Significance
Horizontal gene exchange across species boundaries is considered infrequent relative to vertical inheritance that maintains species coherence. However, haloarchaea living in hypersaline environments take a more relaxed approach to gene exchange. Here we demonstrate that in Deep Lake, Antarctica, haloarchaea exchange DNA between distinct genera, not just species, with some of the DNA being long (up to 35 kb) and virtually 100% conserved. With extremely low cell division rates in the cold (e.g., six generations per year), the remarkable extent of lateral exchange could conceivably homogenize the population. It is therefore equally notable that despite the demonstrated capacity for exchange, different genera are maintained, their coexistence being linked to genomic differences conferring ecotype distinctions that enable niche adaptation.
Keywords: mobile genetic elements, Antarctic haloarchaea, saltern, fragment recruitment, BJ1 virus
Abstract
Deep Lake in Antarctica is a globally isolated, hypersaline system that remains liquid at temperatures down to −20 °C. By analyzing metagenome data and genomes of four isolates we assessed genome variation and patterns of gene exchange to learn how the lake community evolved. The lake is completely and uniformly dominated by haloarchaea, comprising a hierarchically structured, low-complexity community that differs greatly to temperate and tropical hypersaline environments. The four Deep Lake isolates represent distinct genera (∼85% 16S rRNA gene similarity and ∼73% genome average nucleotide identity) with genomic characteristics indicative of niche adaptation, and collectively account for ∼72% of the cellular community. Network analysis revealed a remarkable level of intergenera gene exchange, including the sharing of long contiguous regions (up to 35 kb) of high identity (∼100%). Although the genomes of closely related Halobacterium, Haloquadratum, and Haloarcula (>90% average nucleotide identity) shared regions of high identity between species or strains, the four Deep Lake isolates were the only distantly related haloarchaea to share long high-identity regions. Moreover, the Deep Lake high-identity regions did not match to any other hypersaline environment metagenome data. The most abundant species, tADL, appears to play a central role in the exchange of insertion sequences, but not the exchange of high-identity regions. The genomic characteristics of the four haloarchaea are consistent with a lake ecosystem that sustains a high level of intergenera gene exchange while selecting for ecotypes that maintain sympatric speciation. The peculiarities of this polar system restrict which species can grow and provide a tempo and mode for accentuating gene exchange.
Deep Lake (DL) is an extremely cold and hypersaline environment that has the distinction of being the least productive lake ever recorded (1–3). The lake is a marine-derived system in the Vestfold Hills, East Antarctica, having been isolated from the Southern Ocean by isostatic rebound of the continent ∼3,500 y BP (2, 4, 5) (SI Appendix, Fig. S1). The temperature exceeds 0 °C only in the top few meters for a few summer months per year, and it remains ice-free even in winter when temperatures drop to −40 °C (air) and −20 °C (throughout the lake) (1, 6). Microbial diversity is extremely low, and is dominated by members of the haloarchaea (7). Within this ecosystem, the putative primary producer is the green alga Dunaliella sp., which is found at low biomass concentrations (8–10).
From DL, Halorubrum lacusprofundi, the first member of the Archaea domain isolated from a cold environment (11), has been formally described (12). In the laboratory, H. lacusprofundi grows across a wide range of temperatures from −1 °C to 42 °C, with fastest growth rate occurring at ∼33 °C (12, 13). It is capable of using a number of different carbon sources including glucose, mannose, acetate and ethanol, providing it with a seemingly versatile heterotrophic metabolism (12). Aside from studies that have noted the ability of H. lacusprofundi, and a recent isolate, tADL (14), to form aggregates and biofilms at either high or low temperatures (13, 15, 16), the only studies addressing adaptive responses are those linking the production of unsaturated diether membrane lipids to cold adaptation in H. lacusprofundi (3, 17).
Various mechanisms that alter genetic composition have been reported in haloarchaea, including archaeal viruses, conjugative plasmids, genome rearrangements mediated by transposons, and significant levels of gene exchange via the formation of heterodiploids followed by homologous recombination (18–25). All these studies have been confined to temperate or tropical systems, not polar environments.
For some bacteria, the relationship between recombination frequency and sequence divergence appears to be log-linear (26, 27) but may be as much as two orders of magnitude higher in haloarchaea, such as for Haloferax volcanii and Haloferax mediterranei, which readily undergo cell fusion and DNA exchange (25, 28). Forces driving speciation in microbes include niche adaptation, selective sweeps, genetic drift, recombination, and geographic isolation (29). However, it is unclear how these forces would maintain species homogeneity or bring about lineages when gene flow is high, as is the case in haloarchaea. Although selection is important for speciation, it has been argued that the rate of genomic recombination may be a key determinant when considering sympatric speciation. Modeling has shown that high recombination rates are necessary to generate a new species when an ecological trait inferring increased fitness is controlled by many adaptive loci (30). However, these same high levels of recombination may also increase the number of intermediate (suboptimal) genotypes and reduce “completeness” of speciation. The dynamics of this sympatric evolution hypothesis have not been examined in real populations.
The DL system is geographically very isolated, its organismal growth rates are very low, and the combination of cold plus hypersalinity may be expected to promote the physical process of DNA transformation to levels that exceed those of warm hypersaline environments. In this context, we examined the genomes of four recent haloarchaeal isolates and metagenome data from DL to assess genome variation and patterns of gene exchange and derive an understanding of how this unique Antarctic lake community evolves.
Results
DL Microbial Community Composition.
