Significance
Replacing traditional methods for genetic testing of inheritable disorders with next-generation sequencing (NGS) will reduce the cost of genetic testing and increase the information available for the patients. NGS will become an invaluable resource for the patient and physicians, especially if the sequencing information is stored properly and reanalyzed as bioinformatics tools and annotations improve. NGS is still at the early stages of development, and it is full of false-positive and -negative results and requires infrastructure and specialized personnel to properly analyze the results. This paper will explain our experience with an adult population, our bioinformatics analysis, and our clinical decisions to assure that our genetic diagnostics were accurate to detect carrier status and serious medical conditions in our volunteers.
Keywords: molecular medicine, disease prediction, whole exome sequencing
Abstract
Next-generation sequencing (NGS) is commonly used for researching the causes of genetic disorders. However, its usefulness in clinical practice for medical diagnosis is in early development. In this report, we demonstrate the value of NGS for genetic risk assessment and evaluate the limitations and barriers for the adoption of this technology into medical practice. We performed whole exome sequencing (WES) on 81 volunteers, and for each volunteer, we requested personal medical histories, constructed a three-generation pedigree, and required their participation in a comprehensive educational program. We limited our clinical reporting to disease risks based on only rare damaging mutations and known pathogenic variations in genes previously reported to be associated with human disorders. We identified 271 recessive risk alleles (214 genes), 126 dominant risk alleles (101 genes), and 3 X-recessive risk alleles (3 genes). We linked personal disease histories with causative disease genes in 18 volunteers. Furthermore, by incorporating family histories into our genetic analyses, we identified an additional five heritable diseases. Traditional genetic counseling and disease education were provided in verbal and written reports to all volunteers. Our report demonstrates that when genome results are carefully interpreted and integrated with an individual’s medical records and pedigree data, NGS is a valuable diagnostic tool for genetic disease risk.
Sequencing the whole genome of patients with genetic disorders has become reality since the sequencing of the first individual human in 2007 (1). Further advances in massively parallel DNA sequencing are reducing the price of sequencing an entire genome or exome. The quality and speed of sequencing and analyzing a personal genome are improving at an unprecedented pace, making possible the introduction of next-generation sequencing (NGS) into the clinic on a research basis (2–7). Advancements in NGS have stimulated international research initiatives to identify genetic links to rare disorders in children, with an average diagnostic success of 20–25% and the discovery of new disease–gene associations (8–12).
The rapidly increasing number of aging adults in our society will place unprecedented demands on the health care system. To provide adults with a healthy longevity we need to develop a system to identify genetic risk and apply early intervention on pathology progression. In this report, we decided to sequence the whole exomes of a healthy adult cohort of 81 volunteers and evaluate the value of applying NGS in combination with medical history and pedigree data. In this report we plan to address three main questions. (i) What genetic discoveries need to be provided to the volunteers? (ii) What is the practical value of delivering this information to volunteers? (iii) What are the challenges and barriers to the adoption of this powerful technology into medical practice?
The individual genetic reports yield helpful medical risk information, suggesting that population sequencing of asymptomatic adults may prove to be valuable and useful. We provided to the participants, under our institutional review board, genetic risk findings from the analyses and genetic counseling to discuss their results.
Results
Categories of Variants to Report to Patients.
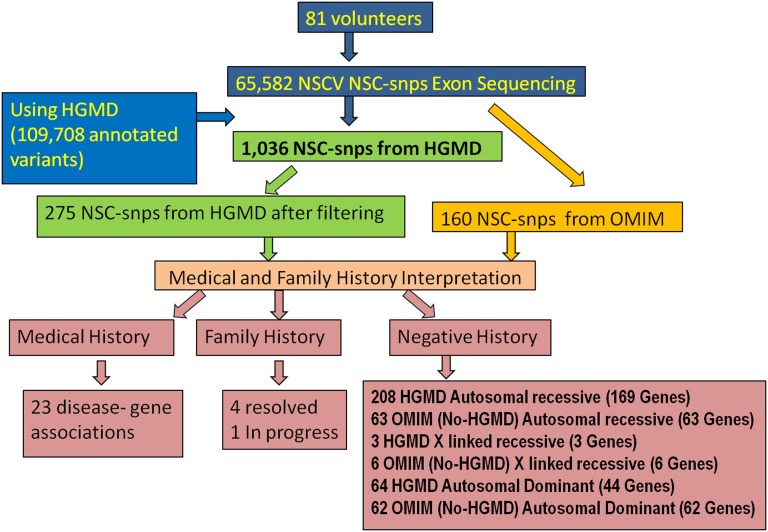
Variants obtained from our workflow (described in Fig. 1) were reported using three categories. Our first variant category consists of variants identified in an individual where the alleles are found in Human Genome Mutation Database (HGMD) (13, 14) and labeled disease-causing mutations (DM). These alleles also were required to be rare [<1% allele frequency in 6,500 exomes from the National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project (15) and the 1,000 Genomes Project Genomes (16, 17)] and predicted to be damaging to protein function by two of three predictions algorithms [Polyphen 2.0 (18), Sift (19–24), and MutationTaster (25)] using Database of Human Non-synonymous SNVs and their functional predictions and annotations (dbNSFP) (26) as described in Fig. 2. The genome sequence data of each volunteer were reviewed and interpreted, taking into account personal medical history, a three-generation pedigree with family history of diseases, and bioinformatics analysis. The medical history of each volunteer in this cohort was rich with detail because each had a private physician used for annual examinations, and in some cases, disease therapy. Fig. 3 summarizes the results of our pipeline: we recruited 81 nonrelated volunteers and sequenced their genomic DNA using exome sequencing. We detected 65,582 unique nonsynonymous coding variants (nscv). Every nscv was interrogated for human inherited disease mutations using the HGMD (13, 14) database from Biobase (DM category consisting of 109,708 variations). We were able to detect 1,036 HGMD (13, 14) DM variations. After using the filters described in Fig. 2, the number was reduced to 275 pathogenic variants. We identified in our cohort 208 autosomal recessive (AR) alleles (169 genes), 64 autosomal dominant (AD) alleles (44 genes), and three X-linked recessive (XLR) alleles (3 genes). These data resulted in an average of 3.5 disease allele reports per volunteer.
