Significance
Our results imply a direct impact of the p38–MAP kinase-activated protein kinase 2 (MK2) kinase pathway on the cellular response to replicative stress. In this situation, MK2 activity determines the decision between replication fork stalling and translesion synthesis. In the absence of MK2 activity, even the otherwise essential checkpoint kinase Chk1 becomes dispensable for S phase progression and cell survival. Moreover, MK2 represents a determinant of cancer cell sensitivity toward nucleoside analogue treatment.
Abstract
DNA damage can obstruct replication forks, resulting in replicative stress. By siRNA screening, we identified kinases involved in the accumulation of phosphohistone 2AX (γH2AX) upon UV irradiation-induced replication stress. Surprisingly, the strongest reduction of phosphohistone 2AX followed knockdown of the MAP kinase-activated protein kinase 2 (MK2), a kinase currently implicated in p38 stress signaling and G2 arrest. Depletion or inhibition of MK2 also protected cells from DNA damage-induced cell death, and mice deficient for MK2 displayed decreased apoptosis in the skin upon UV irradiation. Moreover, MK2 activity was required for damage response, accumulation of ssDNA, and decreased survival when cells were treated with the nucleoside analogue gemcitabine or when the checkpoint kinase Chk1 was antagonized. By using DNA fiber assays, we found that MK2 inhibition or knockdown rescued DNA replication impaired by gemcitabine or by Chk1 inhibition. This rescue strictly depended on translesion DNA polymerases. In conclusion, instead of being an unavoidable consequence of DNA damage, alterations of replication speed and origin firing depend on MK2-mediated signaling.
Replicative stress is a consequence of nonperfect DNA replication, resulting in DNA damage response (DDR) signaling. In contrast to the DDR induced by double strand breaks, our current understanding of replicative stress is still far from complete. However, replicative stress constitutes a limiting factor in cancer cell proliferation (1) and a major mechanism of chemotherapy, and thus merits detailed understanding.
Exogenous damage can enhance replicative stress. UV irradiation forms cross-links between DNA bases at any stage of the cell cycle, but damage is strongly enhanced when the cell tries to use such DNA as a template for replication. Nucleoside analogues, such as gemcitabine or cytarabine, perturb replication by being incorporated into nascent DNA strands, and/or by inducing an imbalance of nucleoside pools. Hence, a deeper understanding of how nucleoside analogues help to eliminate cancer cells can only be achieved through knowledge of how these cells respond to replicative stress.
One way to avoid replicative stress consists in the avoidance of replication itself. Along this line, nongenotoxic activation of p53 induces G1 or G2 arrest that leads to profound resistance toward gemcitabine and UV irradiation (2, 3). This prompted us to ask more generally whether replicative stress represents merely a function of DNA damage before or during S phase, or whether it also depends on the activity of cellular signaling pathways. Indeed, the factors Chk1 and Wee1 are required to avoid replicative stress, and their knockdown induces a severe DDR (4, 5). We were now asking whether some factors can also act in a reverse fashion, provoking a more profound DDR and possibly cell death in response to misincorporations and other conditions that lead to replicative stress. Such mediators of detrimental outcome would contribute to the radiation sensitivity and chemosensitivity of cells.
In this study, we have performed a siRNA screen, interrogating all known human kinases as to their contribution to the cellular response upon UV. We have identified MAP kinase-activated protein kinase 2 (MK2) as a major mediator of this response. MK2 suppresses replication fork progression and conversely enhances the firing frequency of new replication origins in the presence of replicative stress. It dampens translesion synthesis (TLS)-dependent and ongoing replication while promoting stalling of the replication fork.
Results
MK2 Is a Determinant of the DDR and Cell Survival upon UV Irradiation in Vitro and in Vivo.
To obtain a comprehensive overview of kinases that are involved in the DDR to UV, we performed a systematic siRNA-based screen, using a collection of siRNAs targeting all known human kinases and their components. Accumulation of phosphorylated H2AX (termed γH2AX) was used as a readout for the DDR. The human osteosarcoma cell line U2OS was chosen as it has a low level of spontaneous DNA damage and has been extensively used for studies of the DDR to UV before (4, 6–10).
All target genes were ranked according to the sum of the robust z-scores calculated for all three corresponding siRNAs to identify genes that strongly influence H2AX phosphorylation. The results obtained for all siRNAs are displayed in Dataset S1, and a graphical overview is given in Fig. S1A. Knockdown of the kinases Wee1 and Chk1 was found to yield the highest positive z-scores of all targets, in agreement with previous literature (4, 5, 11). At the other end of the scale, the screen identified the kinase MK2 (alias MAPKAPK2) to have the highest negative z-score. Although originally described as a stress response kinase (12, 13), MK2 is also activated in response to UV light irradiation; so far, its function in this context was described to consist in mediating G2 arrest (6–8). The present screen implies a function of MK2 in the phosphorylation of H2AX upon replicative stress, suggesting that the operating range of this kinase in the DDR goes far beyond the control of G2/M transition.