From biomass filter fractionated (20–3.0, 3.0–0.8, 0.8–0.1 µm) from four depths of DL (5, 13, 24, and 36 m), ∼10 Gb of metagenome data (∼3 Gb 454 Titanium all depths; ∼7 Gb Illumina 24 m) and ∼0.5 million universal small subunit (SSU) rRNA gene pyrotag sequences were analyzed (SI Appendix, Table S1). Genome sequences were also analyzed for four DL isolates: tADL [described as “Halohasta litchfieldiae” (14), 1 replicon 3.33 Mb], DL31 (undescribed genus, 3 replicons 3.64 Mb), H. lacusprofundi (3 replicons 3.69 Mb), and DL1 (Halobacterium, 2 replicons 3.16 Mb) (SI Appendix, Table S2). Full descriptions of all data analyzed in this study are provided in SI Appendix.
Phylogenetic reconstructions using SSU rRNA gene sequences cluster all four DL isolates within the family Halobacteriaceae (Archaea; Euryarchaeota; Halobacteria; Halobacteriales; Halobacteriaceae) (SI Appendix, Fig. S2). H. lacusprofundi and DL1 cluster within the described genera Halorubrum and Halobacterium, respectively. tADL is positioned in a cluster along with uncultured clone sequences from other aquatic hypersaline environments, while DL31 forms a discrete cluster containing only clone sequences originating from DL. A phylogenetic distance matrix derived from SSU rRNA genes of the four DL isolates shows they are all ∼85% similar to each other (SI Appendix, Table S3).
From SSU rRNA pyrotag data overall community complexity is very low, as was previously reported (7), and community structure is highly dominated by a few operational taxonomic units (OTUs) (SI Appendix, Figs. S2 and S3). The top 10 OTUs represent ∼90% of the total sequence abundance (SI Appendix, Fig. S2 and Table S4). The four DL isolates accounted for a total of ∼72%, averaged across four depths, with the rank and estimated abundance being tADL, first, 43.5%; DL31, second, 18.2%; H. lacusprofundi, fourth, 9.9%; DL1, 17th, 0.3%. The haloarchaeal community composition differs greatly from other hypersaline environments in the world where Haloarcula spp., Haloferax volcanii, Haloquadratum walsbyi, and Halobacterium salinarum are typically found (but are absent in DL) (31–33).
SSU rRNA gene pyrotag data, and fragment recruitment (FR) read depth of 454 and Illumina data, all gave similar values for the relative proportions of just the four DL isolates averaged across the three filters: tADL 51–58%, DL31 18–30%, H. lacusprofundi 14–16%, DL1 0.4–3% (SI Appendix, Fig. S4). A somewhat lower representation of tADL and higher representation of low-abundance species was evident in the bottom waters of the lake (SI Appendix, Figs. S2, S3, and S5), where a shallow depression may allow cells to settle. The highest level of variation in community composition related to filter size (SI Appendix, Figs. S5 and S6). The most consistent difference across all lake depths was the higher partitioning of DL31, H. lacusprofundi and DL1 cells in the 0.8- and 3.0-µm fractions relative to the 0.1-µm fraction, compared with the relatively even distribution of tADL across all size fractions (SI Appendix, Fig. S6).
From the analysis of eight million metagenomic 454 reads against all available complete and draft genomes, the four DL genomes recruited the largest number of reads (94.4% of all recruited reads) and 28.3% of all reads. Similarly, as a proportion of Illumina reads, the four DL genomes represented 50% of all reads. The lower rate of recruitment for 454 may be explained by the technology’s longer read length and therefore increased rejection of partial degenerate sequence alignment set by the stringent minimum alignment coverage (98%). The unassigned metagenome data could derive from viruses and/or eucarya that are not in the National Center for Biotechnology Information database (SI Appendix).
Characteristics of the Nine Replicons of the Four Genomes.
Secondary replicons in DL31, H. lacusprofundi and DL1 had consistently lower codon adaptation (CAI) and codon bias (CBI) indexes (34, 35) than the primary replicons. Additionally, the primary replicons of tADL and DL1 possessed extended regions with low CAI/CBI. The low CAI/CBI regions often coincided with strong variations in FR and increased density of mobile elements (Fig. 1 A and B and SI Appendix, Fig. S7).
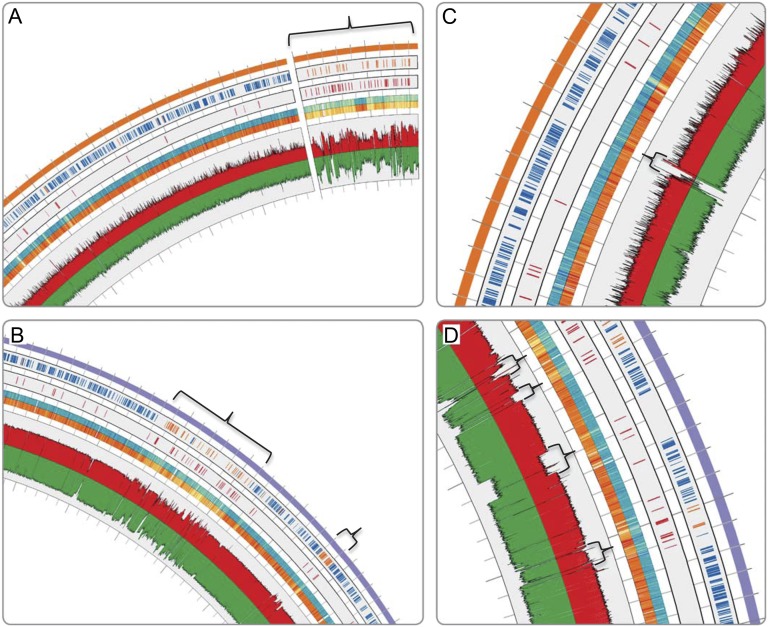
Fig. 1.
