Significance
Coprolites provide unique insights into the plant taxa consumed over a discrete time period by extinct herbivores and have typically been used to reconstruct the diets of single herbivore species. Through ancient DNA, pollen, and plant macrofossil analyses of 51 coprolites deposited by four species of extinct herbivore (the large avian moa of New Zealand) in a single rock shelter, we show the potential for coprolites to also resolve broader paleoecological questions around niche partitioning of extinct sympatric herbivore species and prehistoric herbivore community structure. Such information can help in our understanding of late Quaternary ecosystem functioning and the ecological consequences of prehistoric extinctions, as well as helping to inform rewilding efforts.
Abstract
Knowledge of extinct herbivore community structuring is essential for assessing the wider ecological impacts of Quaternary extinctions and determining appropriate taxon substitutes for rewilding. Here, we demonstrate the potential for coprolite studies to progress beyond single-species diet reconstructions to resolving community-level detail. The moa (Aves: Dinornithiformes) of New Zealand are an intensively studied group of nine extinct herbivore species, yet many details of their diets and community structuring remain unresolved. We provide unique insights into these aspects of moa biology through analyses of a multispecies coprolite assemblage from a rock overhang in a montane river valley in southern New Zealand. Using ancient DNA (aDNA), we identified 51 coprolites, which included specimens from four sympatric moa species. Pollen, plant macrofossils, and plant aDNA from the coprolites chronicle the diets and habitat preferences of these large avian herbivores during the 400 y before their extinction (∼1450 AD). We use the coprolite data to develop a paleoecological niche model in which moa species were partitioned based on both habitat (forest and valley-floor herbfield) and dietary preferences, the latter reflecting allometric relationships between body size, digestive efficiency, and nutritional requirements. Broad ecological niches occupied by South Island giant moa (Dinornis robustus) and upland moa (Megalapteryx didinus) may reflect sexual segregation and seasonal variation in habitat use, respectively. Our results show that moa lack extant ecological analogs, and their extinction represents an irreplaceable loss of function from New Zealand’s terrestrial ecosystems.
Large herbivores are integral components of terrestrial ecosystems, playing a major role in shaping the structure and composition of vegetation communities and providing vital ecosystem functions such as seed dispersal (1, 2). Over the last 50,000 y, large herbivore populations on most landmasses have either gone extinct or been decimated (3), often with significant consequences for local vegetation communities (1, 4, 5). Paleodietary data for extinct large herbivores, and an understanding of how prehistoric large herbivore communities were structured, are therefore important for assessing the full ecological impact of Late Quaternary extinctions. Such information can provide a baseline for attempts to restore the lost ecological functions once provided by extinct herbivores (6). Coprolites provide the strongest potential resource for directly studying the diets and community structure of extinct species. Coprolites of extinct large herbivores have been found in dry caves across the world (7–12), but for most, the herbivore’s identity has only been inferred from associated skeletal remains. Identification of large herbivore coprolites using ancient DNA (aDNA) has thus far been restricted to moa (9) and a limited number of ground sloth (Nothrotheriops shastensis) samples (13). However, this approach offers the possibility of detecting hidden diversity within a coprolite deposit (9), thereby allowing the diets and community structure of entire guilds of sympatric large herbivores to be reconstructed.
The New Zealand moa (Aves: Dinornithiformes) provide an ideal group on which to test the suitability of coprolite analyses to reconstruct the paleoecology of an extinct large herbivore guild. The recent (13th century AD) human settlement of New Zealand (14) means moa fossil remains (including coprolites, bones, feathers, and eggshell) are relatively common and well preserved. The moa were the most diverse radiation within the New Zealand endemic avifauna and played a key role as the largest herbivores in New Zealand’s prehuman terrestrial ecosystems. Nine species, in six genera, occurred in New Zealand (15), with body masses ranging from tens to hundreds of kilograms (16). Due to their recent extinction, moa biology remains highly relevant to understanding current ecosystems in New Zealand (17, 18). Three main themes persist in scientific literature relating to moa ecology and are often mirrored in studies of extinct herbivore paleoecology around the world. First, there has been significant debate about the role of moa herbivory in the evolution of plant structural (e.g., wire-plant syndrome) and developmental (e.g., heteroblasty) characteristics in the New Zealand flora (19–25). Second, there is increasing interest in the legacy that moa extinction may have had on indigenous vegetation community structure and composition (18, 26) based on a hypothesis that reduced herbivory may have caused broadleaf shrubs to flourish in forest understorys, resulting in severely reduced regeneration of podocarp conifers (27). Third, the degree to which introduced herbivores, particularly ungulates, are acting as ecological replacements for moa is the subject of ongoing debate (17, 18, 27, 28).
All three themes rely on a good understanding of moa diets and their ecological niches. However, despite studies on numerous gizzard content samples and coprolites (29–33), many aspects of moa diets remain poorly understood. The exact nature of dietary differences that allowed the apparent sympatry of certain moa species is one such mystery. Multiple moa species [up to six (34)] often occur together within single fossil bone deposits (35–37). Radiocarbon dating and stratigraphic contexts at many sites suggest that the species coexisted, rather than being temporally segregated (35, 38–40). Significant interspecific variation in body size and cranial and beak morphology (41), points to strong dietary niche partitioning between moa species and seems to provide a mechanism to explain the apparent sympatry. However, dietary analyses using only gizzard content samples and coprolites have indicated moa were generalist herbivores and have only provided limited evidence for interspecific diet differences (9, 29, 31–33). We believe this apparent lack of niche partitioning reflects inadequate reconstruction of diets in these studies. Currently, studies that have examined the diets of sympatric moa species (9, 29, 31) have used only plant macrofossils (seeds, twigs, and leaf fragments). This approach may significantly bias the representation of plant remains towards those that are resistant to gizzard mastication and digestion (e.g., twigs and small hard seeds); lead to a significant proportion of samples without identifiable remains (9); and leave much of the diet unexplained.
