardiac arrhythmia (Fig. 1 H and I and Movies S1 and S2). An explanation for this phenotype might be alterations in cell–cell contacts. However, localization of cadherins, adherent junction proteins, was not affected (Fig. S2B), and electron microscopic analyses further support that cell–cell contacts are maintained in R-Gpr126−/− hearts (Fig. S2C). Mitochondria in cardiomyocytes (Fig. 2 A and B) and endocardial cells (Fig. S2D) of R-Gpr126−/− hearts exhibited a more complex shape, less well developed cristae, and electron-dense precipitates. In addition, a large number of lipid droplets (triglycerides) were accumulated in the R-Gpr126−/− cardiac cells, whereas glycogen deposits (Gly) were more frequent in WT heart (Fig. 2 B and C), indicative of aberrant energy metabolism. Lipid staining with oil red O confirmed the lipid accumulation in R-Gpr126−/− hearts. In contrast, lipid droplets were detected neither in WT hearts nor in neural tissue of WT and R-Gpr126−/− mice with this technique (Fig. S2 E and F). By electron microscopy, only a few lipid droplets were detected in the WT heart. Taken together, our data indicate that Gpr126 is required for proper trabeculation, contraction, and energy metabolism of the heart.
Fig. 2.
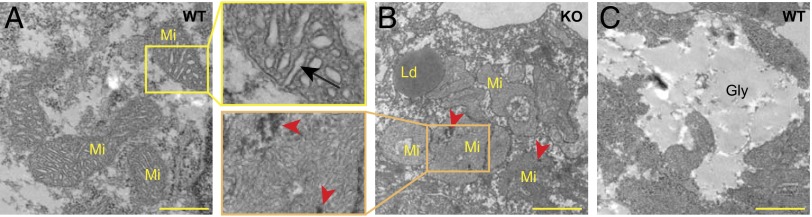
R-Gpr126−/− E11.25 cardiomyocytes contain defective mitochondria. (A–C) Transmission electron micrographs displaying the ultrastructural details of cardiomyocytes in WT (A and C) and KO (B) hearts. (A) Mitochondria (Mi) with well-developed cristae (arrow) in WT cardiomyocytes. (B) KO cardiomyocytes contain abundant lipid droplets (Ld) and mitochondria with a more complex morphology, less well developed cristae, and electron-dense precipitates (arrowheads). (C) Glycogen (Gly) in WT cardiomyocytes. (Scale bar: 1 µm.)
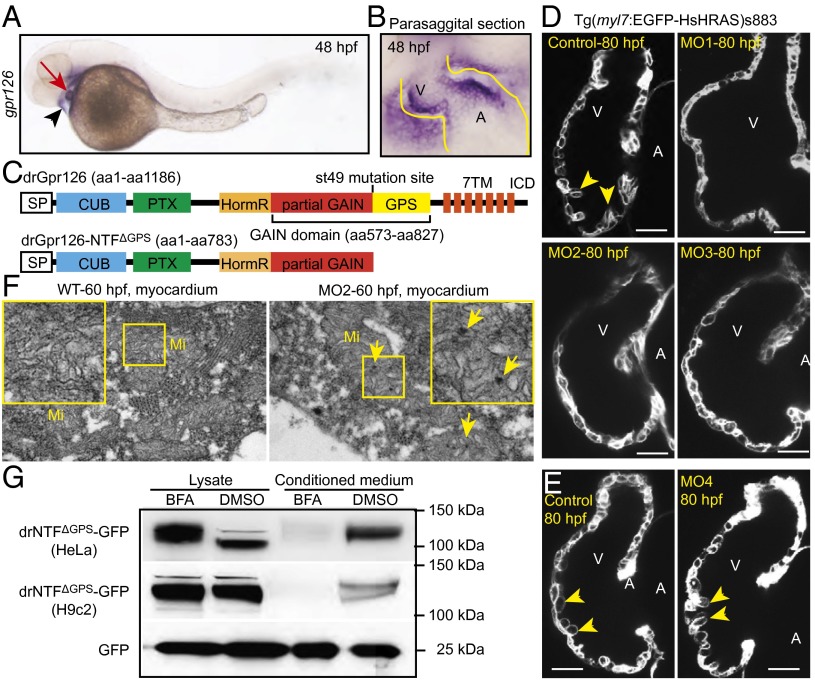
The presented and published data clearly demonstrate that Gpr126 is required for proper cardiac and PNS development. This raises the question of whether Gpr126 plays the same role in both systems, despite very different cellular phenotypes. A recent study showing that the zebrafish mutant line gpr126st49 exhibits a myelination phenotype and ear edema but no heart phenotype suggests that Gpr126 exhibits organ-specific signaling mechanisms (9). ISH demonstrated that in zebrafish, gpr126 is expressed in both cardiac chambers and in the pericardium at 48 h postfertilization (hpf) (Fig. 3 A and B). RT-PCR and sequencing analyses using whole embryos detected four Gpr126 isoforms that differ only in the N-terminal extracellular domain (Fig. S3). However, gpr126st49 zebrafish mutants exhibited no pericardial edema or other cardiac abnormalities at 6 d postfertilization (dpf), the latest time point examined (Fig. S4A), although they did exhibit ear edema, as previously reported (Fig. S4B) (9).