Sectors of circular genome plots highlighting specific features of DL haloarchaea. (A–D) Circos plot (49), outside to inside: first annulus: replicon backbones tADL (purple), H. lacusprofundi (orange); second annulus: core (blue), noncore (orange); third annulus: IS (red); fourth annulus: CAI (yellow–blue), CBI (yellow–orange) heatmaps, deeper color indicates more adapted; fifth annulus: read depth by gsMapper reference mapping (red), FR-hit fragment recruitment (green), log scale y axis. (A and B) Bracketed regions mark low CAI/CBI, high noncore gene content, ISs and FR coverage variability. (C and D) Bracketed regions mark stretches of low FR coverage.
A total of 894 completely conserved ortholog clusters shared across 17 haloarchaeal genomes (including the four DL genomes) were used to define core vs. noncore gene content. Core genes represented metabolism (32%), information storage and processing (30%), poorly characterized (15%), general function prediction only (13%), and cellular processes and signaling (11%) (SI Appendix, Fig. S8), similar to the classes reported for prior analyses that did not include the four Antarctic haloarchaea (36). Core genes were mainly located on the DL primary replicons (Fig. 1 and SI Appendix, Fig. S7). They included genes expected to be efficiently expressed (37), and they had high CAI/CBI indexes. The selective pressure of haloarchaeal genes was considered both within Antarctic genomes, and between Antarctic and non-Antarctic genomes. Single-copy ortholog pairs were identified between tADL and H. lacusprofundi (within population) and tADL and Haloferax volcanii (between populations). Based on the ratio of the number of non-synonymous substitutions per non-synonymous site (Ka) to the number of synonymous substitutions per synonymous site (Ks) (Ka/Ks) values (38), the core gene content for all haloarchaea tested was predicted to be under a similar level of purifying selection.
To explore what persistent contribution secondary replicons might make to the gene repertoire of DL haloarchaea, 68 ortholog groups were selected that possessed at least one member from each secondary replicon of the DL isolates and also one from the primary replicon of tADL. This set was regarded as conserved but potentially noncore gene content and was diagnostic for determining what gene functions were represented on secondary replicons, and where those genes from DL31, H. lacusprofundi, and DL1 resided in the single tADL replicon. The largest ortholog groups were associated with insertion sequences (ISs) and had sizes 72 (ISH3), 27 (ISH4), and 19 (ISH6), which were much greater than the mean of 9 or median of 7. The next most abundant ortholog groups included COG functional groups for transcription (K); replication, recombination, and repair (L); defense mechanisms (V); inorganic ion transport (P); intracellular trafficking (U); and cell cycle, division, and partitioning (D) (SI Appendix, Fig. S9).
Using the ISsaga database (39), a total of 489 matches to ISs were identified across the four genomes, with three families (ISH3, IS200/IS605, and IS5) comprising 65% of all predictions (SI Appendix, Table S5). The density of ISs was noticeably higher in secondary replicons and in distinct regions of the tADL and DL1 primary replicons, which possessed low CAI/CBI indexes (Fig. 1 A and B). The secondary replicon with the lowest density was >4 times the density of any primary replicon, and the density in tADL was >2.5 times higher than the next dense primary replicon. A comparison with 119 other species showed tADL is ranked second in number of complete ISs behind Sulfolobus solfataricus P2, which has considerably more than any other in the ISsaga database.
Dominance and Niche Adaptation.
The unique genomic features of tADL that may contribute to its dominance include a single replicon, and genes for gas vesicles, bacteriorhodopsin, and polyhydroxyalkanoate (PHA) biosynthesis. It also possesses a higher number of predicted ATP-binding cassette transporters for carbohydrates (six), and possesses multiple glycerol kinase orthologs (first step in glycerol breakdown) and a large number of regulatory genes (e.g., signal transduction). Thus, tADL appears to have a highly saccharolytic (carbohydrate degrading) “high energy” metabolism that can respond to changing substrate availability, with glycerol as a preferred substrate. Gas vesicles provide buoyancy that facilitates upward motion, and particularly for slow-growing organisms can allow more efficient vertical migrations than swimming by flagella (40). Buoyancy may facilitate tADL getting to the surface in the summer, thereby allowing light-driven bacteriorhodopsin to generate energy, and faster growth rates to occur in the warmer water. This reasoning is consistent with tADL abundance being somewhat lower in the deepest point (36 m) of the lake (SI Appendix, Figs. S5 and S6). Additionally, surplus carbon and energy could be stored as PHA, and mobilized for biomass production when other limiting substrates become available. The other genomes each have specific characteristics indicative of niche adaptation: DL31 is orientated toward proteolytic (protein-degrading) metabolism of particulate matter rich in protein; DL1 targets amino acids and lacks an ability to use glycerol; H. lacusprofundi has comparatively few over- or underrepresented COGs associated with metabolism, indicating it may prefer to target a broad range of substrates rather than having a specialized metabolism (SI Appendix).
The whole DL metagenome was assembled and organisms assigned to contigs in a two component space of GC content and mean read depth (SI Appendix, Figs. S10 and S11). tADL, DL31 and H. lacusprofundi represented three of four clusters (DL1 was not represented due to its low abundance). An unaccounted for fourth cluster with higher read depth than DL31 or H. lacusprofundi, comprised 52 large contigs (>15 kb) totaling 1.89 Mb, that had highest sequence similarity to tADL: average nucleotide identity (ANI), 0.802 (1.08 Mbp aligning); tetranucleotide use deviation regression, 0.959; NUCMER average identity 85.2%, 492 gaps, total 806,411 bp; CONTIGuator 48 of 52 contigs aligned, total 1.82 Mbp (Fig. 2 and SI Appendix, Fig. S12). From 1,889 predicted full-length protein-coding genes (no detectable 16S rRNA gene), 873 genes (46% of the total and 76% of assigned orthologous groups) were core genes typical of a primary replicon, whereas 40 genes (2%) were assigned as noncore genes. In view of the similarity to tADL and knowledge from SSU rRNA gene data that no other abundant haloarchaea exist in DL, the additional genome may represent a phylotype of tADL that has a significantly different genome to the tADL isolate and is designated the “tADL-related fifth genome” (Fig. 2 and SI Appendix, Fig. S12).