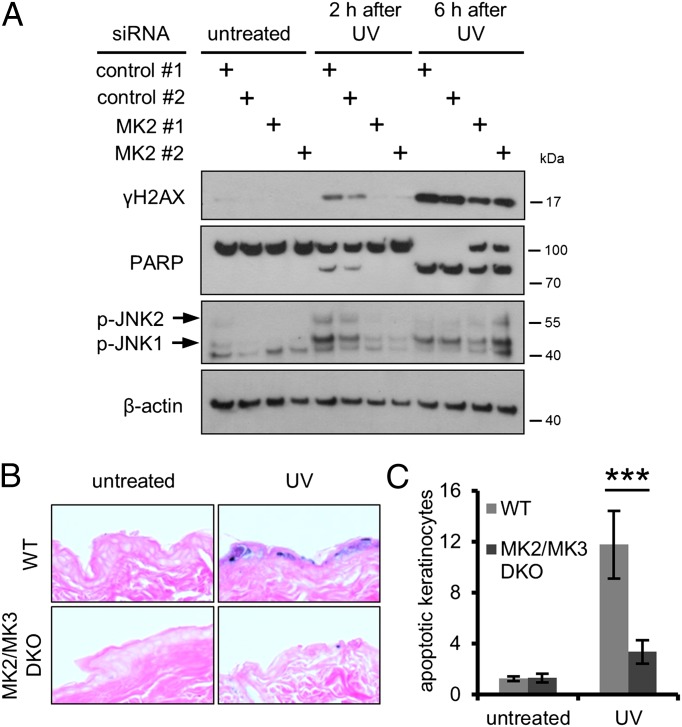
Immunoblot analysis confirmed that MK2 depletion impaired irradiation-induced H2AX phosphorylation (Fig. 1A and Fig. S1B). Importantly, the UV-induced accumulation or removal of DNA lesions was unaltered by MK2 depletion (Fig. S1C). Cleavage of PARP was lowered upon MK2 knockdown (Fig. 1A), indicating reduced caspase activity. Phosphorylation of the stress-responsive JNK1 and JNK2 was diminished upon MK2 knockdown as well. In clonogenic assays, removing MK2 enhanced survival without compromising cell proliferation, whereas Mdm2 knockdown, used as a positive control, reduced colony formation in untreated cells but improved cell survival after exposure to UV light (Fig. S1D). The latter can be attributed to the described protective effect of temporary Mdm2 removal and p53 activation against certain kinds of DNA damage (2, 3). Thus, MK2 depletion not only interfered with UV-induced H2AX phosphorylation but also reduced cellular stress signaling and apoptosis.
Fig. 1.
MK2 depletion reduces UV-induced H2AX phosphorylation and cell death in vitro and in vivo. (A) MK2 knockdown impairs H2AX phosphorylation and apoptosis in U2OS cells upon UV irradiation. Cells were depleted of MK2, exposed to 20 J/m2 UV-C, and harvested at indicated time points after irradiation. Cell lysates were analyzed by immunoblotting. (B and C) MK2/MK3 DKO mice display reduced apoptosis in skin after UV exposure. (B) Backs of MK2/MK3 DKO (n = 6) and WT mice (n = 5) were exposed to 250 mJ/cm2 UV-B irradiation or left untreated. In addition, five WT mice were not irradiated. Mice were euthanized 24 h after irradiation, and skin samples were processed for immunohistochemistry and stained with eosin (red) and for TUNEL (blue). Representative images are shown. (C) Quantification of TUNEL-positive cells per millimeter epidermis from entire tissue sections from animals treated as in B (***P = 0.0003).
A pharmacological MK2 inhibitor (MK2 III) (14) reduced the levels of UV-induced γH2AX compared with control cells (Fig. S1E), confirming the results obtained with MK2 siRNA (Fig. 1A). MK2 inhibition did not influence the formation or removal of UV-induced DNA lesions (Fig. S1F).
To explore the in vivo situation, we made use of mice with genetic ablation of MK2 alone or in combination with its relative MK3. MK2 and MK3 are closely related, and it has been proposed that MK3 can compensate for the loss of MK2 in MK2 KO mice (15). We assessed the consequences of UV-induced DNA damage in the skin of MK2/MK3 double KO (DKO) mice, MK2 single KO mice, and WT animals, identifying apoptotic keratinocytes by TUNEL. Although skin samples from UV-irradiated WT animals displayed strong TUNEL staining, hardly any TUNEL-positive cells were detected in samples from UV-irradiated MK2/MK3 DKO animals (Fig. 1 B and C) or MK2 single KO mice (Fig. S1G). These findings demonstrate that MK2 is required for UV-induced cell death in vivo.
MK2 Activity Promotes DNA Damage Signaling and Slows Down DNA Replication While Enhancing Origin Firing in Response to Gemcitabine.