Fig. 3.
Gpr126 knockdown leads to cardiac abnormalities in zebrafish. (A) Lateral view of a 48-hpf zebrafish embryo after whole-mount in situ hybridization showing gpr126 expression in heart (red arrow) and pericardium (black arrowhead). (B) Parasagittal section confirming cardiac expression of gpr126. (C) Schematic representation of zebrafish full-length Gpr126 (drGpr126) and its NTF part up to the st49 mutation site (drGpr126-NTFΔGPS). (D and E) Confocal sections of hearts from control- and morpholino-injected Tg(myl7:EGFP-HsHRAS)s883 embryos at 80 hpf. In full-length Gpr126-depleted animals (MO1-3) but not in Gpr126-CTF–depleted (MO4) morphants, trabeculation (yellow arrowheads) is perturbed. (Scale bar: 20 µm.) (F) Transmission electron micrographs at 60 hpf reveal that morphants contain elongated Mi with more branched cristae and electron dense precipitates (arrows) compared with WT siblings. (G) Western blot analysis of lysates and conditioned medium from cells overexpressing C-terminal GFP-tagged drGpr126-NTFΔGPS or GFP. Note that both GFP-tagged NTFΔGPS (predicted band size: 113 kDa) and GFP (predicted band size: 27 kDa) were detected in conditioned medium. However, secretion of GFP-tagged NTFΔGPS but not GFP was inhibited by BFA (a blocker of classical trans-Golgi secretory pathway). A, atrium; V, ventricle.
The gpr126st49 mutant allele carries a single point mutation that introduces a stop codon immediately before the GPS motif (Fig. 3C and Fig. S4C) (9). gpr126 mRNA transcripts could be detected in gpr126st49 mutants by RT-PCR (Fig. S4D), as previously shown by in situ hybridization (9). This suggests that Gpr126-NTFΔGPS might be expressed as a functional truncated protein in gpr126st49 mutants. To test our hypothesis, we injected a splicing inhibitory morpholino (MO4) into WT zebrafish, which binds to the mutation site in gpr126st49 mutants and the adjacent intron (Fig. S4E) (9). MO4 binding caused an insertion of a small fragment of the intron (23 bp) (Fig. S4 E and F), resulting in alternatively spliced gpr126 mRNA of all four endogenous splice variants, which encode only Gpr126-NTFΔGPS. As gpr126st49 mutants, MO4 morphants did not exhibit pericardial edema (Fig. S4G) but did exhibit ear edema (Fig. S4H) and abnormalities in posterior lateral line nerve (PLLn) myelination, as assessed by reduced expression of the Schwann cell transcription factor krox20/egr2 and myelin basic protein (mbp) (Fig. S4 I and J) (11, 12). Collectively, our data suggest that Gpr126-CTF is not crucial for heart development but is necessary for PNS myelination.
To determine the physiological functions of full-length Gpr126 in zebrafish heart development, we used one translation-inhibitory (MO1) and two splicing-inhibitory (MO2 and MO3) morpholino oligos (Fig. S5A). MO2 injection resulted in a 144-bp-long intron insertion (Fig. S5B), and MO3 injection resulted in a 166-bp-long exon deletion causing frame shifts (Fig. S5C). Injection of these morpholinos did not cause any obvious defect until 48 hpf. At 54 hpf, we observed mild accumulation of blood at the sinus-venosus (Fig. S5D); however, cardiac cycle was normal in morphant hearts (Fig. S5E). At 75 hpf, morphants exhibited prominent pericardial edema, but most of the morphant hearts contracted regularly (Fig. S5 F and G). In zebrafish, cardiac trabeculation initiates at 72 hpf (13). Detailed morphological analysis at 80 hpf showed that trabeculation was inhibited in the full-length Gpr126-depleted animals (Fig. 3D) but not in MO4 morphants in which the Gpr126-NTFΔGPS was not targeted (Fig. 3E). Electron microscopic analysis at 60 hpf showed a milder cardiac phenotype than R-Gpr126−/− mouse hearts with elongated, more branched mitochondria and electron-dense precipitates compared with WT siblings. However, we observed neither glycogen nor triglyceride deposition in WT or morphant hearts (Fig. 3F). All MOs caused ear edema, confirming that the MOs used disrupted gpr126 (Fig. S5H). Taken together, our data demonstrate that Gpr126 is required for ventricular trabeculation and mitochondria development in zebrafish. Our data further support our hypothesis that Gpr126 function is organ-specific.