Fig. 2.
HIR shared between the four DL genomes. first annulus: genome backbones color-coded; second annulus: tADL-like fifth genome fragments aligned to the tADL genome (purple); third annulus: ISs (red); fourth annulus: read depth by gsMapper reference mapping (red), FR-hit fragment recruitment (green), log scale y axis; internal lines: shared HIR of ≥ 99.8% nucleotide identity and > 5 kb length.
The FR histograms for replicons of tADL, DL31, and H. lacusprofundi (Fig. 1 C and D) revealed discrete regions of low coverage (DL1 was not investigated due to its overall low coverage). Across the three genomes the gene content of the low coverage zones had signatures of: (i) mobile elements including viruses (e.g., BJ1), plasmids (e.g., conjugation) and ISs/transposons; (ii) cell wall synthesis and modification; (iii) metabolism and transport (e.g., amino acids, nitrogen, sugars, iron, phosphate, peptides, urea, and nucleosides); (iv) tRNA modification; and (v) duplicated sets of genes. The low coverage of these regions probably reflects the existence of subpopulations whose genomes do not contain these regions. Although some of the regions probably reflect the movement of mobile elements, others may denote ecotype differences with varying gene content conferring phenotypic properties linked to differential responses to temperature (e.g., cell wall changes and tRNA modification) and/or resource availability (e.g., metabolism and transport).
Genome Variation Detected by FR.
FR to the DL1 primary replicon (mean read depth: 2.5) was well above average in an 80 kb region (63% of assigned reads; mean read depth: 54.2) suggesting that the majority of recruiting reads were not derived from DL1 cells. Gene content included many genes that matched to the haloarchaeal BJ1 virus (41), and genes involved in DNA processing and expression. Regions matching to BJI were also present in H. lacusprofundi (Fig. 1C) and DL31, including regions of lower than average FR. The homologous regions between DL1, H. lacusprofundi and DL31 were rearranged, and ISs were abundant within intervening nonaligning sequences of the homologous regions in H. lacusprofundi.
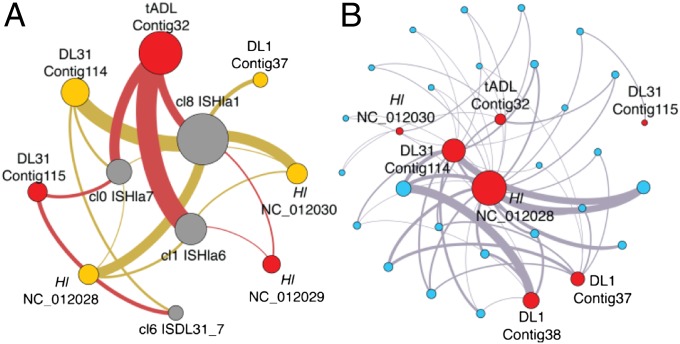
FR to the four DL genomes also revealed short, highly degenerate regions (4.8% of all recruited reads), representing 15 unique sequence clusters: 14 ISs and a conserved haloarchaeal hypothetical gene. A bipartite network of degenerate cluster to replicon was constructed to visualize the prevalence of each cluster within the community, with nodes and edge size (width of interconnecting lines) revealing the relative contribution of each cluster in a specific replicon. The four largest clusters [cl8 (ISHla1), cl1 (ISHla6), cl0 (ISHla7), and cl6 (ISDL31_7; newly defined in DL)] are highly connected across all four genomes (Fig. 3A), and although no cluster is fully associated with all nine replicons, 2 IS clusters (cl8 and cl0) are fully associated at the genome level. tADL, H. lacusprofundi, and DL31 are the most highly interconnected, with tADL possessing 31% of all edges in the network and accommodating the community majority of ISs. Secondary replicons from DL31 and H. lacusprofundi possess ∼twofold more edges than their primary replicons.
Fig. 3.
Association networks for DL features. Bipartite networks for features identified within the four DL haloarchaeal genomes, using Fruchterman–Reingold layout. (A) Nonredundant sequences identified from short highly degenerate regions (gray nodes; read depth ∼2,000) and their containing replicons (red nodes primary replicons, orange nodes secondary replicons), where node radius scales with weighted degree and edge weights are proportional to degenerate region frequency. For clarity, the figure has been filtered to exclude weak (low weight) edges (e.g., between DL1 and cl0 and cl6). (B) HIR (identity > 99.8%, length > 5 kb) (blue nodes) and their containing replicon (red nodes) where node radius scales linearly with weighted degree, and edge weights are proportional to region length.
The prevalence of each IS element within the community was visualized using a bipartite graph of IS to host genome, where nodes and edge size is relative to the number of ISs found within the target genome (SI Appendix, Fig. S13). The out-degree distribution of IS nodes follows an 80/20 power-law (R2 = 0.97) with the top three nodes ISHla1 (38.4%), ISHla6 (20.7%), and ISHla7 (14.6%) possessing 73.7% of all edges. The distribution of occupancy of the 15 unique sequence clusters also follows a power-law (R2 = 0.95) with the largest four clusters comprising 80% of extracted sequences (Fig. 3A). Community composition (as assessed by SSU rRNA genes) also follows a power-law (R2 = 0.972).
Gene Exchange of Long High-Identity Regions.
A striking feature of the four DL genomes was the sharing of numerous long (up to 34.9 kb), high-identity regions (HIR) that share ∼100% nucleotide identity (Fig. 2): 30 regions (>5 kb) shared across seven of the nine replicons and 13 regions (>10 kb) shared between six replicons. Experimental validation by PCR amplification and sequencing confirmed the presence of HIR in their respective genomes (SI Appendix). Represented as a network with replicons as nodes and edges weighted by summation of region lengths >5 kb (Fig. 3B), the most significant node was for replicon NC_012028 of H. lacusprofundi; ranked first both by normalized weighted degree (0.374) and normalized betweenness centrality (0.567). The top three most significant edges (normalized weights: 0.323, 0.182, 0.161) are intergenomic links from H. lacusprofundi NC_012028 to DL31:Contig114, DL1:Contig38, and DL1:Contig37. From a genomic perspective, H. lacusprofundi and DL1 each share regions (>5 kb) with all three other genomes, and DL31 and tADL share regions with two others (Fig. 2).