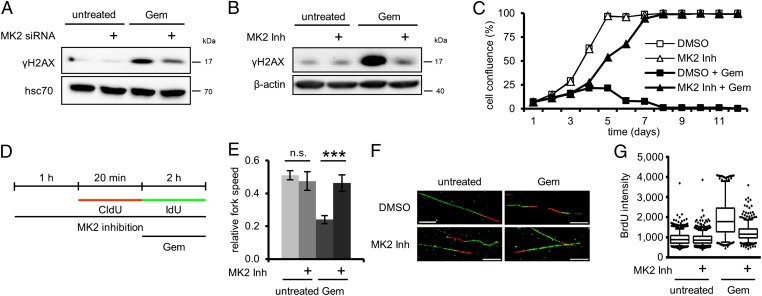
UV exhibits its cytotoxic potential by interfering with various cellular processes. The biggest challenge for cells, however, is arguably imposed by the UV-induced DNA lesions that interfere with DNA replication (16). To test whether MK2 has a function in S phase, we substituted UV with the nucleoside analogue gemcitabine (2′,2′-difluorodeoxycytidine). Gemcitabine interferes with DNA replication and can induce DDR and cell death (17–19). Both MK2 knockdown (Fig. 2A) and inhibition (Fig. 2B) reduced the γH2AX levels in gemcitabine-treated cells (Fig. S2 A–C show control experiments), arguing that kinase-independent activities of MK2 are not sufficient for its contribution to γH2AX accumulation. These results were also reflected by an increased viability of cells treated simultaneously with gemcitabine and MK2 inhibitor compared with gemcitabine alone (Fig. 2C). We previously reported that a reduced responsiveness of cells toward gemcitabine can result from the induction of a G1 arrest by activation of p53 (2). Furthermore, as mentioned, recent publications suggested that MK2 acts as a regulator of the cell cycle (6, 8). However, whereas treatment with the pharmacological Mdm2-antagonist Nutlin-3 (20), used as a positive control, severely reduced the number of cells in S phase, MK2 inhibition did not affect cell cycle progression in otherwise unperturbed cells (Fig. S2D). This demonstrates that the protective effect of MK2 depletion and inhibition is not caused by changes in cell cycle regulation. Rather, we hypothesized that MK2 activity might affect the efficiency of DNA replication in the context of genotoxic stress. Indeed, inhibition of MK2 improved DNA replication in the presence of gemcitabine, as measured by BrdU incorporation (Fig. S2E). This indicated that MK2 is required to compromise DNA replication upon genotoxic stress.
Fig. 2.
Gemcitabine-induced H2AX phosphorylation and reduced replication fork speed depend on MK2. (A and B) H2AX phosphorylation following gemcitabine treatment requires MK2. (A) Cells were depleted of MK2 and treated with 100 nM gemcitabine for 20 h or left untreated. Cell lysates were analyzed by immunoblotting. (B) Cells were treated with 100 nM gemcitabine for 24 h or left untreated and simultaneously treated with MK2 inhibitor or DMSO, followed by immunoblot analysis. (C) Cell survival after gemcitabine treatment is improved by MK2 inhibition. On day 1, cells were treated with 100 nM gemcitabine and MK2 inhibitor or DMSO for 24 h. Cell confluence was determined daily for 12 d by light microscopy. (D–F) MK2 inhibition rescues reduced replication fork speed caused by gemcitabine. (D) Labeling protocol for DNA fiber analysis of replication fork speed. Cells were pretreated with MK2 inhibitor or DMSO for 1 h and throughout the experiment. Cells were then pulse labeled with 5-chloro-2′-deoxyuridine (CldU) for 20 min, followed by IdU for 2 h and simultaneous exposure to 400 nM gemcitabine. CldU and IdU were detected by using specific primary antibodies and secondary antibodies in red and green, respectively. (E) Average relative replication fork speed (ratio of length of IdU-labeled vs. length of CldU-labeled tracks) in cells treated as in D in dependence of gemcitabine and MK2 inhibition (n = 3; ***P = 0.0002). (F) Representative images of fibers treated as in D. (G) MK2 inhibition reduces ssDNA accumulation upon gemcitabine treatment. BrdU-labeled cells were treated with 300 nM gemcitabine and MK2 inhibitor or DMSO for 24 h. Cells were fixed and processed for ssDNA quantification by immunofluorescent detection of accessible BrdU without DNA-denaturing treatment. Image is representative of three independent replicates.
Neither UV irradiation nor gemcitabine treatment was capable of inducing complete nuclear export of MK2, whereas osmotic stress, as observed previously (21), did lead to cytoplasmic MK2 accumulation (Fig. S3A). This argues that, in the context of genotoxic stress, MK2 function is not limited to the cytoplasm.
To test whether MK2 exerts its role in the DDR during DNA replication, we assessed the impact of MK2 inhibition on H2AX phosphorylation in cells that were already in S phase at the time of treatment. For this, we pulse-labeled cells with 5-ethynyl-2′-deoxyuridine (EdU) before gemcitabine treatment. This allowed us to compare the effect of MK2 inhibition on the whole cell population with its impact on cells that are in the process of replicating their DNA (i.e., EdU-positive cells) when treated with gemcitabine. As expected, the MK2 inhibitor reduced gemcitabine-induced γH2AX accumulation when analyzing the whole cell population (Fig. S3B). Importantly, the same held true when the population was gated for EdU-positive cells (Fig. S3C). Hence, inhibition of MK2 impairs the DDR during replication.
Replication stress manifests itself as a decrease in the speed of replication forks (22). At the same time, cells react with an increase in replication origin firing (23, 24). We used DNA fiber assays (25) to assess the role of MK2 in the regulation of replication fork speed and origin firing in response to gemcitabine treatment. (The raw data for all fiber assay results presented are provided in Dataset S2.) The labeling protocol is depicted in Fig. 2D. As expected, the average replication fork speed was heavily reduced by gemcitabine treatment (Fig. 2E; Fig. 2F shows representative fiber images and Fig. S4B shows absolute fork speeds). Strikingly, however, this effect was completely rescued in the presence of MK2 inhibitor, also immediately evident from the distribution of fork speeds in histograms (Fig. S4A). We observed the same effect when replacing the inhibitor by MK2 siRNA (Fig. S4C). The low relative fork speed even in untreated samples probably arises from increased stochastic fork stalling as a result of long 5-iodo-2′-deoxyuridine (IdU) labeling times. It approximates 1 when the IdU label is reduced to 20 min (Fig. S4D).
