Significance
The small GTPase ADP ribosylation factor (Arf) 6 anchors to the plasma membrane, where it coordinates actin filament polymerization to remodel the membrane-associated cytoskeleton in diverse cell processes, yet how Arf6 directs actin polymerization is unknown. By reconstituting membrane-associated actin assembly, we find that Arf6 assembles actin via the WAVE regulatory complex (WRC). In contrast to other Arf family members, which directly bind and activate WRC, Arf6 signaled to WRC indirectly by recruiting the Arf1 activator ARNO to the membrane. Remarkably, we demonstrate this mechanism is hijacked by the pathogen Salmonella that usurped Arf6 and WRC to invade human host cells and establish intracellular infection. This study describes a mechanism for Arf6-driven actin polymerization.
Keywords: Rac1, lamellipodia, nucleation-promoting factor, infection
Abstract
ADP ribosylation factor (Arf) 6 anchors to the plasma membrane, where it coordinates membrane trafficking and cytoskeleton remodelling, but how it assembles actin filaments is unknown. By reconstituting membrane-associated actin assembly mediated by the WASP family veroprolin homolog (WAVE) regulatory complex (WRC), we recapitulated an Arf6-driven actin polymerization pathway. We show that Arf6 is divergent from other Arf members, as it was incapable of directly recruiting WRC. We demonstrate that Arf6 triggers actin assembly at the membrane indirectly by recruiting the Arf guanine nucleotide exchange factor (GEF) ARNO that activates Arf1 to enable WRC-dependent actin assembly. The pathogen Salmonella usurped Arf6 for host cell invasion by recruiting its canonical GEFs EFA6 and BRAG2. Arf6 and its GEFs facilitated membrane ruffling and pathogen invasion via ARNO, and triggered actin assembly by generating an Arf1–WRC signaling hub at the membrane in vitro and in cells. This study reconstitutes Arf6-dependent actin assembly to reveal a mechanism by which related Arf GTPases orchestrate distinct steps in the WRC cytoskeleton remodelling pathway.
ADP ribosylation factor (Arf) GTPases are best known for their roles in vesicle and organelle trafficking (1). Class I and II Arfs (Arf1, Arf3, Arf4, and Arf5) are found predominantly in and around the Golgi apparatus. In contrast, the more divergent Class III Arf (Arf6) operates almost exclusively at the plasma membrane (2). Consistent with its localization, Arf6 has been heavily implicated in trafficking events at the cell surface, including the regulation of endocytosis and exocytosis (1). In particular, Arf6 and its guanine nucleotide exchange factors (GEFs) are believed to be pivotal to the recycling of endosomes and receptors to and from the plasma membrane (3, 4). Arf6 also has a clear role in cortical cytoskeleton rearrangement (5). This is strongly supported by evidence of Arf6 and Rac1 (Ras-related C3 botulinum toxin substrate) interplay (6), exemplified by Arf6 recruitment of the Rac1 GEF Kalirin, Arf6 promotion of Rac1 activation, and lamellipodia formation (7–9).
Rac1 is required for generation of lamellipodia that lead to membrane ruffles and macropinocytosis (10). Rac1 is thought to achieve this by activating the WRC, which comprises WAVE (WASP family veroprolin homolog), Abi (abl-interactor 1), Cyfip (cytoplasmic FMR1 interacting protein), Nap1 (NCK-associated protein 1), HSPC300 (heat shock protein C300), or their homologs (10–12). We recently established that Rac1 was not sufficient for WRC recruitment to the membrane and its activation, which instead requires direct binding by Rac1 and an Arf GTPase (13). This in vitro Arf activity could be supplied by multiple isoforms including Arf1 and Arf5, but only Arf1 facilitated WRC-dependent lamellipodia formation and macropinocytosis of the bacterial pathogen Salmonella into human host cells (13–15). Intriguingly, Arf6 also promoted Salmonella invasion (14), and, given the capability of Arfs to modulate WRC, it seems likely that Arf6 also recruits and activates the WRC at the plasma membrane. However, despite mounting evidence that Arf6 remodels the cytoskeleton, a molecular mechanism by which Arf6 drives Arp2/3-dependent actin assembly has not been resolved.
Results
Arf6-Mediated Actin Assembly via the WAVE Regulatory Complex.
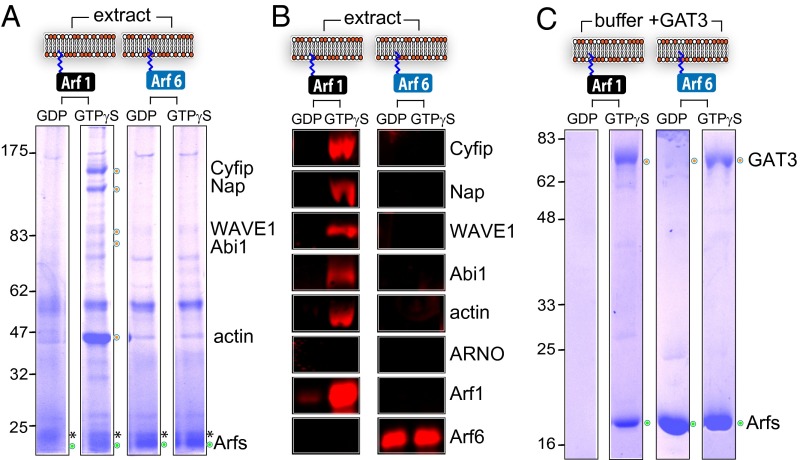
To address how Arf6 mediates actin assembly, we first examined Arf6 recruitment of WRC to the membrane. Purified myristoylated Arf6 and control Arf1 were each loaded with GDP or GTPγS (a nonhydrolysable analog of GTP that maintains activation) before anchoring to silica beads coated with a phospholipid bilayer (membrane platforms) composed of equal amounts of phosphatidylinositol and phosphatidylcholine. The Arf membrane platforms were each incubated in brain cell extract before being isolated and extensively washed and the recruited proteins analyzed by Coomassie blue-stained SDS/PAGE (Fig. 1A). Although control active Arf1 (Fig. 1A, green circle) recruited WRC components Cyfip, Nap1, WAVE1, and Abi1 (Fig. 1A, orange circles), neither inactive (i.e., GDP) nor active (i.e., GTPγS) Arf6 recruited the WRC. Immunoblotting confirmed the presence of recombinant Arf GTPases and that Arf1 but not Arf6 recruited the WRC (Fig. 1B). The low abundance of membrane GDP-bound Arf1 observed (Fig. 1 A and B) was expected, as inactive Arf1 has lower affinity for the membrane than active Arf1 (16). We have previously demonstrated that WRC activation triggers formation of actin comet tails at the membrane (13). Here, actin was observed only on Arf1GTPγS membrane platforms (Fig. 1 A and B), confirming that Arf6 does not mediate actin polymerization in extract. To verify that our recombinant Arf6 was functional, Arf platforms were each incubated in buffer containing purified GAT3 (+GAT3), a domain of GGA3 with homology to GGAs and TOM1 that binds to active Arfs (17). In contrast to inactive GDP-bound isoforms, active control Arf1 and Arf6 recruited GAT3 (Fig. 1C), demonstrating that the Arf6 protein was functional. These experiments show that, unlike Arf1, Arf6 does not directly recruit WRC to the membrane.
