Significance
The opening of P2X receptor channels by extracellular ATP underlies purinergic signaling in many tissues. Here we use computational and functional approaches to study helix interactions within the transmembrane domain of P2X receptors. Our results suggest that the intersubunit crevices observed in the X-ray structure of detergent-solubilized ATP-bound receptors are nonnative but confirm helix interactions within individual subunits observed in both apo and ATP-bound receptors and identify a hot spot within a narrow internal region where the gating and permeation properties can be readily tuned.
Keywords: pore-opening mechanism, transmembrane intersubunit crevices
Abstract
P2X receptor channels open in response to the binding of extracellular ATP, a property that is essential for purinergic sensory signaling. Apo and ATP-bound X-ray structures of the detergent-solubilized zebrafish P2X4 receptor provide a blueprint for receptor mechanisms but unexpectedly showed large crevices between subunits within the transmembrane (TM) domain of the ATP-bound structure. Here we investigate both intersubunit and intrasubunit interactions between TM helices of P2X receptors in membranes using both computational and functional approaches. Our results suggest that intersubunit crevices found in the TM domain of the ATP-bound crystal structure are not present in membrane-embedded receptors but substantiate helix interactions within individual subunits and identify a hot spot at the internal end of the pore where both the gating and permeation properties of P2X receptors can be tuned. We propose a model for the structure of the open state that has stabilizing intersubunit interactions and that is compatible with available structural constraints from functional channels in membrane environments.
P2X receptor channels are a family of trimeric cation-selective channels that are activated by extracellular ATP (1, 2). These ligand-gated ion channels are expressed in many tissues, including the central and peripheral nervous systems and the immune system, where they play a range of important roles in sensory signaling and inflammation (1, 3).
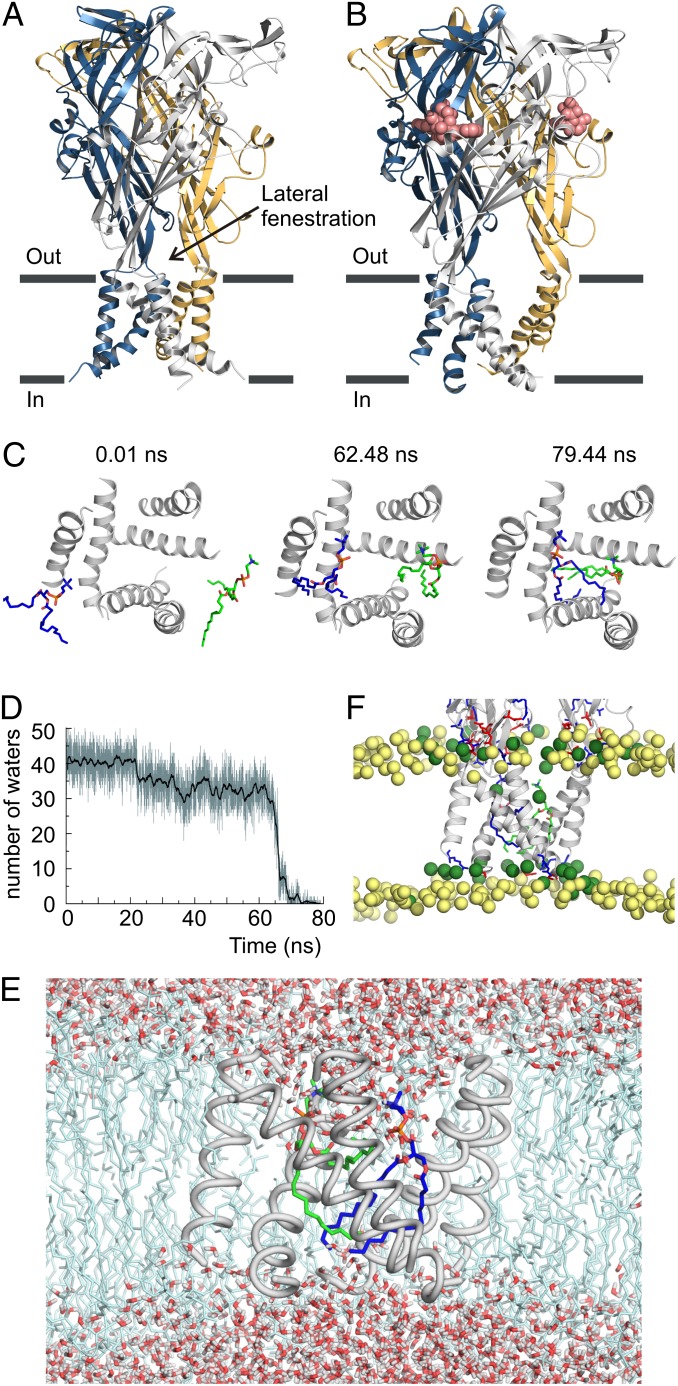
Recent X-ray structures of the zebrafish P2X4 (zfP2X4) receptor in apo and ATP-bound forms (Protein Data Bank ID codes 4DW0 and 4DW1, respectively) have revealed the molecular design of these proteins (2, 4, 5) and have provided valuable information on how ATP binding triggers the opening of the transmembrane (TM) pore (Fig. 1 A and B). ATP binds to a cleft between subunits within the large extracellular domain, inducing cleft closure and an accompanying lateral flexing of the β-sheet connecting the extracellular domain to the TM domain (5). The apo structure of the zfP2X4 receptor reveals that the TM1 helix is positioned peripheral to the TM2 helix and that the TM2 helix occludes the pore at the central axis within the outer half of the membrane (4, 5). In accord with this structure, accessibility studies show that the TM2 helix lines the aqueous pore and that the residues forming the gate are positioned within the occlusion in the apo structure (6–8). Lateral fenestrations within the extracellular domain provide a path for ions to enter and exit the extracellular vestibule positioned above this TM2 occlusion, and these fenestrations are thought to change conformation in response to ATP binding (9, 10). The ATP-bound structure shows that pore opening involves widening of the extracellular vestibule and an iris-like expansion of the pore (5). Intersubunit interactions within the TM domain in the apo structure are limited to the gate region of TM2 (5), and thus the pore expansion observed in the ATP-bound structure leave the three subunits essentially devoid of intersubunit interactions within the membrane (Fig. 1 A and B). In effect, the lateral fenestrations that are positioned above the outer leaflet of the membrane in the apo structure have expanded dramatically to encompass most of the TM domain.
Fig. 1.
MD simulations of the ATP-bound zfP2X4 structure in a membrane environment. (A) Backbone ribbon representation of the X-ray structure of apo zfP2X4 receptor viewed parallel to the membrane with individual subunits colored blue, gold, and gray. (B) Backbone ribbon representation of the X-ray structure of ATP-bound zfP2X4 receptor. ATP is shown as red spheres. (C) Snapshots of the TM domain viewed from the extracellular side at various time points during the restrained MD simulation in a DMPC bilayer. TM helices are shown as gray backbone ribbon representations, and two DMPC lipid molecules are shown as stick representations with carbons in green and blue. Lipids began entering the intersubunit crevices at 62.48 ns and fully occupied the ion conduction pore at 79.44 ns. (D) Plot showing the number of water molecules within a 13-Å–deep section of the pore spanning residues A344–I355. (E) Snapshot of the MD simulation at 79.44 ns with water molecules and lipids shown. Note the absence of water molecules in the section spanning residues A344–I355. (F) Snapshot of the TM domain viewed parallel to the membrane at 79.44 ns in the MD simulation. Blue and red sticks on the protein represent positively and negatively charged residues, respectively. Yellow spheres are phosphorous atoms of bulk lipid molecules, and green spheres are phosphorous atoms of lipids around the receptor. In the lower leaflet, phosphates around the receptor are drawn toward the middle of the bilayer because of the hydrophobic mismatch between the TM domain and the bilayer.
Many of the conformational rearrangements predicted from the X-ray structures of the zfP2X4 receptor are consistent with structural constraints obtained from functional studies on the protein in a membrane environment. For example, ATP binding to the cleft and subsequent cleft closure were predicted to occur based on proximity tethering (11), normal mode analysis (12), and both metal bridging and spectroscopic studies (13, 14). In addition, the accessibility of both methanethiolsulfonate compounds and metals (Ag+ and Cd2+) to cysteine residues introduced into TM1 and TM2 (6–8) is consistent with the expansion of the external pore predicted from the zfP2X4 structures (Fig. S1A). However, metal bridges engineered into the internal region of the TM2 helix (7, 8) suggest that the internal pore narrows as the channel opens, a feature that is not evident in the ATP-bound zfP2X4 structure (Fig. S1B). Moreover, the large crevices between subunits within the TM domain of the ATP-bound structure are unprecedented in membrane proteins and would be expected to destabilize the open state of the protein when it is embedded in a lipid membrane (15). One proposal is that lipids might occupy these crevices and serve to stabilize the structure of the open state (5). The goal of the present study was to investigate the structure of the open state of P2X receptors in a membrane environment using both computational and functional approaches and to assess the validity of the proposed mechanism of ATP activation and pore opening. Our results suggest that the absence of intersubunit interactions in the ATP-bound structure is not representative of the native structure but that intrasubunit interactions within the TM domain are faithfully captured in both apo and ATP-bound crystal structures of detergent-solubilized P2X receptors. Our results also demonstrate that the internal end of the pore plays a crucial role in tuning both the gating and permeation properties of P2X receptors.