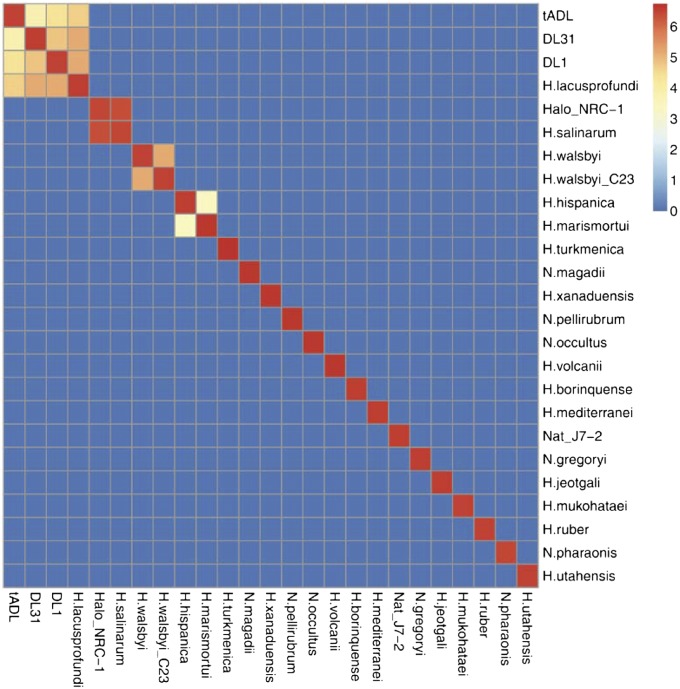
To assess whether HIR were shared between other haloarchaea, an all-vs.-all analysis was performed between 25 closed haloarchaeal genomes, plotting their ANI (SI Appendix, Fig. S14) or total extent of HIR (Fig. 4). The median global ANI was 71.3%, similar to the median for just the four DL haloarchaea (73.1%). However, only nine of the 300 combinations of genome pairs contained HIR (identity >99%, length, >2 kb) (Fig. 4). Three pairs were from closely related Halobacterium (salinarum, NRC-1: >99% ANI), Haloquadratum (walsbyi: 98.7% ANI) and Haloarcula (hispanica, marismortui: 90.6% ANI). The remaining six genome pairs represented the full complement of possible pair-wise combinations of DL haloarchaea. To assess whether the DL-specific HIR were present in haloarchaea from other hypersaline environments, FR was performed using the >10-kb DL HIR against 15 saltern metagenomes (11 Chula Bay, 4 Santa Pola; 2.8 million reads). Incomplete coverage was obtained for alignment identities above 70% from six samples, with the reads primarily mapping to ISs (SI Appendix, Fig. S15). Above 99% alignment identity, no reads mapped. The analyses demonstrate that DL HIR are only represented in DL genome and metagenome data. Comparative analyses using matching genome/metagenome data from other hypersaline environments will need to be performed to determine whether HIR intergenera exchange is unique to DL.
Fig. 4.
HIR analysis of haloarchaeal genomes. Heatmap with rows and columns ordered by minimum variance for the extent of shared HIR (identity > 99.8%, length > 5 kb) between 25 closed haloarchaeal genomes.
The >10-kb HIRs tend to be flanked on either side, and in many cases both sides, by transposase genes, or genes annotated as integrase, resolvase, endonuclease, or in a few cases, hypothetical. The 654 genes completely contained within HIR (>5 kb) consisted of 6 (<1%) core and 180 (27.5%) noncore genes. By ArCOG assignment, 380 (58.1%) were poorly characterized, 177 (27.1%) information storage and processing, 60 (9.2%) cellular processes and signaling, and 37 (5.7%) metabolism. The majority (63%) assigned to cellular processes and signaling were associated with defense mechanisms (V).
SNP Mutations.
The high mean Illumina read depths for the main replicons of tADL (530), DL31 (301), and H. lacusprofundi (112) enabled fixed SNPs (i.e., those present in the isolate genomes but occurring in <10% of metagenome population) to be confidently evaluated (minimum read depth 20, variant frequency above 0.9). The SNPs had a typical trait of being orientated toward transitions (A ↔ G, C ↔ T), and equivalent representation in intergenic or non/synonymous coding regions (SI Appendix, Fig. S7 and Table S6). For tADL, the most highly assigned class of orthologous groups was general function prediction only (R), followed by energy production and conversion (C) (SI Appendix, Table S7). This result contrasts with highest SNP numbers for signal transduction genes (T) and transcriptional regulation genes (K) in Leptospirillum populations in acid mine drainage biofilms (42).
Recombination events would also contribute to the apparent level of SNPs. The population genetic parameters of scaled rates (measured over the effective population and not just per generation per nucleotide) for mutation (θ) and recombination (ρ) were estimated at 95% confidence intervals (SI Appendix). The ratio of rates (ρ/θ) suggests that genetic variability within tADL (3.37, 3.86) occurs at a ∼fourfold lower rate by mutation than recombination, whereas for primary replicons in DL31 (0.79, 0.90) and H. lacusprofundi (0.99, 1.13), and their secondary replicons (SI Appendix), recombination and mutation are approximately equivalent sources of variability. Although the genome variation occurring via ISs and HIR provides information about the extent of genome variation in the DL haloarchaea, these ratio values inform about the relative contribution of recombination and mutation.