Fig. 1.
Arf6 interaction with the WAVE regulatory complex. (A) Proteins recruited from extract to phospholipid bilayers by Arf1 and Arf6. Silica beads coated with phospholipid bilayers (membrane platforms depicted in cartoon) anchored with inactive GDP-bound or active GTPγS-bound myristoylated Arf1 or Arf6 (green circles) isolated from extract. Recruited proteins (labeled at right and marked with orange circles) were extracted and analyzed by SDS/PAGE and Coomassie blue staining. Molecular weight markers are in kilodaltons (Left). Asterisk marks a recruited extract protein migrating above recombinant Arfs. (B) Parallel immunoblotting of samples from A with indicated antibodies (Right). (C) Recruitment of Arf binding domain (GAT, orange circles) of GGA3 to membrane platforms with anchored Arf1 or Arf6 (green circle) in buffer.
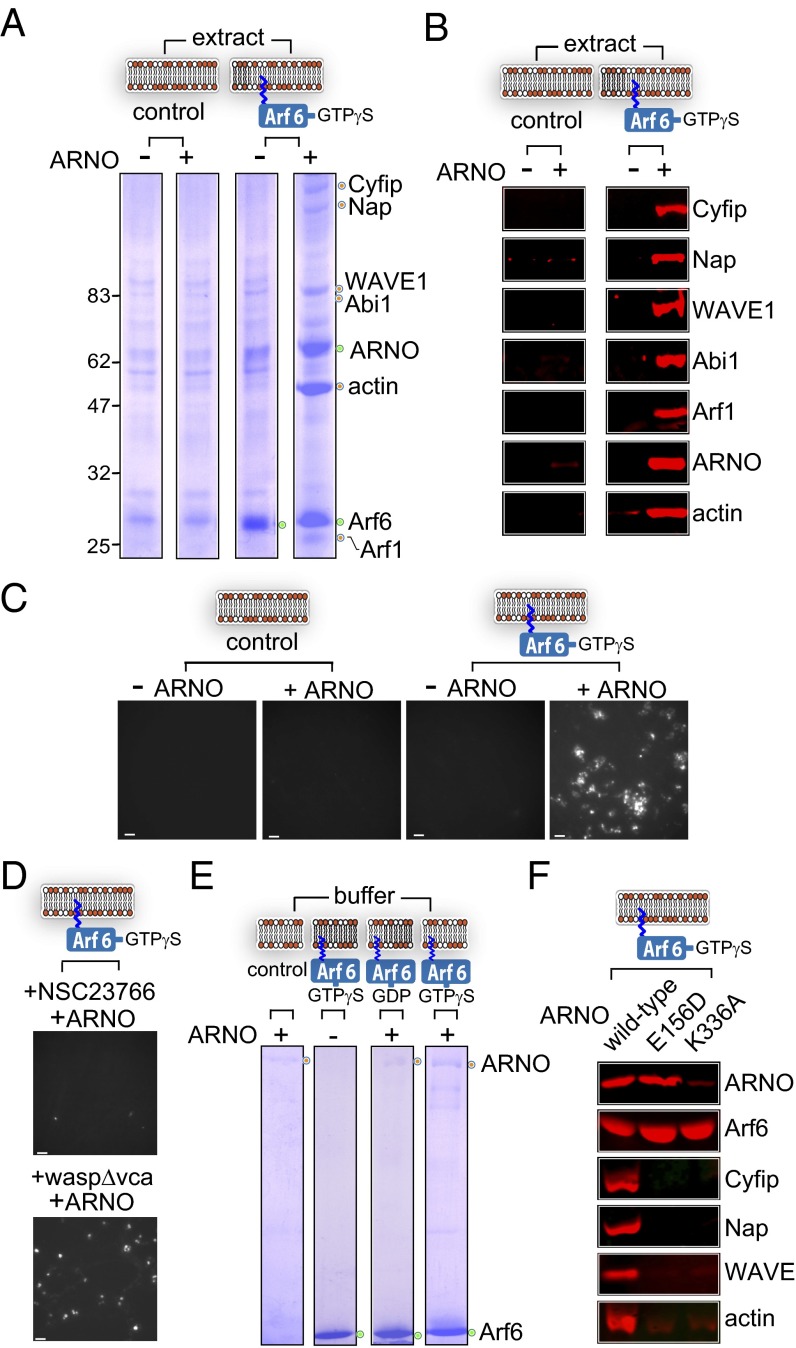
We next sought to address an indirect role for Arf6 in WRC-mediated actin assembly. Arf6 is known to act upstream of Arf1 activation by mediating plasma membrane localization of the Arf GEF ARNO (Arf nucleotide binding site opener) via interaction with its pleckstrin homology (PH) domain (18, 19). Immunoblotting showed that ARNO was recruited by neither Arf1 nor Arf6 membrane platforms (Fig. 1B). However, this is likely because the level of ARNO in the extract was too low to be detected with our antibodies (Fig. S1A). To overcome this barrier, membrane platforms alone (control) or those with anchored Arf6GTPγS were incubated in extract with or without addition of recombinant ARNO. Recruitment of ARNO was clearly evident on Arf6, but not control, membrane platforms (Fig. 2A, green circle). Arf6-dependent recruitment of ARNO triggered Arf1 localization to the membrane and the recruitment of WRC (Fig. 2A, orange circles). Immunoblotting of the extracted proteins confirmed Arf6-mediated recruitment of ARNO, Arf1, and the WRC (Fig. 2B). Arf6-driven actin assembly (Fig. 2A, orange circle) was also apparent on the membrane in the presence of ARNO (Fig. 2A). This was confirmed by parallel immunoblotting (Fig. 2B), and visualized by fluorescence imaging in extract containing rhodamine-labeled actin (Fig. 2C), in which actin polymerization at the membrane was observed only when Arf6 was incubated in extract containing ARNO. In our extract, actin assembly can achieved by activating N-WASP (neuronal wiskott-Aldrich syndrome protein) or WRC, and so WRC activation can be determined by visualizing actin assembly when the N-WASP pathway is inhibited (14). N-WASP–inhibited extract (waspΔvca) had no effect on Arf6-driven actin assembly in the presence of ARNO, but this was abolished by the Rac1 inhibitor NSC23766, showing that Arf6 polymerizes actin filaments via Rac1 and WRC (Fig. 2D). These experiments show that Arf6 uses ARNO to trigger Rac1-dependent actin polymerization via WRC.