Results
Molecular Dynamics Simulations of the ATP-Bound Structure of zfP2X4.
To evaluate how the large intersubunit crevices observed in the X-ray structure of the ATP-bound zfP2X4 receptor impact receptor function, we performed molecular dynamics (MD) simulations with the receptor incorporated into a hydrated 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) bilayer, restraining the receptor and ATP to the X-ray structure while allowing solvent and lipids to move. At the outset of the simulation, the pore was heavily hydrated, compatible with the formation of an ion permeation pathway. However, over time, two lipids diffused into the intersubunit crevices and ultimately entered the pore (Fig. 1C). Shortly after the entrance of the first lipid into the pore at ∼62.5 ns, the number of water molecules in the pore dropped sharply (Fig. 1D). The expulsion of water from the pore facilitated the entrance of the second lipid molecule. In the end, a 13-Å–deep section of the pore became devoid of water (Fig. 1E). We also ran a simulation in which the structural restraint on the receptor was maintained for the first 12.2 ns to allow the lipids, water, and ions to equilibrate with the membrane-embedded receptor and then was released for the duration of the simulation. Again, we observed that two lipids entered the pore (the first at ∼43.8 ns), resulting in dehydration of a large section of the pore.
We also observed a pronounced hydrophobic mismatch between the receptor TM domain and the DMPC lipid bilayer in both the restrained and unrestrained MD simulations. Lipids in the lower leaflet of the bilayer adjacent to the receptor moved toward the center of the bilayer (relative to the more distant lipids) to match the short hydrophobic thickness of the TM domain (Fig. 1F). The short hydrophobic thickness results from the relatively large tilt angle of the TM2 helices with respect to the membrane normal. It is worth noting that DMPC is a relatively short lipid with only 14 carbons on each acyl chain. Longer acyl chains in other common lipids would have increased further the extent of hydrophobic mismatch between the membrane and the TM domain.
These simulations led us to conclude that the large intersubunit crevices observed in the ATP-bound structure of the detergent-solubilized zfP2X4 receptor are not compatible with ion conduction and thus likely represent a nonnative feature. In addition, the length of the TM domain in the X-ray structure may be inconsistent with the thickness of native lipid membranes.
Identification of Candidate Metal Bridges Between TM1 and TM2 Helices in P2X Receptors.
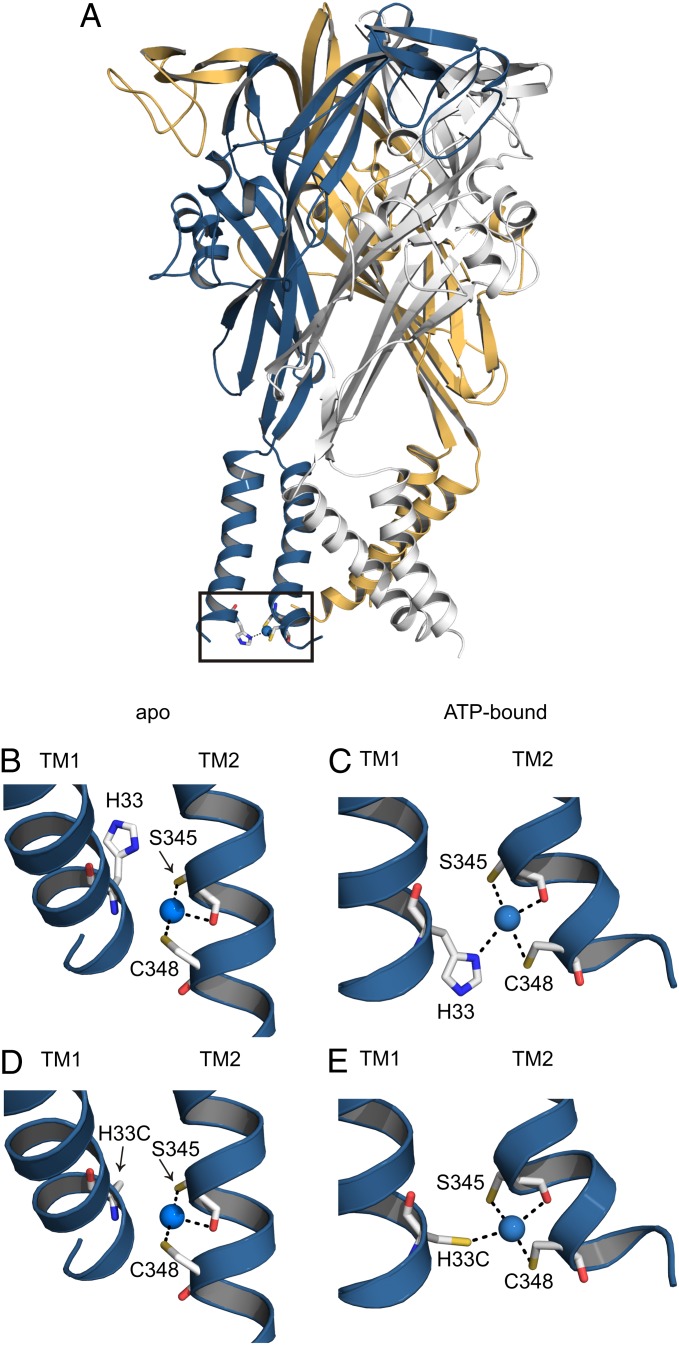
To define interactions within individual subunits of the TM domain, we inspected the interface between TM1 and TM2 in the zfP2X4 X-ray structures to identify positions where state-dependent metal bridges might be engineered. In a previous study on a rat P2X2 (rP2X2) construct in which a native Cys in TM2 was mutated to threonine (C348T), we found that introducing the S345C mutation in TM2 resulted in an inhibitory metal bridge involving H33 in TM1 (7). This region of TM1 and TM2 was resolved in the structure of apo zfP2X4 spans G32–L361 (corresponding to G30–L353 in rP2X2); however, the first TM1 residue resolved in the ATP-bound structure was R36, corresponding to R34 in rP2X2. To evaluate metal bridges involving this intracellular region of TM1 and TM2, we created models for metal coordination in both apo and ATP-bound zfP2X4. To include the position corresponding to H33 in P2X2, we grafted the first turn of the TM1 helix from the apo structure onto the ATP-bound structure, extending the TM1 N terminus to G32 (G30 in rP2X2).
We evaluated these zfP2X4 models to determine whether a state-dependent metal bridge for Cd2+ could be formed at the intrasubunit interface between TM1 and TM2 and found a promising candidate that forms when the N353C mutant (S345C in rP2X2) is introduced in a background that retains the native C356 in TM2 (C348 in rP2X2) and either His or Cys at N35 in TM1 (H33 in rP2X2) (Fig. 2). This predicted metal-coordination site is unusual in that it involves two positions on the TM2 helix that are one helical turn apart and therefore should coordinate Cd2+ in either closed or open states (including a contribution from the backbone carbonyl of N353), but the additional contribution of either His or Cys in TM1 could occur only in the open state (Fig. 2 B–E). The relative motions of TM1 and TM2 between the ATP-bound and apo zfP2X4 structures include a 45° rotation, so that in the apo structure the His or Cys in TM1 cannot readily contribute to bridging Cd2+ with the two Cys residues in TM2 (Fig. 2 B and D).
Fig. 2.
Identifying potential metal bridges between TM1 and TM2 helices. (A) The ATP-bound zfP2X4 receptor with individual subunits colored blue, gold, and gray. A modeled metal bridging site is displayed in the blue subunit involving H33, S345C, and C348 (numbering based on rP2X2). (B) Magnified view of the modeled metal bridging site in the apo zfP2X4 receptor containing the H-C-C bridge, where Cd2+ is coordinated by S345C and C348 in TM2 and H33 in TM1 is rotated away from the S345C/C348 bridge with its nearest Nδ atom 5.2 Å from Cd2+. (C) Magnified view of the modeled metal bridging site in the ATP-bound zfP2X4 receptor containing the H-C-C bridge, where Cd2+ is coordinated by S345C and C348 in TM2 and H33 in TM1. (D) Magnified view of the modeled metal bridging site in the apo zfP2X4 receptor containing the C-C-C bridge, where Cd2+ is coordinated by S345C and C348 in TM2 and H33C in TM1 is rotated away from the S345C/C348 bridge with its S atom 6.3 Å from Cd2+. (E) Magnified view of the modeled metal bridging site in the ATP-bound zfP2X4 receptor containing the C-C-C bridge, where Cd2+ is coordinated by S345C and C348 in TM2 and H33C in TM1.
Metal Bridges Potentiate ATP-Activated Currents in rP2X2 Receptors.
