Significance
Distributed stem cells (DSCs), which continuously divide asymmetrically to replenish mature tissue cells, adopt a special form of mitotic chromosome segregation. Chromosome segregation is nonrandom instead of random. DSCs cosegregate the set of sister chromosomes with the older of the two template DNA strands used for semiconservative DNA replication during the preceding S phase. Neither the responsible molecular mechanisms nor the cellular function of nonrandom segregation are known. Here, we report evidence that immortal strand chromosomes have a higher level of cytosine 5-hydroxymethylation than mortal chromosomes, which contain the younger DNA template strands. We propose that asymmetric chromosomal 5-hydroxymethylation is a key element of a cellular mechanism by which DSCs distinguish older DNA template strands from younger ones.
Keywords: xanthine, tricarboxylic acid cycle, alpha-ketoglutarate, p53, IMPDH II
Abstract
Immortal strands are the targeted chromosomal DNA strands of nonrandom sister chromatid segregation, a mitotic chromosome segregation pattern unique to asymmetrically self-renewing distributed stem cells (DSCs). By nonrandom segregation, immortal DNA strands become the oldest DNA strands in asymmetrically self-renewing DSCs. Nonrandom segregation of immortal DNA strands may limit DSC mutagenesis, preserve DSC fate, and contribute to DSC aging. The mechanisms responsible for specification and maintenance of immortal DNA strands are unknown. To discover clues to these mechanisms, we investigated the 5-methylcytosine and 5-hydroxymethylcytosine (5hmC) content on chromosomes in mouse hair follicle DSCs during nonrandom segregation. Although 5-methylcytosine content did not differ significantly, the relative content of 5hmC was significantly higher in chromosomes containing immortal DNA strands than in opposed mitotic chromosomes containing younger mortal DNA strands. The difference in relative 5hmC content was caused by the loss of 5hmC from mortal chromosomes. These findings implicate higher 5hmC as a specific molecular determinant of immortal DNA strand chromosomes. Because 5hmC is an intermediate during DNA demethylation, we propose a ten-eleven translocase enzyme mechanism for both the specification and maintenance of nonrandomly segregated immortal DNA strands. The proposed mechanism reveals a means by which DSCs “know” the generational age of immortal DNA strands. The mechanism is supported by molecular expression data and accounts for the selection of newly replicated DNA strands when nonrandom segregation is initiated. These mechanistic insights also provide a possible basis for another characteristic property of immortal DNA strands, their guanine ribonucleotide dependency.
Although distributed stem cells (DSCs) are universally essential for normal tissue function, health, and longevity (1–5), understanding of the cellular mechanisms responsible for their unique tissue functions is quite limited. In particular, the mechanisms responsible for the defining properties of DSCs—asymmetric self-renewal (ASR) and nonrandom sister chromatid segregation—are unknown.
ASR is a special subclass of asymmetric cell division by which DSCs continuously renew mature differentiated tissue cells (6–8). During ASR, DSCs divide asymmetrically with retention of their own stem cell phenotype while simultaneously producing nonstem sisters that are precursors for short-lived tissue-specific differentiating cell lineages. This long-lived role of DSCs in the cell kinetics architecture of renewing tissues is the basis for the hypothesis that they are the predominant cells of origin for tumors induced by gene mutations (9, 10).
Nonrandom sister chromatid segregation is tightly associated with ASR (11, 12). During mitosis, asymmetrically self-renewing DSCs (aDSCs) continuously cosegregate to themselves the set of mitotic chromosomes that contain the older of the two parental template DNA strands. By retaining the same set of template DNA strands over many successive ASR divisions, long-lived DSCs are proposed to reduce their rate of accrual of carcinogenic mutations by 100–1,000-fold compared with their shorter-lived differentiating progeny cells, which retain unrepaired and misrepaired replication errors as a consequence of random segregation (9, 13). Nonrandom segregation also may be an important factor contributing to tissue aging. Accrued chemical damage in the cosegregated template DNA strands, called “immortal DNA strands” (9), may compromise the function and viability of tissue DSCs (1). The immortal DNA strands themselves also might organize epigenomic regulators that are responsible for maintaining the DSC fate (3).
