Significance
Recognition and repair of DNA lesions are critical for maintaining genomic stability and reducing the generation of mutations that lead to cancer development. A mechanism of chromatin alteration is described here that is essential for repair of DNA double-strand breaks. The histone chaperone, nucleolin, is recruited to sites of DNA breaks via binding to RAD50 of the MRE11-NBS1-RAD50 complex, where it removes histone proteins H2A and H2B from the nucleosome at the break site. This nucleosome disruption allows DNA repair proteins to access the DNA lesion and facilitate repair of the DNA break. Clinical implications of these findings include the identification of new targets, such as nucleolin, that could be modulated to enhance sensitivity of tumor cells to radiation therapy.
Keywords: chromatin remodeling, DNA damage, I-PpoI, nucleosome disassembly
Abstract
Recruitment of DNA repair factors and modulation of chromatin structure at sites of DNA double-strand breaks (DSBs) is a complex and highly orchestrated process. We developed a system that can induce DSBs rapidly at defined endogenous sites in mammalian genomes and enables direct assessment of repair and monitoring of protein recruitment, egress, and modification at DSBs. The tight regulation of the system also permits assessments of relative kinetics and dependencies of events associated with cellular responses to DNA breakage. Distinct advantages of this system over focus formation/disappearance assays for assessing DSB repair are demonstrated. Using ChIP, we found that nucleosomes are partially disassembled around DSBs during nonhomologous end-joining repair in G1-arrested mammalian cells, characterized by a transient loss of the H2A/H2B histone dimer. Nucleolin, a protein with histone chaperone activity, interacts with RAD50 via its arginine-glycine rich domain and is recruited to DSBs rapidly in an MRE11-NBS1-RAD50 complex-dependent manner. Down-regulation of nucleolin abrogates the nucleosome disruption, the recruitment of repair factors, and the repair of the DSB, demonstrating the functional importance of nucleosome disruption in DSB repair and identifying a chromatin-remodeling protein required for the process. Interestingly, the nucleosome disruption that occurs during DSB repair in cycling cells differs in that both H2A/H2B and H3/H4 histone dimers are removed. This complete nucleosome disruption is also dependent on nucleolin and is required for recruitment of replication protein A to DSBs, a marker of DSB processing that is a requisite for homologous recombination repair.
Human genomes are constantly challenged by DNA-damaging agents from both endogenous and exogenous sources. The cellular responses to DNA damage are critically important in maintaining genomic integrity and preventing tumorigenesis. Initial steps in the cellular response to DSBs involve an early recruitment of the MRE11-NBS1-RAD50 (MRN) complex to the DSB site and activation and recruitment of the ataxia telangiectasia mutated (ATM) kinase resulting in the activation of multiple signaling pathways (1, 2). Because DNA is packaged tightly in eukaryotic cells in highly complex chromatin structures, the ability of repair proteins to access sites of DNA damage and facilitate repair of the damage in the context of these complex structures requires chromatin-remodeling activities (3, 4). The simplest level of DNA packaging in chromatin is at the level of DNA wrapped around nucleosomes, octamers of histones H2A, H2B, H3, and H4 (3). Although disruption of nucleosomes has been reported during the process of DNA-excision repair (5) and DNA DSB repair (6), little is known about the molecular nature of this disruption, the factors that mediate it, or its functional role in the DNA repair process.
Alterations of chromatin structure in general and nucleosome structures in particular are also required during the process of gene transcription. A variety of chromatin remodelers, including switch/sucrose nonfermentable (SWI/SNF) and the facilitates chromatin transportation (FACT) complex, have been shown to promote transcription by removing H2A/H2B histone dimers, resulting in a partial nucleosome disruption (7–9). Increasing evidence suggests that chromatin remodelers such as FACT and SWI/SNF may also play a role in DNA repair (10, 11). However, the characteristics of nucleosome remodeling induced by DNA damage and its role in DNA repair remain to be elucidated. Interestingly, nucleolin, a nucleolus-specific protein, has been recently shown to possess a FACT-like histone chaperone activity as it was required for the removal of H2A/H2B histone dimers and facilitated transcription through the nucleosome in vitro (12). Nucleolin consists of an N-terminal domain containing highly acidic regions, four RNA-binding domains, and a C-terminal RGG domain that is rich in arginine, glycine, and phenylalanine and was shown to interact with ribosomal proteins (13, 14). As with the FACT complex, nucleolin did not require the presence of any additional chromatin-remodeling factors for this activity (12). However, despite the emerging role in transcription, a function of nucleolin in chromatin remodeling after DNA damage and its role in DNA repair remain unknown.
To study factors affecting repair of DNA breaks in mammalian cells as well as the protein dynamics that occur at and around the breaks, we previously developed a DNA breakage system using the homing endonuclease I-PpoI, which generates DSBs at defined endogenous sites in the human genome, allowing us to assess DNA repair and monitor protein recruitment to DSBs directly by ChIP (6). However, the system had some technical limitations, such as a relatively slow induction of DSBs and a suboptimal ability to tightly control DNA cutting by the endonuclease. Here, we report an improved I-PpoI system with the addition of a “destabilization domain” (dd) to the protein sequence, resulting in a tight regulation of I-PpoI endonuclease expression, a rapid induction of DSBs, and an ability to assess DSB repair with kinetics similar to those reported after ionizing irradiation. We also adapted the system to both retroviral and lentiviral expression vectors suitable for transduction of a variety of human and mouse cells. Using this improved system, we demonstrate that nucleosome disruption occurs transiently at the DSB, characterized by the loss of only the H2A/H2B dimer in G1-arrested cells and loss of all four core histones in cycling cells. Nucleolin facilitates nucleosome disruption in chromatin surrounding a DSB, and nucleolin-dependent nucleosome disruption is a necessary step for recruitment of DSB repair factors and for efficient DNA repair. Furthermore, recruitment of nucleolin to the DSB is dependent on the MRN complex and results from an interaction of the RGG domain of nucleolin with the RAD50 protein.
Results
Improved System for Controlled Induction of DNA Breaks and Measuring Repair and Protein Dynamics at DSBs.