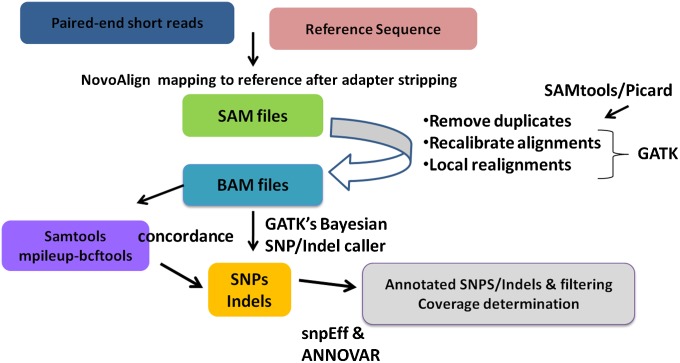
Fig. 1.
Workflow for processing NGS data. Raw sequencing data are aligned against the reference sequence using Novoalign software from NovoCraft. SAM files are preprocessed using SAMtools and Picard to create BAM files and remove duplicates. The Genome Analysis Toolkit (GATK) is then used to recalibrate the alignments, perform local realignments, and identify SNPs and indels. Finally, SnpEff and ANNOVAR are used to annotate variants.
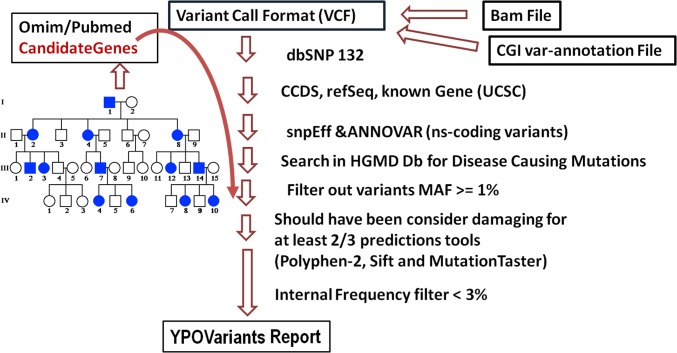
Fig. 2.
Pipeline to generate variants reports. Every variant in the variant call format file is annotated using spnEff and ANNOVAR; nonsynonymous coding variants are annotated using the commercial version of the HGMD database. (Left) Our selection of variants by the creation of a personalized candidate gene list using medical history and family history for each volunteer. Mutations with a minor allele frequency of >1% are removed using frequencies from the NHLBI exome sequencing project (ESP), 1,000 Genomes Project. Variants that are consider benign by two of three predictions tools are removed (using dbNSFP). Finally, we remove variants that are present in our cohort more than three times.
Fig. 3.
Summary of results. The flowchart provides the number of variants from each step of the pipeline described in Fig. 2.
The approach for a second category of variants consisted of creating a personalized list of candidate genes from Online Mendelian Inheritance in Man (OMIM) (27, 28) known to be associated with the disorders reported in the medical literature. We detected 131 alleles (131 genes) using this approach. Each one of these variants provided a potential causation for the volunteer’s disorders. Each one of the variations obtained from this approach passed our stringent pipeline. This approach added on average another 2.0 disease alleles per volunteer report.
The third approach used a family history to create a personalized list of candidate genes from OMIM (27, 28), and as before, we compared our list of candidate genes with the disorders reported in the family history.
Before reporting an allele to the volunteer, we reviewed the original publications that support the pathogenicity of all of the alleles (HGMD) and/or the evidence associating the gene with the disorder (OMIM). At this time, all three abovementioned categories of investigation were reported in full recognition; some would be found to be non–disease-producing alleles as databases improve and functional assays complement informatics predictions. We have updated clinical reports as these data emerged and counseled the patients on the options for reducing or eliminating the disease risk.
Disease Genes Identified in the Cohort.
Table S1 summarizes our disease associations. Matching personal medical records to personal genome reports was informative. We elected to report findings as disease–gene associations instead of reporting findings as diagnostic because we did not included in our study traditional “surrogate markers” (analytes, proteins, and imaging) for the confirmation of a disease diagnosis. We considered potentially causative findings to be those mutations that are predicted to be damaging in addition to being reported in either HGMD (13, 14) or OMIM (27, 28) databases. These mutations are considered to be “need to know” and are reported to volunteers. There was identification of associations for vascular disease and/or hypercholesterolemia in five individuals related to LDL receptor (LDLR) alleles. LDLR mutations are causative of early onset autosomal dominant coronary artery disease (CAD) and manifest hypercholesterolemia (29, 30). Three individuals were taking statins related to their hypercholesterolemia. Two individuals were not under care but had history of personal hypercholesterolemia and in one case a son with hypercholesterolemia.