Fig. 2.
Arf6 mediates WAVE-dependent actin assembly on the membrane via ARNO. (A) Arf6 driven actin assembly in the presence of ARNO. Proteins recruited (orange circles) to control and Arf6GTPγS-anchored membrane platforms (depicted in cartoon) from extract with (+) or without (−) purified ARNO (recombinant proteins marked by green circles). Molecular weight markers are in kilodaltons (Left). (B) Parallel immunoblotting of samples from A with indicated antibodies (Right). (C) Fluorescence microscopy of rhodamine-actin assembly on the control and Arf6GTPγS-anchored membrane platforms in extract with (+) or without (−) purified ARNO. (Scale bars: 10 µm.) (D) Rhodamine-actin assembly on Arf6GTPγS-anchored membrane platforms in extract containing purified ARNO and an inhibitor of Rac1 (NSC23766) or N-WASP (waspΔvca). (Scale bars: 10 µm.) (E) Arf6 interaction with purified ARNO at the membrane in buffer. Control membrane platforms, or platforms anchored with Arf6GDP or Arf6GTPγS (green circles) were incubated in buffer alone (−) or buffer containing diluted recombinant ARNO (+), marked by orange circles. (F) Arf6-mediated recruitment of ARNO in extract. Arf6GTPγS-anchored membrane platforms isolated from extract containing purified WT, GEF mutant (E156D), or Arf6-binding mutant (K336A) variants and analyzed by immunoblotting with indicated antibodies (Right).
We next sought to determine how Arf6 recruits ARNO. To resolve whether Arf6 recruited ARNO via direct interaction, Arf6GDP and Arf6GTPγS membrane platforms were incubated in buffer with or without recombinant ARNO (Fig. 2E). Unlike Arf6 membrane platforms in buffer alone, ARNO (Fig. 2E, orange circles) was observed at control and Arf6GDP membrane platforms. This recruitment likely resulted from the presence of the ARNO PH domain that has affinity for the membrane (1). Nevertheless, ARNO recruitment was enhanced when Arf6GTPγS was at the membrane, showing that Arf6 recruits ARNO directly. ARNO catalyses nucleotide exchange most effectively on Arf1, in cells and in vitro (18, 20). Arf6 can recruit ARNO to the membrane via binding its PH domain (18), but can also be a substrate of the GEF activity (21). To examine ARNO recruitment and relate this to ARNO-triggered actin assembly in extract, we examined Arf6 membrane platforms in extract supplemented with WT ARNO, a catalytically inactive (CI) GEF mutant (E156D), or a variant harboring a mutation (K336A) in the PH domain that abolishes Arf6 binding, rendering ARNO inactive (18, 19, 22) (Fig. 2F). Recruitment of WT and GEF-mutant variants of ARNO were equivalent, but recruitment of the Arf6-binding mutant was reduced. Immunoblotting showed that WRC recruitment and actin assembly by Arf6 membrane platforms was dependent on ARNO GEF activity and Arf6 interaction with the ARNO PH domain. Thus, Arf6 acts upstream of Arf1 in WRC-dependent actin assembly by directing ARNO localization to the membrane.
Arf6 Facilitates Salmonella Invasion via ARNO.
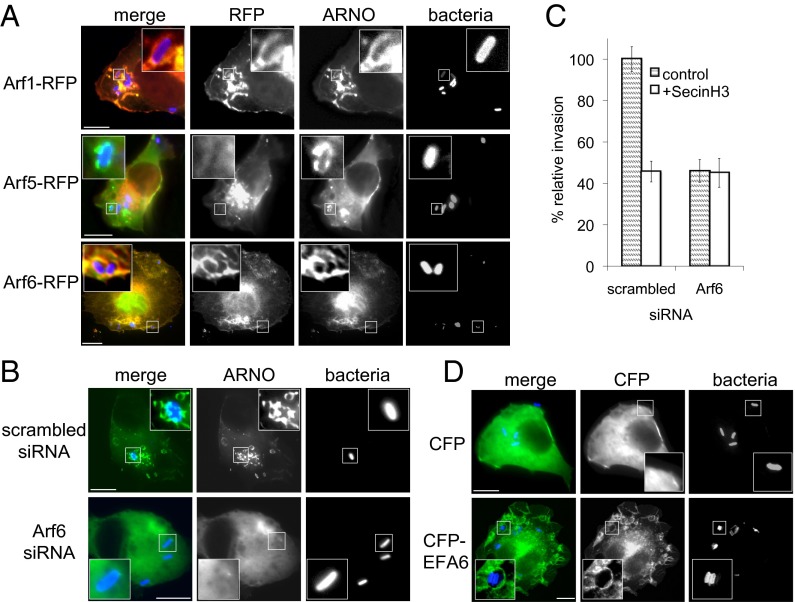
As ARNO is known to generate actin-rich macropinosomes that facilitate Salmonella invasion via Arf1 and the WRC (14), we sought to determine whether Arf6 influences this invasion pathway. To examine localization of Arf6 and ARNO during Salmonella infection, HeLa cells coexpressing CFP-tagged ARNO and either RFP-tagged Arf6, positive control Arf1, or negative control Arf5 were infected with Salmonella and examined by fluorescence microscopy (Fig. 3A). In contrast to Arf5 that has no role in Salmonella invasion (14), Arf1 and Arf6 localized at ARNO-enriched Salmonella-induced macropinosomes (Fig. 3A, Insets). As Arf6 recruited ARNO to membrane platforms, we examined ARNO localization in HeLa cells depleted of Arf6 (quantified in Fig. S1 B and C) by siRNA transfection 72 h before infection (Fig. 3B). In scrambled negative control siRNA-treated cells, ARNO was recruited to invading Salmonella that triggered generation of ARNO-containing macropinosomes, but ARNO recruitment and macropinosome formation were blocked by Arf6 depletion (exemplified by Fig. 3B, Insets). To control for off-target gene knockdown following Arf6 siRNA transfection, we examined ARNO macropinosome formation in Arf6-depleted cells coexpressing Arf6-RFP and CFP-ARNO (Fig. S1F). Generation of macropinosomes (Fig. S1F, arrows) was observed in Salmonella-infected cells expressing Arf6 and ARNO but not ARNO alone.