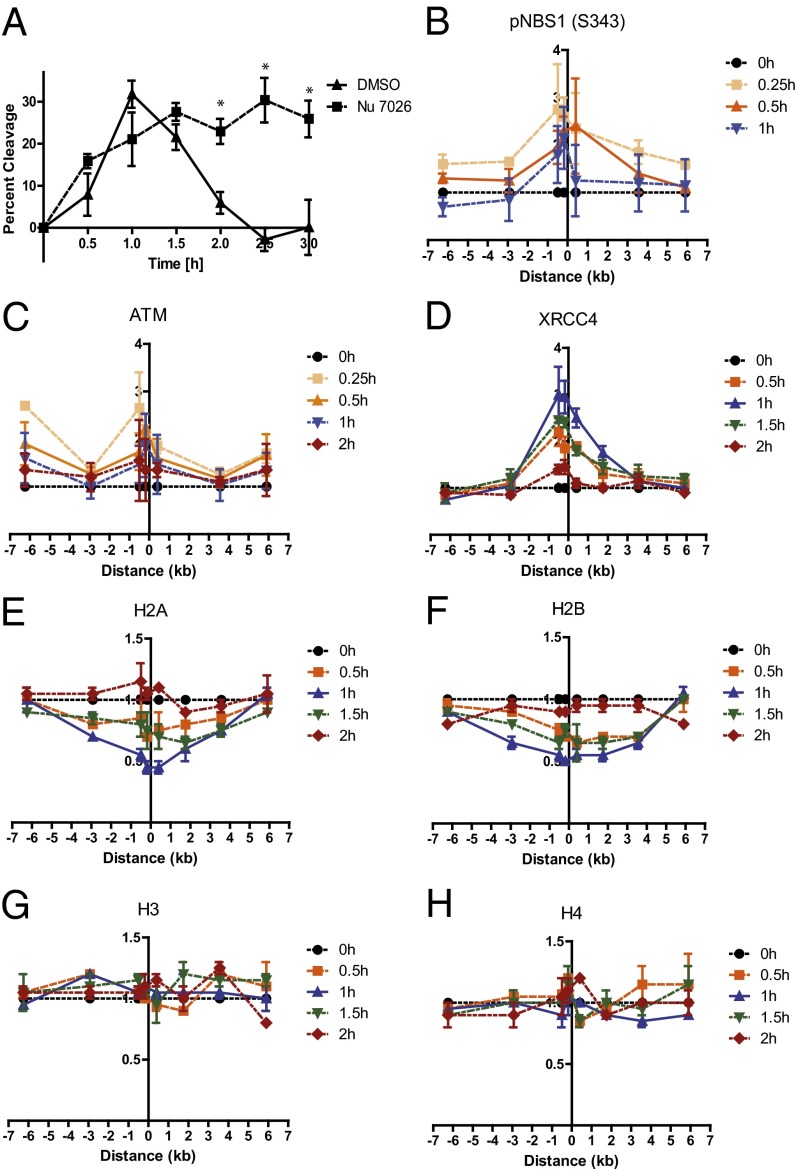
We previously reported a system using the homing endonuclease, I-PpoI, to induce DSBs in cells at defined genomic loci and used this system to monitor the recruitment of DNA repair proteins to the sites of DNA breaks (6). To provide tighter regulation of the endonuclease activity, we added a destabilization domain (dd) (15) to the construct, allowing tighter temporal control of I-PpoI expression (see Fig. S1 for details). This tightly regulated system permitted the generation of stably transfected cell lines amenable to subsequent genetic manipulation as needed for mechanistic studies. To avoid complexities of simultaneously evaluating both homologous recombination (HR) and nonhomologous end-joining (NHEJ) repair processes, cells were arrested in the G1 phase of the cell cycle, in which NHEJ should be the predominant DSB-repair mechanism. DSBs were induced rapidly in MCF7 cells stably expressing ddI-PpoI, reaching maximal levels 1 h after the addition of 4-hydroxytamoxifen (4-OHT), and repair was largely completed 1 h later (Fig. 1A). Notably, these repair kinetics are similar to those reported for DSBs induced by ionizing radiation, in which repair is largely completed within 1 h after irradiation (16). Addition of NU7026, a small-molecule inhibitor of the critical NHEJ factor DNA-PK, inhibited DSB repair (Fig. 1A), confirming that the system is quantitating NHEJ repair. Importantly, ddI-PpoI induction led to detectable activation of the DNA-damage response (DDR) pathway, assessed by ATM activation and phosphorylation of downstream substrates (Fig. S2A). Because I-PpoI target sites exist in all eukaryotic genomes, this system can be used in many different model systems. We adapted the system to immortalized mouse embryonic fibroblasts (MEFs) (Fig. S2 B and C) and developed a lentiviral dd-IPpoI expression cassette (Fig. S2 D and E). Recruitment, egress, and modification of proteins at and around DSBs can be assessed with this system by use of ChIP as demonstrated for phospho-NBS1 (Ser343) and ATM (Fig. 1 B and C). High-resolution mapping of protein dynamics, which can be assessed with a selective choice of PCR primers amplifying selected sequences in the vicinity of the DSB, demonstrated the recruitment of NHEJ factor XRCC4 directly to the site of the DSB but not to the surrounding regions (Fig. 1D). XRCC4-binding kinetics mimicked those of DNA breakage and repair, peaking 1 h after DSB induction and disappearing 1 h later.
Fig. 1.
Monitoring repair, DDR protein dynamics at a DSB, and a partial nucleosome disruption in G1 using a tightly regulated ddI-PpoI system. (A) DNA repair measured by quantitative real-time PCR spanning the unique I-PpoI cleave site at chromosome 1 in MCF7 cells stably expressing ddI-PpoI. The DNA-PK inhibitor Nu7026 was added 1 h before 4-OHT and was present during treatment and recovery. Time indicated is hours after the addition of 4-OHT. Data are shown as mean ± SEM of three independent experiments. *P < 0.05. (B–D) ChIP assay showing the recruitment of (B) pNBS1 (S343), (C) ATM, and (D) XRCC4 to the I-PpoI–induced DSB at chromosome 1 in MCF7 cells stably expressing ddI-PpoI. Time indicated is hours after the addition of 4-OHT. The I-PpoI cleavage site on chromosome 1 is located at distance 0. Data are shown as mean ± SEM of two independent experiments. The y-axis displays the fold change in relative occupancy normalized to the control. (E–H) Partial nucleosome disruption occurs in G1-arrested cells. ChIP of (E) histone H2A, (F) histone H2B, (G) histone H3, and (H) histone H4 in MCF7 cells stably expressing ddI-PpoI as described in B. Cells were cultivated in medium containing 0.1% FBS for 24 h before DSB induction.