There were four volunteers detected with risk genes for diabetes mellitus (31–34). Two of the individuals were under therapy for diabetes 2, whereas two additional volunteers had elevated fasting blood sugars and were being followed by their physicians for further analytes measurements. There were two individuals with morbid obesity (body mass index of 32 and 37 kg/m2) who carried an MC4R allele associated with pediatric obesity and rare heterozygotic adults (35, 36). Two ophthalmologic disease/gene associations were identified. The childhood brittle corneal syndrome type 1 occurred in a volunteer who had undergone successful corneal transplant and carried a putative compound heterozygosity in ZNF469 (37). One volunteer was under care for macular dystrophy and carried an ABCA4 allele (38). One sterile male volunteer was found to have an insertion in gene USP26 (known to be responsible for infertility in men) (39). Associations for melanoma and breast cancer were identified. The two patients with melanoma carried different gene allele associations: GRIN2A and BAG4 (40–42). Two volunteers diagnosed with breast cancer had different allele associations in BRCA2 (43, 44). Single cases of early onset prostate (LRP2) (45) and follicular thyroid cancer (TPR) cancer were identified (46, 47). A volunteer with nonsyndromic deafness was found to have risk alleles in two genes associated with autosomal dominant (AD) deafness and had a three-generation positive family history of deafness (48). In each case, the volunteer was instructed to inform their physician and was requested to confirm the genomic allele identification in a Clinical Laboratory Improvement Amendments (CLIA)-certified laboratory, even when each reported allele had been sequenced twice in independent studies. The finding provided information for personal and family risk counseling not possible before gene association.
Incorporation of Three-Generation Pedigrees into the Genetic Analyses.
The three-generation pedigree medical information was analyzed to identify those volunteer families who warranted additional genetic study. Table S2 lists those genetic disorders identified by pedigree/familial medical history. In each case, the volunteer was counseled for the family risk and encouraged to contact at risk family members who may benefit from focused genetic studies. Three of the families have reported that they have had their familial genetic diagnosis resolved at this time [paraganglioma (49), Prader-Willi syndrome (50, 51), and ankylosing spondylitis (AS) (52)]. One additional family is under study [Tourette syndrome (53)]. Additional familial disease risks were identified by history for atrial fibrillation (AR), bicuspid aortic valve (BAV), dyslexia (AR), Fabry’s (XLR), gall stones (AD), and myotonic dystrophy (anticipation AD). Success with this approach was productive but not universally accepted because disease/gene resolution requires interaction with interested and motivated family members.
Table S3 provides a sampling of the recessive risk alleles. They constitute the majority of the observed alleles. Of the 160 offspring of the 81 volunteers, no children were affected with these disorders. All volunteers indicated their families were complete, and thus, no spousal genetic studies were recommended, but information was proposed to be provided to reproductive age descendants. Many of the genes identified are part of prenatal carrier screens and/or newborn state-sponsored screening programs [phenylketonuria, maple syrup urine disease, cystic fibrosis, Niemann-Pick disease, Gaucher disease, factor V Leiden thrombophilia, medium-chain acyl-CoA dehydrogenase (MCAD) deficiency]. Undoubtedly, NGS will expand the number of nonunreported disease alleles and scope of genes studied for couples in the pregnancy setting. The Beyond Batten Disease Foundation of Austin, TX (54), has this goal.
Table S4 shows that a category of high concern was the identification of XLR disease risk alleles among our female volunteers. One volunteer had an affected son (isolated case) with Fabry disease that was diagnosed before our study. There were four disease alleles identified, each listed in HGMD (13, 14). There was no family history of these disorders found in the three-generation pedigree of each. All were counseled to have their test confirmed and daughters studied in a CLIA-certified laboratory given the high disease risk (50% for men). Three men in our study had alleles predicted from the OMIM (27, 28) disease database to be causative for cutis laxus, Duchenne muscular dystrophy, congenital nystagmus, and hemophilia A, illustrating the challenge of predicting damaging mutations bioinformatically. None had the disorders. Counseling and family study were individualized for each disease risk. Volunteers were made aware of database errors in the reports.
Tables S5–S10 provide a third category that is very problematic, the AD group. The allele identification is as previously described, but counseling is more difficult because of variation in severity and time onset. For this age group of volunteers, the interest was high because disease prevention was frequently expressed as a goal in the face-to-face counseling meetings. A poststudy survey also reflected this objective. We focused in this paper on the three major causes of death in the United States: cancer, cardiovascular disease, and neurodegenerative disease. In our analysis of each volunteer, we reviewed the genomic and family data.
Table S5 lists the breast cancer risk results. There were 12 volunteers found to have breast cancer risk alleles of genes BRCA1, BRCA2, PALB2, RAD51C, and RAD50. Two volunteers with BRCA2 risk alleles were diagnosed with breast cancer. One man carried a premature chain termination mutation and has a first-degree relative with breast cancer (50s). A third volunteer had a frame shift mutation (high-risk allele) but not found to have breast cancer. All alleles were predicted to be damaging. Eight volunteers had first-degree relatives with breast cancer, whereas four had a negative family history of disease. All were advised to seek confirmation via a CLIA-certified laboratory. One patient with an HGMD (13, 14) allele was confirmed but predicted to be “neutral” by a commercial laboratory. All were counseled regarding the need for regular mammograms and gynecological examinations and were requested to inform their physician of this research risk allele identification.
Table S6 displays the colon cancer alleles. There was no disease incidence of colon cancer in this group with the exception of one volunteer with a positive dysplastic polyp biopsy. Five volunteers had a positive family history of colon cancer. Five volunteers had no family history of disease. All were advised to obtain confirmatory CLIA-certified laboratory diagnosis and advise their physician of the research allele identification. Of the 10 volunteers, many had undergone colonoscopy as part of their health care.
Table S7 includes all of the remaining type of cancers. Two volunteers diagnosed with melanomas were found to have different disease gene risk alleles. We identified 10 volunteers with prostate risk alleles. One volunteer reported a diagnosis of prostate cancer at age 55 while the other nine volunteers reported no familial history of the disease. Genetic counseling for cancer risk required the greatest counseling time. The concepts of the two-hit hypothesis (55) and “somatic mutations” (56) were difficult to grasp for the volunteers, even when we discussed the subject in great detail during the education session. All volunteers were provided information regarding standard of practice approaches for early detection of the respective cancer.