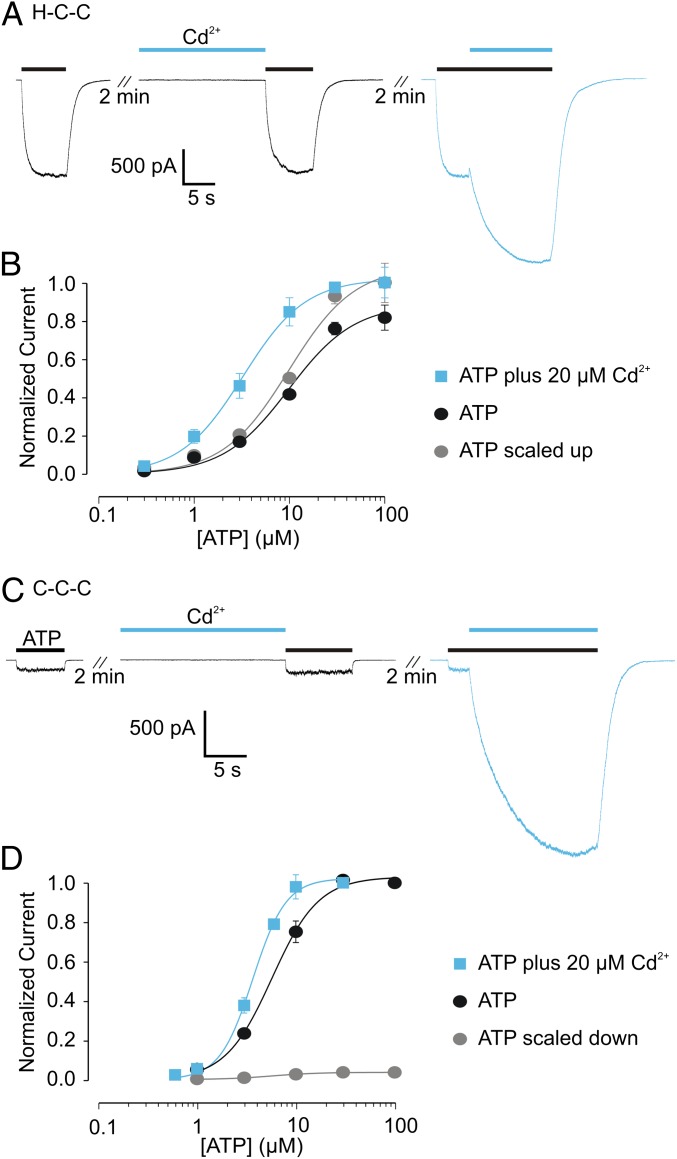
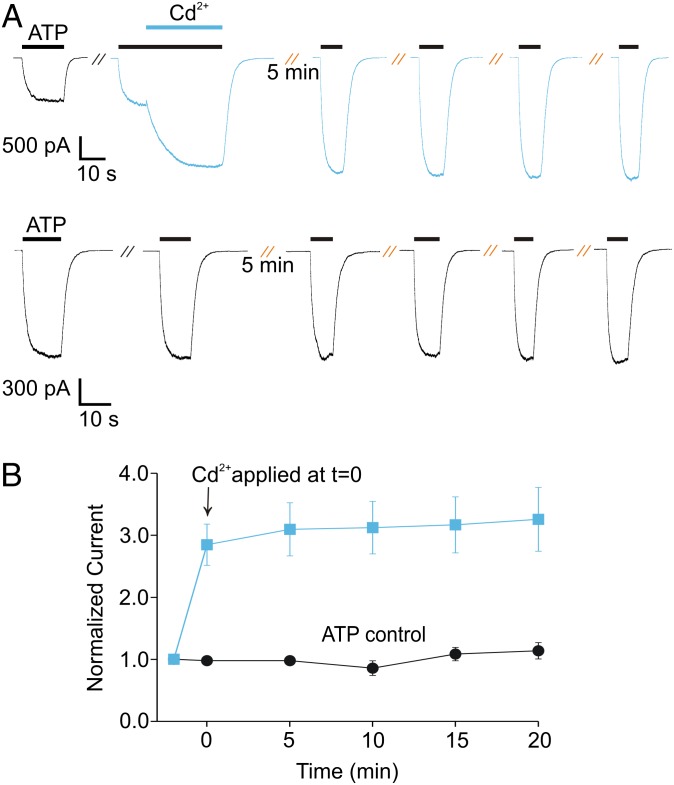
To evaluate this predicted metal coordination site, we initially generated two constructs of the rP2X2 receptor, one containing a native H33, S345C, and a native C348 (designated H-C-C), and the other containing H33C, S345C, and a native C348 (designated C-C-C). When expressed in HEK cells, both constructs gave rise to ATP-activated macroscopic currents and exhibited concentration–response relations comparable to those in the wild-type rP2X2 receptor (Fig. 3 and Table S1) (6). For both constructs, application of external Cd2+ was without effect when applied in the absence of ATP (Fig. 3 A and C), as expected from both structural (4, 5) and functional (6–8) evidence that the gate in P2X receptors is positioned external to this engineered bridging site, and thus closure of the gate in the absence of ATP would prevent Cd2+ from reaching the bridging site. However, when Cd2+ was applied externally in the presence of ATP, the metal produced robust potentiation of ATP-activated currents (Fig. 3 A and C). In the case of the H-C-C site, Cd2+ potentiated currents at an EC20 concentration of ATP by 2.3 ± 0.2-fold (n = 11); for the C-C-C site, Cd2+ potentiated currents at an EC20 concentration of ATP by 48.7 ± 6.6-fold (n = 7). When the concentration-dependence for activation by ATP was examined in the presence and absence of Cd2+, we observed a shift in the EC50 for ATP activation to lower ATP concentrations in both constructs (Fig. 3 B and D). In addition, Cd2+ produced an increase in the maximal current activated at saturating ATP concentrations; for the H-C-C construct that increase was 25 ± 9% (n = 5), and for the C-C-C construct it was 28.1 ± 3.8-fold (n = 4) (Fig. 3 B and D). The collective results demonstrate that both engineered constructs contain Cd2+-binding sites that can be occupied in an open (conducting) state of rP2X2 receptors. The quantitative differences between the effects of Cd2+ on the H-C-C and C-C-C constructs might indicate that formation of the two types of bridges alters the gating and permeation properties of P2X2 receptors to different degrees, or they may indicate that the mutations introduced to form metal bridges have distinct effects on the properties of P2X receptor channels. Nevertheless, the effects of Cd2+ on shifting the EC50 for activation by ATP and increasing the maximal current are consistent with occupancy of the Cd2+ site leading to stabilization of an open state of the channel.
Fig. 3.
Two metal bridges that potentiate ATP-activated currents. (A) Representative whole-cell currents for the H-C-C bridging construct in rP2X2 receptor channels. EC20 concentrations of ATP (3 µM; black bars) and Cd2+ (20 µM; blue bars) were applied as shown. (B) Concentration–response relationships for activation of rP2X2 H-C-C by ATP in the absence and presence of 20 µM Cd2+. A control concentration of ATP (3 µM) was applied to each cell, followed by a test concentration of ATP before the additional application of 20 µM Cd2+. Currents measured with test concentrations of ATP (before and after Cd2+) were normalized to the current amplitude measured at the control concentration of ATP to generate the ATP-alone (black data points and curve) and ATP plus Cd2+ (blue data points and curve) relations. The ATP relation then was scaled (gray data points and curve) to show the shift in EC50. Data points are mean for three to seven measurements, and error bars indicate SEM. (C) Representative whole-cell recordings for the C-C-C bridging construct in rP2X2 receptor channels. EC20 concentrations of ATP (2.6 µM; black bars) and Cd2+ (20 µM; blue bars) were applied as shown. (D) Concentration–response relationships for activation of rP2X2 C-C-C by ATP in the absence and presence of 20 µM Cd2+. A different protocol was used in the C-C-C construct because the large magnitude of potentiation necessitated the activation of currents in the absence of Cd2+ that were too small to be reliably measured. For this construct, concentration–response relationships were generated independently for ATP alone (n = 4) or for ATP plus 20 µM Cd2+ in the external solution (n = 3). The scaled relationship (gray data points and curve) was obtained by measuring fold potentiation at saturating [ATP] in a separate set of experiments (n = 4). Error bars indicate SEM. The Hill equation was fit to the data, and parameters are given in Table S1.
Biophysical Characterization of Metal Bridges in rP2X2 Receptors.