Characterization of Nucleosome Disruption at DSBs.
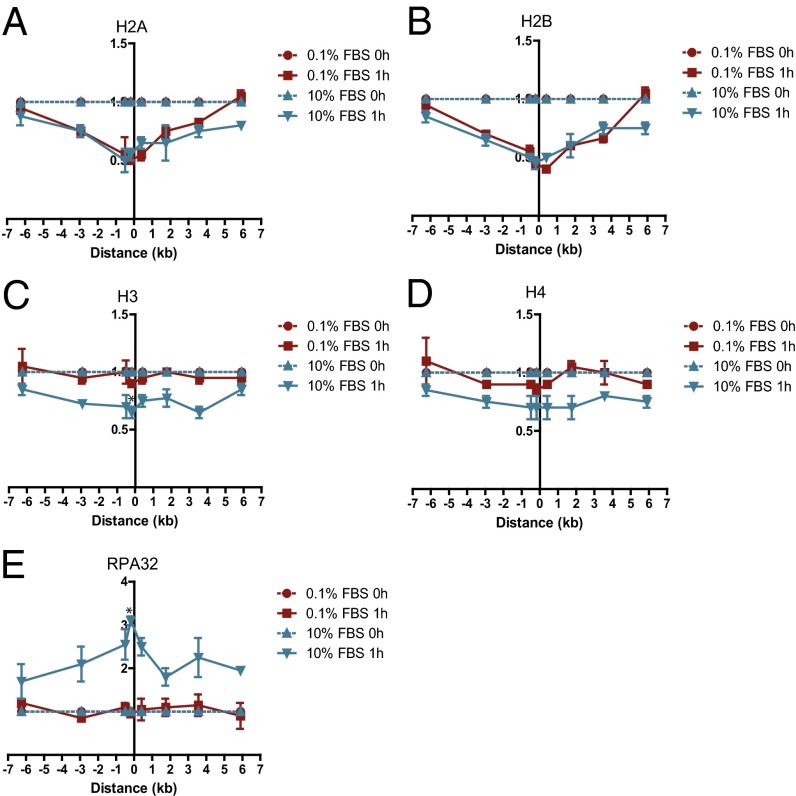
We previously reported that nucleosome disruption occurs at DSBs, assessed by loss of H2B protein at the DSB (6), but the kinetics and extent of nucleosome disruption and the fate of the other core histones has not been elucidated in mammalian cells. Eviction of the H2A/H2B dimer was detectable within 30 min after DSB induction, reaching a maximum at 1 h and returning to baseline 2 h after induction (Fig. 1 E and F), correlating with the kinetics of DSB repair and recruitment of XRCC4 to the break site (Fig. 1 A and D). Nucleosome disruption occurred over a region of ∼3 kb or less on each side of the DSB. Interestingly, the other core histones, H3 and H4, did not show any change in occupancy after DSB induction (Fig. 1 G and H). Thus, only a partial disruption of nucleosomes is associated with DSB induction and NHEJ repair in G1-arrested cells. Because a report using an exogenously introduced HO endonuclease site had suggested that histone H3 also was displaced near DSBs in asynchronous murine cells (17), we postulated that complete, rather than partial, nucleosome disruption might occur as part of the HR repair process that occurs in the S and G2/M phases of the cell cycle. In contrast to the nucleosome disruption in G1-arrested cells involving only the H2A/H2B histone dimer, the levels of all four histones decreased measurably surrounding a DSB in cycling cells (which still include a significant percentage of G1 cells) (Fig. 2 A–D). Further, the complete nucleosome disruption in cycling cells was associated with DNA end resection at the DSB as indicated by recruitment of RPA32, a subunit of the replication protein A (RPA), an event that was absent in G1-arrested cells (Fig. 2E).
Fig. 2.
Complete nucleosome disruption occurs at the DSB in asynchronous cells. ChIP of (A) histone H2A, (B) histone H2B, (C) histone H3, (D) histone H4, and (E) RPA32 in MCF7 cells stably expressing ddI-PpoI as described in Fig. 1B cultivated in serum containing 0.1% or 10% FBS for 24 h before DSB induction. *P < 0.05.
Role of Nucleolin in Nucleosome Disruption.
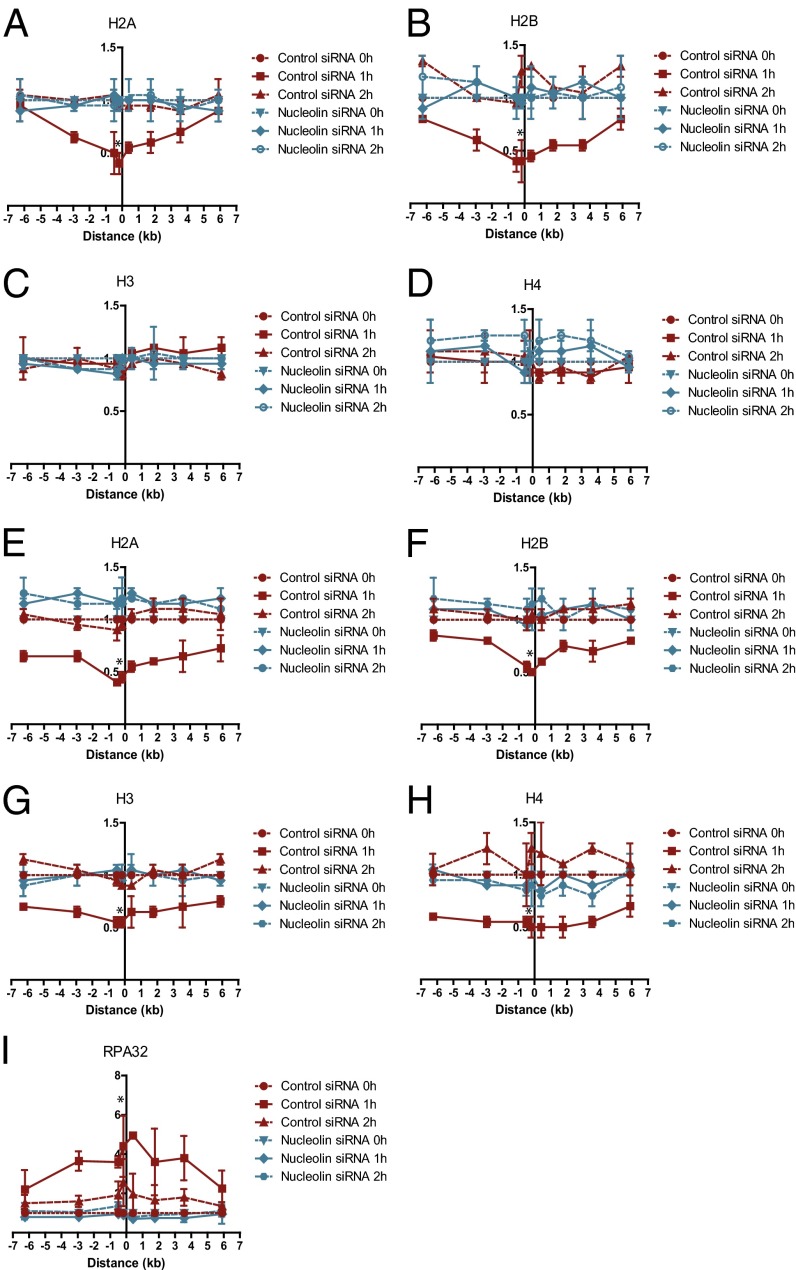
The temporary and partial disruption of the nucleosome at the DSB in G1-arrested cells was reminiscent of the nucleosome disassembly that has been reported during the process of gene transcription (7). Because nucleolin has been shown to function as a histone chaperone with a FACT-like activity in vitro and to facilitate histone eviction from the nucleosome during transcription (12), we investigated a possible role for nucleolin in nucleosome disruption following the induction