Recent advances in paleovegetation studies have shown how increased information can be obtained through combining a number of complimentary proxies (42–44). Although such multiproxy analyses of coprolites can provide a more detailed and nuanced understanding of diet (42), thus far, they have only been applied to single species at any one site. Here, we analyze the pollen, plant aDNA, and plant macrofossil content of a multispecies coprolite assemblage excavated from a single rock overhang at Daley’s Flat, in the Dart River valley, South Island, New Zealand (9). The 0.8-km2 forested sampling locality contains many avalanche boulders, which are commonly >20 m diameter. An earlier study at this location used moa aDNA analysis to attribute the depositor species for 20 coprolites, but reported only macrofossil contents from these (9). In this study, we reexcavated the site and collected an additional 31 coprolites, which, along with the 20 species-identified coprolites from the earlier study, were analyzed for plant macrofossil, pollen, and plant aDNA. This combination of three independent diet proxies and species attribution applied to all 51 coprolites has allowed us to examine the habitats and diets of four sympatric moa species (representing four genera and three families) in greater detail than has previously been attempted for any extinct group of sympatric herbivores. From these, we construct a model of niche partitioning by four of New Zealand’s large prehistoric avian herbivores.
Results
Moa aDNA.
Using aDNA analysis, we attributed 51 coprolites (Table S1) to four moa species: South Island giant moa (Dinornis robustus), n = 21; upland moa (Megalapteryx didinus), n = 19; heavy-footed moa (Pachyornis elephantopus), n = 8; little bush moa (Anomalopteryx didiformis), n = 3. For species known to exhibit phylogeographic structuring (M. didinus, P. elephantopus, and A. didiformis), the coprolite aDNA sequences all fell within the appropriate local clades (Fig. S1). Variation in haplotypes and radiocarbon ages (Table S1) suggest a minimum of 22 individuals are represented by the coprolite assemblage: D. robustus, n ≥ 9; M. didinus, n ≥ 8; P. elephantopus, n ≥ 3; A. didiformis, n ≥ 2. A. didiformis has not previously been reported from coprolite and bone assemblages at the site (9).
Plant aDNA.
Moa diets differed significantly based on coprolite plant aDNA assemblages (Fig. 1) [A. didiformis excluded due to low sample size: permutational multivariate ANOVA (PERMANOVA) F′2,18 = 3.83; P < 0.0001]. Notably abundant plant taxa (mean percentage of DNA sequences per coprolite of a particular moa species > 20%) include Nothofagus (D. robustus), Rubiaceae (A. didiformis, D. robustus, and M. didinus), and Polygonaceae (P. elephantopus and M. didinus) (Dataset S1).
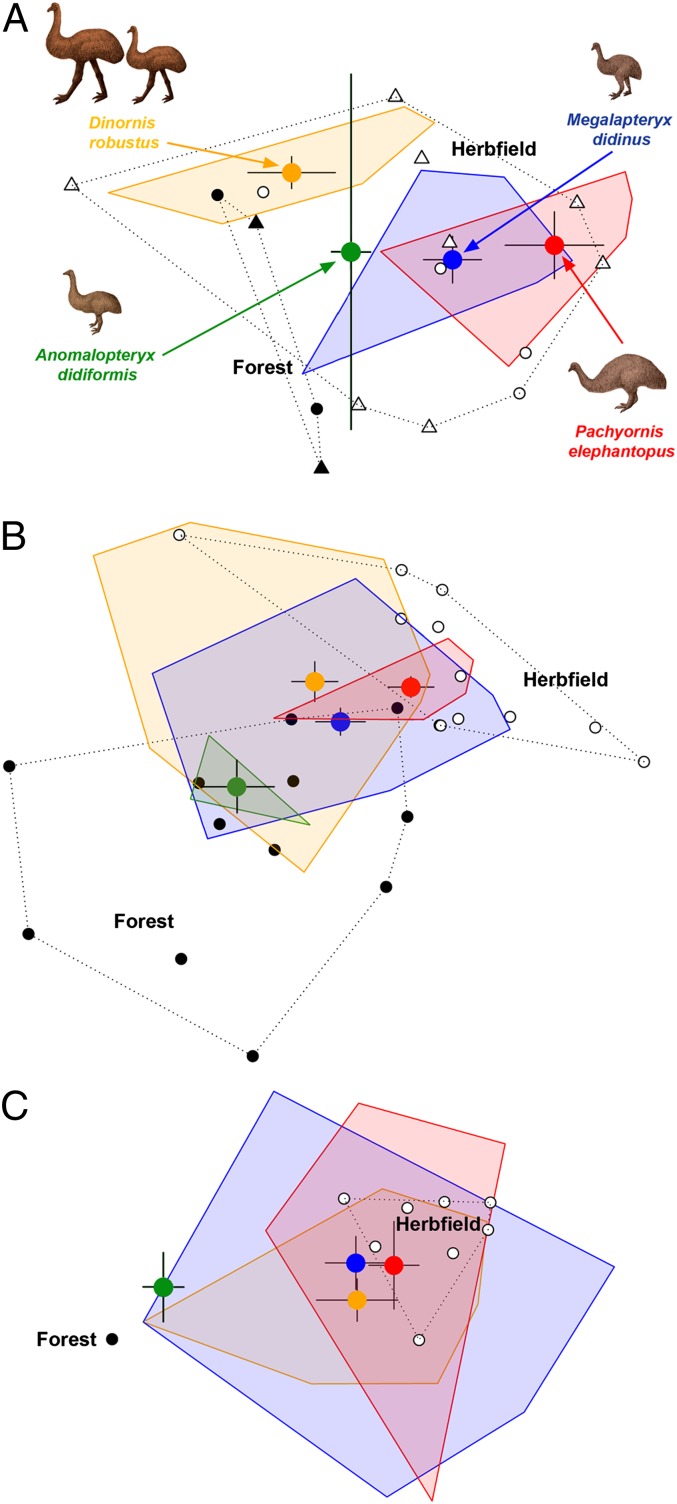
Fig. 1.