Discussion
The DL community is dominated by haloarchaea that exhibit signatures of very high intergenera gene exchange. Secondary replicons and large portions of the primary replicons of tADL and DL1 are characterized by markers of high genomic volatility: high noncore gene content, low CAI/CBI indexes, high numbers of ISs compared with most microbial species, long stretches with markedly lower or higher than average FR, and shared HIR (Figs. 1 and 2). The type of genome variation within the DL haloarchaea has specific characteristics: (i) SNPs are selectively neutral across all four genomes, but recombination is a greater source of genomic variability than mutation in tADL, and ISs (but not HIR) have most connectivity to tADL (Figs. 1–3); (ii) compared with other haloarchaea, the presence of shared intergenera HIR is unprecedented. There is also strong genomic evidence for sympatric speciation in DL: Genomic characteristics of tADL not only describe specific features of competitiveness, but distinctions between all four DL genera are indicative of niche adaptation. This appears to extend to the level of phylotypes, with low coverage FR regions and “the nature of the tADL-related fifth genome” providing evidence that subpopulations exist and fulfill roles in some level of niche adaptation (i.e., ecotypes). The extent and nature of the genome variation that has occurred among the DL haloarchaea raises important questions about the evolutionary path to community composition, species abundance and niche partitioning in this Antarctic lake. Below we consider this:
Power-law distributions can imply a scale-free network, whereby the number of links to nodes provides a measure of node fitness (43), and the abundance of transposons has been linked to cold adaptation (44–46). The fact that tADL has the highest abundance and is highly connected to ISs (Fig. 3A and SI Appendix, Fig. S13) would be consistent with free exchange of ISs promoting a fitness benefit. However, HIR distribution does not follow a power-law and the H. lacusprofundi secondary replicon has the largest number of connections (Fig. 3B), not tADL. It could be argued that HIR are more disruptive to tADL because it contains a single replicon. For that matter, even high IS density is confined primarily to a 400-kb region in tADL (1.150–1.550 Mb). However, it is possible that the “traffic” of ISs and HIR offers no selective advantage (or fitness value). The high contribution of recombination relative to point mutations for other haloarchaea (e.g., ref. 21), and the capacity (in the laboratory) to exchange and recombine fragments >100 kb between genomes differing by ∼86% ANI (25), have been described. In fact, it has previously been suggested that homogenization of haloarchaeal communities could, in theory, override mutations occurring within lineages thereby precluding sympatric speciation from occurring in haloarchaeal dominated hypersaline environments (25).
It is possible that Antarctic DL offers an extreme example of where gene exchange occurs between phylogenetically disparate haloarchaea. Based on laboratory growth rates for H. lacusprofundi (12), haloarchaeal generation times equate to ∼6 generations per year, 100-fold fewer generations annually than that recently described for acid mine drainage biofilms (42). In fact, the number of generations is likely to be considerably less in the bulk of the lake that remains perennially cold, compared with the top few meters where temperatures can elevate above 0 °C (6, 9, 10). Genetic transformation may also be facilitated by the inherently cryogenic stabilizing conditions that are not unlike the cold CaCl2 conditions used for artificial transformation. Biofilm forming characteristics of prominent members of the DL community may also assist gene exchange (13, 15, 16). Importantly, given the very low generation rate of DL haloarchaea, genetic sweeps and establishment of new strains by vertical inheritance is expected to occur very slowly. An approximate calculation shows that it would take a single cell ∼10 y to fully colonize the lake: 60 generations at 6 generations per y to achieve 7 × 1017 total cells, equating to the lake volume of ∼7 × 1012 mL × 105 cells per mL. In reality, competition and cell death would substantially extend this time. Therefore, parallel recombination events would appear to be occurring throughout the lake between individual cells (strains, species, genera), with sympatric speciation being maintained by virtue of ecotype distinctions enabling niche differentiation.
A precedent for Antarctic ecosystem distinctiveness is known for Ace Lake, where a population of essentially clonal green sulfur bacteria, C-Ace, dominates the oxycline (47, 48) (SI Appendix). C-Ace is predicted to have evolved dominance through mechanisms allowing phage evasion linked to a growth response controlled by the annual polar light cycle (48). In DL, by restricting the nature of species that can grow and compete in the lake and providing conditions that naturally promote gene exchange, gene exchange events have become relatively frequent and fixed in the population, with the process becoming an important driver of haloarchaeal community evolution.
Materials and Methods
Metagenomic analyses were performed by filtering DL water (depths: 5, 13, 24, and 36 m) by sequential filtration through a 20-µm prefilter onto 3.0-, 0.8-, 0.1-µm filters. Metagenome sequencing and genome sequencing for tADL, DL31, H. lacusprofundi, and DL1 was performed at the US Department of Energy (DOE) Joint Genome Institute. Full details of materials and methods and full descriptions of Results and Discussion are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Nico Wanandy for extracting DNA from filters, Yan Liao for assistance with PCR and sequencing of HIR, Aaron Darling for helpful discussions, Philip Johnson for guidance on the use of Population genetic Inference In Metagenomics, Jon Magnuson and Jerry Jenkins for access to the draft Dunaliella salina Culture Collection of Algae and Protozoa 19/18 genome sequence, and Tassia Kolesnikow for comments on the manuscript. We also thank the Editor and reviewers who provided very insightful and constructive feedback. This work was supported by the Australian Research Council and the Australian Antarctic Science program and undertaken with the assistance of resources provided at the National Computational Infrastructure National Facility systems at the Australian National University through the National Computational Merit Allocation Scheme supported by the Australian Government. The work conducted by the US DOE Joint Genome Institute is supported by the Office of Science of the US DOE under Contract DE-AC02-05CH11231. M.D.S. is grateful for the support by the Max Planck Society, and particularly D. Oesterhelt (Department of Membrane Biochemistry, Max Planck Institute).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The genome data reported in this paper have been deposited in the National Center for Biotechnology Information BioProject database, www.ncbi.nlm.nih.gov/bioproject [accession nos. PRJNA53493 (tADL), PRJNA58807 (HI), PRJNA72619 (DL31), and PRJNA75121 (DL1)]; and the metagenome data have been deposited in the Genomes OnLine database, www.genomesonline.org [accession nos. Gs0000582 (13m_0.1), Gs0000585 (24m_0.8), Gs0000586 (SSU rRNA pyrotag data), Gs0000587 (24m_0.1), Gs0000592 (36m_pooled), Gs0000594 (24m_3.0), and Gs0000595 (5mRS_0.1)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1307090110/-/DCSupplemental.