Nonrandom segregation of mitotic sister chromatids was discovered in experiments with cultured mouse fetal fibroblasts (14) and the root tips of legumes and wheats (15, 16). More recently, nonrandom segregation has been described in a diverse range of mammalian species and normal (3, 4, 11, 12, 17–27) and cancerous (28–30) tissue types. Thus, far, these studies have shed only limited light on the nature of the responsible molecular mechanisms. In particular, the molecular basis for the specification and maintenance of immortal DNA strands during nonrandom segregation remains unknown nearly a half century after their discovery (16).
As proposed initially (9), we have confirmed that nonrandom segregation occurs specifically when DSCs adopt an ASR program (4, 11, 12). Five specific cellular proteins—p53, inosine 5′ monophosphate dehydrogenase type II (EC 1.2.1.14) (12), left-right dynein (25), histone H2A.Z (3), and PIM-1 kinase (26)—have been identified as playing a role in nonrandom chromosome segregation mechanisms. In addition, we have shown that guanine ribonucleotide precursors and high cell density are physiological regulators of ASR and nonrandom segregation (4, 12, 31, 32). However, so far, none of these factors has led to an understanding of how DSCs achieve nonrandom segregation.
Two recently discovered properties of the histone H2A variant H2A.Z motivated us to investigate the 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) content of immortal and mortal chromosomes in mouse hair follicle DSCs undergoing nonrandom segregation. H2A.Z chromosomal detection shows a reciprocal relationship with 5mC sites (33), and H2A.Z is molecularly masked on the mortal set of chromosomes in nonrandomly segregating DSCs (3). These analyses reveal that immortal chromosomes, which contain immortal DNA strands, have a significantly higher level of 5hmC modification. The identification of this chromosomal 5hmC asymmetry suggests a molecular mechanism that can account for immortal DNA strand specification and maintenance as well as their long-enigmatic guanine ribonucleotide dependency (12, 32).
Results and Discussion
Detection of 5hmC Chromosomal Asymmetry.
Previously described mouse hair follicle DSC strains, expanded in culture via the suppression of asymmetric cell kinetics, were used for these studies (3, 4). Previously, we discussed the derivation of these cell strains, given that other laboratories have not detected nonrandom segregation in mouse hair follicles in skin-tissue sections (4). The cells have an intrinsic high frequency of ASR and nonrandom segregation. However, when their medium is supplemented with the purine base xanthine (Xn), they show a marked shift to symmetric self-renewal (SSR) and random segregation (4). This Xn sensitivity of ASR and nonrandom segregation are important properties for the detection and analysis of these DSC-specific functions.
The relative 5mC or 5hmC content of sister nuclei or sister chromosome sets in 3C5 mouse hair follicle aDSCs was evaluated in three different previously described (2–4) assay formats (Fig. 1): sister pair (SPr), cytochalasin D-arrested binucleated cells (CD), and mitotic cell assays. The three assays are complementary, with the CD and mitotic cell assays ensuring exact sister–sister comparisons for interphase nuclei and segregating chromosome sets, respectively. In all three assays, sister nuclei or sister chromosome sets were evaluated by in situ indirect immunofluorescence (ISIF) analyses after cells were grown for 24 h under conditions that promoted either nonrandom segregation (Xn-free) or random segregation (Xn-supplemented). The CD assay required an additional 14-h period of CD arrest.
Fig. 1.