of DSBs. Knockdown of nucleolin completely abrogated the partial nucleosome disruption surrounding a DSB (Fig. 3 A–D and Fig. S3A) and inhibited DSB religation (Fig. 4A), indicating that nucleosome disruption plays a crucial role in DSB repair. Nucleolin knockdown did not affect the amount of DSBs induced by ddI-PpoI (Fig. S3B) or ATM-dependent DDR signaling (Fig. S3D). Unexpectedly, nucleolin depletion also abrogated the displacement of the four core histones and RPA32 recruitment to the DSB in cycling cells (Fig. 3 E–I) without significantly affecting the cell-cycle distribution (Fig. S3C).
Fig. 3.
Nucleolin is required for nucleosome disruption surrounding a DSB. (A–D) Nucleolin knockdown abrogates partial nucleosome disruption in G1. ChIP of (A) histone H2A, (B) histone H2B, (C) histone H3, and (D) histone H4 in MCF7 cells stably expressing ddI-PpoI as described in Fig. 1B cultivated in medium containing 0.1% FBS for 24 h before DSB induction. (E–I) Nucleolin knockdown abrogates complete nucleosome disruption in cycling cells. ChIP of (E) histone H2A, (F) histone H2B, (G) histone H3, (H) histone H4, and (I) RPA32 in MCF7 cells stably expressing ddI-PpoI as described in Fig. 1B that were cultivated in medium containing 10% FBS for 24 h before DSB induction. Cells were transfected with nontargeting control siRNA or nucleolin-targeting siRNA. *P < 0.05.
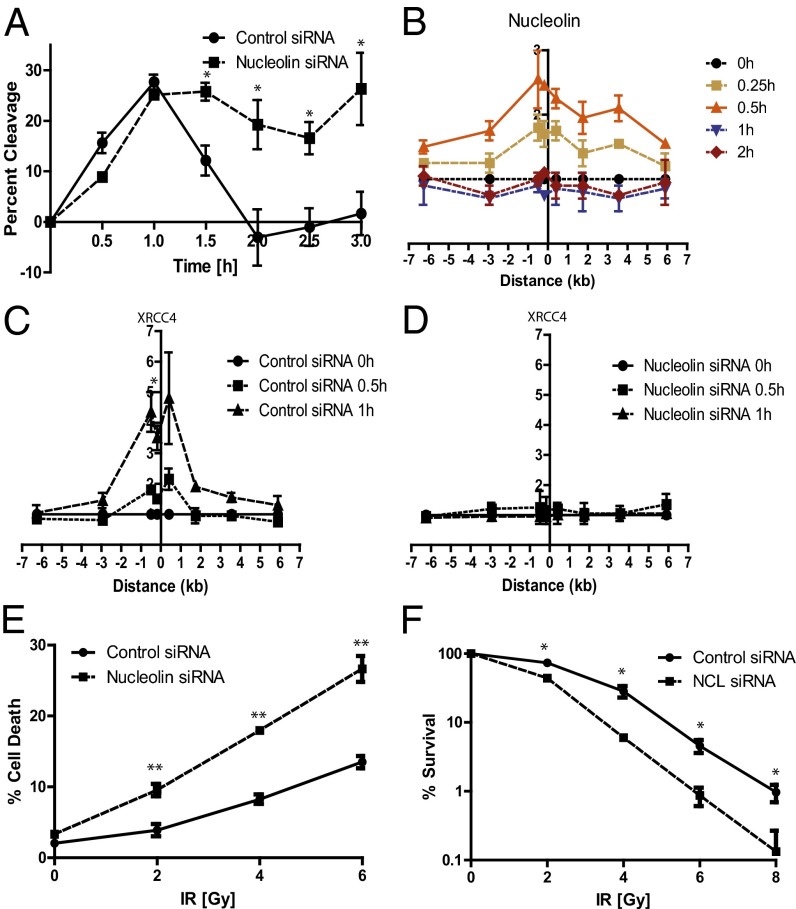
Fig. 4.
Partial nucleosome disruption is required for recruitment of XRCC4 to the DSB and efficient DNA repair. (A) Nucleolin knockdown inhibits DSB repair. Repair assay in MCF7 cells stably expressing ddI-PpoI as described in Fig. 1A. Cells were transfected with nontargeting control siRNA or nucleolin-targeting siRNA. (B) Nucleolin is recruited rapidly to the DSB. ChIP of nucleolin in MCF7 cells stably expressing ddI-PpoI as described in Fig. 1B that were cultivated in medium containing 0.1% FBS for 24 h before DSB induction. (C and D) Nucleolin knockdown prevents XRCC4 recruitment to the DSB. ChIP of XRCC4 in MCF7 cells stably expressing ddI-PpoI as described in Fig. 1B cultivated in medium containing 0.1% FBS for 24 h before DSB induction. Cells were transfected with nontargeting control siRNA (C) or nucleolin-targeting siRNA (D). (E and F) Nucleolin knockdown increases radio-sensitivity. (E) Quantification of cell death in p53-null HCT116 cells 96 h after ionizing radiation. Cell death was determined by quantifying the SubG1 cellular fraction. (F) Cell survival as determined by clonogenic assay in p53-null HCT116 cells after indicated doses of ionizing radiation. Data are shown as mean ± SEM of three independent experiments. *P < 0.05; **P < 0.01.