Table S8 lists all of the affected volunteers with cardiomyopathies (57). Five volunteers had a medical history of cardiac dysrhythmia with identified risk alleles. One younger (50s) volunteer had first-degree relatives requiring pacemakers and carried two risk alleles. Three volunteers had either stent placements or bypass procedures related to CAD. Each was in their 70s.
Table S9 lists the 11 volunteers who had no apparent disease but had a positive family history of tachycardia, sudden death, and CAD and carried risk alleles. We provide this experience to broaden alertness to both genetic causation and risk of disease for adult-onset cardiovascular disease (58). Of the alleles listed in Tables S8 and S9, 13 alleles were found in HGMD (13, 14). We advised volunteers to inform their physicians of these results for their long-term clinical care.
In Table S10, we listed the results for adult-onset neurodegenerative diseases. Our findings were limited but of high interest to the cohort. It was frequently asked by volunteers if they had Alzheimer’s risk. We summarize our findings for Alzheimer’s and Parkinson risk alleles (59, 60). The genes included APOE, APP, PSEN1, MAPT, EIF461, GBA, GIGYF2, LRRK2, PARK2, PM20D1, and SNCA. There were nine volunteers with HGMD (13, 14) listed risk alleles. Of these, two had a positive family history of Parkinson disease and one with Alzheimer’s disease. One of the PARK2 alleles occurred in a volunteer who provided a history of three second-degree relatives in a sibship affected with disease. The reminder had no family history of either disease. There were 25 alleles predicted to be damaging. One is a frameshift allele. None of these volunteers had a family history of disease.
Discussion
Exome Sequencing Is Limited.
The full spectrum of disease mutation identification is not satisfied by exome sequencing alone because large deletions, copy number variations (CNVs), and triplet repeats are not reliably identified at this time. Furthermore, exon capture relies on probe design. For example, the discovery of the MAGEL2 mutation in our Prader-Willi patient was made using whole genome sequencing (WGS) from complete genomics and missed by exome capture because of high GC content (51). The accuracy of coding allele identifications was, however, quite high and thus of great utility as a genome screening approach. CGI (61) sequencing produced higher coverage than exome sequencing; data for CNV, large deletions, and regulatory elements will have utility as we analyze previously labeled “junk” DNA for disease causation (62). There is also the issue of our limited knowledge of disease alleles within the databases. One of our biggest challenges for the interpretation of human genomes is the lack of gene annotations and the errors in databases. Our knowledge base for human disorders is small. There are only ∼100,000 pathogenic variants in the HGMD (13, 14) database and a fraction of them have errors. If we do not use annotated variants but instead gene annotations as our source of information, we can calculate the fraction of knowledge that we can use at this time. For example, the number of genes associated with human disorders reported by HGMD (13, 14), OMIM (27, 28), UniProtKB (63), Gene Atlas (64), etc. is 4,622. From the 4,622 genes, only 1,955 genes have high-quality data because they are part of the GeneTest (65) database. GeneTest (65) is a database originally created by the National Center for Biotechnology Information to track all of the laboratories worldwide that offer a genetic test for a gene. With this information, we know that the fraction of genes that we can use for the interpretation of a human genome of a successful high-quality whole exome or whole genome dataset is ∼7–18% when using the high confidence set of 1,955 genes or a set of 4,622 genes. Despite these limitations, this report documents the utility for disease associations and risk.
During the last few years, the field of NGS has developed a large number of tools that make it easier to handle the analysis of reads, variant calling, functional prediction, and annotation (66). There are also large publicly available datasets of healthy individuals that can be used as controls that can be used to remove technology specific errors or filter out common polymorphisms. As we begin to use whole genome sequencing at an increasing depth, we are discovering more variants, so these public datasets are becoming increasingly important for quality control and filtering of variants in smaller projects. One of the main limitations is the lack of access to public and private genome and exome variants. There are thousands of datasets, but the majority are inaccessible to the scientific community. We recognize the existence of the 1,000 Genomes project, the NHLBI Exome Sequencing Project (ESP), Exome variant server, and the 69 sets of whole genomes from CGI (15–17, 67). However, we need larger datasets from very carefully phenotyped patients to assist in the interpretation of the variants in our patients. The million genome project of the US Department of Veterans Affairs (68) has the potential to provide such data, as well as private health plans considering adaptation of genome sequencing.
Genetic Discoveries Provided to Volunteers.
There are several approaches to disclose the results to volunteers. Groups like Patel et al. use the statistics and epidemiology approach in reporting the polygenic risk assessment using common SNPs that have been previous associated with genetic disorders from genome-wide association studies (69). The PGP-10 project uses an automated tool or Genome Environment Trait Evidence (GET-Evidence) system, wich is a system that is collaboratively edited (70). For this project, we decided to focus on reporting only high-quality variants that are rare in the population and considered damaging by two of three commonly used predictions algorithms. In addition, the variant has to be either reported in HGMD under category DM or the gene has to have been previously associated with a genetic disorder (OMIM). The group of volunteers consisted of adults with complete medical and family history so we personalized the reports as described in Fig. 2 to specifically try to identify molecular explanations for the maladies reported in their medical or family history. This approach generated reports that were easy to explain and accepted by the patients during the genetic counseling session.
Medical Histories and Family Pedigrees Complement Sequencing Results.