We next asked whether MK2 inhibition also affects origin firing (Fig. S4E shows an overview of the relevant replication structures; labeling protocol is in Fig. S4F). Indeed, the marked increase in origin firing upon gemcitabine treatment was also reversed by inhibition of MK2 (Fig. S4G; Fig. S4H shows representative images of fibers). The frequency of other replication structures remained largely untouched by MK2 inhibition (Fig. S4I).
We further hypothesized that MK2 might also be required for the gemcitabine-induced accumulation of ssDNA. ssDNA may accumulate as a result of perturbed replication and can be readily detected by immunofluorescent staining for exposed BrdU (5). Indeed, we found that cells in which MK2 was inhibited accumulated less ssDNA in response to gemcitabine treatment than control cells (Fig. 2G). Furthermore, we observed a strong correlation between ssDNA accumulation and H2AX phosphorylation (Fig. S2F).
In summary, upon gemcitabine-induced replicative stress, MK2 inhibition restores the replication fork speed while it decreases excess origin firing and premature replication termination. Thus, the cell is capable of adapting its replication not only in response to the presence of a toxic nucleoside analogue, but also as a function of MK2 activity.
MK2 Is Required for DNA Damage Signaling and Replication Impairment upon Inhibition or Depletion of Chk1.
DNA replication, even in unperturbed cells, is subject to tight regulation. Upon randomly occurring replication errors, the cell activates an intra–S-phase checkpoint, which is mainly mediated by ATR and Chk1 (26-29). Chk1 depletion or inhibition leads to the abrogation of this checkpoint (30) and has deleterious effects, ranging from increased replication initiation to DNA breakage and cell death (11, 31). Thus, Chk1 inhibition or depletion induces replicative stress. We therefore tested whether the genotoxic effects of Chk1 inhibition or depletion also depended on MK2.
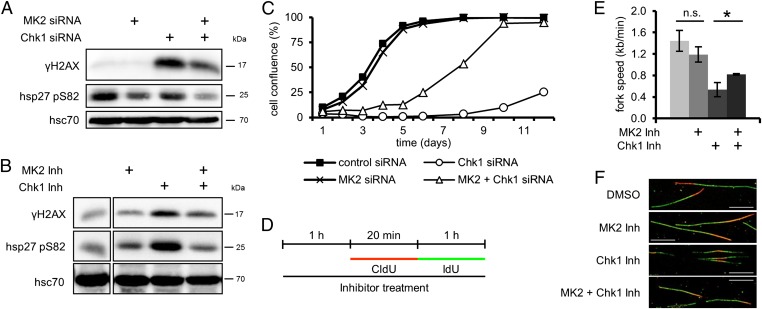
Chk1 knockdown or inhibition with the pharmacological inhibitor SB218078 (32) (called “Chk1 inhibitor” from here on) strongly enhanced H2AX phosphorylation (Fig. 3 A and B; Fig. S1B shows depletion efficiencies), in line with previous observations (4, 5, 30). Importantly, MK2 depletion or inhibition reduced this effect. Phosphorylation of Hsp27, an MK2 substrate (33), served as a readout for MK2 activity. Furthermore, the previously reported (34) increase in gemcitabine-induced H2AX phosphorylation by Chk1 depletion also depended on MK2 (Fig. S5A).
Fig. 3.
The genotoxic effects caused by depletion or inhibition of Chk1 depend on MK2. (A and B) H2AX phosphorylation upon depletion or inhibition of Chk1 is reduced by codepletion or inhibition of MK2. (A) Cells were depleted of MK2 and Chk1 and harvested 48 h later, and cell lysates were analyzed by immunoblotting. (B) Cells were treated with MK2 inhibitor, Chk1 inhibitor (Chk1 Inh), or DMSO for 12 h, and analyzed by immunoblotting. (C) Cell proliferation after Chk1 depletion is improved by codepletion of MK2. Cells were depleted of MK2 and/or Chk1 and reseeded. At 24 h later, measurement was started (day 1). Cell confluence was measured on subsequent days. Averages of three replicates are shown. (D–F) MK2 inhibition improves the reduced replication fork speed following Chk1 inhibition. (D) Labeling protocol for DNA fiber analysis. Cells were pretreated with MK2 inhibitor, Chk1 inhibitor, or DMSO for 1 h and then pulse-labeled with CldU for 20 min and IdU for 1 h in the continuous presence of inhibitors. CldU and IdU were detected by immunofluorescence in red and green, respectively. (E) Average replication fork speed in dependence of Chk1 and MK2 inhibition. The length of CldU tracks of ongoing forks was used to calculate the replication fork speed (n = 3; *P = 0.0226). (F) Representative images of fibers from cells treated as in D. (Scale bar: 10 µm.)
Depletion of Chk1, as expected (5, 35), resulted in an increase in the fraction of cells in S phase (Fig. S5 B and C), but this was accompanied by reduced EdU incorporation (Fig. S5D), indicating that cells depleted of Chk1 accumulate in S phase but stop DNA replication. Simultaneous knockdown of MK2 reduced the accumulation of cells in S phase (Fig. S5 B and C), arguing that MK2 is required for intra–S-phase arrest in response to Chk1 depletion.
Finally, although Chk1-depleted cells displayed a strongly reduced proliferation rate and only started to recover approximately 1 wk after the knockdown, cells codepleted of Chk1 and MK2 showed only a mild decrease in proliferation (Fig. 3C).
In conclusion, MK2 is needed for the DDR and the accumulation of cells in S phase upon elimination of Chk1. Hence, Chk1 is not strictly required for S phase progression. Rather, MK2 mediates a block in DNA synthesis when Chk1 is absent.