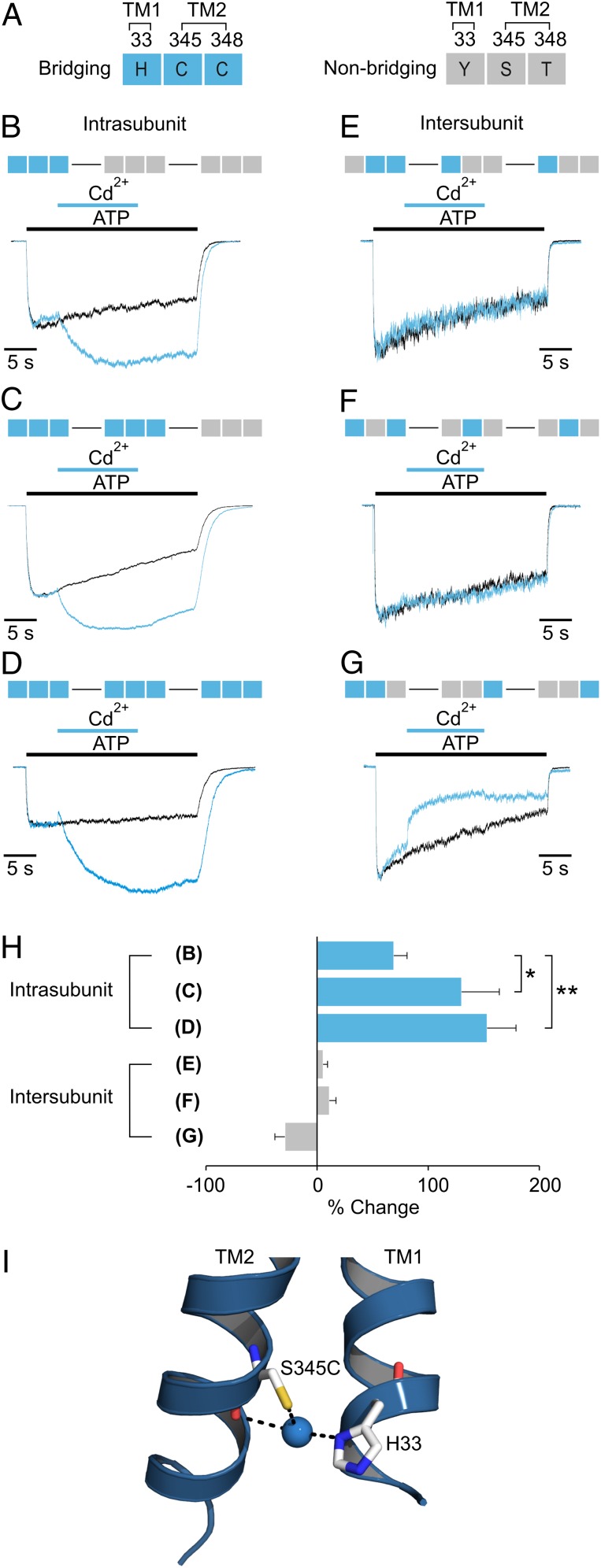
One prediction concerning the crystal structures of the apo and ATP-bound zfP2X4 receptors is that these engineered metal sites should be contained within individual subunits. To test this possibility, we constructed concatenated trimers containing different combinations of bridging and nonbridging residues at the three relevant positions (33, 345, and 348, rP2X2 numbering; Fig. 4A). When all three bridging residues (H-C-C) were present in only a single subunit within a trimer, we observed readily detectable potentiation by external Cd2+ when applied in the presence of ATP, and the extent of potentiation increased further when a second or third subunit contained all three bridging residues (Fig. 4 B–D and H). In contrast, we observed either no effect or reversible inhibition when Cd2+ was applied to three concatenated constructs where bridging residues were placed in different subunits (Fig. 4 E–H). The one instance in which we observed inhibition is consistent with a previously reported inhibitory bridge involving H33 and S345C (7) (H-C), which in our metal coordination model of the apo zfP2X4 receptor could form when the native C348 is not present (Fig. 4I). Additionally, concatenated constructs containing subunits with one, two, or three subunits containing a Cys at position 345 showed that the extent of Cd2+ inhibition increases with the number of mutant subunits (7), suggesting that this inhibitory bridge occurs within individual subunits. The present experiments with control concatamers (Fig. 4 E–H) reveal that the inhibitory bridge forms only when H33 and S345C are present in the same subunit, a finding that is supported further by the demonstration of an inhibitory disulfide bridge between H33C and S345C within individual subunits (16). We therefore conclude that both the potentiating H-C-C and inhibitory H-C metal bridges between TM1 and TM2 occur within individual subunits, consistent with the expectations from the apo and ATP-bound X-ray structures.
Fig. 4.
Subunit relationships of metal bridges. (A) Legend for concatenated subunit constructs. Each rectangle (made up of three squares) corresponds to one subunit of the trimer, and the residues at the three bridging positions (33, 345, and 348) are represented by their one-letter amino acid code in each square of the rectangle. Bridging residues (H33, S345C, and C348) are colored blue; nonbridging residues (H33Y, S345, and C348T) are colored gray. (B–D) Concatenated H-C-C subunit constructs with one, two, or three subunits containing all three residues necessary for bridging. Control currents activated by ATP alone (black trace) or ATP plus Cd2+ (blue trace) are superimposed for comparison. (E–G) Concatenated H-C-C subunit constructs with the three residues necessary for bridging split between subunits to test for intersubunit bridges. In each construct, one of the three bridging residues was removed from subunit 1 and placed in the two adjacent subunits. Control currents activated by ATP alone (black trace) or ATP plus Cd2+ (blue trace) are superimposed for comparison. (H) Bar graph summarizing the effects of Cd2+ on each concatameric construct (n = 3–4). Measurements of statistical significance are based on unpaired Student t tests; *P ≤ 0.05, **P ≤ 0.005. See Table S1 for concentration–response relations for each construct. EC 50 concentrations of ATP and 20 µM Cd2+ were used for each construct. (I) Modeling of a Cd2+ bridge between S345C and H33 in the 3T construct of rP2X2 receptor (7) using the X-ray structure of the apo zfP2X4 receptor. In 3T rP2X2 a native Cys at 348 and two additional Cys residues in the C and N termini were mutated to Thr. Bridging residues S345C and H33 (rP2X2 numbering) are shown for only one subunit, with Cd2+ represented as a blue sphere. The side view shown here is from the side opposite that depicted in Fig. 2.
A unique feature of the H-C-C and C-C-C bridges is that full coordination of Cd2+ involving residues in TM1 and TM2 would be predicted to occur in the open state, but partial and potentially stable coordination involving S345C and C348 could occur even in the closed state. In both apo and ATP-bound structures, these TM2 residues are equivalently positioned on the same face of the helix, and there is precedence in other proteins in which two residues on one face of an α-helix form a strong metal-binding site (17–19). In experiments in which both Cd2+ and ATP were removed following potentiation of ATP-activated currents, we observed that both H-C-C and C-C-C constructs could close (Fig. 3 A and C), indicating either that Cd2+ rapidly dissociates from the channel or that the bridge does not lock the channel open per se, as was observed previously for the internal gate region of the Shaker Kv channel (20). To determine whether Cd2+ remains stably associated with the P2X2 receptor after ATP unbinds and channels close, we used a protocol in which Cd2+ was applied together with ATP to potentiate channel activity; once the channel closed after both Cd2+ and ATP were removed, we repeatedly challenged the cell with ATP alone to assess how long channel activity would remain potentiated (Fig. 5A). In experiments with the H-C-C construct, we observed little evidence of recovery from Cd2+-mediated potentiation even 20 min after the removal of Cd2+ (Fig. 5), demonstrating stable interaction of the metal with the closed channel. Using this protocol for the C-C-C construct, we also observed stable potentiation 15 min after removal of Cd2+.
Fig. 5.
Cd2+ remains tightly bound in the closed state. (A) Representative whole-cell recordings measuring Cd2+ dissociation from closed channels for the H-C-C construct. Control traces show no change in ATP-activated current over time. In both experiments, there was a 2-min delay between the first and second ATP applications. (B) Plot of fractional potentiation as a function of time for experiments such as that shown in A. Data points are mean ± SEM (n = 3–4).