Ordination hulls for Dart River Valley moa coprolites based on (A) plant aDNA assemblages (n = 23 coprolites); (B) pollen assemblages (n = 51 coprolites); and (C) plant macrofossil assemblages (n = 33 coprolites). Colors represent the same moa species across all plots. Centroids for moa coprolites are shown (large colored circles) with SE bars. A habitat gradient is indicated by the position of plant taxa that are either restricted to forest (black circle) or open herbfield/grassland (white circle) habitats. In A, plant taxa that are probably forest (black triangles) and probably herbfield/grassland (white triangle) are also shown.
DNA sequences from the coprolites resolved 26 plant taxa (8 to genera; 2 to subfamily; 9 to family; 5 to higher orders; 2 unranked clades; Dataset S1; Table S2). There was significant agreement (83.5% of sequences) in the identities provided by the two analytical methods (phylogenetic analysis based on GenBank plant sequences vs. sequences from a local species dataset), with identities of just 0.76% of sequences in disagreement between methods. For 15.8% of the sequences, one method was able to assign an identity where the other method did not. Overall, the plant taxa identified by aDNA were consistent with the coprolite pollen and macrofossil assemblages and the local extant flora (45).
Pollen.
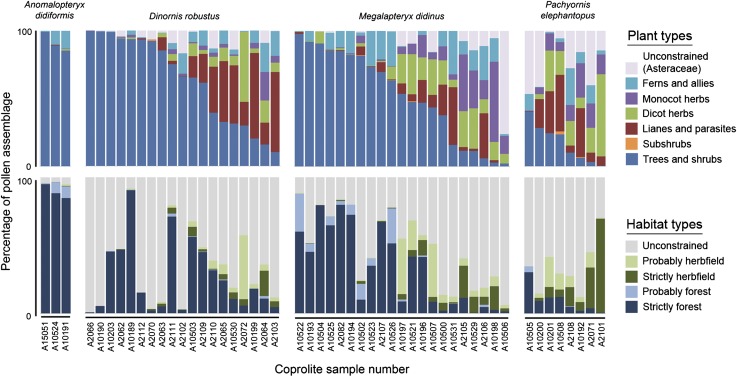
Fifty-eight pollen and spore taxa were identified in the coprolites, with a mean diversity of 14.6 taxa per coprolite (Dataset S2). The assemblage included plant taxa diagnostic of both forest and open valley-floor herbfield habitats (Fig. 2). Moa diets also differed significantly based on coprolite pollen assemblages (Fig. 1) (A. didiformis excluded due to low sample size: PERMANOVA F′2,45 = 2.17; P = 0.0181).
Fig. 2.
Summary moa coprolite pollen assemblages from Daley’s Flat rock avalanche, Dart River Valley.
Plant Macrofossils.
At least 19 plant taxa were identified by macrofossil remains in the coprolites (Dataset S3). No new taxa in addition to those previously reported from Dart River moa coprolites (9) were identified. The coprolite plant macrofossil assemblages did not differ significantly between moa species (Fig. 1C) (A. didiformis excluded due to low sample size: PERMANOVA F′2,31 = 1.01; P = 0.4424).
Radiocarbon Dating.
Seven radiocarbon dates were obtained for coprolites and fall within the range of ages previously obtained for moa bones and coprolites from the deposit (39). In total, 20 of the coprolites have been radiocarbon dated (Table S3) and show that all four moa species inhabited the area at the same time between ∼1000 and 1400 AD (Fig. S2). The earlier limit of this range reflects the formation of the rock avalanche deposit, which correlates with a known rupture of the Alpine Fault (39). The later limit of the range reflects the local extinction of moa.
Discussion
Moa Niche Partitioning.
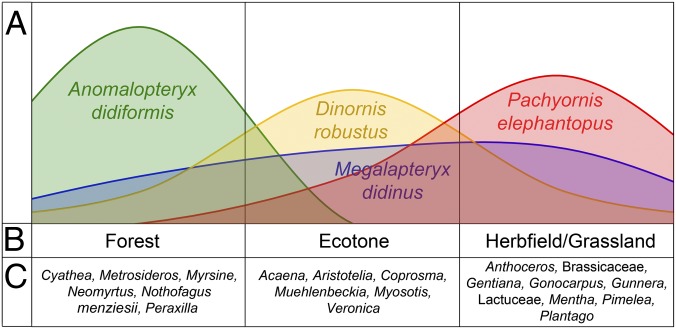
We propose an ecological niche model for the four sympatric moa species at Daley’s Flat based on the content of the analyzed coprolites and previous studies of moa diet. In the model, moa are partitioned based on both habitat and dietary preferences. The most significant niche partitioning was observed between A. didiformis and P. elephantopus. The ordination hulls representing the habitat and dietary niche spaces of these species did not overlap for any of the proxies (Figs. 1 and 3). In both the plant aDNA and pollen assemblages, A. didiformis was biased toward forest species and P. elephantopus toward open herbfield plants (Figs. 1 and 3). Niche partitioning between these two moa species is supported by evidence from other studies (16, 46, 47). Pollen and macrofossil analyses of two coprolites (46) from near Lake Wakatipu, ∼60 km south of Daleys Flat, indicate that A. didiformis was a forest herbivore, whereas pollen analysis of two P. elephantopus coprolites from the Kawarau and Clutha River gorges, ∼80 and 110 km south of Daley’s Flat, respectively (47), indicate that this species was predominantly a herbfield grazer. The different habitat preferences of A. didiformis and P. elephantopus may explain the rarity of bone deposits containing contemporaneous remains of these two species. Their coexistence at Daley’s Flat was facilitated by the close proximity of both forest and herbfield communities.
Fig. 3.
Habitat niche partitioning between four sympatric moa species in the Dart River Valley, based on pollen content of coprolites. (A) Proportion of coprolites assigned to habitat class. (B) Habitat class. (C) Representative pollen types.
D. robustus and M. didinus had much broader habitat preferences than A. didiformis and P. elephantopus. We suggest that there are alternative explanations for the broad habitat ranges of D. robustus and M. didinus.