The utility of genome data was significantly enhanced when integrating standard medical care features of personal and family disease diagnosis. The significant number of 23 disease associations in all likelihood represents a bias of our volunteers to seek answers to their personal disease history. This observation may hold a key to how we obtain maximal use of genome sequencing—sequence the disease index cases. Our experience would suggest a high value for that utilization. This approach has been clearly documented to be successful for pediatric genetic disorders but not exploited for adult-onset disease. The practical value of this study is summarized in Tables S1 and S2 and fell into two general categories: (i) new knowledge of the genetic risk and heritability for themselves and family; and (ii) options for therapy (CAD) or imaging (cancer) for personal and extended family care. By using the medical and family history, we were able to clarify the genetic risk in 6 of the 81 cases. One of the cases yielded a new discovery of a gene associated with Prader-Willi syndrome, which is described in another paper (51).
Prenatal vs. Adult Genetic Screening.
The technology and this report beg the question of whether we are prepared to offer adult disease risk screening. Currently, prenatal and newborn screening for a selected set of frequently occurring disease alleles (not genome sequencing) is a standard of practice. There are questions that deserve medical and ethical review before adult screening becomes a standard of practice. First, for reproductive and newborn diagnosis, typically only actionable childhood diseases are explored, which respects the future autonomy of the child and preserves her right to an open future (71, 72). Because adult screening decisions would be made by an autonomous individual for her own health decisions, broader conceptions of utility, including personal utility, need to be considered (73). It is a clear and simple decision to provide patients with actionable genetic information from a WES study; on the other hand, it is challenging and it raises a difficult ethical question to decide what to do with incidental genetic findings that are not actionable and could lead to physiological distress to the patient (e.g. APO-E for Alzheimer disease). Despite this ethical dilemma our group of volunteers elected to receive information even if the genetic information might not be actionable. Only 3% of the volunteers were uncertain about receiving nonactionable information (SI Poststudy Survey).
Volunteer Response to Clinical Reports.
From our poststudy survey, we found that 72% of the responders reported speaking with their physician about their results. This raises important questions about whether nongeneticists are adequately prepared to counsel patients based on WES results and whether such follow-up will lead to iatrogenic harm or unjustified use of health care resources (74). Twenty-five percent reported changing their behaviors because of the results, which is surprising given that previous reports found no significant behavior change resulting from adult risk screening in a direct-to-consumer setting (75). Despite that all of the participants were clearly informed that their results originated from two independent sequencing experiments and that we advised them to have their results clinically validated in a CLIA-certified laboratory, 78% reported that they did not have the results confirmed. This low percentage of confirmatory results from the volunteers raises the question of whether it is sufficient to counsel research participants to have results clinically confirmed or if investigators should be required to confirm results before disclosure.
It was apparent for some volunteers that they were seeking information related to familial diseases. Resolution of these questions required family member interest and motivation because, in all cases, we had sequenced the nonrisk family member. We followed up each case with a referral to a qualified genetics program with diagnostic capacity for the suspected genetic disease.
Our efforts to analyze cancer, cardiovascular, neurodegenerative, and obesity/diabetes risk were successful but needed considerable education/counseling to avoid confusion over risk vs. diagnosis. Second, there are standard of care options for those with risk alleles for cancer, cardiovascular disease, and diabetes for disease modification or early diagnosis. Thus, sequencing serves as a new screening risk detection approach toward the objective of improved health. It is expected that genomic studies will increase surveillance studies (e.g., colonoscopy, gynecologic examinations, mammograms, cardiovascular markers and scanning studies) but has the possibility of more precisely identifying the patients who may benefit from disease prevention surveillance.
The area of adult-onset neurologic disorders is an increasing concern worldwide as our population ages, thus exposing disease incidence not seen earlier. The genetic disease discoveries are limited. Confirmatory diagnostics such as image analysis and biomarkers/surrogate markers are just emerging, and prevention therapeutic options are nonexistent. Although one might question the utility of screening for these disorders at this time, the experience with Huntington disease (76) screening taught valuable lessons on how to proceed with studying and counseling families at risk. Furthermore, there are new therapeutic trials in disease prevention for Alzheimer’s (58) and Parkinson disease based on the genetic cause of disease. These clinical trials use genetic diagnosis to select participants, which is also a successful approach in cancer drug development (77–79).
Barriers to the Adoption of Genetic Screening via Sequencing.
Although the above comments would present the case for the value of adult genetic screening via whole genome sequencing, there are major issues to be addressed. In our opinion, the least is sequencing technology and cost. Bioinformatics focused on the practical extraction of medical relevant/actionable data are a challenge. We relied heavily on HGMD alleles for “need to know” information to patients. This approach is flawed in three ways: (i) databases contain errors; (ii) highly validated disease databases are scattered, private, and limited; and (iii) the future will provide more disease risk alleles by sequencing than by patient reports in the literature. Our current limitation for interpretation of a genome is not the quality of the data of the coverage of the genome but our disease knowledge database. R. Cotton’s Human Variome Project (62) together with Beijing Genome Institute are proposing to create a highly validated disease allele database.
New technological advances such as structure-based prediction of protein–protein interactions on a genome wide scale (80), 3D structure of protein active and contact sites (81), high-throughput functional assays of damaging alleles (81–83), and new approaches that combine analytes, metabolomics and genetic information from a single individual (84) are just a few examples of the new technologies that will help us to generate better interpretation of genomic data.
The delivery of the genome risk information will need to be carried out by a new cadre of physicians and counselors skilled in medicine, genetics, and education/counseling. These experts will need to integrate into medical care as well as has been done for newborn screening, prenatal diagnosis, and newborn genetic disease diagnosis.
The approach of adult screening is in its early phase but from our data appears very promising. We conclude that the genomic study of adults deserves intensified effort to determine if “need to know” genome information has the utility for improved quality of health for our aging population.
Materials and Methods
The oversight of this research was under two institutional review boards: (i) HSC-IMM-08-0641 (University of Texas Health Science Center at Houston) and (ii) H-30710 (Baylor College of Medicine).
Cohort Description.