Because the FACT complex also can remove the H2A/H2B dimer from the nucleosome (7), we sought to verify whether it is involved in nucleosome disruption at the DSB, perhaps interacting functionally with nucleolin. However, knocking down structure-specific recognition protein 1 (SSRP1), a subunit of FACT that affects its chromatin-remodeling activities (7, 18), marginally reduced nucleosome disassembly (Fig. S4), suggesting that nucleolin can remove the H2A/H2B dimer from the nucleosome directly rather than mediating nucleosome disruption as a part of the FACT complex. Further, we hypothesized that because nucleolin binds H2A/H2B, but not the H3/H4 dimer, in vitro, it is unlikely that it can remove all four core histones from the nucleosome in proliferating cells. Rather, the H3/H4 dimer may be removed by a different histone chaperone with specificity for H3/H4 dimer in a step that is dependent on the initial H2A/H2B eviction by nucleolin. Indeed, we found that simultaneous knockdown of human isoforms A and B of the histone chaperone anti-silencing function 1 (ASF1), which can evict histones H3 and H4 during transcription (19, 20), attenuated the removal of the H3/H4 dimer in cycling cells without affecting the removal of H2A/H2B (Fig. S5). Both isoforms were knocked down simultaneously to avoid a masked effect by possible redundancies of their function.
Consistent with the involvement of nucleolin in nucleosome disassembly, the recruitment of nucleolin to the break site temporally preceded the eviction of the H2A/H2B histone dimer (Figs. 4B and 1 E and F). Nucleolin was released from the DSB site less than 1 h after DSB induction, occurring simultaneously with the removal of the H2A/H2B dimer from the nucleosome and prior to repair. Since knockdown of nucleolin inhibited both disruption of the nucleosome and DNA repair, we investigated whether nucleolin knockdown affects the recruitment of repair factors to the DSB. Depletion of nucleolin abrogated the recruitment of the NHEJ repair factor XRCC4 to the DSB (Fig. 4 C and D), demonstrating that nucleolin plays a critical role in DNA repair by promoting partial nucleosome disruption and permitting subsequent recruitment of DNA repair factors. Consistent with this model, down-regulation of nucleolin enhanced the sensitivity to ionizing radiation in p53-null HCT116 cells (chosen to avoid modulation of ionizing radiation responses resulting from altered p53 responses caused by nucleolin depletion) (21) (Fig. 4 E and F).
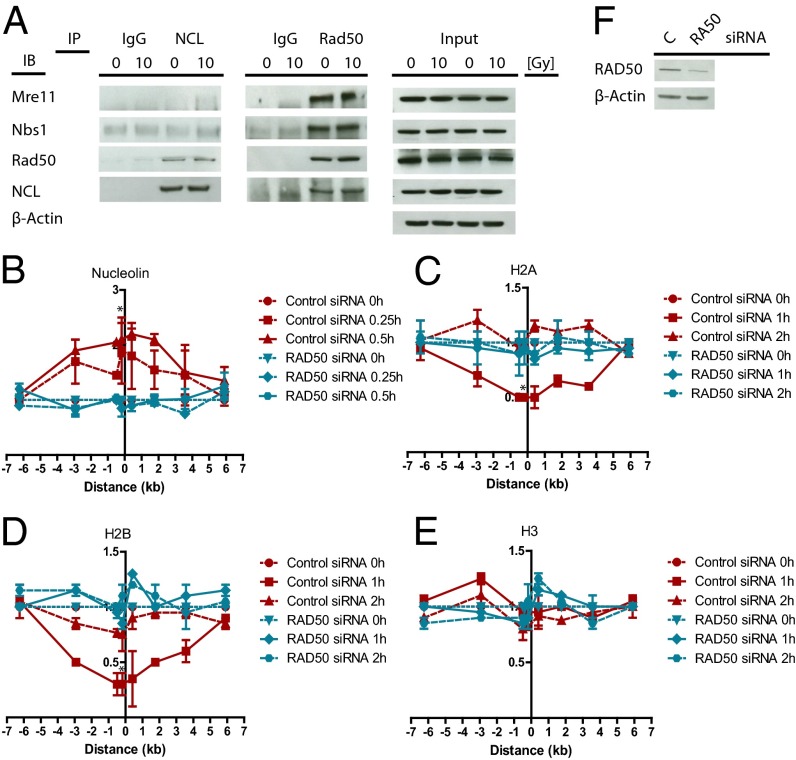
MRN-Dependent Recruitment of Nucleolin to the DSB.