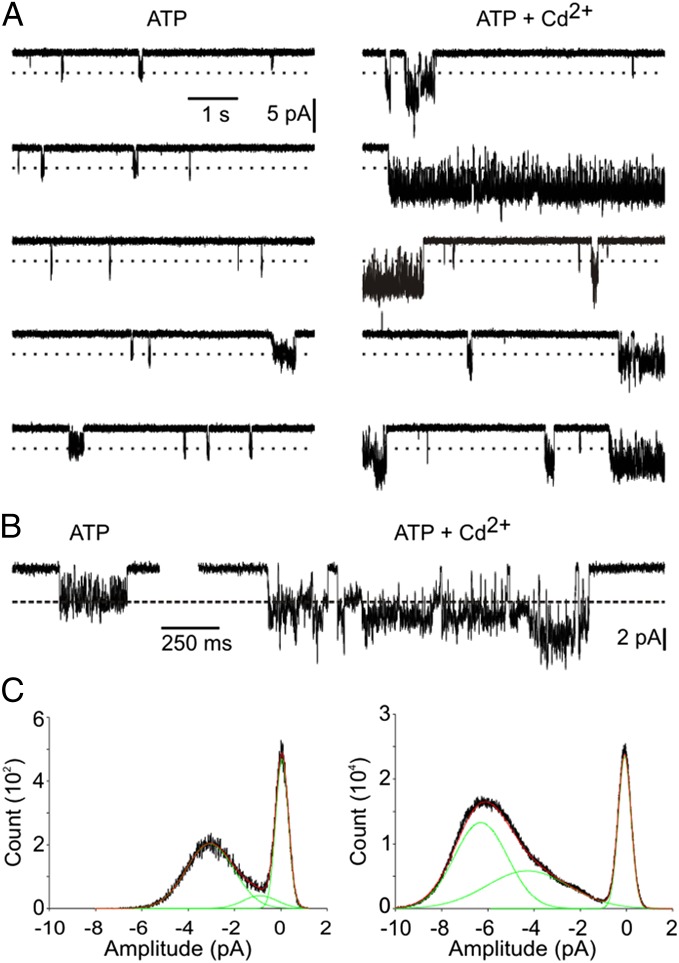
To investigate the mechanism of Cd2+ potentiation further, we undertook single-channel recordings. Membrane patches in the outside-out configuration of the patch-clamp technique were isolated from cells expressing the C-C-C construct because these channels produced the most robust potentiation of ATP-activated currents by Cd2+. Analysis was conducted on three patches in which channel activity was observed in the presence of ATP but not in its absence. Two additional patches exhibited qualitatively similar results but were excluded from the analysis because infrequent channel openings in the absence of ATP precluded the definitive identification of ATP-activated currents. Channels activated by 0.3 μM ATP fluctuated rapidly among multiple current levels, with a predominant level of 3.1 ± 0.1 pA at −120 mV (Fig. 6, Left). The addition of 2 μM Cd2+ resulted in dramatic increases in both the open probability and unitary current amplitudes of the channel (Fig. 6, Right). The predominant current amplitude in the single-channel current distribution increased approximately twofold in the presence of Cd2+, whereas individual burst events, defined as channel openings not interrupted by a close event exceeding 50 ms in duration, increased 7.5 ± 1.8-fold. The maximum observed burst duration in ATP alone was 471 ms, whereas in the presence of Cd2+ the longest measurable burst lasted 6,698 ms. The effect of Cd2+ on burst duration is an underestimation, because bursts of channel activity during which a second channel also became active were excluded from the analysis, and such instances predominantly occurred during long bursts of channel activity. The observed effect of Cd2+ on burst duration indicates that the bridge formed by Cd2+ stabilizes open states. The effect of Cd2+ on the unitary conductance of the channel is intriguing, because it suggests that the metal bridge either stabilizes channel conformations infrequently visited by the channel in its absence or that the metal bridge leads to a conformational change that results in an approximate doubling of the current at all channel sublevels.
Fig. 6.
Metal bridges stabilize open states. (A) Representative outside-out single-channel recordings for the C-C-C construct. (Left) Thirty seconds of continuous recording in the presence of 0.3 µM ATP. (Right) Thirty seconds of continuous recording from the same patch after switching to a solution containing 0.3 µM ATP and 2 µM Cd2+. The dotted line is placed at −3 pA as a visual aid. (B) Expanded traces of single bursts of channel activity in ATP alone (Left) and in ATP plus Cd2+ (Right), demonstrating subconductance states. (C) All-points histograms from three patches for ATP alone (Left) and ATP plus Cd2+ (Right). In each case, the sum of three Gaussian distributions, corresponding to one closed-state level (red curves) and two open-state levels (green curves) were fit to the data. When the three patches were analyzed separately, the Gaussian distributions for the open levels had amplitudes of −1.1 ± 0.3 pA and −3.1 ± 0.1 pA for ATP alone and −3.4 ± 0.7 pA and −6.3 ± 0.4 pA in the presence of Cd2+.
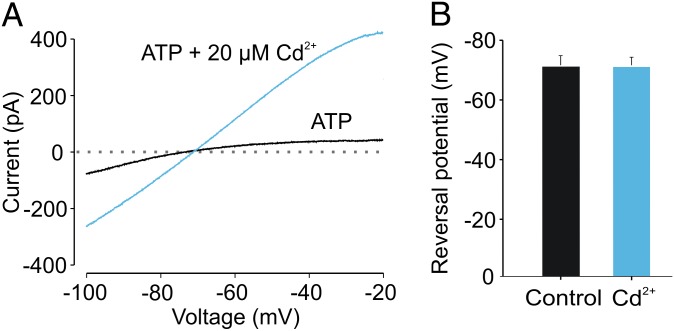
We also explored whether the conducting states stabilized by the C-C-C bridge might correspond to the dilated state of P2X receptors that gives rise to a change in the relative permeability of Na+ to N-methyl-d-glucamine (NMDG+) (21–23). Macroscopic current–voltage (I–V) relationships were obtained using voltage ramps in whole-cell recordings in which Na+ was the primary internal cation and NMDG+ was the primary external cation. In previous reports with related ionic conditions and protocols, the zero-current potential shifted in the positive direction in the continuous presence of ATP as the relative permeability of Na+ to NMDG+ changes (21–23). When Cd2+ was applied to the C-C-C construct, we observed large increases in ATP-activated macroscopic current for both inward and outward limbs of the I–V relationship, with no detectable change in the zero-current potential (Fig. 7). These results indicate that the relative permeability of Na+:NMDG+ for the rP2X2 receptor does not change when Cd2+ forms a bridge between TM1 and TM2, and thus the state stabilized by the bridge does not correspond to the proposed dilated state of P2X receptor channels.
Fig. 7.
The Cd2+ metal bridge does not alter the relative permeability of Na+ to NMDG+. (A) Representative whole-cell ATP-activated current–voltage relationships in response to voltage ramps for the C-C-C construct in the absence (blue trace) and presence (black trace) of Cd2+. EC20 [ATP] or EC20 [ATP] plus 20 µM Cd2+ were applied externally. The currents shown are the net current activated by ATP alone or by ATP in the presence of Cd2+. Each current trace is the average of five consecutive voltage ramps from −100 to +40 mV (ramp duration was 2.3 s with 1-s intervals between ramps) after subtracting the current in the absence of ATP. (B) Bar graph displaying the average zero-current potential measured from three cells.
A Structural Model for the Open State of zfP2X4 Receptors.
The results presented thus far suggest that the intrasubunit interactions observed in both the apo and ATP-bound structures are remarkably consistent with our engineered metal bridges between TM1 and TM2. However, the ATP-bound structure of the zfP2X4 receptor is nonnative in that intersubunit interactions are largely absent within the TM domain. To generate a model for the open state that contains intersubunit interactions but retains intrasubunit helix–helix interactions, we developed a conformational search algorithm to explore alternate quaternary arrangements of the TM2 helices (see Methods, Fig. S2). TM1 then was introduced into each subunit using the same relative positioning with respect to TM2 as in the ATP-bound X-ray structure, and finally the extracellular ATP-binding domain was grafted after superimposing the TM domain. The resulting model was chosen because it is as close as possible to the X-ray structure of the ATP-bound zfP2X4 receptor but introduces additional intersubunit interactions to stabilize the TM domain. Consistent with both accessibility (6–8) and metal coordination (7, 8) data, our model features expansion of the external pore and narrowing of the internal pore (Fig. S1 B and C).
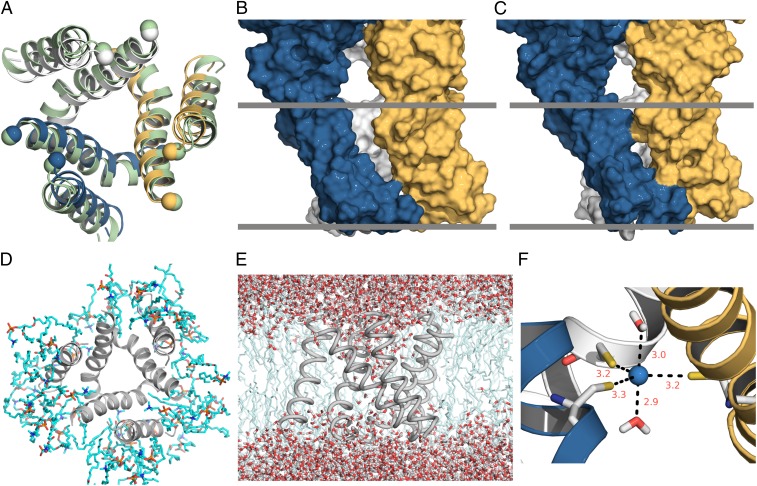
In this model, relative to the X-ray structure, the TM2 helices are translated 1.3 Å toward the central axis, the twist of the TM2 three-helix bundle is increased by 10°, and each TM2 helix is rotated 5° counterclockwise (as viewed from the top) around its helical axis, creating a much more tightly packed intersubunit interface (Fig. 8 A–C and Fig. S2). We performed MD simulations on this model, similar to those performed on the ATP-bound X-ray structure (Fig. 1), and observed that lipid molecules were excluded from the pore (Fig. 8D), which remained well-hydrated throughout the simulation (Fig. 8D). The addition of one turn of the helix in TM1 and a slight reduction in the tilt angle of TM2 result in a modest improvement in hydrophobic mismatch (Fig. S3A). The pore diameter of our model is somewhat less than that of the X-ray structure, especially toward the internal end (Fig. S3 B and C); however, it still is 4.7 Å wide at its narrowest point and therefore should support permeation of physiologically relevant cations such as Na+, K+, or Ca2+. Although we found that formation of a Cd2+ bridge in the C-C-C construct does not alter the relative permeability of Na+ to NMDG+, this channel clearly permeates NMDG+ at voltages negative to −70 mV (Fig. 7A). Corey–Pauling–Koltun (CPK) models of NMDG+ can fit into a box with dimensions of 5 × 6.4 × 12 Å (24), providing an upper limit on the dimensions necessary for permeation. The actual pore diameter required to support permeation would be lower, because the structures of both the organic cation and the protein will fluctuate on a rapid timescale. Thus, it seems reasonable to conclude that the diameter of the pore in our open-state model is compatible with NMDG+ permeation.