References
- 1.Campbell PJ. Primary productivity of a hypersaline Antarctic lake. Aust J Mar Freshwater Res. 1978;29(6):717–724. [Google Scholar]
- 2.Gibson JAE. The meromictic lakes and stratified marine basins of the Vestfold Hills, East Antarctica. Antarct Sci. 1999;11(2):175–192. [Google Scholar]
- 3.Cavicchioli R. Cold-adapted archaea. Nat Rev Microbiol. 2006;4(5):331–343. doi: 10.1038/nrmicro1390. [DOI] [PubMed] [Google Scholar]
- 4.Zwartz D, Bird M, Stone J, Lambeck K. Holocene sea-level change and ice-sheet history in the Vestfold Hills, East Antarctica. Earth Planet Sci Lett. 1998;155(1-2):131–145. [Google Scholar]
- 5.Wilkins D, et al. Key microbial drivers in Antarctic aquatic environments. FEMS Microbiol Rev. 2012;37(3):303–335. doi: 10.1111/1574-6976.12007. [DOI] [PubMed] [Google Scholar]
- 6.Ferris JM, Burton HR. The annual cycle of heat content and mechanical stability of hypersaline Deep Lake, Vestfold Hills, Antarctica. Hydrobiologia. 1988;165(1):115–128. [Google Scholar]
- 7.Bowman JPJ, McCammon SAS, Rea SMS, McMeekin TAT. The microbial composition of three limnologically disparate hypersaline Antarctic lakes. FEMS Microbiol Lett. 2000;183(1):81–88. doi: 10.1111/j.1574-6968.2000.tb08937.x. [DOI] [PubMed] [Google Scholar]
- 8. Akiyama M (1981) Plankton and bottom deposits of Lake Funazoko-ike in Skarvs Nes, Antarctica. Shimane University Education Department Lett 9:1975.
- 9. Tominaga H, Fukui F (1981) Saline lakes at Syowa Oasis, Antarctica. Hydrobiologia 81-82(1):375–389.
- 10.Wright SW, Burton HR. The biology of Antarctic saline lakes. Hydrobiologia. 1981;81-82:319–338. [Google Scholar]
- 11.Cavicchioli R. Archaea—timeline of the third domain. Nat Rev Microbiol. 2011;9(1):51–61. doi: 10.1038/nrmicro2482. [DOI] [PubMed] [Google Scholar]
- 12.Franzmann PD, et al. Halobacterium lacusprofundi sp. nov., a halophilic bacterium isolated from Deep Lake, Antarctica. Syst Appl Microbiol. 1988;11(1):20–27. [Google Scholar]
- 13.Reid IN, et al. Terrestrial models for extraterrestrial life: Methanogens and halophiles at Martian temperatures. Int J Astrobiol. 2006;5(2):89–97. [Google Scholar]
- 14.Mou YZ, et al. Halohasta litorea gen. nov. sp. nov., and Halohasta litchfieldiae sp. nov., isolated from the Daliang aquaculture farm, China and from Deep Lake, Antarctica, respectively. Extremophiles. 2012;16(6):895–901. doi: 10.1007/s00792-012-0485-5. [DOI] [PubMed] [Google Scholar]
- 15.Fröls S. Archaeal biofilms: Widespread and complex. Biochem Soc Trans. 2013;41(1):393–398. doi: 10.1042/BST20120304. [DOI] [PubMed] [Google Scholar]
- 16.Fröls S, Dyall-Smith M, Pfeifer F. Biofilm formation by haloarchaea. Environ Microbiol. 2012;14(12):3159–3174. doi: 10.1111/j.1462-2920.2012.02895.x. [DOI] [PubMed] [Google Scholar]
- 17.Gibson JAE, et al. Unsaturated diether lipids in the psychrotrophic archaeon Halorubrum lacusprofundi. Syst Appl Microbiol. 2005;28(1):19–26. doi: 10.1016/j.syapm.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 18.Torsvik T, Dundas ID. Bacteriophage of Halobacterium salinarium. Nature. 1974;248(450):680–681. doi: 10.1038/248680a0. [DOI] [PubMed] [Google Scholar]
- 19.DasSarma S, RajBhandary UL, Khorana HG. High-frequency spontaneous mutation in the bacterio-opsin gene in Halobacterium halobium is mediated by transposable elements. Proc Natl Acad Sci USA. 1983;80(8):2201–2205. doi: 10.1073/pnas.80.8.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenshine I, Tchelet R, Mevarech M. The mechanism of DNA transfer in the mating system of an archaebacterium. Science. 1989;245(4924):1387–1389. doi: 10.1126/science.2818746. [DOI] [PubMed] [Google Scholar]
- 21.Papke RT, Koenig JE, Rodríguez-Valera F, Doolittle WF. Frequent recombination in a saltern population of Halorubrum. Science. 2004;306(5703):1928–1929. doi: 10.1126/science.1103289. [DOI] [PubMed] [Google Scholar]
- 22.Legault BA, et al. Environmental genomics of “Haloquadratum walsbyi” in a saltern crystallizer indicates a large pool of accessory genes in an otherwise coherent species. BMC Genomics. 2006;7:171. doi: 10.1186/1471-2164-7-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papke RT, et al. Searching for species in haloarchaea. Proc Natl Acad Sci USA. 2007;104(35):14092–14097. doi: 10.1073/pnas.0706358104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhodes ME, Spear JR, Oren A, House CH. Differences in lateral gene transfer in hypersaline versus thermal environments. BMC Evol Biol. 2011;11:199. doi: 10.1186/1471-2148-11-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naor A, Lapierre P, Mevarech M, Papke RT, Gophna U. Low species barriers in halophilic archaea and the formation of recombinant hybrids. Curr Biol. 2012;22(15):1444–1448. doi: 10.1016/j.cub.2012.05.056. [DOI] [PubMed] [Google Scholar]