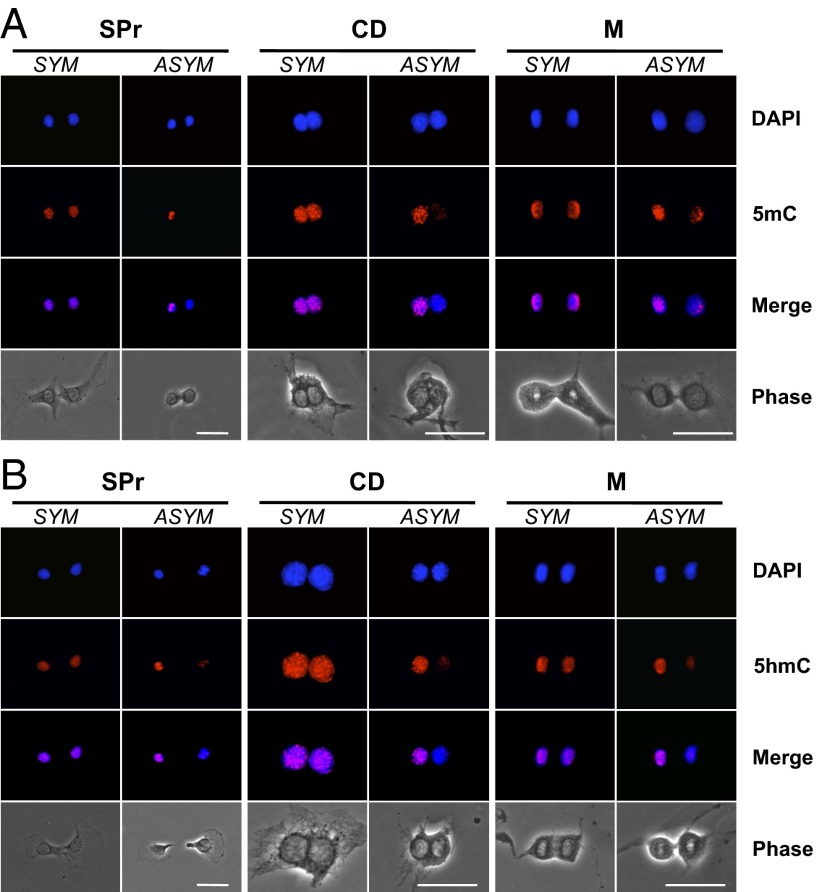
Chromosomal patterns of 5mC and 5hmC content in mouse hair follicle DSCs under conditions that promote nonrandom sister chromatid segregation. Shown are epifluorescence and phase-contrast micrographs of 3C5 cells grown under Xn-free conditions to promote ASR with nonrandom segregation. Fluorescence micrographs compare nuclear DNA fluorescence (DAPI), in situ immunofluorescence images with either anti-5mC antibodies (A) or anti-5hmC antibodies (B), and overlaid images (Merge in A and B). Examples of symmetric (SYM) and asymmetric (ASYM) patterns of chromosomal 5mC and 5hmC content from SPr, CD, and mitotic cell analyses are compared. (Scale bars, 25 microns.)
5mC and 5hmC content was determined in ISIF analyses with specific antibodies (Materials and Methods and Fig. S1). In both 5mC (Fig. 1A) and 5hmC (Fig. 1B) analyses, examples of symmetric and asymmetric fluorescent sister patterns were observed. After quantification of the fluorescence intensities of the members of paired sister nuclei and sister chromosome sets individually, asymmetry was defined as a ≥50% difference between sister nuclei or sister chromosome sets. The frequency of 5mC asymmetry was inconsistent across the three assay formats and was not significantly affected by addition of Xn (Fig. 2 A–C). In contrast, the frequency of 5hmC asymmetry was consistently higher than 5mC asymmetry in all three assays and was reduced significantly by addition of Xn (Fig. 2 D–F). In Xn-free medium, the frequency of 5hmC asymmetry was 22, 40, and 40%, respectively for SPr, CD, and mitotic cell assays. However, in Xn-supplemented medium it decreased to 5, 17, and 13%, respectively (P = 0.04, 0.08, and 0.039, respectively).
Fig. 2.