Genetic analyses of bones have revealed extreme reversed sexual dimorphism in D. robustus, with females weighing up to almost three times as much as males (48, 49). In browsing herbivores, ecological segregation is significantly correlated with increasing sexual body size dimorphism (50). It would be expected that an unbiased coprolite deposit would include specimens from both males and females (as both are present in the bone assemblage from Daley’s Flat) and therefore capture this ecological segregation. Multiple attempts to amplify nuclear DNA from the Dart River moa coprolites following established methods (48, 49) were unsuccessful for all but one specimen (female P. elephantopus, A10192), likely reflecting a much lower abundance of nuclear DNA in coprolites compared with ancient bone. However, allometry of digestive organ capacity and food retention times mean that large herbivores are able to use lower quality food items than small herbivores, which have shorter retention times and require foods of higher nutritional value (51). Based on these relationships, we suggest that D. robustus coprolites with a higher proportion of forest plant species may have been deposited by females, whereas those toward the herbfield end of the spectrum represent males. Our hypothesis that female D. robustus browsed within forests is supported by 10 gizzard content samples from the Pyramid Valley swamp (northeast South Island), which are dominated by macrofossils of forest trees and shrubs (29). The Pyramid Valley bone deposit is heavily biased toward female individuals [95% of skeletons determined by nuclear DNA (52)]. Nothofagus aDNA sequences were prevalent in some of the D. robustus coprolites from the Dart. Twigs and leaves of these trees would be considered low-quality food for herbivores (53, 54). Foliar nutrient differentiation may explain the sympatry of moa species within the forest understory; the large female D. robustus fed on low-quality fibrous plant matter, and the smaller A. didiformis and M. didinus browsed a wide range of leaves from fast-growing deciduous, parasitic, and nitrogen-fixing plants that have higher leaf nitrogen and protein concentrations (54) (as indicated by aDNA of Loranthaceae, Malvaceae, Coriaria, and Polygonaceae; Dataset S1).
With no discernible sexual dimorphism in M. didinus (55), we would not expect ecological segregation between sexes to be as pronounced as in D. robustus. The relatively small body size of M. didinus may have allowed it to coexist with the larger moa species in each habitat type by allowing it to use different food resources. Alternatively, adaptations that allowed specialization in montane and alpine zones (16) may have included seasonal variation in habitat use. An example of this can be seen in an extant New Zealand avian herbivore, the South Island takahe (Porphyrio hochstetteri), which feeds in alpine grasslands during summer and in forest understories during winter when snow cover prevents access to the grasses (56).
Our proposed model of moa partitioning based on habitat and dietary preferences explains the observed differences and similarities between species diets and reflects preferred feeding sites. Within habitats, we suggest moa species were partitioned based on different body sizes, digestive capabilities, nutritional requirements, and dominant types of plant tissues consumed. However, the home ranges of each moa species probably encompassed a broader span of habitat types than were present in their preferred feeding territories. Both forest and herbfields on frosty river terraces are in close proximity at Daley’s Flat, and all four moa species probably ranged between both of these habitat types to some degree. Forest would have provided shelter during adverse weather, protection from predation by their only predator, Haast’s eagle (Aquila moorei), and sites for roosting and nesting. Further testing of the model could include development of methods to increase the potential for nuclear aDNA to be retrieved from the coprolites to distinguish the sex of the depositing bird and studies to determine useful local indicators of seasonality (e.g., short period flowering taxa in pollen assemblages).
Ecological Consequences of Moa Extinction.
Late Quaternary extinctions of megafauna are likely to have had major consequences for terrestrial ecosystems across the globe (1–3). The extinction of moa could have affected New Zealand ecosystems through altering vegetation composition and structure, regeneration patterns, and fire frequency (17). By defining the niches of different moa species, we can progress beyond treating moa as a single entity and examine the possible ecological implications of extinction for each species separately. Our results provide evidence for three sympatric guilds of moa in the Dart River Valley. Extinction of the grazer guild (dominated by P. elephantopus) would have resulted in reduced seed dispersal for herbs with small indehiscent fruits (e.g., Pratia and Wahlenbergia) and altered the composition of turf communities (57). Loss of the tall browser guild (including D. robustus) may have caused changes to forest understories and canopy openness in short forest. Extinction of the short browser guild (including A. didiformis and M. didinus) may have had the most effect on increasing the density of forest understories (through postextinction flourishing of previously browsed tree and shrub species) and regeneration patterns (through reduced consumption of seedlings and increased shading). Testing these hypotheses within modern vegetation communities would be complicated due to forest modification by introduced mammalian herbivores. The full diversity of plant taxa represented in the moa coprolites is now largely restricted to “island gardens,” which exist on the large boulders that are sufficiently tall and steep-sided to exclude browsing ungulates. A detailed examination of the ecological change that followed moa extinction will likely require high stratigraphic resolution analyses of forest soil profiles (18).
It has been suggested that some extant herbivores could act as ecological surrogates for moa (18). Detailed information on the ecology of extinct species is critical for determining appropriate ecological surrogates. Our study shows that no extant herbivore taxa (including other large ratites such as emu and ostrich) could replace the entire range of feeding ecologies exhibited by moa. The extinction of moa resulted in the irreplaceable loss of ecological function in New Zealand’s ecosystems.
Coprolites and Herbivore Paleoecology.