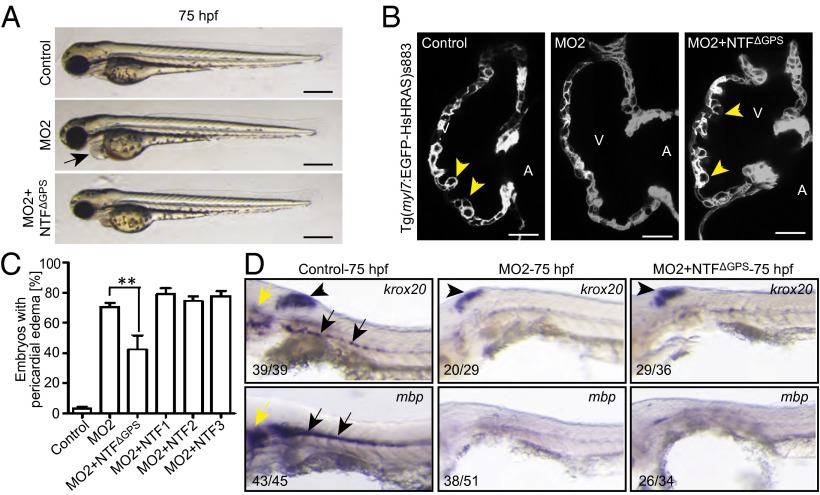
To directly test whether Gpr126-NTFΔGPS acts independently of Gpr126-CTF during heart development, we generated C-terminal GFP-tagged full-length Gpr126 (drGpr126-GFP) and Gpr126-NTFΔGPS (drGpr126-NTFΔGPS-GFP, cloned up to the st49 mutation) (Fig. S6 A and B). Overexpression of these fusion proteins and GFP (Fig. S6C, empty cloning vector) in HeLa cells indicate that NTFΔGPS and CTF both localize to the endoplasmic reticulum, whereas GFP localized predominantly to the nucleus (Fig. S6D). Overexpression of drGpr126-GFP but not drGpr126-NTFΔGPS-GFP resulted in membrane localization, suggesting that the CTF part of Gpr126 can translocate to the membrane but that the NTFΔGPS part by itself cannot. Furthermore, overexpression of drGpr126-NTFΔGPS-GFP in HeLa or cardiac H9c2 cells (14) resulted in secretion of drGpr126-NTFΔGPS-GFP (predicted size: 113 kDa) (Fig. S6 E and F). This secretion was inhibited by Brefeldin A (BFA), a blocker of the classical trans-Golgi secretory pathway (Fig. 3G) (15, 16). These data suggest that the mutant drGpr126-NTFΔGPS can be secreted from the cell as it has been shown for human and mouse Gpr126-NTF (2). Surprisingly, we have detected by Western blotting a drGpr126-NTFΔGPS-GFP band of over 100 kDa. Previously, Moriguchi et al. (2) demonstrated that human and mouse Gpr126-NTFs contain two cleavage sites: the GPS and the S2 cleavage sites. Cleavage of a C-terminal–fused NTF at the S2 site should result in a signal corresponding to around 60 kDa (C-terminal GFP fused) that we could not detect, suggesting that in zebrafish the NTF of Gpr126 is not cleaved at S2 (Fig. S6 E and F). This is further supported by amino acid sequence alignment of zebrafish, human, and mouse Gpr126 revealing that zebrafish Gpr126 contains the GPS cleavage site but lacks eight amino acids at the S2 cleavage site (Fig. S7). Collectively, our data indicate that the NTFΔGPS part of the st49 mutant form of drGpr126 can function as an independent, stable, secreted protein. Therefore, we injected capped drgpr126-NTFΔGPS mRNA together with MO2 and assayed for rescue of the heart phenotype. MO2 morphants were characterized by pericardial edema, lack of trabeculation, and reduced expression of mbp and krox20/egr2 at the PLLn. Injection of 100 pg of capped drgpr126-NTFΔGPS mRNA encoding amino acids 1–783 (variant 1) rescued the pericardial edema and ventricular trabeculation phenotype in a subset of the MO2 morphants (Fig. 4 A–C). In contrast, krox20/egr2 or mbp expression was not rescued in cardiac-rescued morphants at 75 hpf (Fig. 4D and Fig. S8A). No obvious phenotype was observed when drgpr126-NTFΔGPS mRNA was injected alone. To explore the domain-specific functions of the drGpr126-NTF in heart development, we coinjected mRNAs, which encode subfragments of NTF, with MO2 (drgpr126-NTF1, lacks GAIN domain, amino acid 1–572; drgpr126-NTF2, lacks GAIN and HormR binding domain, amino acid 1–504; and drgpr126-NTF3, lacks GAIN, HormR binding, and PTX domain, amino acid 1–149) (Fig. S8B). None of these mRNAs rescued the cardiac phenotype caused by MO2-mediated depletion of Gpr126 (Fig. 4C). Although we cannot definitively show that the injected mRNAs encoding subfragments of NTF are properly expressed and secreted due to experimental limitations, our data are consistent with the hypothesis that the drGpr126-NTF without the GPS motif is required for zebrafish heart development.
Fig. 4.