The cohort consists of members and spouses in the Houston Chapter of the Young Presidents Organization (YPO) (85). The entire description of the cohort can be found in SI Materials and Methods.
WES Sequencing.
Standard NGS was performed using illumina HighSeq; an extended explanation can be found in SI Materials and Methods.
Sequencing Analysis.
Fig. 1 illustrates our pipeline, and Fig. 2 describes our pipeline to detect known pathogenic variations. Additional details can be found in SI Materials and Methods.
Counseling.
Genome counseling was conducted by a board-certified internist and medical geneticist by both individual meetings and two written summaries over a period of 12 mo. Additional information can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by the Cullen Foundation for Higher Education and the Governing Board of the Greater Houston Community Foundation. The funding organizations made the awards to the University of Texas Health Science Center at Houston and Baylor College of Medicine. C.T.C. was the principal investigator of both grants.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1315934110/-/DCSupplemental.
References
- 1.Levy S, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5(10):e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bamshad MJ, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12(11):745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 3.Tabor HK, Berkman BE, Hull SC, Bamshad MJ. Genomics really gets personal: How exome and whole genome sequencing challenge the ethical framework of human genetics research. Am J Med Genet A. 2011;155A(12):2916–2924. doi: 10.1002/ajmg.a.34357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lander ES. Genome-sequencing anniversary. The accelerator. Science. 2011;331(6020):1024. doi: 10.1126/science.1204037. [DOI] [PubMed] [Google Scholar]
- 5.Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470(7333):187–197. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- 6.Biesecker LG, Burke W, Kohane I, Plon SE, Zimmern R. Next-generation sequencing in the clinic: Are we ready? Nat Rev Genet. 2012;13(11):818–824. doi: 10.1038/nrg3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hennekam RC, Biesecker LG. Next-generation sequencing demands next-generation phenotyping. Hum Mutat. 2012;33(5):884–886. doi: 10.1002/humu.22048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Anonymous Finding of rare disease genes in Canada (forge Canada). Available at http://www.genomebc.ca/portfolio/projects/health-projects/finding-of-rare-disease-genes-in-canada-forge-canada/. Accessed September 19, 2013.
- 9.Gahl WA, et al. The National Institutes of Health undiagnosed diseases program: Insights into rare diseases. Genet Med. 2012;14(1):51–59. doi: 10.1038/gim.0b013e318232a005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gahl WA, et al. The National Institutes of Health undiagnosed diseases program: Insights into rare diseases. Genet Med. 2012;14(1):51–59. doi: 10.1038/gim.0b013e318232a005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gahl WA, Tifft CJ. The NIH undiagnosed diseases program: Lessons learned. JAMA. 2011;305(18):1904–1905. doi: 10.1001/jama.2011.613. [DOI] [PubMed] [Google Scholar]
- 12.Koenekoop RK, et al. Finding of Rare Disease Genes (FORGE) Canada Consortium Mutations in NMNAT1 cause Leber congenital amaurosis and identify a new disease pathway for retinal degeneration. Nat Genet. 2012;44(9):1035–1039. doi: 10.1038/ng.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stenson PD, et al. (2012) The Human Gene Mutation Database (HGMD) and its exploitation in the fields of personalized genomics and molecular evolution. Curr Protocol Bioinform 39:1.13.1-1.13.20. [DOI] [PubMed]
- 14.Stenson PD, et al. The Human Gene Mutation Database: 2008 update. Genome Med. 2009;1(1):13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Anonymous NHLBI exome sequencing project (ESP) exome variant server. Available at http://evs.gs.washington.edu/EVS/. Accessed September 19, 2013.
- 16.Clarke L, Zheng-Bradley X, et al. The 1000 Genomes Project: Data management and community access. Nat Methods. 2012;9(5):459–462. doi: 10.1038/nmeth.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abecasis GR, et al. 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 20. Sim NL, Kumar P, et al. (2012) SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res 40(Web Server issue):W452–W457. [DOI] [PMC free article] [PubMed]
- 21.Hu J, Ng PC. Predicting the effects of frameshifting indels. Genome Biol. 2012;13(2):R9. doi: 10.1186/gb-2012-13-2-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11(5):863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu Rev Genomics Hum Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- 25.Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7(8):575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 26.Liu X, Jian X, Boerwinkle E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat. 2011;32(8):894–899. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Anonymous Online Mendelian Inheritance in man OMIM. Available at http://omim.org. Accessed September 19, 2013.
- 28. Anonymous NCBI OMIM Online Mendelian Inheritance in Man. Available at http://www.ncbi.nlm.nih.gov/omim. Accessed September 19, 2013.