Our study demonstrates the potential for multiproxy studies of coprolite assemblages to reconstruct the community structure of extinct herbivores, particularly for closely-related taxa where ecological niche differences may not be obvious. Three important points should be considered when using coprolite assemblages to study extinct herbivores. First, the importance of using aDNA to identify herbivore taxa, rather than inferring this solely from associated skeletal remains, was shown by the identification of coprolites from a moa species not represented in the bone assemblage (A. didiformis). Second, with larger sample sizes, the ability to assess the full dietary breadth of generalist herbivore species is greatly improved. Third, by combining information obtained using different diet proxies, it is possible to refine the likely identity of the most important diet taxa. For example, Polygonaceae aDNA sequences occurred in 73.9% of the analyzed coprolite samples (including M. didinus, P. elephantopus, and D. robustus), and were the dominant sequences obtained from some M. didinus and P. elephantopus coprolites (Dataset S1). Muehlenbeckia pollen was common in coprolites from M. didinus, P. elephantopus, and D. robustus (Dataset S2) and provides a likely generic identity for the Polygonaceae aDNA. Seeds of Muehlenbeckia axillaris were identified in some coprolites. This species is currently common throughout the Daley’s Flat herbfield and is the taxa most likely represented in the coprolite content. Similarly, Rubiaceae aDNA sequences occurred in 82.6% of the analyzed samples and were present in coprolites from all four moa species. Coprosma seeds and pollen in the coprolites provide a likely generic identity for the Rubiaceae sequences. Importantly, our study further highlights the benefit of using multiple proxies to reveal information about diets from coprolites (42). As was reported by Wood et al. (9), there was no significant difference between moa species based on coprolite macrofossil content. However, we showed here that plant aDNA and pollen analyses can tease apart important interspecific differences.
Our study demonstrated that multiproxy analyses of coprolites can provide a detailed picture of extinct herbivore diets, including information on specific diet items and niche breadth. It also shows that coprolite analyses can be used to reconstruct lost community structure, in addition to single species diet reconstructions. Increased application of multiproxy analyses and sample identification using DNA will mean coprolite analyses can begin to shed new light on Late Quaternary large herbivore communities across the globe.
Materials and Methods
Study Site and Samples.
The coprolites were excavated from beneath an overhanging rock on the Daley's Flat rock avalanche deposit (44.542°S, 168.385°E) in the Dart River Valley, South Island, New Zealand. Wood et al. (9, 39) provided a summary of the geology, vegetation, and climate of the area. Fifty-one coprolites were analyzed. These coprolites included 20 coprolites identified to species and sampled for plant macrofossil in a previous study (9) (sample numbers A2062–A2112; these were analyzed here for pollen and plant aDNA assemblages), and 31 excavated in July 2010 (sample numbers A10189–A10531; these were analyzed for pollen, plant macrofossils, and aDNA).
Subsampling.
Newly excavated coprolites were subsampled in a still-air perspex hood, the surfaces of which were cleaned between samples with 10% (wt/vol) decon, 2% (wt/vol) bleach, and ethanol. Approximately 2–3 mm was removed from the outer surface of each coprolite by scraping with a scalpel blade, and the freshly exposed surface was UV irradiated for 15 min to destroy any potential exogenous environmental contamination. The irradiated coprolites were bisected (Fig. S3), and subsamples (0.4–1.0 g) for pollen and aDNA analysis were taken from the interior. Additional interior samples for radiocarbon dating and to be kept as voucher specimens were also taken, and the remainder of each coprolite was analyzed for macrofossils.
aDNA Extraction, Amplification, and Sequencing.
aDNA extractions, amplification, and sequencing were performed at the Australian Centre for Ancient DNA following the methods described by Wood et al. (9, 46). Multiple extraction and PCR control blanks were included. Newly excavated coprolites were identified using the primers MoaCR262F and MoaCR294R (9) to amplify a species-diagnostic 31-bp fragment of moa mitochondrial control region. The identities of A. didiformis coprolites were verified using the Andi/Eucu primers, which were designed to distinguish coprolites of this species from those of Euryapteryx curtus (46). Plant aDNA was amplified (95-bp fragment of the rbcL gene using primers h1aF and h2aR) and cloned following the methods described by Wood et al. (42).
Moa aDNA Sequence Analysis.
Sequences obtained from the coprolites were aligned with 131 reference moa mitochondrial control region sequences representing all nine moa species (obtained from GenBank). The alignment was assessed using ModelGenerator (58) and analyzed in BEAST 1.6.1 (59) using a HKY + G model.
Plant aDNA Sequence Analysis.
The total dataset of 526 clones (Table S2) was collapsed to 178 unique sequences that were analyzed using two complimentary methods. First, clone sequences were compared with sequences in GenBank using the Bayesian Statistical Assignment Package (60). Second, the clone sequences were aligned with 220 reference rbcL sequences, including sequences from extant plant taxa native to the Dart River Valley, and selected sequences from GenBank to help constrain order and family level branches. Sequences were aligned using Seaview (61), assessed using ModelGenerator (58), and analyzed using PhyML 4.3.0 (62) (with HKY85 model, six gamma categories, and tree topology search for the best of nearest neighbor interchange (NNI) and subtree pruning and regrafting (SPR)). Sequences that could be resolved only to order or higher taxonomic clades were omitted from statistical analyses, except where no subordinate taxa within the order were resolved (e.g., Oxalidales). Sequences identified as Fabaceae and Proteaceae were also excluded from analyses, as suspected contamination (SI Text).
Microscopic Analyses.
Subsamples for macrofossil analysis were rehydrated in 0.5% trisodium phosphate for several weeks and then sieved on a 0.25-mm mesh. Residues were examined at 10–40× magnification. Subsamples for pollen analysis were prepared by heating in potassium hydroxide (10 min), treatment with hydrochloric acid, heavy-liquid flotation (lithium polytungstate; specific gravity, 2.2), acetolysis, staining, and mounting on microscope slides following standard preparation methods (63). Pollen concentrations were quantified by adding tablets containing a known number of exotic Lycopodium spores to samples before preparation. A minimum of 215 pollen/spores were counted from each sample. Pollen and spore nomenclature follows the standard New Zealand system (64).
Radiocarbon Dating.
Coprolite subsamples were Accelerator Mass Spectrometry (AMS) radiocarbon dated at the Waikato Radiocarbon Dating Laboratory, New Zealand. Radiocarbon ages were calibrated using the ShCal04 curve (65) in OxCal 4.1 (66).
Statistical Methods.
Compositional overlap between the diets of moa species were assessed using all three proxies (pollen, plant macrofossil, and aDNA).