NTFΔGPS mRNA rescues the trabeculation phenotype in Gpr126-depleted zebrafish. (A) Lateral view of control-, MO2-, or MO2+NTFΔGPS mRNA-injected embryos at 75 hpf. MO2 injection resulted in pericardial edema (arrow). (B) Confocal sections of the hearts from control-, MO2-, and MO2+NTFΔGPS mRNA-injected Tg(myl7:EGFP-HsHRAS)s883 embryos at 80 hpf. In Gpr126-depleted animals, trabeculation (yellow arrowheads) is perturbed. Note that this phenotype is rescued by NTFΔGPS mRNA injection. (C) Quantitative analysis of rescue experiments of coinjections of MO2 and mRNAs encoding NTFΔGPS or NTF subfragments. Mean ± SEM. (D) Whole-mount in situ hybridization demonstrating krox20/egr2 and mbp expression in the otic vesicle (yellow arrow) and PLLn Schwann cells (black arrow) and krox20/egr2 expression in the hindbrain (arrowhead) at 75 hpf. In contrast, expression of krox20/egr2 and mbp is down-regulated in PLLn of MO2-mediated full-length Gpr126-depleted embryos. Coinjection of NTFΔGPS mRNA failed to rescue the PLLn myelineation phenotype in the cardiac phenotype-rescued animals. **P < 0.01.
To assess whether the secreted NTFΔGPS functions in a cell-autonomous or paracrine fashion, we performed a Gpr126-NTFΔGPS binding in situ using purified mouse Gpr126-NTFΔGPS fused at its C terminus with the Fc fragment of murine IgG2b subclass (mFc) (mGpr126-NTFΔGPS-mFc). Subsequent immunofluorescence staining for cardiomyocytes revealed that mGpr126-NTFΔGPS-mFc specifically binds to cardiomyocytes and pericardial cells (Fig. S9). Collectively, our data suggest that during heart development, Gpr126-NTF functions independently of Gpr126-CTF, presumably in a paracrine fashion. This is in contrast to the PNS, where CTF signaling is essential for myelination (9).
Discussion
Taken together, our data provide evidence of tissue- and domain-specific adhesion GPCR function. We confirm the previous finding that Gpr126 deletion causes heart phenotypes in mice causing embryonic lethality (7). Moreover, our data provide additional insights into the cardiac phenotype suggesting that the endocardium communicates through Gpr126 with the myocardium to regulate energy metabolism, contractility, and trabeculation. We hypothesize that the contractility phenotype is a consequence of defective mitochondria because several studies have demonstrated that mitochondrial respiratory dysfunction or the deficiency of lipid transporter in the mitochondrial membrane leads to a metabolic shift from fatty acid oxidation to glycolysis and the accumulation of lipids causing cardiac arrhythmia (17–20). In gpr126-depleted zebrafish hearts, neither lipid accumulation nor cardiac arrhythmia was observed despite abnormal mitochondria. This suggests that early heart morphogenesis in zebrafish relies more on glycolysis than on fatty acid oxidation. In contrast, trabeculation defects, which occur later in development, were observed in both species. It remains to be determined whether trabeculation defects are due to defective mitochondria or represent an independent phenotype.
Our analysis of gpr126 mutants and morphants in combination with rescue experiments and the analysis of two different organ systems provide a first glimpse in the versatile function and signaling properties of adhesion GPCRs using the example of Gpr126 in vertebrates. Our data demonstrate that Gpr126-CTF is not essential for cardiac development, offering an explanation for the lack of a heart phenotype in gpr126st49 zebrafish mutants. In contrast, Gpr126-NTFΔGPS is sufficient to rescue the heart phenotype but not the myelination defect. This suggests that Gpr126 acts as a receptor to drive myelin gene expression in PNS Schwann cells, whereas in the heart, the NTF of Gpr126 acts as a ligand or coreceptor activating an unknown receptor.
Materials and Methods
Ethics Statement.
Animal experiments were approved by the local committee for care and use of laboratory animals (Regierungspräsidium, Darmstadt, Germany, or Washington University School of Medicine) and conform to Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH) (21).
mRNA Expression Screening.
Large mRNA expression screening was performed as described previously (22). Briefly, total RNA of rat cardiac ventricles from different developmental stages was isolated using TRIzol (Invitrogen). The Affymetrix GeneChip RAT230 Expression Set was used for expression analysis.
Zebrafish Maintenance.
Transgenic Tg(myl7:EGFP-HsHRAS)s883 (23), wild-type AB, and gpr126st49 heterozygous zebrafish (Danio rerio) were maintained at standard laboratory condition as described (24). Zebrafish embryos were maintained at 28 °C in E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4, and 0.6 µM methylene blue). For confocal microscopy and in situ hybridization, 0.2 mM 1-phenyl-2-thio-urea (Alfa Aesar) was added to the E3 medium at around 24 hpf to prevent pigmentation.
Genotyping.