Fig. 3.
Arf6 facilitates Salmonella macropinocytosis via ARNO. (A) Localization of Arf1, Arf5, and Arf6 during Salmonella invasion. HeLa cells expressing RFP-tagged Arf1, Arf5, or Arf6 as indicated (Left) together with CFP-tagged ARNO were infected for 10 min with fluorescently labeled WT Salmonella (bacteria). (Insets) Magnified areas. (Scale bar: 8 µm.) (B) Influence of Arf6 depletion on ARNO localization during Salmonella invasion. HeLa cells were transfected with scrambled or Arf6 siRNA 72 h before expression of CFP-tagged ARNO and infection as in A. (Insets) Magnified areas. (Scale bar: 8 µm.) (C) Influence of Arf6 depletion on ARNO-dependent Salmonella invasion. HeLa cells were transfected with scrambled or Arf6 siRNA 72 h before treatment with SecinH3 and infection with Salmonella (10 min) encoding pM975 that expresses GFP inside pathogen-containing vacuoles. Error bars represent ± SEM. (D) Localization of control CFP and CFP-tagged EFA6 in HeLa cells infected for 10 min with fluorescently labeled Salmonella (bacteria). (Insets) Magnified areas. (Scale bar: 8 µm.)
We next sought to address whether Arf6 mediates pathogen invasion via ARNO. We first assessed Salmonella invasion in Arf6-depeleted HeLa cells (Fig. 3C), which was significantlyimpaired by ∼54% relative to scrambled siRNA-treated cells (P < 0.05; n = 3). This impairment was not a result of off-target gene knockdown, as multiple siRNAs targeting Arf6 also reduced invasion (Fig. S1D), and invasion was restored to ∼86% when Arf6-depleted cells expressed Arf6-RFP but not control RFP alone, which remained impaired at ∼51% (Fig. S1E). To demonstrate that Arf6 mediates pathogen invasion via ARNO, Salmonella invasion was determined in Arf6-depeleted HeLa cells treated with a specific inhibitor (SecinH3) of the cytohesin family (23) to which ARNO belongs (Fig. 3C). In scrambled siRNA-treated cells, SecinH3 treatment reduced invasion by ∼55%, which was mirrored in Arf6-depleted cells, revealing no additive effect to that seen with SecinH3 alone, and established that Arf6 acts via ARNO. These results show that Arf6 localizes to Salmonella, where it drives ARNO recruitment and cytoskeleton remodeling to facilitate pathogen macropinocytosis.
EFA6 and BRAG2 Initiate Actin Assembly via Arf1 and WAVE.
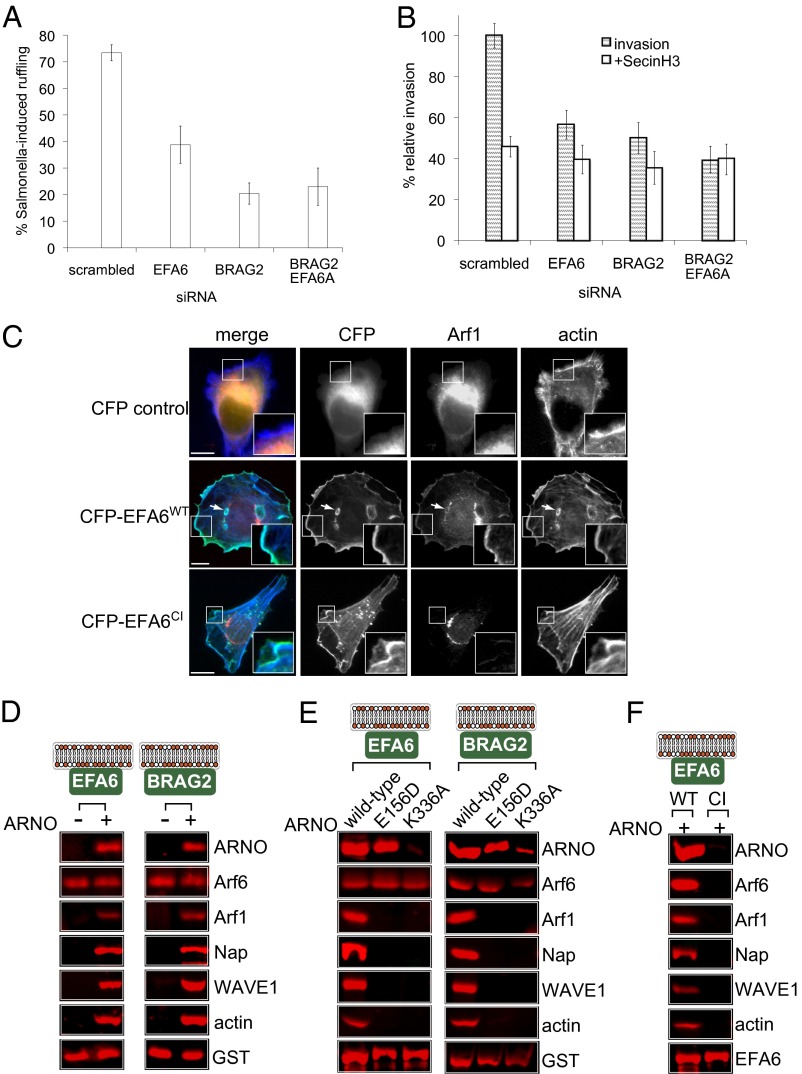
Arf6 function is intimately coupled to the activity of its canonical GEFs EFA6 (exchange factor for Arf6) and BRAG2 (brefeldin-resistant Arf GEF) that localize to the plasma membrane via their PH domains. We therefore examined the localization of CFP-tagged EFA6 (Fig. 3D) and BRAG2 (Fig. S2A) in HeLa cells infected with Salmonella. Each GEF was found enriched on macropinosomes, that often contained engulfed bacteria (Fig. 3D, Insets), whereas control CFP was diffusely localized and not recruited to invasion sites (Fig. 3D). The macropinosomes surrounding engulfed Salmonella were rich in polymerized actin as exemplified in EFA6-expressing cells (Fig. S2B, Insets). To examine the role of Arf6 GEFs in cytoskeleton rearrangement, EFA6 and BRAG2 were individually depleted (quantified in Fig. S1 B and C) by siRNA transfection 72 h before infection with Salmonella and analysis of pathogen-induced membrane ruffling (Fig. 4A). Salmonella triggered invasion ruffles in ∼73% of scrambled siRNA-treated cells, but this was significantly reduced to ∼37% in EFA6-depleted cells and to ∼20% in BRAG2-depleted cells (P < 0.05; n = 3). This impairment in membrane ruffling was mirrored in Salmonella invasion into host cells, which was