Data tranformations.
Macrofossil data were quantitative, ordinal, and categorical. To preserve the relative differences between samples, we converted all data to an ordinal scale as follows: 0 (absent), 1 (a single seed/leaf present), 2 (2–5 seeds/leaves), 3 (>5 seeds/leaves). This approach effectively compensates for the effect of consumption of fruits containing abundant seeds (e.g., Gaultheria). Pollen data were weighted according to pollen dispersal characteristics and production quantities. This accounts for potential environmental-input biases in coprolite pollen assemblages and provides a better representation of taxa on which the birds were feeding (42). Pollen counts were divided by the product of values (64) for pollination mode (wind = 3; animal = 1), pollen production (prolific = 3; moderate = 2; low = 1), and dispersability [regional (>5 km) = 3; local (5–∼0.25 km) = 2; restricted (<∼0.25 km) = 1]. A list of possible plant species accounting for each pollen/spore type was created from Moar et al. (64) and refined to include just those present at the study site using a local plant species list of Mark (45). The distribution of these taxa within the vegetation communities at Daley’s Flat was used to score each pollen/spore type as being restricted to either forest or herbfield or present in both.
Analyses.
We used nonmetric multidimensional scaling (NMS) to optimally arrange the compositional data from each coprolite in multidimensional space. Pollen data were log transformed while preserving zero values. NMS was analyzed using metaMDS in the vegan library of R with Bray’s distance measure, 9,999 permutations, and default settings following McCune et al. (67). Two dimensions (axes) minimized the stress of the NMS solution, and more dimensions only marginally reduced stress further (68). Sample scores (coprolites) were plotted for each moa species, and the area of the hull containing samples from a moa species was calculated to estimate niche breadth. Overlap among niches was also calculated from these hulls. We estimated statistical differences in composition with a nonparametric PERMANOVA using adonis in the vegan library with default settings, Bray’s distance measure, and 9,999 permutations. Pollen data were log transformed before analysis.
Plotting niches.
We calculated the total percentage of forest and herbfield pollen types in each coprolite. These percentages were fitted as environmental vectors to the NMS ordination using envfit in the vegan library of R. Both vectors were highly significant (P < 0.001) and were plotted as smoothed surfaces across the ordination using thinplate splines in gam (generalized additive models) implemented by ordisurf (vegan library of R). Coprolites were categorized according to these surfaces as containing >40% forest and <5% herbfield pollen types; >30% herbfield and <10% forest pollen types; and an intermediate category reflecting the ecotone between forest and herbfield. The number of coprolites for each moa species in each of these three categories was expressed as a proportion of the total number of coprolites for each moa species, and these proportions were plotted to represent niche position and overlap (Fig. 3).
Supplementary Material
Acknowledgments
We thank the Otago Museum for providing samples, the New Zealand Department of Conservation for permitting further excavation at the Dart River Valley coprolite site, and G. Rattray for preparation of pollen samples. We also thank J. Austin, J. Soubrier, I. Dickie, and M. McGlone, who provided advice and assistance with various aspects of this study. Funding was provided by the Royal Society of New Zealand Marsden Fund and the Australian Research Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. C.L. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1307700110/-/DCSupplemental.
References
- 1.Johnson CN. Ecological consequences of Late Quaternary extinctions of megafauna. Proc Biol Sci. 2009;276(1667):2509–2519. doi: 10.1098/rspb.2008.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hansen DM, Galetti M. Ecology. The forgotten megafauna. Science. 2009;324(5923):42–43. doi: 10.1126/science.1172393. [DOI] [PubMed] [Google Scholar]
- 3.Burney DA, Flannery TF. Fifty millennia of catastrophic extinctions after human contact. Trends Ecol Evol. 2005;20(7):395–401. doi: 10.1016/j.tree.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 4.Gill JL, Williams JW, Jackson ST, Lininger KB, Robinson GS. Pleistocene megafaunal collapse, novel plant communities, and enhanced fire regimes in North America. Science. 2009;326(5956):1100–1103. doi: 10.1126/science.1179504. [DOI] [PubMed] [Google Scholar]
- 5.Rule S, et al. The aftermath of megafaunal extinction: Ecosystem transformation in Pleistocene Australia. Science. 2012;335(6075):1483–1486. doi: 10.1126/science.1214261. [DOI] [PubMed] [Google Scholar]
- 6.Hansen DM. On the use of taxon substitutes in rewilding projects on islands. Menorqui d'Estudis. Recerca. 2010;19:111–146. [Google Scholar]
- 7.Mead JI, Agenbroad LD, Davis OK, Martin PS. Dung of Mammuthus in the Arid Southwest, North-America. Quat Res. 1986;25(1):121–127. [Google Scholar]