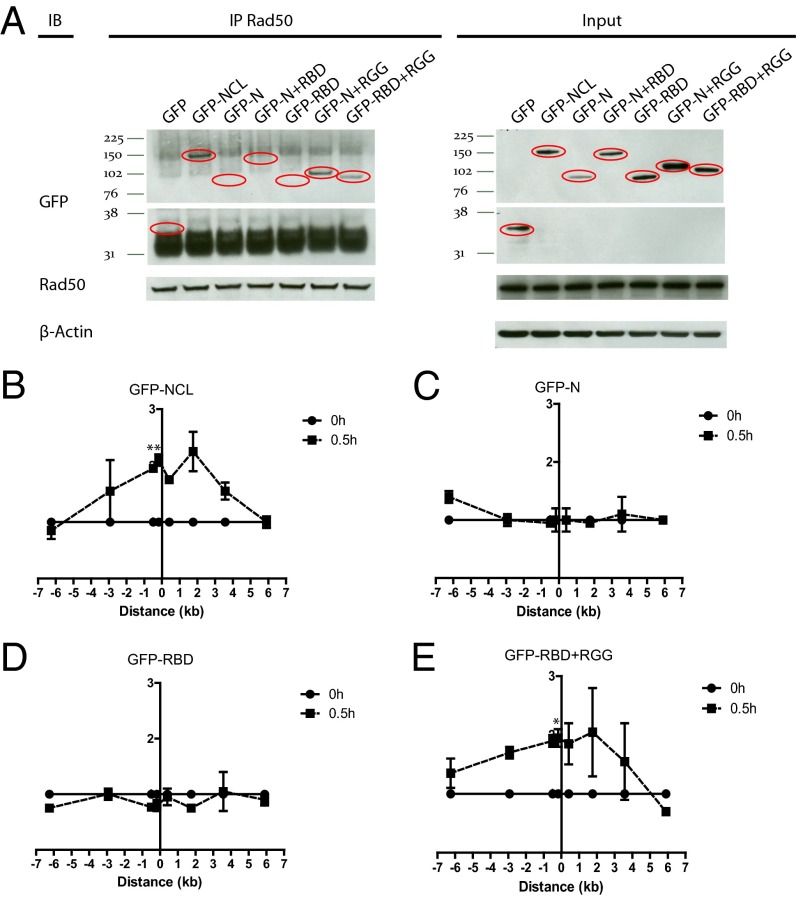
After identifying nucleolin as a chromatin remodeler that is involved in nucleosome disruption at the DSB, we explored the mechanism of nucleolin recruitment to the DSB site. Recruitment of nucleolin is an early event, occurring 15–30 min after DSB induction (Fig. 4B). These similarities with kinetics of the MRN recruitment (Fig. 1B) led us to investigate the potential role of the MRN complex in nucleolin recruitment to the DSB. It is noted that disrupting the MRN complex by knocking down one of the factors prevents the other two MRN complex members from binding to the DSB site (22). NBS1 knockdown significantly impaired the recruitment of nucleolin to the DSB (Fig. S6 A and E) and abolished nucleosome disruption (Fig. S6 B–D), suggesting that the MRN complex facilitates the role of nucleolin in the DSB response. Reciprocal coimmunoprecipitations revealed that nucleolin interacts with RAD50 but not with NBS1 or MRE11 (Fig. 5A). Unexpectedly, this interaction occurred in unstressed cells and persisted after DSB induction. Consistent with a requirement of RAD50 in the recruitment of nucleolin to the DSB, RAD50 knockdown abrogated nucleolin recruitment and, accordingly, the nucleosome disruption (Fig. 5 B–F). Conversely, nucleolin knockdown did not affect the recruitment of NBS1 or RAD50 to the DSB (Fig. S6 F and G). Domain mapping demonstrated that the C-terminal RGG domain of nucleolin is required for interaction with RAD50 (Fig. 6A and Fig. S7A) and for the recruitment of nucleolin to the DSB (Fig. 6 B–E). Overall, these results indicate that nucleolin interacts with RAD50 via the RGG domain and confirm that this interaction is required for the recruitment of nucleolin to the DSB by the MRN complex, where it facilitates nucleosome disruption. Because the N-terminal domain is required for binding to and evicting the H2A/H2B dimer by nucleolin in vitro (12), we tested if the ΔN mutant consisting of the RBD and RGG domains can act as a dominant-negative mutant, thus inhibiting nucleosome disruption. Overexpression of the RBD+RGG mutant impaired the eviction of the H2A/H2B dimer compared with wild-type nucleolin (Fig. S7 B–E) indicating that nucleolin must bind directly to histone proteins for nucleosome disruption.
Fig. 5.
Nucleolin interacts with RAD50 and is recruited to the DSB in an MRN complex-dependent manner to facilitate nucleosome disruption. (A) Nucleolin constitutively interacts with RAD50. (A) Coimmunoprecipitation of nucleolin and MRE11, NBS1, RAD50, and a reciprocal coimmunoprecipitation of RAD50 with nucleolin in untreated MCF7 cells or in MCF7 cells 30 min after 10 Gy ionizing radiation. IB, immunoblot; IP, immunoprecipitation. Normal rabbit IgG was used as control. RAD50 knockdown inhibits nucleolin recruitment to the DSB (B) and nucleosome disruption (C–E). ChIP of (B) nucleolin, (C) histone H2A, (D) histone H2B, and (E) histone H3 in MCF7 cells stably expressing ddI-PpoI as described in Fig. 1B. Cells were transfected with nontargeting control siRNA or RAD50-targeting siRNA. *P < 0.05. (F) Western Blot showing a down-regulation of RAD50 expression by siRNA. IR, ionizing radiation.
Fig. 6.
The C-terminal RGG domain of nucleolin is responsible for the interaction with RAD50 and the recruitment of nucleolin to the DSB. (A) Nucleolin interacts with RAD50 via its RGG domain. Coimmunoprecipitation of RAD50 and indicated nucleolin-deletion mutants in MCF7 cells transiently transfected with GFP or GFP-tagged wild-type nucleolin or nucleolin-deletion mutants. (B–E) The RGG domain is required for nucleolin recruitment to the DSB. ChIP showing recruitment of wild-type nucleolin or indicated nucleolin-deletion mutants in MCF7 cells stably expressing ddI-PpoI as described in Fig. 1B that were transiently transfected with wild-type nucleolin or nucleolin-deletion mutants. Anti-GFP antibody was used to precipitate GFP-tagged wild-type nucleolin and nucleolin-deletion mutants. *P < 0.05, **P < 0.01.
Discussion
We have developed a tightly regulated, rapidly inducible system for studying protein dynamics and DNA repair at sites of DNA breakage in eukaryotic cells. The development of facile, quantitative assessments of DSB repair in mammalian cells has been a long-standing challenge. Pulsed-field gel electrophoresis and comet assays detect broken DNA ends, but they tend to be cumbersome or require relatively high doses of DNA breakage for quantitation and do not provide site-specificity of repair. Resolution of γH2AX or other immunofluorescent foci involving posttranslational modifications of proteins have been used recently as markers of repair (23, 24). Although useful, they are surrogate markers in that they assess the removal of posttranslational modifications rather than directly assessing the religation of broken DNA. Therefore, manipulation of factors that affect protein modifications such as protein phosphatases can affect foci resolution without affecting DNA repair (16). Like Kinner et al. (16), we found that DSB induction by ddI-PpoI leads to the generation of γH2AX foci and that treatment with a phosphatase inhibitor blunts the resolution of the γH2AX foci (Fig. S8 A and B). Inhibition of the phosphatase PP2A also delayed the resolution of the γH2AX foci induced by the topoisomerase II inhibitor etoposide (Fig. S8 C and D). However, PCR assays demonstrated that repair of the ddI-PpoI–induced DSBs was not altered by treatment with the phosphatase inhibitor (Fig. S8A). Therefore, this endonuclease system provides a facile, direct, and quantitative approach to measuring DSB repair in mammalian cells, whereas monitoring the resolution of of γH2AX foci can be affected by factors that modulate the reversal of damage-induced protein modifications instead of those affecting DSB religation.