reduced by ∼44% in EFA6-depleted cells and ∼50% in BRAG2-depleted cells (P < 0.05; n = 3; Fig. 4B). These observations were not a result of off-target gene knockdown, as multiple siRNAs targeting BRAG2 or EFA6 also inhibited membrane ruffling and invasion (Fig. S1G). Relative to individual Arf GEF knockdowns, no significant effect on Salmonella-induced ruffling and pathogen invasion was observed when BRAG2 and EFA6 were depleted simultaneously (Fig. 4 A and B), demonstrating that both GEFs are required. These results show that the recruited GEFs EFA6 and BRAG2 facilitate cytoskeleton remodelling by activating Arf6 to promote pathogen invasion into host cells.
Fig. 4.
Arf6 GEFs facilitate Salmonella invasion via Arf1-ARNO-WAVE. (A) Influence of Arf6 GEF depletion on Salmonella ruffle formation. HeLa cells were transfected with scrambled, EFA6, or BRAG2 siRNA, or with EFA6 or BRAG2 siRNA together, 72 h before infection with WT Salmonella (10 min). Error bars represent ± SEM. (B) Influence of Arf6 GEF depletion on ARNO-driven Salmonella invasion. HeLa cells were transfected with siRNA as in A 72 h before treatment with SecinH3 and infection with Salmonella (10 min) encoding pM975 that expresses GFP inside pathogen-containing vacuoles. Error bars represent ± SEM. (C) Localization of RFP-tagged Arf1 in HeLa cells coexpressing control CFP alone, CFP-tagged WT, or CI EFA6. HeLa cells were stained with Alexa Fluor 594–phalloidin to visualize actin. (Insets) Magnified areas. Arrows indicate intracellular vacuoles. (Scale bar: 8 µm.) (D) Actin assembly on the membrane driven by Arf6 GEFs in the presence of ARNO. Proteins recruited to EFA6 and BRAG2-associated membrane platforms (depicted in cartoons) from extract with (+) or without (−) purified ARNO analyzed by immunoblotting with indicated antibodies (Right). Anti-GST was used to detect GST-tagged EFA6 and BRAG2. (E) Actin assembly by Arf6 GEFs in the presence of CI ARNO (E156D) and the ARNO mutant defective for Arf6-binding (K336A). Proteins recruited to EFA6 and BRAG2-associated membrane platforms (depicted in cartoons) from extract containing indicated ARNO derivatives, analyzed by immunoblotting as in D. (F) Actin assembly via CI EFA6. Proteins recruited to WT and CI EFA6 from extract containing purified WT ARNO (+), analyzed by immunoblotting as in D.
To develop our hypothesis that Arf6 modulates the cytoskeleton via ARNO, we compared Salmonella invasion into EFA6- and BRAG2-depleted cells in the presence or absence of SecinH3 (Fig. 4B). In contrast to scrambled siRNA-treated cells, no additional reduction on invasion was observed when EFA6- and BRAG2-depleted cells were incubated with SecinH3, showing that Arf6 GEFs act via ARNO. To confirm that Arf6 GEFs act upstream of WRC by recruiting Arf1, we investigated Arf1-RFP localization in HeLa cells coexpressing CFP-tagged WT EFA6 or a CI variant carrying the GEF domain mutation E242K (24) (Fig. 4C). In control cells or cells expressing CI EFA6, no Arf1 recruitment to the plasma membrane was observed (Fig. 4C, Insets) and Arf1 was only found at the Golgi apparatus. In contrast, Arf1 was recruited by EFA6, which colocalized with Arf1 at the actin-rich cell cortex (Fig. 4C, Insets). Arf1 was also observed on EFA6-positive actin-rich intracellular vacuoles (Fig. 4C, arrows) that are generated following sustained Arf6 activation (25). Consistent with a role for EFA6 in recruiting Arf1 to the cell cortex via ARNO, both EFA6 and ARNO colocalized at actin assembly sites at the plasma membrane (Fig. S2C, Insets).