- 29.Huijgen R, Kindt I, Defesche JC, Kastelein JJ. Cardiovascular risk in relation to functionality of sequence variants in the gene coding for the low-density lipoprotein receptor: A study among 29,365 individuals tested for 64 specific low-density lipoprotein-receptor sequence variants. Eur Heart J. 2012;33(18):2325–2330. doi: 10.1093/eurheartj/ehs038. [DOI] [PubMed] [Google Scholar]
- 30.Boekholdt SM, et al. Association of LDL cholesterol, non-HDL cholesterol, and apolipoprotein B levels with risk of cardiovascular events among patients treated with statins: A meta-analysis. JAMA. 2012;307(12):1302–1309. doi: 10.1001/jama.2012.366. [DOI] [PubMed] [Google Scholar]
- 31.Waeber G, et al. The gene MAPK8IP1, encoding islet-brain-1, is a candidate for type 2 diabetes. Nat Genet. 2000;24(3):291–295. doi: 10.1038/73523. [DOI] [PubMed] [Google Scholar]
- 32.Mosca L, et al. Genetic variability of the fructosamine 3-kinase gene in diabetic patients. Clin Chem Lab Med. 2011;49(5):803–808. doi: 10.1515/CCLM.2011.133. [DOI] [PubMed] [Google Scholar]
- 33.da Silva Xavier G, et al. Per-arnt-sim (PAS) domain-containing protein kinase is downregulated in human islets in type 2 diabetes and regulates glucagon secretion. Diabetologia. 2011;54(4):819–827. doi: 10.1007/s00125-010-2010-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MacDonald PE, Rorsman P. Per-arnt-sim (PAS) domain kinase (PASK) as a regulator of glucagon secretion. Diabetologia. 2011;54(4):719–721. doi: 10.1007/s00125-011-2072-1. [DOI] [PubMed] [Google Scholar]
- 35.O’Rahilly S. Human genetics illuminates the paths to metabolic disease. Nature. 2009;462(7271):307–314. doi: 10.1038/nature08532. [DOI] [PubMed] [Google Scholar]
- 36.van den Berg L, et al. Melanocortin-4 receptor gene mutations in a Dutch cohort of obese children. Obesity (Silver Spring) 2011;19(3):604–611. doi: 10.1038/oby.2010.259. [DOI] [PubMed] [Google Scholar]
- 37.Al-Owain M, Al-Dosari MS, Sunker A, Shuaib T, Alkuraya FS. Identification of a novel ZNF469 mutation in a large family with Ehlers-Danlos phenotype. Gene. 2012;511(2):447–450. doi: 10.1016/j.gene.2012.09.022. [DOI] [PubMed] [Google Scholar]
- 38.Fritsche LG, et al. A subgroup of age-related macular degeneration is associated with mono-allelic sequence variants in the ABCA4 gene. Invest Ophthalmol Vis Sci. 2012;53(4):2112–2118. doi: 10.1167/iovs.11-8785. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J, et al. [Polymorphism of Usp26 correlates with idiopathic male infertility] Zhonghua Nan Ke Xue. 2012;18(2):105–108. [PubMed] [Google Scholar]
- 40.Wei X, et al. NISC Comparative Sequencing Program Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet. 2011;43(5):442–446. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Howell PM, Jr, Li X, Riker AI, Xi Y. MicroRNA in melanoma. Ochsner J. 2010;10(2):83–92. [PMC free article] [PubMed] [Google Scholar]
- 42.Xi Y, et al. Global comparative gene expression analysis of melanoma patient samples, derived cell lines and corresponding tumor xenografts. Cancer Genomics Proteomics. 2008;5(1):1–35. [PMC free article] [PubMed] [Google Scholar]
- 43.Nelson HD, Huffman LH, Fu R, Harris EL. U.S. Preventive Services Task Force Genetic risk assessment and BRCA mutation testing for breast and ovarian cancer susceptibility: Systematic evidence review for the U.S. Preventive Services Task Force. Ann Intern Med. 2005;143(5):362–379. doi: 10.7326/0003-4819-143-5-200509060-00012. [DOI] [PubMed] [Google Scholar]
- 44. Anonymous National Cancer Institute BRCA1 and BRCA2. Available at http://www.cancer.gov/cancertopics/factsheet/Risk/BRCA. Accessed September 19, 2013.
- 45.Holt SK, et al. Association of megalin genetic polymorphisms with prostate cancer risk and prognosis. Clin Cancer Res. 2008;14(12):3823–3831. doi: 10.1158/1078-0432.CCR-07-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frank-Raue K, et al. Prevalence and clinical spectrum of nonsecretory medullary thyroid carcinoma in a series of 839 patients with sporadic medullary thyroid carcinoma. Thyroid. 2013;23(3):294–300. doi: 10.1089/thy.2012.0236. [DOI] [PubMed] [Google Scholar]
- 47.Mak HH, et al. Oncogenic activation of the Met receptor tyrosine kinase fusion protein, Tpr-Met, involves exclusion from the endocytic degradative pathway. Oncogene. 2007;26(51):7213–7221. doi: 10.1038/sj.onc.1210522. [DOI] [PubMed] [Google Scholar]
- 48.Ruel J, et al. Impairment of SLC17A8 encoding vesicular glutamate transporter-3, VGLUT3, underlies nonsyndromic deafness DFNA25 and inner hair cell dysfunction in null mice. Am J Hum Genet. 2008;83(2):278–292. doi: 10.1016/j.ajhg.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Hulsteijn LT, Dekkers OM, Hes FJ, Smit JW, Corssmit EP. Risk of malignant paraganglioma in SDHB-mutation and SDHD-mutation carriers: A systematic review and meta-analysis. J Med Genet. 2012;49(12):768–776. doi: 10.1136/jmedgenet-2012-101192. [DOI] [PubMed] [Google Scholar]
- 50.Jiang Y, Tsai TF, Bressler J, Beaudet AL. Imprinting in Angelman and Prader-Willi syndromes. Curr Opin Genet Dev. 1998;8(3):334–342. doi: 10.1016/s0959-437x(98)80091-9. [DOI] [PubMed] [Google Scholar]
- 51.Schaaf CP, et al. Truncating mutations of MAGEL2 cause Prader-Willi phenotypes and autism. Nat Genet. 2013 doi: 10.1038/ng.2776. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rashid T, Ebringer A. Gut-mediated and HLA-B27-associated arthritis: An emphasis on ankylosing spondylitis and Crohn’s disease with a proposal for the use of new treatment. Discov Med. 2011;12(64):187–194. [PubMed] [Google Scholar]
- 53.Deng H, Gao K, Jankovic J. The genetics of Tourette syndrome. Nat Rev Neurol. 2012;8(4):203–213. doi: 10.1038/nrneurol.2012.26. [DOI] [PubMed] [Google Scholar]
- 54. Anonymous Beyond Batten Disease Foundation. Available at http://beyondbatten.org/. Accessed September 19, 2013.