Zebrafish larvae were genotyped for the st49 mutation by a DCAPs assay, as described (9). The following primers were used to amplify genomic DNA: 5′-TAGAGATTGCACATTTGGATTA-3′ and 5′-ATCGGAACACACCAAACAGG-3′. PCR products were digested with MseI, resulting in a smaller fragment for the mutant allele. Genomic DNA of R-Gpr126 mice was extracted from tail biopsies using the ISOLATE Genomic DNA Mini Kit (Bioline). Genotyping PCR was performed using the common reverse primer 5′-GAGAACTGGAAAGGAAAGGTGAGTGCAGC-3′ binding WT and targeted alleles at intron 2 and the forward primers 5′-TGTGTTACCTGTGTTCCTCTCTCAGGTAG-3′ binding wild-type gpr126 at exon2–intron2 junction and 5′-TCGTGCTTTACGGTATCGCCGCTCCCGATT-3′ binding the neomycin resistance cassette, yielding products of 829 bp and 530 bp, respectively.
In Situ Hybridization.
Mice or zebrafish embryos were harvested at indicated time points and fixed overnight in 4% (wt/vol) formaldehyde at 4 °C. ISH was performed following standard protocols as described (22). For the riboprobe against zebrafish gpr126 and mouse Gpr126 mRNA, 271-bp (zebrafish gpr126, 5′-TGTCATGGTCAGCTCAGAGG-3′ and 5′-CAGTGTTGGAGGCAGAGTGA-3′) and 692-bp (mouse Gpr126, 5′-CTCCGATAACCTGGGGAAAT-3′ and 5′-TTCTTGGGGTTCTCCTCTCA-3′) fragments were amplified by PCR using cDNA from 2-dpf zebrafish and embryonic day E12.5 mouse hearts, respectively, and cloned into the pGEM-T Easy vector (Promega) to create pGEMTeasy-drgpr126 and pGEMTeasy-mGpr126. Zebrafish embryos embedded after ISH [17% (wt/vol) gelatin in PBS] were fixed with 4% (wt/vol) PFA in PBS (overnight at room temperature), sectioned (80 µm) using a vibratome (Leica), and mounted (Kaiser’s glycerol gelatin; Merck) on microscope slides. To examine the cell type-specific expression of Gpr126, we performed multiplex ISH (QuantiGene ViewRNA In Situ Hybridization kit, Affymetrix) on 5-µm thin paraffin sections through the heart of E11.5 mouse embryos following the manufacturer’s instructions. Probe set against Gpr126 was 1,226 bp to 2,226 bp of NM_001002268. Probe set against cardiomyocyte-specific Actc1 was 1,213 bp to 1,390 bp of NM_009608.
Cloning of gpr126 and gpr126-NTFΔGPS and Microinjection.
A 3,558-bp fragment of zebrafish full-length gpr126 (NM_001163291.1, variant 1) (5′-ATGATTTCGTTCATCAGTGGTC-3′ and 5′-TTATTGCAGGGTACTATCTGCATTAC-3′); a 2,349-bp fragment of zebrafish gpr126-NTFΔGPS (sense 5′-CACCATGATTTCGTTCATCAGTGGTC-3′ and antisense 5′-CTGTTACTCCAAATGTGCAATCTCTA-3′); a 1,716-bp fragment of gpr126-NTF lacking the GAIN domain (sense 5′-ATGATTTCGTTCATCAGTGGTC-3′ and antisense 5′-TTATGAAACCGTAATGCTGGTTAT-3′); a 1,512-bp fragment of gpr126-NTF lacking GAIN and HormR binding domain (sense 5′-ATGATTTCGTTCATCAGTGGTC-3′ and antisense 5′-TTATAAGTTCACGGTAACACTGTC-3′); and a 447-bp fragment of zebrafish NTF-gpr126 lacking GAIN, HormR binding, and PTX domain (sense 5′-ATGATTTCGTTCATCAGTGGTC-3′ and antisense 5′-TTACACTTGCTTGTAACTGATATG-3′) were amplified by PCR using cDNA from 2-dpf embryos. The PCR products were cloned into pGEM-T Easy vector (Promega). Next, fragments were subcloned into pCS2+ vector (named pCS2+-drgpr126-NTFΔGPS, pCS2+-drgpr126-NTF1, pCS2+-drgpr126-NTF2, and pCS2+-drgpr126-NTF3) or pGFP [referred to as drgpr126-NTFΔGPS-GFP (C-terminal GFP tagged) and drgpr126-GFP (C-terminal GFP tagged)]. Capped mRNA of drgpr126-NTFΔGPS fragments were synthesized after linearization (NotI) using the mMESSAGE mMACHINE SP6 In Vitro Transcription Kit (Ambion). Single-cell–stage embryos were injected into the cell or yolk (≤3 nL; PV820 Injector; World Precision Instruments) with morpholinos [gpr126 MO1, 5′-ACCGACCACTGATGAACGAAATCAT-3′; gpr126 MO2, 5′-GTCTGATTGAGCTGTAAAACATCAA-3′; gpr126 MO3, 5′-AGCTCTGTACCAAGACATATGATGA-3′; and gpr126 MO4, 5′-CGGGTTGGACTCAAGACGATAG-3′ (Gene Tools)] and/or capped mRNA in nuclease-free water. Control injection was with 0.05% Phenol Red (Sigma). For the in situ protein binding assay a 2,310-bp fragment of mouse Gpr126-NTF without GPS motif was amplified (sense 5′-AAAAAGAATTCATGATGTTTGACACTCTCGGG-3′ and antisense 5′-AAAAAGAATTCATGATGCACTTCCTGTGTTCT-3′) by PCR using the mouse Gpr126 clone in a pcDps vector (a gift from I. Liebscher, University of Leipzig, Leipzig, Germany). Subsequently, the fragment was subcloned (EcoRI) in a modified pCDNA3.1(+) vector (25) to generate a fusion protein containing a C-terminal Fc fragment of murine IgG2b subclass (mFc) referred to as mGpr126-NTFΔGPS-mFc.