Fig. 8.
A structural model for the open state of P2X receptors. (A) Comparison of the TM domain between the X-ray structure for the ATP-bound zfP2X4 receptor (shown in light green) and the open-state model (with individual subunits colored in blue, gold, and gray) viewed from the external side of the membrane. The Cα atoms of the C terminus (residue G56) of TM1 and the N terminus (residue I335) of TM2 are shown as spheres. (B and C) Interfaces between adjacent subunits for the ATP-bound zfP2X4 receptor and the open-state model, respectively, depicted in a surface rendering. Hypothetical boundaries of the lipid bilayer are represented by gray bands. (D) Snapshot of the TM domain of the open-state model and its adjacent lipid molecules in the restrained MD simulation, viewed from the external side of the membrane. In contrast to observations in the simulations of the ATP-bound zfP2X4 receptor, no lipid molecules entered the pore in simulations of the open-state model. (E) Continuous water chain in the pore of the model. (F) Cd2+ coordination by the L351C trimer and two water molecules.
In the simulation of the open-state model, the side chains of L351 alternate between two rotamers, one of which positions the side chain toward the inter-TM2 interface, and the other with the side chain projected into the pore. Interestingly, this residue is equivalent to V343 in the rP2X2 receptor, where substitution of Cys results in the formation of a stable Cd2+ bridge that blocks the channel (7). This Cd2+ bridge forms rapidly (rate constant = 106 M/s) in the presence of ATP, suggesting that bridging occurs in the highly populated open state, and requires Cys in all three subunits, implying that it forms at the central axis of the pore (7). L351 in the X-ray structure of the ATP-bound zfP2X4 receptor cannot coordinate Cd2+ because of the larger diameter of the pore at this position (L351 Cβ–Cβ distances of 11.2 Å; Fig. S1B). However, in our open-state model L351C residues in the alternative rotamer, together with two water molecules, can coordinate Cd2+ in the triagonal bipyramidal geometry (Fig. 8F).
Discussion
The objective of the present study was to evaluate both intersubunit and intrasubunit interactions within the TM domain of P2X receptors using both computational and functional approaches. A key motivation for this work was the unexpected absence of intersubunit interactions in the recent X-ray structure of the ATP-bound zfP2X4 receptor, which raises the possibility that the structure is distorted (15) and brings into question the proposed mechanism of ATP activation. Indeed, our MD simulations suggest that lipid molecules can diffuse through the intersubunit crevices to occupy and dehydrate the ion permeation pathway (Fig. 1), a scenario that is incompatible with ion conduction. As noted previously (15), the predominance of hydrocarbon side chains in helical transmembrane domains implicates nonspecific van der Waals and weak electrostatic interactions as the main intraprotein forces for tertiary and quaternary structural stability. In native membranes, the hydrophobic environment provided by the acyl chains of lipid molecules enhances helix–helix packing and helps define the hydrophobic dimension. The lack of such a hydrophobic environment in the crystalline lattice is likely the reason for the poor intersubunit packing of the TM1 and TM2 helices and the apparent hydrophobic mismatch in the X-ray structure of the ATP-bound zfP2X4 receptor. Such distortions may be particularly problematic for membrane proteins such as P2X receptors, which contain large extracellular domains that provide most of the crystal contacts, together with small transmembrane domains that contain only six helices in the trimer (15).
Although the intersubunit crevices are a striking feature of the ATP-bound X-ray structure, only relatively modest structural rearrangements are required to form an intersubunit interface within the membrane. In our open-state model of the zfP2X4 receptor, intersubunit interactions within two regions of the TM domain were created by small rotations and translation of the TM2 helix toward the central axis (Fig. 8). The first interface is within the internal region of the TM domain, where residues on three turns of the TM2 helix (L351, I355, and W358) from one subunit make contact with L346, A347, and V354 on the adjacent TM2 helix. The second is within the external end of the TM domain, where Y45 in TM1 contacts L340 in TM2 of the adjacent subunit. These contacts do not completely seal off the pore from the surrounding lipid membrane but diminish the crevices to openings or portals (Fig. 8C), similar to what has been seen in the KcsA potassium channel (25) and the NavAb voltage-activated sodium channel (26). In addition, our model exhibits a modest improvement in hydrophobic mismatch (Fig. S3A).
We also investigated the intrasubunit interactions depicted in both the apo and ATP-bound X-ray structures (Fig. 2), and in this case we identified metal bridges that are fully compatible with those structures. Our results demonstrate that S345C and C348 in TM2 and H/C33 in TM1 form a robust intrasubunit Cd2+ bridge that stabilizes the open state of the rP2X2 receptor (Figs. 3–6). The X-ray structures position the equivalent of S345C and C348 on one face of the TM2 helix where they can coordinate Cd2+ in either the apo or ATP-bound states (Fig. 2), consistent with our functional results demonstrating that Cd2+ remains stably associated with the channel in the closed state (Fig. 5). Those structures also show that ATP binding triggers a relative rotation of the TM1/TM2 interface so that residue 33 in TM1 also can participate in coordinating the metal. Thus, our metal-bridging results substantiate intrasubunit helix interactions and the intrasubunit motions between TM1 and TM2 upon ATP binding that are depicted in the X-ray structures of the zfP2X4 receptor. When considered together with the modest modifications that are required to decrease the size of the intersubunit crevices, these results support the proposed structural mechanism by which ATP binding leads to an iris-like opening of P2X receptor channels (5). In our structural model for the open state, this structural rearrangement would also be accompanied by a narrowing of the internal pore to create intersubunit interactions between the TM2 helices (Fig. 8).
The metal bridges that we engineered between TM1 and TM2 also identify an internal region of the TM domain that is particularly sensitive to modification, where the activity of the channel can be tuned readily (Figs. 3–6). The relative motions between TM1 and TM2 in this region are not very large (Fig. 2), but occupancy of the engineered metal-binding site has profound consequences for both the gating and conduction properties of P2X receptors. In the case of the C-C-C bridge, Cd2+ occupancy results in an increase in burst duration, suggesting that the bridge stabilizes the open state. We also observed a substantial increase in mean unitary conductance (Fig. 6), implying that this local region plays a key role in determining the permeation properties of P2X receptor channels. The internal pore in our open-state model of the zfP2X4 receptor is the narrowest region of the pore, and it contains the most extensive intersubunit interactions, providing an initial picture of the structure of this important region of P2X receptor channels. Formation of intersubunit interfaces with the internal region of TM2 is consistent with observations that this region of TM2 is particularly sensitive to mutations (6, 27–29), and the model is fully compatible with metal bridges that have been described for the TM domain of rP2X2 receptor channels. These bridges include an intersubunit metal bridge formed at the threefold axis by V343C (Fig. 8F) (7), stable metal bridges formed by D349C (8), as well as the metal bridges presented here that are located at the intrasubunit interface between the TM1 and TM2 helices. Refinement of this working model to reduce further the hydrophobic mismatch, while retaining the constraints imposed by the metal bridges discussed here, will help to identify key intersubunit interfaces involved in channel gating.
Methods
Evaluation of the X-Ray Structure for ATP-Bound zfP2X4 Receptor by MD Simulations.
Before being embedded in a lipid bilayer for MD simulations, the X-ray structure of ATP-bound zfP2X4 receptor was preprocessed as follows. Using symmetry operations, the monomer entry in ATP-bound zfP2X4 receptor was replicated into a threefold symmetric trimer. The trimer was energy minimized for 10,000 steps while the Cα atoms and the heavy atoms of the ATP molecules and the surrounding residues (N296, R298, K316, K70, K72, and T189) were restrained in their original positions with a force constant of 10 kcal−1⋅mol−1⋅Å−2.
A preequilibrated DMPC lipid bilayer was obtained from CHARMM-GUI (30), replicated, and trimmed to generate a membrane with 398 lipid molecules solvated by 32,187 water molecules. The preprocessed structure of the zfP2X4 receptor then was inserted into the membrane, with lipid molecules within 1 Å and water molecules within 3 Å removed. Sodium and chloride ions were added to neutralize the system and provide a physiological concentration of salt. The resulting system contained the receptor along with 332 lipid molecules, 32,004 water molecules, 93 sodium ions, and 90 chloride ions (corresponding to a salt concentration of 0.15 M). The system preparation was done using VMD (31) and its Solvate and Ionize plugins.
The system was energy minimized for 10,000 steps and then was simulated for 79.44 ns, with the restraints stated above, but the restraining force constant was reduced to 1 kcal−1⋅mol−1⋅Å−2 after the first 21.98 ns. A second simulation was started from the snapshot at 12.15 ns of the restrained simulation and was run for 53.66 ns. In this simulation, all the previous restraints were released. However, to ensure that the ATP molecules stayed in the binding pockets, distance restraints were introduced between pairs of atoms involved in ATP–zfP2X4 receptor salt bridges and hydrogen bonds (5).