- 8.James HF, Burney DA. The diet and ecology of Hawaii's extinct flightless waterfowl: Evidence from coprolites. Biol J Linn Soc Lond. 1997;62(8):279–297. [Google Scholar]
- 9.Wood JR, et al. Coprolite deposits reveal the diet and ecology of the extinct New Zealand megaherbivore moa (Aves, Dinornithiformes) Quat Sci Rev. 2008;27(27-28):2593–2602. [Google Scholar]
- 10.Kropf M, Mead JI, Anderson RS. Dung, diet, and the paleoenvironment of the extinct shrub-ox (Euceratherium collinum) on the Colorado Plateau, USA. Quat Res. 2007;67(1):143–151. [Google Scholar]
- 11.Ghosh R, et al. Ovi-caprid dung as an indicator of paleovegetation and paleoclimate in northwestern China. Quat Res. 2008;70(2):149–157. [Google Scholar]
- 12.Marcolino CP, dos Santos Isaias RM, Cozzuol MA, Cartelle C, Dantas MAT. Diet of Palaeolama major (Camelidae) of Bahia, Brazil, inferred from coprolites. Quat Int. 2012;278:81–86. [Google Scholar]
- 13.Poinar HN, et al. Molecular coproscopy: Dung and diet of the extinct ground sloth Nothrotheriops shastensis. Science. 1998;281(5375):402–406. doi: 10.1126/science.281.5375.402. [DOI] [PubMed] [Google Scholar]
- 14.Wilmshurst JM, Anderson AJ, Higham TFG, Worthy TH. Dating the late prehistoric dispersal of Polynesians to New Zealand using the commensal Pacific rat. Proc Natl Acad Sci USA. 2008;105(22):7676–7680. doi: 10.1073/pnas.0801507105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bunce M, et al. The evolutionary history of the extinct ratite moa and New Zealand Neogene paleogeography. Proc Natl Acad Sci USA. 2009;106(49):20646–20651. doi: 10.1073/pnas.0906660106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Worthy TH, Holdaway RN. The Lost World of the Moa. Christchurch, New Zealand: Canterbury Univ Press; 2002. [Google Scholar]
- 17.Lee WG, Wood JR, Rogers GM. Legacy of avian-dominated plant-herbivore systems in New Zealand. N Z J Ecol. 2010;34(1):28–47. [Google Scholar]
- 18.Forsyth DM, Wilmshurst JM, Allen RB, Coomes DA. Impacts of introduced deer and extinct moa on New Zealand ecosystems. N Z J Ecol. 2010;34(1):48–65. [Google Scholar]
- 19.Atkinson IAE, Greenwood RM. Relationships between moas and plants. N Z J Ecol. 1989;12(Suppl):67–96. [Google Scholar]
- 20.Lee D, Gould K. Three birds with one stone: Moas, heteroblasty and the New Zealand flora. New Phytol. 2009;184(2):282–284. doi: 10.1111/j.1469-8137.2009.03025.x. [DOI] [PubMed] [Google Scholar]
- 21.Bond WJ, Lee WG, Craine JM. Plant structural defenses against browsing birds: A legacy of New Zealand's extinct moas. Oikos. 2004;104(3):500–508. [Google Scholar]
- 22.Darrow HE, Bannister P, Burritt DJ, Jameson PE. Are juvenile forms of New Zealand heteroblastic trees more resistant to water loss than their mature counterparts? NZ J Bot. 2002;40(2):313–325. [Google Scholar]
- 23.Day JS. Light conditions and the evolution of heteroblasty (and the divaricate form) in New Zealand. N Z J Ecol. 1998;22(1):43–54. [Google Scholar]
- 24.Darrow HE, Bannister P, Burritt DJ, Jameson PE. The frost resistance of juvenile and adult forms of some heteroblastic New Zealand plants. NZ J Bot. 2001;39(2):355–363. [Google Scholar]
- 25.Cooper A, Atkinson IAE, Lee WG, Worthy TH. Evolution of the moa and their effect on the New Zealand flora. Trends Ecol Evol. 1993;8(12):433–437. doi: 10.1016/0169-5347(93)90005-A. [DOI] [PubMed] [Google Scholar]
- 26.Tanentzap AJ, Lee WG, Monks A. Increased nitrogen cycling facilitates native forest regeneration: Potential for restoring extinct ecological processes? Ecol Appl. 2013;23(1):36–45. doi: 10.1890/12-0247.1. [DOI] [PubMed] [Google Scholar]
- 27.Batcheler CL. Moa browsing and vegetation formations, with particular reference to deciduous and poisonous plants. N Z J Ecol. 1989;12(Suppl):57–65. [Google Scholar]
- 28.Duncan K, Holdaway R. Footprint pressures and locomotion of moas and ungulates and their effects on the New-Zealand indigenous biota through trampling. N Z J Ecol. 1989;12(Suppl):97–101. [Google Scholar]
- 29.Burrows CJ, McCulloch B, Trotter MM. The diet of moas based on gizzard contents samples from Pyramid Valley, North Canterbury, and Scaifes Lagoon, Lake Wanaka, Otago. Rec Cant Mus. 1981;9(6):309–336. [Google Scholar]
- 30.Wood JR. Moa (Aves: Dinornithiformes) nesting material from rockshelters in the semi-arid interior of South Island, New Zealand. J R Soc N Z. 2008;38(3):115–129. [Google Scholar]
- 31.Wood JR. Moa gizzard content analyses: Further information on the diets of Dinornis robustus and Emeus crassus, and the first evidence for the diet of Pachyornis elephantopus (Aves: Dinornithiformes) Rec Cant Mus. 2007;21:27–39. [Google Scholar]
- 32.Horrocks M, D'Costa D, Wallace R, Gardner R, Kondo R. Plant remains in coprolites: Diet of a subalpine moa (Dinornithiformes) from southern New Zealand. Emu. 2004;104(2):149–156. [Google Scholar]
- 33.Gregg D. Holocene stratigraphy and moas at Pyramid Valley, North Canterbury, New Zealand. Rec Cant Mus. 1972;9(3):151–158. [Google Scholar]
- 34. Hutton FW (1895) On the leg bones of Meionornis from Glenmark. Trans Proc N Z Institute 29:557–560.