We directly assessed the effects of MK2 in the context of Chk1 inhibition by DNA fiber assays. Fig. 3D shows the labeling protocol. Chk1 inhibition strongly decreased replication fork speed (Fig. 3E and Fig. S6A; Fig. 3F shows representative images of fibers) and increased origin firing (Fig. S6B), in line with previous reports (10, 36, 37). Simultaneous treatment with MK2 inhibitor, however, improved the fork speed and rescued enhanced origin firing almost completely. Again, the frequency of other replication structures remained largely unchanged (Fig. S6C).
Thus, upon Chk1 impairment, MK2 suppresses DNA replication while enhancing origin firing.
TLS Is Required for the Rescue of Gemcitabine-Induced Replication Impairment by MK2 Inhibition.
Our findings raise the question of how MK2 blocks DNA replication, and how replication is rescued upon inhibition of MK2, despite the continuous presence of gemcitabine.
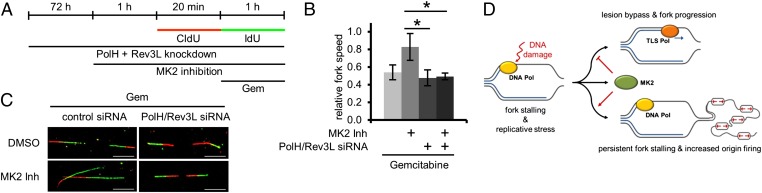
We hypothesized that TLS does not reach its full activity in the presence of MK2, but that it overcomes gemcitabine-induced lesions when MK2 is inactivated. In such a scenario, one would predict that MK2 inhibition can no longer rescue the gemcitabine-induced block in replication when TLS is impaired. To test this, we depleted cells of polymerase (Pol) η, the TLS polymerase previously associated with gemcitabine-induced TLS (38), and Rev3L, the catalytic subunit of Pol ζ, which is specialized to synthesize DNA from a distorted DNA duplex (39) (Fig. S7A). We then assessed replication fork speed upon gemcitabine treatment and MK2 inhibition as before (Fig. 4A). The removal of Pol η and Rev3L did not grossly affect replication in the absence of gemcitabine (Fig. S7 B and D; Fig. S7C shows absolute fork speeds), underscoring that, in unperturbed cells, TLS is not essential. However, in the absence of these polymerases, MK2 inhibition was no longer capable of reversing the suppression of replication fork speed by gemcitabine (Fig. 4B; Fig. 4C shows representative images of fibers and Fig. S7G shows absolute fork speeds). The changes in fork speed are also documented by the distribution of fork rates in unperturbed (Fig. S7E) and gemcitabine-treated cells (Fig. S7F). Thus, upon the knockdown of these TLS components, gemcitabine slowed down the fork rate regardless of MK2.
Fig. 4.
Rescue of gemcitabine-induced slow replication fork speed by MK2 inhibition depends on TLS. (A) Labeling protocol for DNA fiber analysis. Cells were depleted of PolH and Rev3L. At 72 h later, cells were pretreated with MK2 inhibitor or DMSO for 1 h and then pulse-labeled with CldU for 20 min. Afterward, cells were pulse-labeled with IdU for 1 h and simultaneously exposed to 400 nM gemcitabine. (B) Average relative replication fork speed (ratio of length of IdU-labeled vs. length of CldU-labeled tracks) in gemcitabine-treated cells in dependence of MK2 inhibition and depletion of TLS polymerases PolH and Rev3L (n = 3; *P = 0.0308 and *P = 0.0186, respectively). (C) Representative images of fibers from cells treated with gemcitabine as in A (Scale bar: 10 µm.) (D) A model of how, upon replicative stress, the decision between TLS and replication fork stalling and increased origin firing is regulated by MK2.
Treatment of cells with the ribonucleotide reductase inhibitor hydroxyurea (HU) also led to H2AX phosphorylation, but MK2 inhibition did not influence γH2AX levels or ssDNA accumulation in this context (Fig. S8 A and B). This is in contrast to the role of MK2 in the response to gemcitabine or Chk1 inhibition. We propose that this difference is caused by a reduction of dNTP levels by the inhibition of ribonucleotide reductase through HU, resulting in diminished TLS. This consideration is in accordance with the previously described dependence of Pol η activity on high dNTP levels (40).
We conclude that MK2 inhibition can only rescue the deleterious effects of gemcitabine on DNA replication when the TLS machinery is intact. In agreement with this notion, we found that Pol η phosphorylation can be enhanced by MK2 in vitro (Fig. S8 C and D), suggesting a direct regulatory activity of MK2 on Pol η. These findings imply that MK2 acts as a mediator of the replicative stress response. It appears to block or at least to limit the ability of a cell to overcome replicative stress by TLS. In the absence of MK2 activity, the cells are considerably more tolerant to replicative stress, but this resistance requires TLS.
Discussion
Our results indicate that the kinase MK2 is required for the accumulation of DNA damage upon replicative stress. To our knowledge, MK2 represents the first example of a cellular factor the removal of which enables the continuation of DNA replication under conditions that would normally lead to fork stalling. The removal of CDKs also promotes fork progression; this, however, appears to represent a consequence of decreased origin firing rather than a direct effect on processivity (10). Because gemcitabine directly affects replication processivity and because MK2 inhibition is capable of restoring replication under such conditions, we suggest that MK2 activity primarily determines the decision of fork stalling vs. continuous replication. Thus, replicative stress is not only determined by DNA lesions before or during S phase. Rather, the activity of specific signaling components governs the accumulation of DNA damage during replication. MK2 activity is required for efficient replication blocks; its inhibition leads to a permissive state that allows replication despite disturbances, with the help of TLS (Fig. 4D).