We sought to confirm that, like Arf6, EFA6 and BRAG2 trigger WRC-dependent actin assembly at the membrane through ARNO. Purified GEFs were each associated to membrane platforms and incubated in extract with or without addition of recombinant ARNO (Fig. 4D). Immunoblotting of the membrane-associated proteins showed that EFA6 and BRAG2 in extract alone triggered Arf6 recruitment, but, in the absence of ARNO, cytoskeleton regulators were not recruited. When extract was supplied with ARNO, EFA6 and BRAG2 recruited ARNO, leading to generation of a signaling hub enriched in the WRC and Arf1, which triggered actin assembly at the membrane.
To examine the WRC actin assembly pathway further, we examined signaling by EFA6 and BRAG2 membrane platforms in extract containing WT ARNO, the CI mutant (E156D), or the variant (K336A) defective for Arf6 binding (Fig. 4E). Arf6 was recruited by EFA6 and BRAG2 in all cases. CI (E156D) ARNO was recruited equivalently to WT, whereas recruitment of the Arf6-binding mutant was significantly impaired. These results showed that Arf6 binding is required for recruitment of ARNO by EFA6 and BRAG2. Immunoblotting demonstrated that Arf6 interaction with ARNO and ARNO GEF activity was required for WRC recruitment and actin assembly at the membrane by EFA6 and BRAG2. Consistent with this, and in contrast to WT EFA6 membrane platforms, those harboring the CI EFA6 were incapable of recruiting Arf6 and therefore signaling downstream via ARNO to drive WRC-dependent actin assembly (Fig. 4F).
Discussion
Understanding how multiple signals are integrated at the membrane to orchestrate cytoskeleton rearrangement remains a considerable challenge. We recently showed that Arf and Rac1 GTPases cooperate to directly recruit and activate the WRC at the membrane (13). This demonstrated that Arf GTPases directly modulate nucleation-promoting factors (NPFs, e.g., N-WASP, WRC), which opened up new questions with regard to cytoskeleton control. In contrast to other Arf GTPases, Arf6 is distinguished by its prominent plasma membrane localization (2), where it regulates endosomal recycling and the cortical actin cytoskeleton (1), but the precise means by which Arf6 actually triggers actin assembly and through which NPF was unknown. In this study, we reconstituted Arf6-mediated actin assembly at the membrane and demonstrated that it was driven via WRC. In contrast to other Arf family members, actin polymerization was not mediated by direct interaction with WRC but instead was achieved indirectly through the recruitment of ARNO and subsequent Arf1 activation. This Arf6 pathway was recapitulated in cells in which WRC-dependent membrane ruffling and macropinocytosis of Salmonella was driven through an Arf6 signaling network involving its GEFs EFA6 and BRAG2.
Arf6 has long been implicated in cytoskeleton remodeling, but how this is achieved has been difficult to address as a result of its diverse activities. Arf6 membrane trafficking has been shown to promote plasma membrane localization of Cdc42 and Rac1 (26, 27), and Arf6 stimulates PI4P5K to generate PI(4,5)P2 (28) that works in synergy with Cdc42 for the activation of N-WASP (29). Indeed, we have previously demonstrated that PI(4,5)P2 membrane platforms trigger the formation of actin comet tails in extract via N-WASP (13). By eliminating the contribution of PI(4,5)P2 in our reconstitution experiments, we were able to solely focus on the direct contribution of Arf6 at the membrane showing that Arf6-dependent actin assembly was driven through active Arf and Rac1. These results are consistent with the reported role for Arf6 in Rac1 activation and lamellipodia formation (8, 9, 27).
As Arf6 was unable to directly modulate WRC, the GTPase used the activity of ARNO to mediate actin assembly in vitro and in cells. ARNO recruitment to the plasma membrane and its activation require Arf6 that directly binds the ARNO PH domain, which triggers Arf1 activation (18, 19). Our findings support this mechanism by showing that Arf6 and its GEFs recruit ARNO to the membrane in extract and in cells, stimulating an Arf1-driven pathway, which was further supported by experiments revealing EFA6-mediated localization of Arf1 at the cell cortex.
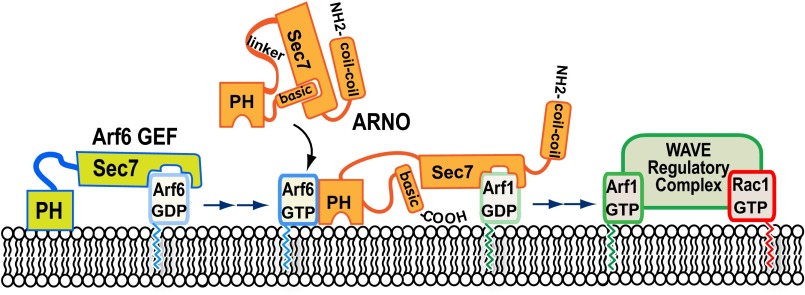
Despite the mechanistics of Arf1 recruitment to the plasma membrane being clear, why a normally Golgi-associated protein is translocated was uncertain (18, 19). The direct activation of WRC by Arf1 explains its recruitment by ARNO, which is supported by the role of Arf1 in lamellipodia formation, and Salmonella-triggered WRC-dependent membrane ruffling and pathogen macropinocytosis (13, 14, 30). This study resolves a new aspect of WRC control by defining how the Arf6 network fits into this pathway (Fig. 5), and highlights the elaborate spatiotemporal small GTPase control mechanisms that underlie actin polymerization at the membrane.
Fig. 5.
Model for Arf6 regulation of the WAVE regulatory complex. Arf6 GEFs such as EFA6 or BRAG activate Arf6. ARNO resides in a soluble autoinhibited conformation, and is recruited to the plasma membrane and activated by Arf6 binding of the ARNO PH domain. ARNO activates Arf1 that collaborates with Rac1 to drive WRC-dependent actin polymerization.
Methods
Plasmids and Recombinant Proteins.