- 35.Worthy TH. The Quaternary fossil avifauna of Southland, South Island, New Zealand. J R Soc N Z. 1998;28(4):537–589. [Google Scholar]
- 36.Worthy TH. Quaternary fossil faunas of Otago, South Island, New Zealand. J R Soc N Z. 1998;28(3):421–521. [Google Scholar]
- 37.Worthy TH, Holdaway RN. Quaternary fossil faunas, overlapping taphonomies, and palaeofaunal reconstruction in North Canterbury, South Island, New Zealand. J R Soc N Z. 1996;26(3):275–361. [Google Scholar]
- 38.Rawlence NJ, et al. New palaeontological data from the excavation of the Late Glacial Glencrieff miring bone deposit, North Canterbury, South Island, New Zealand. J R Soc N Z. 2011;41(3):217–236. [Google Scholar]
- 39.Wood JR, Wilmshurst JM, Rawlence NJ. Radiocarbon-dated faunal remains correlate very large rock avalanche deposit with prehistoric Alpine fault rupture. NZ J Geol Geophys. 2011;54(4):431–434. [Google Scholar]
- 40.Allentoft ME, et al. A molecular characterisation of a newly discovered megafaunal fossil site in North Canterbury, South Island, New Zealand. J R Soc N Z. 2012;42(4):241–256. [Google Scholar]
- 41.Worthy TH, Scofield RP. 21st Century advances in the understanding of moa biology. NZ J Zool. 2012;39(2):87–153. [Google Scholar]
- 42.Wood JR, et al. High-resolution coproecology: Using coprolites to reconstruct the habits and habitats of New Zealand’s extinct upland moa (Megalapteryx didinus) PLoS ONE. 2012;7(6):e40025. doi: 10.1371/journal.pone.0040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jørgensen T, et al. A comparative study of ancient sedimentary DNA, pollen and macrofossils from permafrost sediments of northern Siberia reveals long-term vegetational stability. Mol Ecol. 2012;21(8):1989–2003. doi: 10.1111/j.1365-294x.2011.05287.x. [DOI] [PubMed] [Google Scholar]
- 44.Parducci L, et al. Molecular- and pollen-based vegetation analysis in lake sediments from central Scandinavia. Mol Ecol. 2013;22(13):3511–3524. doi: 10.1111/mec.12298. [DOI] [PubMed] [Google Scholar]
- 45.Mark AF. Vegetation of Mount Aspiring National Park New Zealand. Wellington, New Zealand: National Parks Authority; 1977. [Google Scholar]
- 46.Wood JR, Wilmshurst JM, Worthy TH, Cooper A. First coprolite evidence for the diet of Anomalopteryx didiformis, an extinct forest ratite from New Zealand. N Z J Ecol. 2012;36(2):164–170. [Google Scholar]
- 47.Wood JR, Wilmshurst JM. Pollen analysis of coprolites reveals dietary details of heavy-footed moa (Pachyornis elephantopus) and coastal moa (Euryapteryx curtus) from Central Otago. N Z J Ecol. 2013;37(1):151–155. [Google Scholar]
- 48.Bunce M, et al. Extreme reversed sexual size dimorphism in the extinct New Zealand moa Dinornis. Nature. 2003;425(6954):172–175. doi: 10.1038/nature01871. [DOI] [PubMed] [Google Scholar]
- 49.Huynen L, Millar CD, Scofield RP, Lambert DM. Nuclear DNA sequences detect species limits in ancient moa. Nature. 2003;425(6954):175–178. doi: 10.1038/nature01838. [DOI] [PubMed] [Google Scholar]
- 50.Mysterud A. The relationship between ecological segregation and sexual body size dimorphism in large herbivores. Oecologia. 2000;124(1):40–54. doi: 10.1007/s004420050023. [DOI] [PubMed] [Google Scholar]
- 51.Belovsky GE. Optimal foraging and community structure: The allometry of herbivore food selection and competition. Evol Ecol. 1997;11(6):641–672. [Google Scholar]
- 52.Allentoft ME, Bunce M, Scofield RP, Hale ML, Holdaway RN. Highly skewed sex ratios and biased fossil deposition of moa: Ancient DNA provides new insight on New Zealand's extinct megafauna. Quat Sci Rev. 2010;29(5-6):753–762. [Google Scholar]
- 53.Forsyth DM, Coomes DA, Nugent G, Hall GMJ. Diet and diet preferences of introduced ungulates (Order: Artiodactyla) in New Zealand. NZ J Zool. 2002;29(4):323–343. [Google Scholar]
- 54.Wardle DA, Bonner KI, Barker GM. Linkages between plant litter decomposition, litter quality, and vegetation responses to herbivores. Funct Ecol. 2002;16(5):585–595. [Google Scholar]
- 55.Worthy TH. A re-examination of the genus Megalapteryx. Notornis. 1988;35(2):99–108. [Google Scholar]
- 56.Mills JA, Mark AF. Food preferences of takahe in Fiordland National Park, New Zealand, and effect of competition from introduced red deer. J Anim Ecol. 1977;46(3):939–958. [Google Scholar]
- 57.Korsten AC, Lee WG, Monks A, Wilson JB. Understanding the role of birds in sustaining indigenous turf communities in a lacustrine wetland in New Zealand. N Z J Ecol. 2013;37(2):206–213. [Google Scholar]
- 58.Keane TM, Creevey CJ, Pentony MM, Naughton TJ, Mclnerney JO. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol. 2006;6:29. doi: 10.1186/1471-2148-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214–221. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Munch K, Boomsma W, Huelsenbeck JP, Willerslev E, Nielsen R. Statistical assignment of DNA sequences using Bayesian phylogenetics. Syst Biol. 2008;57(5):750–757. doi: 10.1080/10635150802422316. [DOI] [PubMed] [Google Scholar]
- 61.Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27(2):221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 62.Guindon S, Dufayard JF, Hordijk W, Lefort V, Gascuel O. PhyML: Fast and accurate phylogeny reconstruction by maximum likelihood. Infect Genet Evol. 2009;9(3):384–385. [Google Scholar]
- 63.Moore PD, Webb JA, Collinson ME. Pollen Analysis. Malden, MA: Blackwell Science; 1991. [Google Scholar]
- 64.Moar NT, Wilmshurst JM, McGlone M. Standardising names applied to pollen and spores in New Zealand palynology. NZ J Bot. 2011;49(2):201–229. [Google Scholar]
- 65.McCormac FG, et al. SHCal04 Southern Hemisphere calibration, 0-11.0 cal kyr BP. Radiocarbon. 2004;46(3):1087–1092. [Google Scholar]
- 66.Ramsey CB. Bayesian analysis of radiocarbon dates. Radiocarbon. 2009;51(1):337–360. [Google Scholar]
- 67.McCune B, Grace JB, Urban DL. Analysis of Ecological Communities. Gleneden Beach, OR: MjM Software Design; 2002. [Google Scholar]
- 68.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26(1):32–46. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.