All simulations were performed using NAMD 2.9 (32) with temperature at 310 K and pressure at 1 atm. The Langevin dynamics and Nosé–Hoover Langevin piston methods were used for temperature and pressure coupling, respectively. CHARMM27 protein (33) and CHARMM36 lipid (34) force fields were used. The force field for ATP was from Pavelites et al. (35).
Modeling Cd2+ Bridges Between TM1 and TM2 of the zfP2X4 Receptor.
Putative bridging residues (e.g., N35 and N353) were mutated into Cys or His using the Mutator plugin of VMD. Sidechain dihedral angles of the introduced Cys or His residues were adjusted manually so that their sulfur or nitrogen atoms were at possible positions to coordinate Cd2+. To eliminate possible steric clashes by the mutations, each mutant was energy minimized for 10,000 steps, while Cα atoms were restrained with a force constant of 10 kcal−1⋅mol−1⋅Å−2.
Construction of an Open-State Model of the zfP2X4 Receptor.
The TM2 helix from a single subunit in the energy-minimized ATP-bound zfP2X4 receptor structure was used as the starting point for the conformational search to generate the open-state model. This helix was translated and rotated so that its helical axis was aligned with the z-axis and its center of mass was at the origin of the coordinate system and is called “TM2mono” hereafter. The z-axis represents the membrane normal. All translation and rotation operations were done using VMD scripts.
A series of rotation, translation, and replication was performed on TM2mono to generate an ensemble of conformations of TM2 trimer upon which several filters were applied to select plausible models. The operations on TM2mono were carried out sequentially as follows: (i) rotation around its helical axis (i.e., the z-axis) by an angle ρ; (ii) rotation around the x-axis by an angle θ; (iii) rotation around the z axis by an angle ϕ; (iv) translation along the x-axis by a distance r; (v) replication to create a threefold symmetric TM2 trimer.
We constrained the search space to be centered around the ATP-bound X-ray structure by using the following ranges for the four degrees of freedom: ρ from −30° to 10° with increments of 10°; θ from 25° to 45° with increments of 5°; ϕ from −40° to 80° with increments of 10°; and r from 6–10 Å with increments of 1 Å.
The first filter aimed to tighten the interfaces in the TM2 trimer. We therefore selected models with Cα–Cα distances for neighboring A344 residues and for neighboring L351 residues that were shorter than in the ATP-bound X-ray structure.
The second filter was applied to avoid steric clashes on the one hand and to ensure adequate intersubunit contacts on the other. We found the minimum Cα–Cα distances between neighboring TM2 helices are 4.5 Å in the apo structure and 9.5 Å in the ATP-bound structure. We eliminated any model in which at least one intersubunit Cα–Cα distance was less than 4.5 Å or in which no single intersubunit Cα–Cα distance was less than 9.5 Å.
The third filter was used to select those with adequate pore sizes from the surviving models. For each model, the pore-radius profile along the threefold axis was calculated using the HOLE program (36), and the minimum radius was obtained. Models with minimum pore radius greater than 2 Å were selected.
The final model was chosen, by visual inspections, because the inter-TM2 crevices were filled to the greatest extent. In this model ρ = −10°, θ = 45°, ϕ = 30°, and r = 7 Å. The side chain dihedral angles of L351, V354, and W358 were adjusted to open the pore further and improve the packing in the interfaces. To the final model of the TM2 trimer, we added a TM1 helix in each subunit by using its relative positioning with respect to TM2 as in the ATP-bound X-ray structure. We did so by taking TM1 and TM2 in one subunit of the ATP-bound X-ray structure and then superimposing the TM2 portion on our model, one subunit at a time; the resulting TM1 helices in the three subunits were the desired additions. Finally the ectodomain of the ATP-bound X-ray structure was grafted after the TM domain was superimposed on our model. A restrained MD simulation was run on this open-state model, similar to the simulation performed for the ATP-bound X-ray structure.
Channel Constructs.
Mutations were introduced into an rP2X receptor construct (generously provided by David Julius, University of California, San Francisco) in which C9 and C430 were mutated to threonine (6), a construct termed “2T.” The previously reported inhibitory Cd2+ bridge between H33 and S345C (7) was studied in the 3T background, a construct that also contains the C348T mutation. The primary potentiating Cd2+ bridges studied here involved H33 or H33C in TM1 and both S345C and C348 in TM2 (H-C-C and C-C-C, respectively), and these were studied in the 2T background. Cd2+ also has weak and rapidly reversible potentiating activity on the wild-type rP2X2 receptor channel; external application of 20 µM Cd2+ increases macroscopic currents activated by 15 µM ATP by 15 ± 3% (n = 3), and 100 µM Cd2+ increases ATP-activated currents by 48 ± 8% (n = 3). Although we have not determined the coordinating residues responsible for this weak and rapidly reversible potentiation in the wild-type channel, a simple explanation that would be consistent with the present study is that H33 and C348C can form a weak intrasubunit Cd2+ bridge that stabilizes the open state. Concatamers were constructed as described previously (7, 37, 38) and were confirmed by restriction digests and DNA sequencing. In addition, cell lysates were evaluated with SDS/PAGE and Western blot analysis, which confirmed that the most abundant species of rP2X2 receptors in HEK cells expressing the concatenated trimeric construct corresponds to the molecular weight of a trimer (Fig. S4).
Cell Culture and Transfection.
HEK293 cells were cultured in DMEM supplemented with 10% (vol/vol) FBS and 10 mg/L gentamicin. All cell-culture reagents were obtained from GIBCO. Trypsin-treated HEK293 cells were transiently transfected and seeded onto glass coverslips in six-well plates and were placed in a 37 °C incubator with 95% air and 5% CO2. Transfections were performed using FuGENE6 Transfection Reagent (Promega). P2X receptors were cotransfected with a GFP cDNA construct in pGreen-Lantern (Invitrogen) at ratios varying from 2:1–8:1. Recordings were conducted 24–60 h after transfection.
Western Blot Analysis of Concatenated Constructs.
Cells grown in T25 25-cm2 flasks were transfected with P2X receptor DNA using FuGene 6. Two days after transfection, cells were collected in Dulbecco’s PBS and then were centrifuged at 500 × g for 3 min. The supernatant was discarded after centrifugation, and a lysis buffer containing 1% Triton X-100 and protease inhibitors was used to suspend the cell pellet. Cells in lysis solution were sonicated for 5 s on ice and incubated on ice for 30 min, with vortexing for 5 s every 5 min. The cell lysate then was centrifuged at 13,000 × g for 20 min at 4 °C, and the supernatant was combined with LDS buffer, DTT, and 2-mercaptoethnol. Samples were heated at 95 °C for 5 min and then were centrifuged at 13,000 × g for 2 min. Proteins were separated in a 4–12% NuPage Bis-Tris gel (Invitrogen) using a running buffer containing (in mM): 50 3-(N-morpholino)propanesulfonic acid, 50 Tris base, 3.46 SDS, and 1 EDTA. SeeBlue Plus2 (Invitrogen) was used as the protein molecular-weight marker. Protein in the gel was transferred to nitrocellulose membrane and probed with goat anti-hP2X2 antibody (Santa Cruz).
Electrophysiology.
All experiments were performed in transiently transfected HEK293 cells under voltage-clamp (−60 mV for whole-cell and −120 mV for single-channel) recording using an Axopatch 200B patch-clamp amplifier (Axon Instruments, Inc.) and were digitized on-line using a Digidata 1321A interface board and pCLAMP 10.2 software (Axon Instruments, Inc.). Whole-cell currents were filtered at 5 kHz using eight-pole Bessel filters and were digitized at 20 kHz. Single-channel recordings were filtered initially at 5 kHz, were digitized at 10–50 kHz, and were further filtered at 2 kHz for data analysis and at 1 kHz for presentation.
The whole-cell external solution contained (in mM): 140 NaCl, 5.4 KCl, 2 CaCl2, 0.5 MgCl2, 10 Hepes, and 10 d-glucose, adjusted to pH 7.3 with NaOH. The internal solution contained (in mM): 140 NaCl, 10 EGTA and 10 Hepes, adjusted to pH 7.0 with NaOH. The single-channel external solution contained (in mM): 147 NaCl, 1 CaCl2, 10 Hepes, and 11 d-glucose, adjusted to pH 7.4 with NaOH. The internal solution contained (in mM): 140 NaF, 5 NaCl, 10 EGTA and 10 Hepes, adjusted to pH 7.0 with NaOH. NMDG+ permeability experiments were conducted with an external solution containing (in mM): 150 NMDG+, 10 Hepes, adjusted to pH 7.3 with HCl. The internal solution contained (in mM): 140 NaCl, 10 EGTA, 10 Hepes, adjusted to pH 7.0 with NaOH. Voltage ramps were given from −100 mV to +40 mV. In each experimental condition, five consecutive current traces were averaged, and then the average leak current was subtracted from the average ATP- and ATP plus Cd2+-evoked currents.
Solution exchange was achieved using the Rapid Solution Changer RSC-200 (BioLogic), which has the capacity of switching between nine solutions within 40 ms depending on the size of the cell. ATP and Cd2+ solutions were prepared daily and diluted to the desired concentration in external solution immediately before each experiment.