The following plasmids were generated by Invitrogen Gateway methodology: pET20b-Arf6 encoding the Arf family N-myristoyltransferase site as previously described for Arf1 (13); pET15b-ARNO-2G, pcDNA3-mRFP-Arf1, pcDNA3-mRFP-Arf5, pcDNA3-mRFP-Arf6, peCFP-C1-EFA6, peCFP-C1-EFA6E242K, peCFP-C1-BRAG2524-933, pGEX2T-GGA3-GAT1-313, peYFP-N1-ARNO, and peCFP-C1-ARNO. The following were provided to us: pGEX3X-EFA6 splice variant 2 from Michel Franco (l'Institut de Pharmacologie Moléculaire et Cellulaire, Université Nice Sophia-Antipolis, Valbonne, France) and pM975 from Wolf-Dietrich Hardt (Eidgenössiche Technische Hochschule, Zurich), and pET-arf1 and pBB131 encoding the Arf family N-myristoyltransferase from Martin Spiess (Biozentrum, Basel). GST- and His-tagged proteins were expressed in Escherichia coli Rosetta (Novagen) at 16 °C before affinity purification (14).
Antibodies.
Antibodies were purchased from Abcam (BRAG2, ab122442; Rac1, ab33186; Arf1, ab58578; Arf6, ab81650; ARNO, ab56510; and tubulin, ab7291), Sigma (Abi1, A5106; actin, A2066; Cyfip, P0092; EFA6, SAB1-400227; and Nap1, N3788) or were raised against recombinant peptides in rabbits by Diagnostics Scotland (WAVE1; amino acids 180–241).
Bacterial Strains and Infection of HeLa Cells.
WT Salmonella enterica serovar Typhimurium SL1344 (gift from Jean Guard-Petter, US Department of Agriculture, Athens, GA) were used in all experiments. For fluorescence microscopy and quantification of membrane ruffling, bacteria were washed with PBS solution, conjugated to Alexa Fluor-350 carboxylic acid succinimidyl ester (15 min, 37 °C), washed in Tris, pH 7.4, buffered saline solution, then used to infect HeLa cells (multiplicity of infection of 50). Quantification of membrane ruffling was performed as described previously (14) whereby fixed infected cells were stained with Alexa Fluor 594–phalloidin to visualize actin, the number of fluorescently labeled bacteria in each field of view was counted, and the proportion of those bacteria associated with actin ruffles was determined. To quantify invasion, Salmonella encoding pM975 that expresses GFP via the SPI2 promoter when bacteria are within Salmonella-containing vacuoles (31) were used to infect HeLa cells (10 min) and the number of fluorescent bacteria were counted per cell (∼400 cells per experiment) microscopically. When appropriate, HeLa cells were incubated with 25 µM SecinH3 (Merck). Immunofluorescence microscopy was performed and images assembled as described previously (14). All experiments were performed in triplicate at least three times. Geometric means were calculated, and significance was determined by Student t test or one-way ANOVA followed by a post hoc Dunnett comparison. P <0.05 was considered significant.
Actin-Based Motility by Phospholipid Beads.
Preparation of porcine brain extract, actin-based motility by phospholipid-coated beads, and isolation of bead membrane-associated proteins have been described in detail (13, 14). When indicated, extract or buffer was preincubated with recombinant GAT165–314, NSC23766, and waspΔvca as described (13, 14).
Mammalian Cell Culture and Transfections.
Mammalian HeLa cells (CCL-2; American Type Culture Collection) were routinely cultured in complete growth media consisting of DMEM supplemented with 10% (vol/vol) FBS, 2 mM l-glutamine, 200 μg⋅mL−1 streptomycin, and 100 U⋅mL−1 penicillin (37 °C, 5% CO2). Transient transfection of HeLa cells by microporation was performed by using the Neon Transfection System according to the manufacturer’s instructions (Invitrogen). For RNAi, siRNA from Qiagen against EFA6 (gene accession number NM_002779, Hs_PSD_5 sequence ATCACTCGCATCAATGTAGTA), Hs_PSD_6 sequence ATCGCTAGAGCCAAATGGGAA, BRAG2 (gene accession number NM_001134382, Hs_IQSEC1_4 sequence CCGGAAGAAATTCACCGATGA, Hs_IQSEC1_5 sequence CTGAAGGGTAGCAGTAATGAA), ARF6 (gene accession number NM_001663, Hs_ARF6_5 sequence CAACGTGGAGACGGTGACTTA), Hs_ARF6_7 sequence AAGACCAGTATAGTAAACTTA, or AllStars negative control siRNA (Qiagen) were combined for each individual gene (unless otherwise stated) and transfected into HeLa cells with Oligofectamine transfection reagent (Invitrogen) according to the manufacturer’s instructions. Transfection mixture was replaced after 24 h with complete growth medium, and cells were cultured 72 h in total. RNAi efficiency was determined by EXPRESS One-Step SYBR GreenER quantitative real-time–PCR according to the manufacturer’s instructions (Invitrogen), with actin as a relative control in each case.
Supplementary Material
Acknowledgments
This work was supported by the Wellcome Trust, the Biotechnology and Biological Sciences Research Council, and the Cambridge Isaac Newton Trust.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1311680110/-/DCSupplemental.