TLS appears as a double-edged sword in the determination of cell fate. It represents a convenient mechanism to allow the successful completion of S phase. However, TLS may also lead to the accumulation of small mutations. It is therefore conceivable that mechanisms evolved to limit TLS. The results displayed in this paper suggest that MK2 is an essential component to carry out this TLS control.
MK2 inactivation restored the rate of replication forks that were slowed down by gemcitabine or Chk1 inhibition. However, at the same time, the origin firing frequency was inversely correlated to the replication rate. Although this correlation was observed previously (36, 37, 41), the underlying mechanism remains elusive. This also makes it difficult to decide whether the replication speed or the origin-firing rate is the primary function to be governed by MK2. The fact that TLS is required for the rescue of gemcitabine effects argues in favor of replication efficiency as the primary target of MK2. This further argues that increased origin firing may be triggered by slow or stalled replication forks, as has been suggested earlier (23, 24).
Previous studies have implied MK2 in the arrest of cells in G2, and in a translational block (6, 8). In particular, the arrest in G2 may still represent a result of incomplete DNA replication and is therefore in agreement with our study. An intimate link between the control of DNA replication and mitosis emerges as a common theme, as many factors that were first described to control mitosis later turned out to determine S phase progression as well. Examples include the kinases CDK1 and Wee1, as well as the phosphatases of the Cdc25 family (42–44). Mechanistically, this link is perhaps not surprising, as incompletely replicated DNA easily results in mitotic catastrophe (45, 46).
MK2 was initially described as a mediator of p38-driven signaling, typically seen in inflammatory responses (13, 47–49). We propose that MK2 may integrate inflammation and replicative stress, resulting in a particularly stringent control on replication when cells are exposed to inflammatory stimuli. This would allow further enhancement of the inflammatory response by denying smooth replication to cells under these conditions. Further studies are required to determine whether the deliberate activation of inflammatory responses, in particular p38, may enhance the efficacy of nucleoside analogues in cancer therapy.
Chk1 and MK2 share structural and functional homologies also present in Chk2 (6), leading to the assumption that all three kinases may serve similar purposes, i.e., transmitting the DDR. However, Chk1 and MK2 act in an antagonistic fashion under the conditions presented here. This striking observation may have its roots in a differential activation pattern and/or in different substrate specificities. Although MK2 can undergo nuclear export as part of a stress response, MK2 was mostly retained in the nucleus in response to UV or gemcitabine treatment. We therefore propose that differential interactions with signaling and DNA replication factors, rather than intracellular localization, confer the mechanistic difference between Chk1 and MK2.
The crucial role of MK2 in replication control also raises the question whether MK2 levels in tumor cells may be suitable as predictors of therapeutic outcome when patients with tumors are treated with gemcitabine or other nucleoside analogues. Although this is of interest, it should be considered that the overall response is likely to be determined by the combination of MK2 and the previously identified negative regulators of replicative stress, including Chk1 and Wee1. Additional factors may also be involved, such as the effectors of TLS and of general DNA replication, as well as nucleotide metabolism, cell cycle checkpoints, and upstream regulators of MK2. The targeted manipulation of Chk1 was already reported to increase the efficacy of nucleoside analogues as anticancer agents (50) and is currently being tested in phase I clinical trials (51). It should be noted that the findings presented here suggest that down-regulation or loss of MK2 in tumors likely constitute a source of resistance to Chk1 inhibition. We anticipate that the combined inhibition of Chk1 and Wee1, along with the activation of MK2, may provide a more effective strategy for chemosensitization.
Materials and Methods
The siRNA screen was performed with high content automated cell microscopy. Furthermore, cell culture, immunoblotting, immunofluorescence microscopy, UV irradiation of mice, immunohistochemistry and TUNEL, cell proliferation and clonogenic assays, flow cytometry, DNA fiber assay, quantitative RT-PCR, and in vitro kinase assay were used. The detailed methods are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
This paper is dedicated to the memory of Dr. Ellen Fanning, who was a mentor and a friend. Dr. Fanning's research focus was on DNA replication, the central topic of our paper. We thank Andreas Wodarz (University Medicine Göttingen) for sharing the confocal microscope. This work was supported by the Wilhelm Sander Stiftung; Deutsche José Carreras Stiftung; German Cancer Aid/Dr. Mildred Scheel Stiftung; European Union Sixth Framework Program (Integrated Project Active p53); German Research Foundation (Deutsche Forschungsgemeinschaft); Studienstiftung des Deutschen Volkes (F.K.); Göttingen Graduate School of Neurosciences and Molecular Biosciences (F.K.); and European Union Cooperation in Science and Technology action BM0703 Cancer and Control of Genomic Integrity (to F.K., I.E., T.H., and M.D.). F.K. was a Fellow of the International Max Planck Research School, Master PhD program in Molecular Biology, during this work.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304355110/-/DCSupplemental.