Chromatin-structure modulation clearly has important roles in transcription, replication, and DNA repair (3). In this report, we used our DSB-inducing system to characterize nucleosome disruption at DSBs in mammalian cells and to define its role in DNA repair. We show that a partial nucleosome disassembly characterized by the displacement of the H2A/H2B histone dimer occurs temporarily surrounding a DSB during NHEJ. Notably, recruitment of XRCC4, an NHEJ factor involved in ligase IV-dependent DSB religation (25), to the DSB is limited to the region of nucleosome disassembly. Additionally, XRCC4 recruitment temporally coincides with disruption of the nucleosome, and XRCC4 release from the break site correlates with completion of DNA repair and nucleosome reassembly. Overlapping spatiotemporal kinetics of nucleosome disassembly and reassembly and of DNA repair factor recruitment and release, respectively, reveal a precise orchestration of these events during DNA repair. Still, the question remains whether one or both H2A/H2B dimers are removed from the nucleosome, since the ChIP assay does not allow a precise quantification of the number of evicted histone dimers.
Like the FACT complex, nucleolin is capable of removing the H2A/H2B dimer from the nucleosome, thereby promoting transcription elongation through the nucleosomal DNA in vitro (12). In this report we demonstrate a role for nucleolin in nucleosome disruption at the DSB and show that this chromatin change is required for efficient DNA repair. Interestingly, after a rapid recruitment, nucleolin is released from the DSB simultaneously with the removal of the core histones from the nucleosome, leading us to speculate that nucleolin is acting as a histone chaperone, binding the H2A/H2B dimer and escorting it from the nucleosome. Consistent with this model, a direct interaction between nucleolin and the H2A/H2B dimer has been shown previously (12). A minor effect of a knockdown of SSRP1, a FACT complex subunit, on nucleosome disruption and the inhibitory effect of the overexpression of the RBD+RGG nucleolin mutant that is recruited to the DSB but lacks the ability to bind the H2A/H2B dimer support a direct and potentially autonomous role of nucleolin in nucleosome disassembly at the DSB.
A previously published study reported that the histone H3 level decreases in the proximity of an endonuclease-induced DSB in a murine cell line (17). Indeed, results presented here show that all four core histones can be displaced from the chromatin surrounding DSBs in proliferating cells. Thus, the NHEJ repair pathway, the major DSB repair mechanism in mammalian cells (26), is dependent on nucleolin-mediated partial nucleosome disruption, whereas a nucleolin-dependent complete nucleosome disruption occurs during the S and G2 phases and is associated with DNA resection, a process that is required for homologous strand invasion and HR. The latter finding was unexpected, because nucleolin is a histone chaperone specifically binding to the H2A/H2B histone dimer and, at least in vitro, is incapable of transferring the H3/H4 histone dimer from the nucleosome (12). Possibly, nucleolin initiates the nucleosome disruption process in S/G2 cells by removing the H2A/H2B dimer and serves as a prerequisite for the subsequent removal of the H3/H4 dimer that is facilitated by another histone chaperone, resulting in the complete nucleosome disruption, DSB processing, and facilitation of HR. Interestingly, ASF1A and B seem to be involved in H3/H4 removal. In yeast, Asf1 is involved in nucleosome disruption during transcription (19, 20) but was implicated in nucleosome reassembly following DSB repair (27). However, human ASF1 may have a different function at the DSB, given the different interaction partners of human and yeast ASF1 among histone chaperones (28). It will be important to investigate if ASF1A and B are recruited to the DSB and facilitate the eviction of the H3/H4 dimer as part of larger chromatin-remodeling complexes such as HIR histone cell cycle regulation defective homologue A (HIRA) or chromatin assembly factor 1 (CAF-1).
As we elucidated the mechanism of nucleolin recruitment to the DSB, we found that nucleolin interacts with RAD50, and its recruitment and the subsequent nucleosome disruption are dependent on the functional MRN complex. This finding was not surprising, because recruitment of nucleolin is an early event, and the binding of the MRN complex to the DSB is one of the first events in the DDR. The observation that nucleolin interacts with RAD50 in unstressed cells via the RGG domain suggests that MRN relocates to the DSB, carrying nucleolin to the break site.
A recently published report using reporter constructs suggested that nucleolin may affect DNA repair (29). However, the repair of DSBs in an artificially integrated reporter can be different from the repair of endogenous DNA, and the assays used to implicate nucleolin in nucleosome modulation (determining protein levels in chromatin versus nucleoplasmic fractions by Western blot) were relatively insensitive and lacked specificity for the break site. Further, they concluded that the role of nucleolin in this process resulted from its role in ATM-dependent or Mdc1-dependent signaling pathways. However, our data demonstrate no effect of nucleolin knockdown on ATM-dependent DDR signaling (Fig. S3D) and instead present a more definitively supported mechanism in which nucleolin recruitment to the DSB occurs in an MRN-dependent manner and directly regulates nucleolin-mediated nucleosome disruption, repair-factor recruitment, and efficient DSB repair. Taken together, our results provide insights into the mechanism of chromatin-structure modulation that occurs at the DSB and identify nucleolin as a chromatin-remodeling factor that facilitates nucleosome disassembly and efficient DSB repair.
Experimental Procedures
Cell Culture, Chemicals, and siRNA.