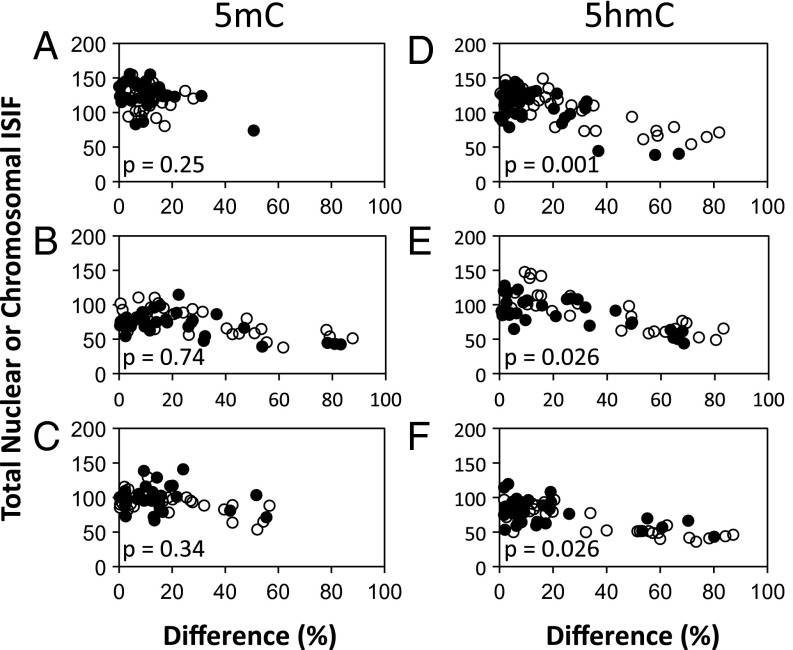
Quantitative analysis of the relationships between the patterns of 5mC or 5hmC chromosomal content and their total nuclear or mitotic chromosome content. The fluorescence intensity of individual paired nuclei or segregating chromosome sets, like those presented in Fig. 1, was quantified by digital imaging epifluorescence microscopy. Fluorescence intensity caused by 5mC or 5hmC antibodies in independent experiments was determined for 3C5 mouse hair follicle DSCs. For each analysis, the percent difference in ISIF intensity between paired nuclei or segregating sets of anaphase/telophase mitotic chromosomes (x-axis) is plotted versus the total ISIF intensity of each pair (y-axis). Increasing percent differences indicate an increasing degree of asymmetry in 5mC or 5hmC chromosomal content. (A and D) Data from nuclei of sister pairs. (B and E) Data from nuclei of cytochalasin D-arrested binucleated cells. (C and F) Data from segregating chromosome sets in anaphase/telophase mitotic cells. Open circles represent Xn-free conditions that promote ASR with nonrandom segregation. Filled circles represent Xn-supplemented conditions that promote SSR and random segregation. p, the Kolmogorov–Smirnov probability that the percent differences shown in the Xn-free and Xn-supplemented distributions would occur by chance.
5hmC asymmetry was caused by decreased 5hmC on one segregating set of chromosomes. This cause was evident from the lower total specific antibody fluorescence of sister cell nuclei, CD-arrested binucleated cells, or mitotic cells that showed 5hmC asymmetry (≥50% along the x-axis) (Fig. 2 D–F) compared with the total specific antibody fluorescence of sister cell nuclei that showed a symmetric 5hmC chromosomal pattern (<50% along the x-axis) (Fig. 2 D–F). We confirmed this interpretation by determining the average fluorescence of the nuclei or chromosome sets with the greater fluorescence of their pair. The average determined from symmetric pairs was compared with the average determined from asymmetric pairs. Across the three assay formats, these two averages differed by only 12–18%. This degree of variance is similar to our observations for the percent differences in nuclear DAPI fluorescence. Therefore, these results indicate that nuclei or chromosome sets with the greater specific antibody fluorescence of their pair had similar 5hmC content in both symmetric and asymmetric pairs. In contrast, the same analysis performed with the nuclei or chromosome sets that had the lower specific antibody fluorescence level in their pairs yielded differences in the range of 46–59%, with the average from the asymmetric pairs being less (P < 0.0001). These data indicate that, on average, asymmetrically self-renewing cells have one set of segregating mitotic chromosomes with ∼50% lower 5hmC, and this reduced level is maintained in the interphase of the asymmetric sister cells produced.
Higher 5hmC Identifies Immortal DNA Chromosomes.