RT-PCR.
RNA was isolated from different developmental stages of rat (E11 to E20, n ≥ 10; P5, P10, and adult, n ≥ 3) or mouse hearts (E9.5 to E18.5, n ≥ 10; P1, P8, and adult, n ≥ 3) and zebrafish embryos (n ≥ 30) at indicated developmental stages, using TRIzol (Invitrogen) or RNAeasy kit (Qiagen). RT-PCR was performed following standard protocols. Primers for mouse were gapdh, 5′-CAGAAGACTGTGGATGGCCC-3′ and 5′-AGTGTAGCCCAGGATGCCCT-3′, and gpr126 variants, 5′-CTCCGATAACCTGGGGAAAT-3′ and 5′-TTCTTGGGGTTCTCCTCTCA-3′. Primers for zebrafish were gapdh, 5′-TGGGTGTCAACCATGAGAAA-3′ and 5′-AACCTGGTGCTCCGTGTATC-3′; gpr126 F1, 5′-CCCTTGTGGTTACTGATGAACA-3′; R1, 5′-GGAACTCATCTGCCTGACAA-3′; F2, 5′-TGTCATGGTCAGCTCAGAGG-3′; R2, 5′-GGTCCTGTTGCTGTTGGAGT-3′; and F3, 5′-TGGCAACTTCACCATCAAAA-3′.
Microscopy.
For bright-field (Leica) and confocal microscopy (Zeiss), tricane-anesthetized embryos were mounted in 1% low–melting-point agarose in E3 medium. The 1.24-μm thin optical sections of the zebrafish heart were imaged on a Zeiss LSM710 confocal microscope, and images were processed by Laser Scanning Microscope Image Browser (Zeiss). For transmission electron microscopy, E11.25 mouse or 3-dpf zebrafish embryos were fixed by immersion in 1.5% (wt/vol) glutaraldehyde/1.5% paraformaldehyde in 0.15 M Hepes buffer (pH 7.3) for at least 24 h. Subsequently, the specimens were osmicated, stained en bloc with uranyl acetate, dehydrated in an ascending ethanol/acetone series, and embedded in epoxy resin. Ultrathin sections from comparable regions of the heart were generated and analyzed using a Zeiss Transmission Electron Microscope 902.
Histology and Immunohistochemical Analysis.
Following standard protocols, E11.25 mouse embryos were embedded in paraffin and sectioned through the heart (5 µm, Microtome; Leica). For histological analysis, Hematoxylin and Eosin staining (AppliChem) were performed on deparaffinized tissue sections. Immunohistochemistry was performed as described (26). In brief, after antigen retrieval, deparaffinized sections or fixed cultured cells were incubated 20 min in blocking solution (5% goat serum/0.2% Tween 20/PBS) and subsequently with primary antibodies [mouse anti-sarcomeric α-actinin, 1:200 (Sigma); rabbit anti-connexin 43, 1:50 (Santa Cruz Biotechnology); and mouse anti-calnexin, 1:100 (BD Biosciences)] for 1 h. Alexa 488- or Alexa 594-conjugated antibodies (1:200; Molecular Probes) were used to detect primary immune complexes. TOPRO3 (Invitrogen) or 4′,6′-diamidino-2-phenylindole (DAPI; Sigma) (0.5 mg/mL PBS) was used to detect DNA. Images were captured using a Leica microscope or a Zeiss LSM 710 confocal laser scanning microscope.
Transfection, Western Blotting, and mGpr126-NTFΔGPS Binding Assay.