References
- 1.Bartkova J, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444(7119):633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 2.Kranz D, Dobbelstein M. Nongenotoxic p53 activation protects cells against S-phase-specific chemotherapy. Cancer Res. 2006;66(21):10274–10280. doi: 10.1158/0008-5472.CAN-06-1527. [DOI] [PubMed] [Google Scholar]
- 3.Kranz D, Dohmesen C, Dobbelstein M. BRCA1 and Tip60 determine the cellular response to ultraviolet irradiation through distinct pathways. J Cell Biol. 2008;182(1):197–213. doi: 10.1083/jcb.200712014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beck H, et al. Regulators of cyclin-dependent kinases are crucial for maintaining genome integrity in S phase. J Cell Biol. 2010;188(5):629–638. doi: 10.1083/jcb.200905059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Syljuåsen RG, et al. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol Cell Biol. 2005;25(9):3553–3562. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manke IA, et al. MAPKAP kinase-2 is a cell cycle checkpoint kinase that regulates the G2/M transition and S phase progression in response to UV irradiation. Mol Cell. 2005;17(1):37–48. doi: 10.1016/j.molcel.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 7.Reinhardt HC, Aslanian AS, Lees JA, Yaffe MB. p53-deficient cells rely on ATM- and ATR-mediated checkpoint signaling through the p38MAPK/MK2 pathway for survival after DNA damage. Cancer Cell. 2007;11(2):175–189. doi: 10.1016/j.ccr.2006.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reinhardt HC, et al. DNA damage activates a spatially distinct late cytoplasmic cell-cycle checkpoint network controlled by MK2-mediated RNA stabilization. Mol Cell. 2010;40(1):34–49. doi: 10.1016/j.molcel.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doil C, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136(3):435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 10.Petermann E, Woodcock M, Helleday T. Chk1 promotes replication fork progression by controlling replication initiation. Proc Natl Acad Sci USA. 2010;107(37):16090–16095. doi: 10.1073/pnas.1005031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gagou ME, Zuazua-Villar P, Meuth M. Enhanced H2AX phosphorylation, DNA replication fork arrest, and cell death in the absence of Chk1. Mol Biol Cell. 2010;21(5):739–752. doi: 10.1091/mbc.E09-07-0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clifton AD, Young PR, Cohen P. A comparison of the substrate specificity of MAPKAP kinase-2 and MAPKAP kinase-3 and their activation by cytokines and cellular stress. FEBS Lett. 1996;392(3):209–214. doi: 10.1016/0014-5793(96)00816-2. [DOI] [PubMed] [Google Scholar]
- 13.Gaestel M. MAPKAP kinases - MKs - two’s company, three’s a crowd. Nat Rev Mol Cell Biol. 2006;7(2):120–130. doi: 10.1038/nrm1834. [DOI] [PubMed] [Google Scholar]
- 14.Anderson DR, et al. Pyrrolopyridine inhibitors of mitogen-activated protein kinase-activated protein kinase 2 (MK-2) J Med Chem. 2007;50(11):2647–2654. doi: 10.1021/jm0611004. [DOI] [PubMed] [Google Scholar]
- 15.Ronkina N, et al. The mitogen-activated protein kinase (MAPK)-activated protein kinases MK2 and MK3 cooperate in stimulation of tumor necrosis factor biosynthesis and stabilization of p38 MAPK. Mol Cell Biol. 2007;27(1):170–181. doi: 10.1128/MCB.01456-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herrlich P, Karin M, Weiss C. Supreme EnLIGHTenment: Damage recognition and signaling in the mammalian UV response. Mol Cell. 2008;29(3):279–290. doi: 10.1016/j.molcel.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karnitz LM, et al. Gemcitabine-induced activation of checkpoint signaling pathways that affect tumor cell survival. Mol Pharmacol. 2005;68(6):1636–1644. doi: 10.1124/mol.105.012716. [DOI] [PubMed] [Google Scholar]
- 18.Ewald B, Sampath D, Plunkett W. Nucleoside analogs: Molecular mechanisms signaling cell death. Oncogene. 2008;27(50):6522–6537. doi: 10.1038/onc.2008.316. [DOI] [PubMed] [Google Scholar]
- 19.Ewald B, Sampath D, Plunkett W. H2AX phosphorylation marks gemcitabine-induced stalled replication forks and their collapse upon S-phase checkpoint abrogation. Mol Cancer Ther. 2007;6(4):1239–1248. doi: 10.1158/1535-7163.MCT-06-0633. [DOI] [PubMed] [Google Scholar]
- 20.Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303(5659):844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 21.Engel K, Kotlyarov A, Gaestel M. Leptomycin B-sensitive nuclear export of MAPKAP kinase 2 is regulated by phosphorylation. EMBO J. 1998;17(12):3363–3371. doi: 10.1093/emboj/17.12.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11(3):208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 23.Ge XQ, Jackson DA, Blow JJ. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev. 2007;21(24):3331–3341. doi: 10.1101/gad.457807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ibarra A, Schwob E, Méndez J. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc Natl Acad Sci USA. 2008;105(26):8956–8961. doi: 10.1073/pnas.0803978105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: Evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140(6):1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feijoo C, et al. Activation of mammalian Chk1 during DNA replication arrest: a role for Chk1 in the intra-S phase checkpoint monitoring replication origin firing. J Cell Biol. 2001;154(5):913–923. doi: 10.1083/jcb.200104099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heffernan TP, et al. An ATR- and Chk1-dependent S checkpoint inhibits replicon initiation following UVC-induced DNA damage. Mol Cell Biol. 2002;22(24):8552–8561. doi: 10.1128/MCB.22.24.8552-8561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miao H, Seiler JA, Burhans WC. Regulation of cellular and SV40 virus origins of replication by Chk1-dependent intrinsic and UVC radiation-induced checkpoints. J Biol Chem. 2003;278(6):4295–4304. doi: 10.1074/jbc.M204264200. [DOI] [PubMed] [Google Scholar]