Next, we investigated relationships between 5hmC asymmetry and chromosomes containing an immortal DNA strand. This analysis was accomplished by serial ISIF (Materials and Methods) for BrdU and 5hmC in our standardized label-retention CD assay designed for maximal detection of immortal DNA strands in cells undergoing nonrandom segregation (4). 3C5 and 5B8 mouse hair follicle DSC strains were labeled with BrdU in Xn-supplemented medium to promote the maximal level of random segregation. This labeling pulse was maintained for approximately one cell cycle (24 h) to produce cells with chromosomes hemisubstituted with BrdU. Thereafter, parallel labeled cultures were either continued in Xn-supplemented medium (maintaining random segregation) or switched to Xn-free medium (promoting nonrandom segregation). Both media were supplemented with excess thymidine as a chase for four to five cell cycles (96 h). After CD arrest, serial ISIF was performed to evaluate the respective BrdU and 5hmC content of sister nuclei. In Fig. 3 these data are presented as scatter plots of the percent difference in anti-BrdU immunofluorescence versus the percent difference in anti-5hmC immunofluorescence for the sister nuclei of each quantified CD-arrested binucleated cell. Chromosomal asymmetry for BrdU or 5hmC was defined as a ≥50% difference between sister nuclei. For BrdU, this degree of difference is indicative of nonrandom segregation, and the sister chromosome sets with significantly greater anti-BrdU immunofluorescence correspond to the immortal sister chromosomes that contain highly BrdU-substituted immortal DNA strands (Fig. 4).
Fig. 3.
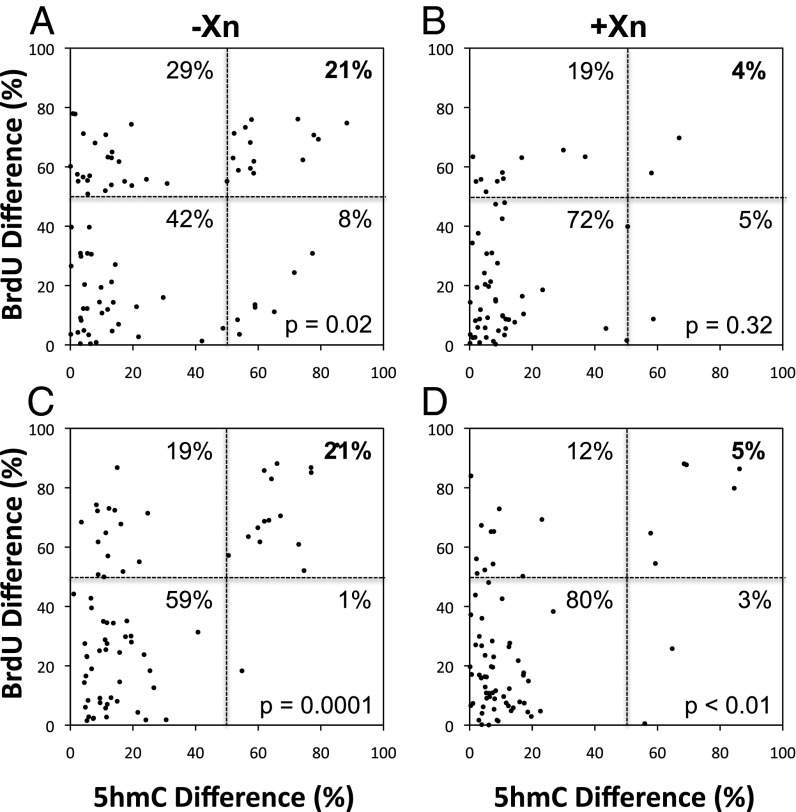
Quantitative analysis of the relationship between nonrandom chromosome segregation and asymmetry of 5hmC chromosomal content in mouse hair follicle DSCs. A label-retention assay with cytochalasin D-arrested binucleated cells was used to identify cells undergoing nonrandom segregation by virtue of the unequal BrdU content of sister nuclei. Parallel assays were performed under Xn-free (−Xn) and Xn-supplemented (+Xn) conditions to promote nonrandom or random segregation, respectively. Both strain 3C5 (A and B) and strain 5B8 (C and D) cells were evaluated. Serial ISIF analyses were performed with distinct anti-BrdU antibodies and anti-5hmC antibodies (Fig. 4). The BrdU and 5hmC immunofluorescence of paired sister nuclei were quantified independently and were used to calculate corresponding percent differences for each marker addressed to each quantified binucleated cell. Differences ≥50% in the values for BrdU and 5hmC correspond to cells undergoing nonrandom segregation and cells with an asymmetric pattern of 5hmC chromosomal content, respectively. Differences <50% in the values for BrdU and 5hmC correspond to cells undergoing random segregation and cells with a symmetric pattern of 5hmC chromosomal content, respectively. The four quadrants formed by the grid lines at 50% difference correspond to cells with the following patterns of chromosome segregation pattern/5hmC chromosomal content: nonrandom:symmetric (Upper Left); random:symmetric (Lower Left); nonrandom:asymmetric (Upper Right); random:asymmetric (Lower Right). The percent values denote the fraction of total quantified binucleated cells in each quadrant. P values indicate levels of confidence, based on Fisher’s two-tailed exact test, that the observed greater fraction of binucleated cells with 5hmC chromosomal asymmetry among cells undergoing nonrandom segregation, compared with cells undergoing random segregation, did not occur by chance.