- 26.Zawadzki P, Roberts MS, Cohan FM. The log-linear relationship between sexual isolation and sequence divergence in Bacillus transformation is robust. Genetics. 1995;140(3):917–932. doi: 10.1093/genetics/140.3.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fraser C, Hanage WP, Spratt BG. Recombination and the nature of bacterial speciation. Science. 2007;315(5811):476–480. doi: 10.1126/science.1127573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams D, Gogarten JP, Papke RT. Quantifying homologous replacement of loci between haloarchaeal species. Genome Biol Evol. 2012;4(12):1223–1244. doi: 10.1093/gbe/evs098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papke RT, Gogarten JP. Ecology. How bacterial lineages emerge. Science. 2012;336(6077):45–46. doi: 10.1126/science.1219241. [DOI] [PubMed] [Google Scholar]
- 30.Friedman J, Alm EJ, Shapiro BJ. Sympatric speciation: When is it possible in bacteria? PLoS ONE. 2013;8(1):e53539. doi: 10.1371/journal.pone.0053539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benlloch S, et al. Archaeal biodiversity in crystallizer ponds from a solar saltern: Culture versus PCR. Microb Ecol. 2001;41(1):12–19. doi: 10.1007/s002480000069. [DOI] [PubMed] [Google Scholar]
- 32.Ochsenreiter T, Pfeifer F, Schleper C. Diversity of Archaea in hypersaline environments characterized by molecular-phylogenetic and cultivation studies. Extremophiles. 2002;6(4):267–274. doi: 10.1007/s00792-001-0253-4. [DOI] [PubMed] [Google Scholar]
- 33.Oh D, Porter K, Russ B, Burns D, Dyall-Smith M. Diversity of Haloquadratum and other haloarchaea in three, geographically distant, Australian saltern crystallizer ponds. Extremophiles. 2010;14(2):161–169. doi: 10.1007/s00792-009-0295-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bennetzen JL, Hall BD. Codon selection in yeast. J Biol Chem. 1982;257(6):3026–3031. [PubMed] [Google Scholar]
- 35.Sharp PM, Li WH. The codon Adaptation Index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Capes MD, DasSarma P, DasSarma S. The core and unique proteins of haloarchaea. BMC Genomics. 2012;13:39. doi: 10.1186/1471-2164-13-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. von Mandach C, Merkl R. (2010) Genes optimized by evolution for accurate and fast translation encode in Archaea and Bacteria a broad and characteristic spectrum of protein functions. BMC Genomics 11:617. [DOI] [PMC free article] [PubMed]
- 38.Zhang Z, et al. KaKs_Calculator: Calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics. 2006;4(4):259–263. doi: 10.1016/S1672-0229(07)60007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varani AM, Siguier P, Gourbeyre E, Charneau V, Chandler M. ISsaga is an ensemble of web-based methods for high throughput identification and semi-automatic annotation of insertion sequences in prokaryotic genomes. Genome Biol. 2011;12(3):R30. doi: 10.1186/gb-2011-12-3-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walsby AE. Gas vesicles. Microbiol Rev. 1994;58(1):94–144. doi: 10.1128/mr.58.1.94-144.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pagaling E, et al. Sequence analysis of an Archaeal virus isolated from a hypersaline lake in Inner Mongolia, China. BMC Genomics. 2007;8:410. doi: 10.1186/1471-2164-8-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Denef VJ, Banfield JF. In situ evolutionary rate measurements show ecological success of recently emerged bacterial hybrids. Science. 2012;336(6080):462–466. doi: 10.1126/science.1218389. [DOI] [PubMed] [Google Scholar]
- 43.Caldarelli GG, Capocci AA, De Los Rios PP, Muñoz MAM. Scale-free networks from varying vertex intrinsic fitness. Phys Rev Lett. 2002;89(25):258702. doi: 10.1103/PhysRevLett.89.258702. [DOI] [PubMed] [Google Scholar]
- 44.DeLong EF, et al. Community genomics among stratified microbial assemblages in the ocean’s interior. Science. 2006;311(5760):496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- 45.Lauro FM, et al. Large-scale transposon mutagenesis of Photobacterium profundum SS9 reveals new genetic loci important for growth at low temperature and high pressure. J Bacteriol. 2008;190(5):1699–1709. doi: 10.1128/JB.01176-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allen MA, et al. The genome sequence of the psychrophilic archaeon, Methanococcoides burtonii: The role of genome evolution in cold adaptation. ISME J. 2009;3(9):1012–1035. doi: 10.1038/ismej.2009.45. [DOI] [PubMed] [Google Scholar]
- 47.Ng C, et al. Metaproteogenomic analysis of a dominant green sulfur bacterium from Ace Lake, Antarctica. ISME J. 2010;4(8):1002–1019. doi: 10.1038/ismej.2010.28. [DOI] [PubMed] [Google Scholar]
- 48.Lauro FM, et al. An integrative study of a meromictic lake ecosystem in Antarctica. ISME J. 2011;5(5):879–895. doi: 10.1038/ismej.2010.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krzywinski M, et al. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.