- 29.Zachos G, Rainey MD, Gillespie DA. Chk1-deficient tumour cells are viable but exhibit multiple checkpoint and survival defects. EMBO J. 2003;22(3):713–723. doi: 10.1093/emboj/cdg060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao Z, Xue J, Sowin TJ, Rosenberg SH, Zhang H. A novel mechanism of checkpoint abrogation conferred by Chk1 downregulation. Oncogene. 2005;24(8):1403–1411. doi: 10.1038/sj.onc.1208309. [DOI] [PubMed] [Google Scholar]
- 31.Sørensen CS, et al. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol. 2005;7(2):195–201. doi: 10.1038/ncb1212. [DOI] [PubMed] [Google Scholar]
- 32.Jackson JR, et al. An indolocarbazole inhibitor of human checkpoint kinase (Chk1) abrogates cell cycle arrest caused by DNA damage. Cancer Res. 2000;60(3):566–572. [PubMed] [Google Scholar]
- 33.Stokoe D, Engel K, Campbell DG, Cohen P, Gaestel M. Identification of MAPKAP kinase 2 as a major enzyme responsible for the phosphorylation of the small mammalian heat shock proteins. FEBS Lett. 1992;313(3):307–313. doi: 10.1016/0014-5793(92)81216-9. [DOI] [PubMed] [Google Scholar]
- 34.Parsels LA, et al. Gemcitabine sensitization by checkpoint kinase 1 inhibition correlates with inhibition of a Rad51 DNA damage response in pancreatic cancer cells. Mol Cancer Ther. 2009;8(1):45–54. doi: 10.1158/1535-7163.MCT-08-0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forment JV, Blasius M, Guerini I, Jackson SP. Structure-specific DNA endonuclease Mus81/Eme1 generates DNA damage caused by Chk1 inactivation. PLoS ONE. 2011;6(8):e23517. doi: 10.1371/journal.pone.0023517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petermann E, et al. Chk1 requirement for high global rates of replication fork progression during normal vertebrate S phase. Mol Cell Biol. 2006;26(8):3319–3326. doi: 10.1128/MCB.26.8.3319-3326.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maya-Mendoza A, Petermann E, Gillespie DA, Caldecott KW, Jackson DA. Chk1 regulates the density of active replication origins during the vertebrate S phase. EMBO J. 2007;26(11):2719–2731. doi: 10.1038/sj.emboj.7601714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen YW, Cleaver JE, Hanaoka F, Chang CF, Chou KM. A novel role of DNA polymerase eta in modulating cellular sensitivity to chemotherapeutic agents. Mol Cancer Res. 2006;4(4):257–265. doi: 10.1158/1541-7786.MCR-05-0118. [DOI] [PubMed] [Google Scholar]
- 39.Waters LS, et al. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev. 2009;73(1):134–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Washington MT, Johnson RE, Prakash L, Prakash S. The mechanism of nucleotide incorporation by human DNA polymerase eta differs from that of the yeast enzyme. Mol Cell Biol. 2003;23(22):8316–8322. doi: 10.1128/MCB.23.22.8316-8322.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shechter D, Costanzo V, Gautier J. ATR and ATM regulate the timing of DNA replication origin firing. Nat Cell Biol. 2004;6(7):648–655. doi: 10.1038/ncb1145. [DOI] [PubMed] [Google Scholar]
- 42.Hochegger H, et al. An essential role for Cdk1 in S phase control is revealed via chemical genetics in vertebrate cells. J Cell Biol. 2007;178(2):257–268. doi: 10.1083/jcb.200702034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katsuno Y, et al. Cyclin A-Cdk1 regulates the origin firing program in mammalian cells. Proc Natl Acad Sci USA. 2009;106(9):3184–3189. doi: 10.1073/pnas.0809350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Domínguez-Kelly R, et al. Wee1 controls genomic stability during replication by regulating the Mus81-Eme1 endonuclease. J Cell Biol. 2011;194(4):567–579. doi: 10.1083/jcb.201101047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol. 2009;11(6):753–760. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- 46.Ichijima Y, et al. DNA lesions induced by replication stress trigger mitotic aberration and tetraploidy development. PLoS ONE. 2010;5(1):e8821. doi: 10.1371/journal.pone.0008821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stokoe D, et al. MAPKAP kinase-2; a novel protein kinase activated by mitogen-activated protein kinase. EMBO J. 1992;11(11):3985–3994. doi: 10.1002/j.1460-2075.1992.tb05492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kotlyarov A, et al. MAPKAP kinase 2 is essential for LPS-induced TNF-alpha biosynthesis. Nat Cell Biol. 1999;1(2):94–97. doi: 10.1038/10061. [DOI] [PubMed] [Google Scholar]
- 49.Winzen R, et al. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 1999;18(18):4969–4980. doi: 10.1093/emboj/18.18.4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garrett MD, Collins I. Anticancer therapy with checkpoint inhibitors: What, where and when? Trends Pharmacol Sci. 2011;32(5):308–316. doi: 10.1016/j.tips.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 51.Ma CX, Janetka JW, Piwnica-Worms H. Death by releasing the breaks: CHK1 inhibitors as cancer therapeutics. Trends Mol Med. 2011;17(2):88–96. doi: 10.1016/j.molmed.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.