To generate the concentration–response relationships, a reference concentration of ATP was applied before applying a test concentration, as previously described (6). The Hill equation was fit to the data according to
where I is the normalized current at a given concentration of ATP, Imax is the maximum normalized current, EC50 is the concentration of ATP ([ATP]) producing half-maximal currents, and n is the Hill coefficient.
Analysis of the duration of single-channel bursts was conducted in pCLAMP 10.2 (Axon Instruments, Inc.). Only patches in which no channel activity was observed in the absence of ATP were analyzed. All events detected by the software were inspected visually and were rejected if two or more channel openings superimposed. Bursts of single-channel currents were extracted from the original data file and transferred to SigmaPlot 10.0, which was used for generating the all-point histogram. Origin 8.1 was used for fitting the data with Gaussian distributions.
Supplementary Material
Acknowledgments
We thank Andres Jara-Oseguera, Mark Mayer, Miguel Holmgren, Dmitriy Krepkiy, Jeet Kalia, Gilman Toombes, and other members of the K.J.S. laboratory for helpful discussions. This work was supported by the Intramural Research Program of the National Institute of Neurological Disorders and Stroke, National Institutes of Health (NIH) (K.J.S.), NIH Pathway to Independence Award NS070954 (to M.L.), and NIH Grants GM058187 and GM 088187(to H.-X.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1311071110/-/DCSupplemental.
References
- 1.Khakh BS, North RA. Neuromodulation by extracellular ATP and P2X receptors in the CNS. Neuron. 2012;76(1):51–69. doi: 10.1016/j.neuron.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang R, Taly A, Grutter T. Moving through the gate in ATP-activated P2X receptors. Trends Biochem Sci. 2013;38(1):20–29. doi: 10.1016/j.tibs.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Surprenant A, North RA. Signaling at purinergic P2X receptors. Annu Rev Physiol. 2009;71:333–359. doi: 10.1146/annurev.physiol.70.113006.100630. [DOI] [PubMed] [Google Scholar]
- 4.Kawate T, Michel JC, Birdsong WT, Gouaux E. Crystal structure of the ATP-gated P2X(4) ion channel in the closed state. Nature. 2009;460(7255):592–598. doi: 10.1038/nature08198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hattori M, Gouaux E. Molecular mechanism of ATP binding and ion channel activation in P2X receptors. Nature. 2012;485(7397):207–212. doi: 10.1038/nature11010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li M, Chang TH, Silberberg SD, Swartz KJ. Gating the pore of P2X receptor channels. Nat Neurosci. 2008;11(8):883–887. doi: 10.1038/nn.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li M, Kawate T, Silberberg SD, Swartz KJ. Pore-opening mechanism in trimeric P2X receptor channels. Nat Commun. 2010;1:44. doi: 10.1038/ncomms1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kracun S, Chaptal V, Abramson J, Khakh BS. Gated access to the pore of a P2X receptor: Structural implications for closed-open transitions. J Biol Chem. 2010;285(13):10110–10121. doi: 10.1074/jbc.M109.089185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawate T, Robertson JL, Li M, Silberberg SD, Swartz KJ. Ion access pathway to the transmembrane pore in P2X receptor channels. J Gen Physiol. 2011;137(6):579–590. doi: 10.1085/jgp.201010593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samways DS, Khakh BS, Dutertre S, Egan TM. Preferential use of unobstructed lateral portals as the access route to the pore of human ATP-gated ion channels (P2X receptors) Proc Natl Acad Sci USA. 2011;108(33):13800–13805. doi: 10.1073/pnas.1017550108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang R, et al. Agonist trapped in ATP-binding sites of the P2X2 receptor. Proc Natl Acad Sci USA. 2011;108(22):9066–9071. doi: 10.1073/pnas.1102170108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du J, Dong H, Zhou HX. Gating mechanism of a P2X4 receptor developed from normal mode analysis and molecular dynamics simulations. Proc Natl Acad Sci USA. 2012;109(11):4140–4145. doi: 10.1073/pnas.1119546109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lörinczi E, et al. Involvement of the cysteine-rich head domain in activation and desensitization of the P2X1 receptor. Proc Natl Acad Sci USA. 2012;109(28):11396–11401. doi: 10.1073/pnas.1118759109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang R, et al. Tightening of the ATP-binding sites induces the opening of P2X receptor channels. EMBO J. 2012;31(9):2134–2143. doi: 10.1038/emboj.2012.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou HX, Cross TA. Influences of membrane mimetic environments on membrane protein structures. Annu Rev Biophys. 2013;42:361–392. doi: 10.1146/annurev-biophys-083012-130326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang X, et al. Functional Identification of Close Proximity Amino Acid Side Chains within the Transmembrane-Spanning Helixes of the P2X2 Receptor. PLoS ONE. 2013;8(8):e70629. doi: 10.1371/journal.pone.0070629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arnold FH, Haymore BL. Engineered metal-binding proteins: Purification to protein folding. Science. 1991;252(5014):1796–1797. doi: 10.1126/science.1648261. [DOI] [PubMed] [Google Scholar]
- 18.Suh SS, Haymore BL, Arnold FH. Characterization of His-X3-His sites in alpha-helices of synthetic metal-binding bovine somatotropin. Protein Eng. 1991;4(3):301–305. doi: 10.1093/protein/4.3.301. [DOI] [PubMed] [Google Scholar]
- 19.Taraska JW, Puljung MC, Olivier NB, Flynn GE, Zagotta WN. Mapping the structure and conformational movements of proteins with transition metal ion FRET. Nat Methods. 2009;6(7):532–537. doi: 10.1038/nmeth.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holmgren M, Shin KS, Yellen G. The activation gate of a voltage-gated K+ channel can be trapped in the open state by an intersubunit metal bridge. Neuron. 1998;21(3):617–621. doi: 10.1016/s0896-6273(00)80571-1. [DOI] [PubMed] [Google Scholar]
- 21.Browne LE, Compan V, Bragg L, North RA. P2X7 receptor channels allow direct permeation of nanometer-sized dyes. J Neurosci. 2013;33(8):3557–3566. doi: 10.1523/JNEUROSCI.2235-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Virginio C, MacKenzie A, Rassendren FA, North RA, Surprenant A. Pore dilation of neuronal P2X receptor channels. Nat Neurosci. 1999;2(4):315–321. doi: 10.1038/7225. [DOI] [PubMed] [Google Scholar]
- 23.Khakh BS, Bao XR, Labarca C, Lester HA. Neuronal P2X transmitter-gated cation channels change their ion selectivity in seconds. Nat Neurosci. 1999;2(4):322–330. doi: 10.1038/7233. [DOI] [PubMed] [Google Scholar]
- 24.Villarroel A, Burnashev N, Sakmann B. Dimensions of the narrow portion of a recombinant NMDA receptor channel. Biophys J. 1995;68(3):866–875. doi: 10.1016/S0006-3495(95)80263-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature. 2001;414(6859):43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 26.Payandeh J, Gamal El-Din TM, Scheuer T, Zheng N, Catterall WA. Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature. 2012;486(7401):135–139. doi: 10.1038/nature11077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silberberg SD, Li M, Swartz KJ. Ivermectin Interaction with transmembrane helices reveals widespread rearrangements during opening of P2X receptor channels. Neuron. 2007;54(2):263–274. doi: 10.1016/j.neuron.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 28.Silberberg SD, Chang TH, Swartz KJ. Secondary structure and gating rearrangements of transmembrane segments in rat P2X4 receptor channels. J Gen Physiol. 2005;125(4):347–359. doi: 10.1085/jgp.200409221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao L, Broomhead HE, Young MT, North RA. Polar residues in the second transmembrane domain of the rat P2X2 receptor that affect spontaneous gating, unitary conductance, and rectification. J Neurosci. 2009;29(45):14257–14264. doi: 10.1523/JNEUROSCI.4403-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J Comput Chem. 2008;29(11):1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 31. Humphrey W, Dalke A. Schulten K. (1996) VMD: Visual molecular dynamics. J Mol Graph 14(1):33–38. [DOI] [PubMed]
- 32.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mackerell AD, Jr, Feig M, Brooks CL., 3rd Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25(11):1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 34.Klauda JB, et al. Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J Phys Chem B. 2010;114(23):7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pavelites JJ, Gao J, Bash PA, Mackerell AD. A molecular mechanics force field for NAD+, NADH, and the pyrophosphate groups of nucleotides. J Comput Chem. 1997;18(2):221–239. [Google Scholar]
- 36.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14(6):354–360, 376. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 37.Stoop R, et al. Contribution of individual subunits to the multimeric P2X(2) receptor: Estimates based on methanethiosulfonate block at T336C. Mol Pharmacol. 1999;56(5):973–981. doi: 10.1124/mol.56.5.973. [DOI] [PubMed] [Google Scholar]
- 38.Nagaya N, Tittle RK, Saar N, Dellal SS, Hume RI. An intersubunit zinc binding site in rat P2X2 receptors. J Biol Chem. 2005;280(28):25982–25993. doi: 10.1074/jbc.M504545200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.