Fig. 4.
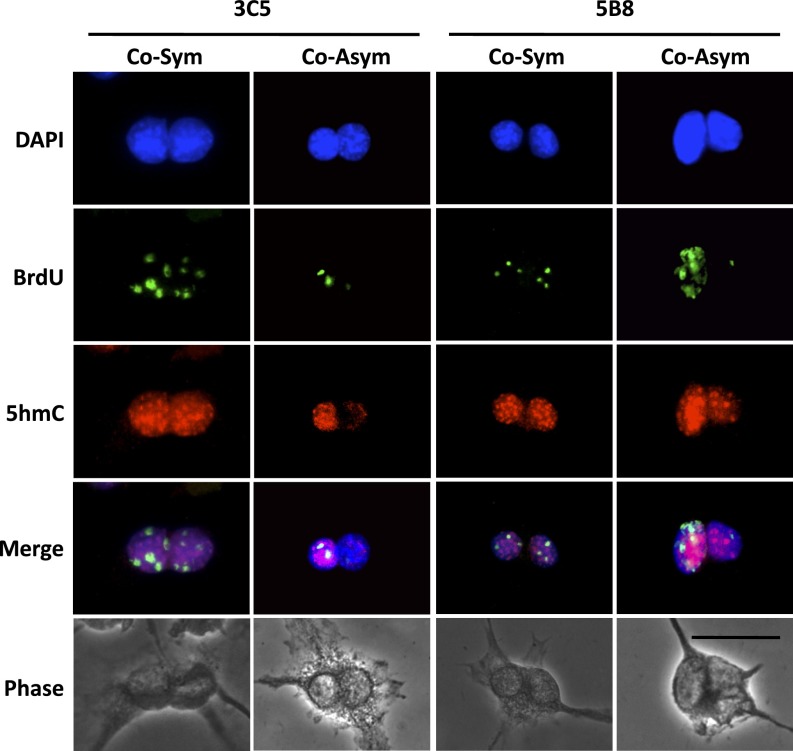
Higher 5hmC content marks immortal DNA chromosomes in mouse hair follicle DSCs undergoing nonrandom segregation. Shown are epifluorescence and phase-contrast micrographs of cytochalasin D-arrested binucleated 3C5 and 5B3 cells at the completion of a BrdU label retention-CD study with culture under Xn-free conditions to promote ASR with nonrandom segregation. Fluorescence micrographs compare nuclear DNA fluorescence (DAPI), serial ISIF images with anti-BrdU and anti-5hmC antibodies, and overlaid images (Merge). Examples of cosymmetric (Co-Sym) and coasymmetric (Co-Asym) patterns of BrdU and 5hmC chromosomal content are shown. (Scale bars, 25 microns.)
In the label-retention CD studies, both the 3C5 and 5B8 cell cultures displayed nonrandom segregation cell fractions and 5hmC asymmetry cell fractions that were reduced significantly by Xn supplementation. For 3C5 cells, the nonrandom segregation fraction decreased from 50 to 23% (P = 0.0014), and the corresponding 5hmC asymmetry fraction decreased from 29 to 9% (P = 0.0081). Similarly, for 5B8 cells, the nonrandom segregation fraction decreased from 40 to 17% (P = 0.0035), and the corresponding 5hmC asymmetry fraction decreased from 22 to 8% (P = 0.022). Under Xn-free conditions, which maximized nonrandom segregation, on average 82% of cells with 5hmC asymmetry also displayed nonrandom segregation. Importantly, 94% of these cells were coasymmetric for BrdU and 5hmC (See examples in Fig. 4, Co-Asym). The coasymmetric pattern of association demonstrated that the higher levels of 5hmC were found on the chromosomes that contained the immortal DNA strands.