H9c2 or HeLa cells (ATCC) were transfected with plasmid DNA using FuGENE or X-tremeGENE 9 DNA transfection reagents (Roche), following the manufacturer’s instructions. Protein was isolated from the cells 48 h after transfection. Briefly, cells in medium were scraped off from the plates, pelleted (300 × g, 3 min, 4 °C), and washed with PBS. Cell pellets were lysed by sonication (six pulses of 5 s each) in lysis buffer [Cell Signaling buffer plus protease inhibitor mix (Roche) and 1 mM PMSF (Sigma)]. Supernantants after centrifugation (17,000 × g, 10 min, 4 °C) were resolved in NuPAGE Novex Bis⋅Tris Gels (Invitrogen), blotted, and analyzed with anti-GFP (1:500; Novus) and anti-Gapdh (1:4,000; Sigma) antibodies. To produce mGpr126-NTFΔGPS-mFc (aa1 to aa770), HEK-293 cells were transfected with plasmid DNA using FuGENE HD DNA transfection reagents (Promega). Cell culture medium was replaced with serum-free DMEM (Sigma) 24 h posttransfection and incubated for another 48 h. Conditioned medium was harvested, centrifuged (600 × g, 15 min), and passed through a 0.45-µm filter. Then, mGpr126-NTFΔGPS-mFc was purified with the Protein A-Sepharose 4 Fast Flow kit (Amersham Biosciences), dialyzed with 10 mM Tris⋅HCl (pH 7.3) containing 100 mM NaCl and 10 mM CaCl2, and concentrated with a Vivaspin column (30-kDa cutoff, 1,640 × g, 15 min, 4 °C; Satorius). For the mGpr126-NTFΔGPS binding assay, freshly prepared 10-µm-thick E11.5 mouse embryo cryosections were permeabilized (PBS/0.5% Triton-X100, 10 min), incubated with purified concentrated protein (overnight, 4 °C), and washed in 0.1% Nonidet P-40/PBS. Subsequently, sections were incubated with rabbit anti-sarcomeric alpha actinin antibody (1:100, 2 h, RT; Abcam), and mFc-tagged mGpr126-NTFΔGPS and primary immune complexes were detected with ALEXA 488- and ALEXA 594-conjugated antibodies (1:200; Molecular Probes), respectively. To detect DNA, DAPI (Sigma) (0.5 mg/mL PBS) was used.
Video Capture, Processing, and Kymograph Analysis.
For zebrafish, 2-dpf embryos were positioned in 1% low-melting-point agarose in E3 medium, and movies were captured with a Sony HDR-SR12 camcorder under a Leica DM6000 B microscope. Movies of beating hearts from E11.25 mouse embryos were taken immediately after isolation from the mother. All movies were formatted by Wondershare or iSkysoft video converter and analyzed with the Image J Kymograph, which allows the measurement of the velocity of moving structures in image time series. The y axis of the kymograph is a time axis. A line in the kymograph parallel to the y axis means there is no movement. A line or peak parallel to the x axis represents one heartbeat.
Data Analysis.
Data are expressed as the mean ± SEM of at least three independent experiments. Statistical significance of differences was evaluated by one-way ANOVA followed by Bonferroni’s post hoc test (GraphPad Prism) or Fisher’s exact test (Microsoft Excel). P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank M. Müller-Boche for excellent fish care; I. Hauck-Schmalenberger, P. Freund, S. Rühl, G. Magdowski, and C. Lichtenberg for technical support; H.-H. Lin for the modified pCDNA3.1 vector; I. Liebscher for the mouse Gpr126 clone; W. S. Talbot for the zebrafish mutant line gpr126st49; and S. Offermanns, D. Stainier, and B. Jungblut for critical reading of the manuscript. We specially thank RIKEN for providing the heterozygous Gpr126 mice for this study. This work was supported by the Alexander von Humboldt Foundation Sofja Kovalevskaja Award (to F.B.E.), the NIH Grant R01 NS079445 (to K.R.M.), and the Faculty Development Program of Government College University Faisalabad (A.S.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304837110/-/DCSupplemental.