References
- 1.Donaldson JG, Jackson CL. ARF family G proteins and their regulators: Roles in membrane transport, development and disease. Nat Rev Mol Cell Biol. 2011;12(6):362–375. doi: 10.1038/nrm3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peters PJ, et al. Overexpression of wild-type and mutant ARF1 and ARF6: Distinct perturbations of nonoverlapping membrane compartments. J Cell Biol. 1995;128(6):1003–1017. doi: 10.1083/jcb.128.6.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Radhakrishna H, Donaldson JG. ADP-ribosylation factor 6 regulates a novel plasma membrane recycling pathway. J Cell Biol. 1997;139(1):49–61. doi: 10.1083/jcb.139.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunphy JL, et al. The Arf6 GEF GEP100/BRAG2 regulates cell adhesion by controlling endocytosis of beta1 integrins. Curr Biol. 2006;16(3):315–320. doi: 10.1016/j.cub.2005.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Myers KR, Casanova JE. Regulation of actin cytoskeleton dynamics by Arf-family GTPases. Trends Cell Biol. 2008;18(4):184–192. doi: 10.1016/j.tcb.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cotton M, et al. Endogenous ARF6 interacts with Rac1 upon angiotensin II stimulation to regulate membrane ruffling and cell migration. Mol Biol Cell. 2007;18(2):501–511. doi: 10.1091/mbc.E06-06-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu B, et al. ADP-ribosylation factor 6 regulates glioma cell invasion through the IQ-domain GTPase-activating protein 1-Rac1-mediated pathway. Cancer Res. 2009;69(3):794–801. doi: 10.1158/0008-5472.CAN-08-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koo TH, Eipper BA, Donaldson JG. Arf6 recruits the Rac GEF Kalirin to the plasma membrane facilitating Rac activation. BMC Cell Biol. 2007;8:29. doi: 10.1186/1471-2121-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palamidessi A, et al. Endocytic trafficking of Rac is required for the spatial restriction of signaling in cell migration. Cell. 2008;134(1):135–147. doi: 10.1016/j.cell.2008.05.034. [DOI] [PubMed] [Google Scholar]
- 10.Bisi S, et al. Membrane and actin dynamics interplay at lamellipodia leading edge. Curr Opin Cell Biol. 2013;25(5):565–573. doi: 10.1016/j.ceb.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Gautreau A, et al. Purification and architecture of the ubiquitous Wave complex. Proc Natl Acad Sci USA. 2004;101(13):4379–4383. doi: 10.1073/pnas.0400628101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miki H, Suetsugu S, Takenawa T. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J. 1998;17(23):6932–6941. doi: 10.1093/emboj/17.23.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koronakis V, et al. WAVE regulatory complex activation by cooperating GTPases Arf and Rac1. Proc Natl Acad Sci USA. 2011;108(35):14449–14454. doi: 10.1073/pnas.1107666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Humphreys D, Davidson A, Hume PJ, Koronakis V. Salmonella virulence effector SopE and Host GEF ARNO cooperate to recruit and activate WAVE to trigger bacterial invasion. Cell Host Microbe. 2012;11(2):129–139. doi: 10.1016/j.chom.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Humphreys D, Liu T, Davidson AC, Hume PJ, Koronakis V. The Drosophila Arf1 homologue Arf79F is essential for lamellipodium formation. J Cell Sci. 2012;125(pt 23):5630–5635. doi: 10.1242/jcs.108092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franco M, Chardin P, Chabre M, Paris S. Myristoylation is not required for GTP-dependent binding of ADP-ribosylation factor ARF1 to phospholipids. J Biol Chem. 1993;268(33):24531–24534. [PubMed] [Google Scholar]
- 17.Dell’Angelica EC, et al. GGAs: A family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J Cell Biol. 2000;149(1):81–94. doi: 10.1083/jcb.149.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen LA, et al. Active Arf6 recruits ARNO/cytohesin GEFs to the PM by binding their PH domains. Mol Biol Cell. 2007;18(6):2244–2253. doi: 10.1091/mbc.E06-11-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stalder D, et al. Kinetic studies of the Arf activator Arno on model membranes in the presence of Arf effectors suggest control by a positive feedback loop. J Biol Chem. 2011;286(5):3873–3883. doi: 10.1074/jbc.M110.145532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macia E, Chabre M, Franco M. Specificities for the small G proteins ARF1 and ARF6 of the guanine nucleotide exchange factors ARNO and EFA6. J Biol Chem. 2001;276(27):24925–24930. doi: 10.1074/jbc.M103284200. [DOI] [PubMed] [Google Scholar]
- 21.Frank S, Upender S, Hansen SH, Casanova JE. ARNO is a guanine nucleotide exchange factor for ADP-ribosylation factor 6. J Biol Chem. 1998;273(1):23–27. doi: 10.1074/jbc.273.1.23. [DOI] [PubMed] [Google Scholar]
- 22.Béraud-Dufour S, et al. A glutamic finger in the guanine nucleotide exchange factor ARNO displaces Mg2+ and the beta-phosphate to destabilize GDP on ARF1. EMBO J. 1998;17(13):3651–3659. doi: 10.1093/emboj/17.13.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hafner M, et al. Inhibition of cytohesins by SecinH3 leads to hepatic insulin resistance. Nature. 2006;444(7121):941–944. doi: 10.1038/nature05415. [DOI] [PubMed] [Google Scholar]
- 24.Franco M, et al. EFA6, a sec7 domain-containing exchange factor for ARF6, coordinates membrane recycling and actin cytoskeleton organization. EMBO J. 1999;18(6):1480–1491. doi: 10.1093/emboj/18.6.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown FD, Rozelle AL, Yin HL, Balla T, Donaldson JG. Phosphatidylinositol 4,5-bisphosphate and Arf6-regulated membrane traffic. J Cell Biol. 2001;154(5):1007–1017. doi: 10.1083/jcb.200103107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balasubramanian N, Scott DW, Castle JD, Casanova JE, Schwartz MA. Arf6 and microtubules in adhesion-dependent trafficking of lipid rafts. Nat Cell Biol. 2007;9(12):1381–1391. doi: 10.1038/ncb1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Osmani N, Peglion F, Chavrier P, Etienne-Manneville S. Cdc42 localization and cell polarity depend on membrane traffic. J Cell Biol. 2010;191(7):1261–1269. doi: 10.1083/jcb.201003091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Honda A, et al. Phosphatidylinositol 4-phosphate 5-kinase alpha is a downstream effector of the small G protein ARF6 in membrane ruffle formation. Cell. 1999;99(5):521–532. doi: 10.1016/s0092-8674(00)81540-8. [DOI] [PubMed] [Google Scholar]
- 29.Rohatgi R, Ho HY, Kirschner MW. Mechanism of N-WASP activation by CDC42 and phosphatidylinositol 4, 5-bisphosphate. J Cell Biol. 2000;150(6):1299–1310. doi: 10.1083/jcb.150.6.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hänisch J, et al. Molecular dissection of Salmonella-induced membrane ruffling versus invasion. Cell Microbiol. 2010;12(1):84–98. doi: 10.1111/j.1462-5822.2009.01380.x. [DOI] [PubMed] [Google Scholar]
- 31.Schlumberger MC, et al. Two newly identified SipA domains (F1, F2) steer effector protein localization and contribute to Salmonella host cell manipulation. Mol Microbiol. 2007;65(3):741–760. doi: 10.1111/j.1365-2958.2007.05823.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.