MCF7 and human foreskin fibroblasts (1070SK) (American Type Culture Collection) were grown in DMEM supplemented with 10% (vol/vol) FBS. MCF7 cells stably expressing ddI-PpoI were grown in DMEM supplemented with 10% (vol/vol) FBS + 1 μg/mL Puromycin (Sigma). MEFs were grown in DMEM supplemented with 10% (vol/vol) FBS, 1% antibiotics, and 4 mM l-glutamine, and 0.05 mM β-mercaptoethanol. 4-OHT (Sigma) was added to a final concentration of 1 μM. Shield-1 (Cheminpharma) was added to a final concentration of 1 μM for 3 h to stabilize the ddI-PpoI fusion protein. NU7026 (Calbiochem) was added to a final concentration of 10 μM. Etoposide (Sigma) was added to a final concentration of 2 μM for 1 h. Thereafter, cells were washed twice with PBS and allowed to recover. Fostriecin (Calbiochem) was added to a final concentration of 100 nM. ON-TARGETplus SMARTpool siRNA toward nucleolin, NBS1, RAD50, SSRP1, ASF1A and ASF1B (Dharmacon) or ON-TARGETplus nontargeting siRNA (Dharmacon) was transfected into cells using Dharmafect-1 transfection reagent (Dharmacon) according to the manufacturer’s protocols to a final concentration of 100 nM.
Antibodies, ChIP, DSB-Repair Assay, Cell-Death Analysis, Immunofluorescence, and Protein Coimmunoprecipitation Assays.
Detailed information about antibodies, ChIP, DSB-repair assay, SubG1, colony formation, immunofluorescence, and protein coimmunoprecipitation assays is supplied in SI Experimental Procedures. Primer sequences for the real-time PCR are listed in Table S1.
Supplementary Material
Acknowledgments
We thank the members of the M.B.K. laboratory for advice and technical support throughout the course of this study, especially Diane Woods and Donald Fleenor. This work was supported by National Institutes of Health Grants CA71387, CA159826, P30CA14236, and P30CA21765 (to M.B.K.); German Research Foundation Grant GO1894/1-1 (to M.G.); and by the American Lebanese Syrian Associated Charities of St. Jude Children’s Research Hospital.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1306160110/-/DCSupplemental.
References
- 1.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421(6922):499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 2.Ciccia A, Elledge SJ. The DNA damage response: Making it safe to play with knives. Mol Cell. 2010;40(2):179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avvakumov N, Nourani A, Côté J. Histone chaperones: Modulators of chromatin marks. Mol Cell. 2011;41(5):502–514. doi: 10.1016/j.molcel.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11(7):481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 5.Nakanishi S, Prasad R, Wilson SH, Smerdon M. Different structural states in oligonucleosomes are required for early versus late steps of base excision repair. Nucleic Acids Res. 2007;35(13):4313–4321. doi: 10.1093/nar/gkm436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berkovich E, Monnat RJ, Jr, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9(6):683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- 7.Belotserkovskaya R, et al. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301(5636):1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 8.Levchenko V, Jackson B, Jackson V. Histone release during transcription: Displacement of the two H2A-H2B dimers in the nucleosome is dependent on different levels of transcription-induced positive stress. Biochemistry. 2005;44(14):5357–5372. doi: 10.1021/bi047786o. [DOI] [PubMed] [Google Scholar]
- 9.Yang X, Zaurin R, Beato M, Peterson CL. Swi3p controls SWI/SNF assembly and ATP-dependent H2A-H2B displacement. Nat Struct Mol Biol. 2007;14(6):540–547. doi: 10.1038/nsmb1238. [DOI] [PubMed] [Google Scholar]
- 10.Park JH, et al. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting gamma-H2AX induction. EMBO J. 2006;25(17):3986–3997. doi: 10.1038/sj.emboj.7601291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu Y, et al. The p400 ATPase regulates nucleosome stability and chromatin ubiquitination during DNA repair. J Cell Biol. 2010;191(1):31–43. doi: 10.1083/jcb.201001160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Angelov D, et al. Nucleolin is a histone chaperone with FACT-like activity and assists remodeling of nucleosomes. EMBO J. 2006;25(8):1669–1679. doi: 10.1038/sj.emboj.7601046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ginisty H, Sicard H, Roger B, Bouvet P. Structure and functions of nucleolin. J Cell Sci. 1999;112(Pt 6):761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- 14.Bouvet P, Diaz JJ, Kindbeiter K, Madjar JJ, Amalric F. Nucleolin interacts with several ribosomal proteins through its RGG domain. J Biol Chem. 1998;273(30):19025–19029. doi: 10.1074/jbc.273.30.19025. [DOI] [PubMed] [Google Scholar]
- 15.Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126(5):995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinner A, Wu W, Staudt C, Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008;36(17):5678–5694. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sunavala-Dossabhoy G, De Benedetti A. Tousled homolog, TLK1, binds and phosphorylates Rad9; TLK1 acts as a molecular chaperone in DNA repair. DNA Repair (Amst) 2009;8(1):87–102. doi: 10.1016/j.dnarep.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 18.Lolis AA, et al. Myogenin recruits the histone chaperone facilitates chromatin transcription (FACT) to promote nucleosome disassembly at muscle-specific genes. J Biol Chem. 2013;288(11):7676–7687. doi: 10.1074/jbc.M112.426718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adkins MW, Howar SR, Tyler JK. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol Cell. 2004;14(5):657–666. doi: 10.1016/j.molcel.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 20.Schwabish MA, Struhl K. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol Cell. 2006;22(3):415–422. doi: 10.1016/j.molcel.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 21.Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123(1):49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 22.Yang YG, et al. Conditional deletion of Nbs1 in murine cells reveals its role in branching repair pathways of DNA double-strand breaks. EMBO J. 2006;25(23):5527–5538. doi: 10.1038/sj.emboj.7601411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodarzi AA, et al. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31(2):167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 24.Ziv Y, et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8(8):870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 25.Christmann M, Tomicic MT, Roos WP, Kaina B. Mechanisms of human DNA repair: An update. Toxicology. 2003;193(1-2):3–34. doi: 10.1016/s0300-483x(03)00287-7. [DOI] [PubMed] [Google Scholar]
- 26.Goodarzi AA, Jeggo P, Lobrich M. The influence of heterochromatin on DNA double strand break repair: Getting the strong, silent type to relax. DNA Repair (Amst) 2010;9(12):1273–1282. doi: 10.1016/j.dnarep.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 27.Chen CC, et al. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell. 2008;134(2):231–243. doi: 10.1016/j.cell.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Koning L, Corpet A, Haber JE, Almouzni G. Histone chaperones: An escort network regulating histone traffic. Nat Struct Mol Biol. 2007;14(11):997–1007. doi: 10.1038/nsmb1318. [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi J, et al. Nucleolin participates in DNA double-strand break-induced damage response through MDC1-dependent pathway. PLoS ONE. 2012;7(11):e49245. doi: 10.1371/journal.pone.0049245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.