References
- 1.Bjarnadóttir TK, et al. The human and mouse repertoire of the adhesion family of G-protein-coupled receptors. Genomics. 2004;84(1):23–33. doi: 10.1016/j.ygeno.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 2.Moriguchi T, et al. DREG, a developmentally regulated G protein-coupled receptor containing two conserved proteolytic cleavage sites. Genes Cells. 2004;9(6):549–560. doi: 10.1111/j.1356-9597.2004.00743.x. [DOI] [PubMed] [Google Scholar]
- 3.Prömel S, et al. The GPS motif is a molecular switch for bimodal activities of adhesion class G protein-coupled receptors. Cell Rep. 2012;2(2):321–331. doi: 10.1016/j.celrep.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaur B, Brat DJ, Devi NS, Van Meir EG. Vasculostatin, a proteolytic fragment of brain angiogenesis inhibitor 1, is an antiangiogenic and antitumorigenic factor. Oncogene. 2005;24(22):3632–3642. doi: 10.1038/sj.onc.1208317. [DOI] [PubMed] [Google Scholar]
- 5.Koh JT, et al. Extracellular fragment of brain-specific angiogenesis inhibitor 1 suppresses endothelial cell proliferation by blocking alphavbeta5 integrin. Exp Cell Res. 2004;294(1):172–184. doi: 10.1016/j.yexcr.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 6.Silva JP, Lelianova V, Hopkins C, Volynski KE, Ushkaryov Y. Functional cross-interaction of the fragments produced by the cleavage of distinct adhesion G-protein-coupled receptors. J Biol Chem. 2009;284(10):6495–6506. doi: 10.1074/jbc.M806979200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waller-Evans H, et al. The orphan adhesion-GPCR GPR126 is required for embryonic development in the mouse. PLoS ONE. 2010;5(11):e14047. doi: 10.1371/journal.pone.0014047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monk KR, Oshima K, Jörs S, Heller S, Talbot WS. Gpr126 is essential for peripheral nerve development and myelination in mammals. Development. 2011;138(13):2673–2680. doi: 10.1242/dev.062224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monk KR, et al. A G protein-coupled receptor is essential for Schwann cells to initiate myelination. Science. 2009;325(5946):1402–1405. doi: 10.1126/science.1173474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glenn TD, Talbot WS. Analysis of Gpr126 function defines distinct mechanisms controlling the initiation and maturation of myelin. Development. 2013;140(15):3167–3175. doi: 10.1242/dev.093401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyons DA, et al. erbb3 and erbb2 are essential for schwann cell migration and myelination in zebrafish. Curr Biol. 2005;15(6):513–524. doi: 10.1016/j.cub.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 12.Pogoda HM, et al. A genetic screen identifies genes essential for development of myelinated axons in zebrafish. Dev Biol. 2006;298(1):118–131. doi: 10.1016/j.ydbio.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 13.Liu J, et al. A dual role for ErbB2 signaling in cardiac trabeculation. Development. 2010;137(22):3867–3875. doi: 10.1242/dev.053736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimes BW, Brandt BL. Properties of a clonal muscle cell line from rat heart. Exp Cell Res. 1976;98(2):367–381. doi: 10.1016/0014-4827(76)90447-x. [DOI] [PubMed] [Google Scholar]
- 15.Rosa P, Barr FA, Stinchcombe JC, Binacchi C, Huttner WB. Brefeldin A inhibits the formation of constitutive secretory vesicles and immature secretory granules from the trans-Golgi network. Eur J Cell Biol. 1992;59(2):265–274. [PubMed] [Google Scholar]
- 16.Tanudji M, Hevi S, Chuck SL. Improperly folded green fluorescent protein is secreted via a non-classical pathway. J Cell Sci. 2002;115(Pt 19):3849–3857. doi: 10.1242/jcs.00047. [DOI] [PubMed] [Google Scholar]
- 17.Choong K, Clarke JT, Cutz E, Pollit RJ, Olpin SE. Lethal cardiac tachyarrhythmia in a patient with neonatal carnitine-acylcarnitine translocase deficiency. Pediatr Dev Pathol. 2001;4(6):573–579. doi: 10.1007/s10024001-0101-7. [DOI] [PubMed] [Google Scholar]
- 18.Hansson A, et al. A switch in metabolism precedes increased mitochondrial biogenesis in respiratory chain-deficient mouse hearts. Proc Natl Acad Sci USA. 2004;101(9):3136–3141. doi: 10.1073/pnas.0308710100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walters AM, Porter GA, Jr, Brookes PS. Mitochondria as a drug target in ischemic heart disease and cardiomyopathy. Circ Res. 2012;111(9):1222–1236. doi: 10.1161/CIRCRESAHA.112.265660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldberg IJ, Trent CM, Schulze PC. Lipid metabolism and toxicity in the heart. Cell Metab. 2012;15(6):805–812. doi: 10.1016/j.cmet.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. National Institutes of Health (1996) Guide for the Care and Use of Laboratory Animals (Natl Inst Health, Bethesda), DHHS Publ No (NIH) 85-23.
- 22.Patra C, et al. Nephronectin regulates atrioventricular canal differentiation via Bmp4-Has2 signaling in zebrafish. Development. 2011;138(20):4499–4509. doi: 10.1242/dev.067454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D’Amico L, Scott IC, Jungblut B, Stainier DY. A mutation in zebrafish hmgcr1b reveals a role for isoprenoids in vertebrate heart-tube formation. Curr Biol. 2007;17(3):252–259. doi: 10.1016/j.cub.2006.12.023. [DOI] [PubMed] [Google Scholar]
- 24. Westerfield M (1993). The Zebrafish Book: a Guide for the Laboratory Use of Zebrafish (Brachydanio rerio). Univ of Oregon Press, Eugene, OR.
- 25.Stacey M, et al. EMR4, a novel epidermal growth factor (EGF)-TM7 molecule up-regulated in activated mouse macrophages, binds to a putative cellular ligand on B lymphoma cell line A20. J Biol Chem. 2002;277(32):29283–29293. doi: 10.1074/jbc.M204306200. [DOI] [PubMed] [Google Scholar]
- 26.Patra C, et al. Silk protein fibroin from Antheraea mylitta for cardiac tissue engineering. Biomaterials. 2012;33(9):2673–2680. doi: 10.1016/j.biomaterials.2011.12.036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.