- 55.Knudson AG. Hereditary cancer: Two hits revisited. J Cancer Res Clin Oncol. 1996;122(3):135–140. doi: 10.1007/BF01366952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nik-Zainal S, et al. Breast Cancer Working Group of the International Cancer Genome Consortium The life history of 21 breast cancers. Cell. 2012;149(5):994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alcalai R, Seidman JG, Seidman CE. Genetic basis of hypertrophic cardiomyopathy: from bench to the clinics. J Cardiovasc Electrophysiol. 2008;19(1):104–110. doi: 10.1111/j.1540-8167.2007.00965.x. [DOI] [PubMed] [Google Scholar]
- 58.Rader DJ, Cohen J, Hobbs HH. Monogenic hypercholesterolemia: New insights in pathogenesis and treatment. J Clin Invest. 2003;111(12):1795–1803. doi: 10.1172/JCI18925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin I, Dawson VL, Dawson TM. Recent advances in the genetics of Parkinson’s disease. Annu Rev Genomics Hum Genet. 2011;12:301–325. doi: 10.1146/annurev-genom-082410-101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Selkoe DJ. Preventing Alzheimer’s disease. Science. 2012;337(6101):1488–1492. doi: 10.1126/science.1228541. [DOI] [PubMed] [Google Scholar]
- 61. Anonymous Complete Genomics Inc. Available at http://www.completegenomics.com. Accessed September 19, 2013.
- 62. Anonymous Human variome project. Available at http://www.humanvariomeproject.org. Accessed September 19, 2013.
- 63. Anonymous UniProtKB. Available at http://www.uniprot.org/uniprot. Accessed September 19, 2013.
- 64. Anonymous Gene atlas. Available at http://www.geneatlas.org/gene/main.jsp. Accessed September 19, 2013.
- 65. Anonymous Genetic Testing Registry (GeneTests). Available at http://www.genetests.org. Accessed September 19, 2013.
- 66. Anonymous SEQanswers. Available at http://seqanswers.com. Accessed September 19, 2013.
- 67. Anonymous 69 genomes data. Available at http://www.completegenomics.com/public-data/69-Genomes/. Accessed September 19, 2013.
- 68. Anonymous The million veteran program. Available at http://www.va.gov/opa/pressrel/pressrelease.cfm?id=2090. Accessed September 19, 2013.
- 69.Patel CJ, et al. Whole genome sequencing in support of wellness and health maintenance. Genome Med. 2013;5(6):58. doi: 10.1186/gm462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ball MP, et al. A public resource facilitating clinical use of genomes. Proc Natl Acad Sci USA. 2012;109(30):11920–11927. doi: 10.1073/pnas.1201904109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. American Academy of Pediatrics Committee on Bioethics (2001) Ethical issues with genetic testing in pediatrics. Pediatrics 107(6):1451–1455. [DOI] [PubMed]
- 72.Davis DS. Genetic dilemmas and the child’s right to an open future. Hastings Cent Rep. 1997;27(2):7–15. [PubMed] [Google Scholar]
- 73. Wolf SM, Lawrenz FP, et al. (2008) Managing incidental findings in human subjects research: Analysis and recommendations. J Law Med Ethics 36(2):219–248. [DOI] [PMC free article] [PubMed]
- 74.McGuire AL, Burke W. An unwelcome side effect of direct-to-consumer personal genome testing: Raiding the medical commons. JAMA. 2008;300(22):2669–2671. doi: 10.1001/jama.2008.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bloss CS, Schork NJ, Topol EJ. Effect of direct-to-consumer genomewide profiling to assess disease risk. N Engl J Med. 2011;364(6):524–534. doi: 10.1056/NEJMoa1011893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wexler NS. Huntington’s disease: Advocacy driving science. Annu Rev Med. 2012;63:1–22. doi: 10.1146/annurev-med-050710-134457. [DOI] [PubMed] [Google Scholar]
- 77.Caskey CT. The drug development crisis: Efficiency and safety. Annu Rev Med. 2007;58:1–16. doi: 10.1146/annurev.med.58.042705.124037. [DOI] [PubMed] [Google Scholar]
- 78.Caskey CT. Using genetic diagnosis to determine individual therapeutic utility. Annu Rev Med. 2010;61:1–15. doi: 10.1146/annurev-med-011209-132719. [DOI] [PubMed] [Google Scholar]
- 79.Miller G. Alzheimer’s research. Stopping Alzheimer’s before it starts. Science. 2012;337(6096):790–792. doi: 10.1126/science.337.6096.790. [DOI] [PubMed] [Google Scholar]
- 80.Zhang QC, et al. Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature. 2012;490(7421):556–560. doi: 10.1038/nature11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Edwards AM, Bountra C, Kerr DJ, Willson TM. Open access chemical and clinical probes to support drug discovery. Nat Chem Biol. 2009;5(7):436–440. doi: 10.1038/nchembio0709-436. [DOI] [PubMed] [Google Scholar]
- 82. Namovic MT, Jarvis MF, Donnelly-Roberts D (2012) High throughput functional assays for P2X receptors. Curr Protocol Pharmacol Jun;Chapter 9:Unit 9.15. [DOI] [PubMed]
- 83.Trivedi S, Liu J, Liu R, Bostwick R. Advances in functional assays for high-throughput screening of ion channels targets. Expert Opin Drug Discov. 2010;5(10):995–1006. doi: 10.1517/17460441.2010.513377. [DOI] [PubMed] [Google Scholar]
- 84.Suhre K, et al. CARDIoGRAM Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477(7362):54–60. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Anonymous Membership criteria YPO. Available at http://www.ypo.org/join-ypo/. Accessed September 19, 2013.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.