It is noteworthy that ∼50% of cells showing evidence of nonrandom segregation had a symmetric pattern of chromosomal 5hmC content (Fig. 3, Upper Left Quadrants). This population of cells was less sensitive to suppression by Xn supplementation than the coasymmetric population (compare Fig. 3, Upper Right Quadrants). The exact nature of these cells is unknown. One likely explanation is that these are cells that spontaneously switched from ASR and nonrandom segregation back to SSR and random segregation late during the label-retention chase. ASR is readily suppressed by increases in cell density (12, 31) that would occur late in these experiments. If this suppression were to occur, the restoration of 5hmC symmetry might occur on a faster time scale than the 24-h period required for complete randomization of previously cosegregating immortal DNA chromosomes. Another possible explanation is that the in vitro conditions are not optimal for achieving the full in vivo efficiency.
Proposed Mechanism for Specification and Maintenance of Immortal DNA Strands.
5hmC modification of chromosomal DNA is a rapidly emerging new molecular factor associated with gene-regulatory events and determinants of varied cell phenotypes (34–36). Recently, 5hmC has been viewed primarily as an intermediate in DNA demethylation (37–40). Based on our observation of an apparently stabilized coasymmetric association of 5hmC with immortal chromosomes, here, we consider its potential to be a major actor in the specification and maintenance of immortal DNA strands in homeostatic, aDSCs in mammalian tissues.
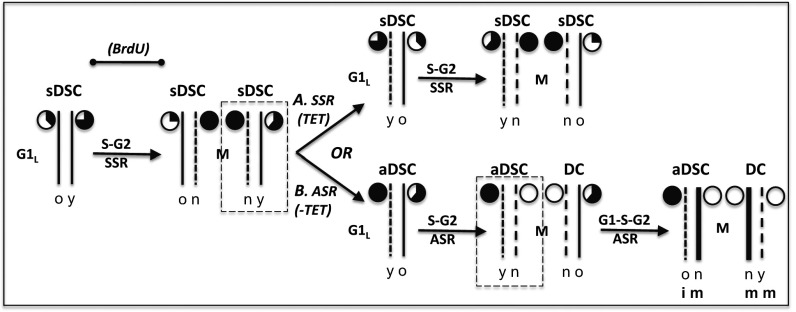
Fig. 5 provides a schematic of the proposed role of 5hmC as a determinant of the specification and maintenance of immortal DNA strands. The essential concept for the model is that the level of 5hmC in DNA strands serves as a molecular indicator of their age. DNA methyltransferases modify DNA in concert with DNA replication. Thereafter, ten-eleven translocase enzymes (TETs) proceed to catalyze the removal of the methyl groups, 5hmC being the first intermediate in the overall demethylation reaction. First, TET enzymes convert 5mC to 5hmC. TET enzymes continue to convert 5hmC into later intermediates that are excised by repair enzymes with restoration of unmodified cytosine (37–40). We suggest that 5hmC generated on newly synthesized DNA strands reaches a maximal level in G2/M phase of the cell cycle (Fig. 5, closed, filled pie charts) but declines during subsequent cell cycles as a result of the completion of demethylation reactions. Thus, in SSR cells (i.e., symmetrically self-renewing DSC, sDSCs) (Fig. 5A), the level of 5hmC in DNA strands is proportional to the amount of time that has passed since the strands were first synthesized (Fig. 5A, partially filled pie charts). This mechanism would cause even randomly segregating sister chromatid pairs to differ in 5hmC content. However, this basal difference is predicted to be relatively small, and its randomization over many independently assorting sister chromosomes would average it out. As a result, randomly segregating chromosome sets overall have nearly equivalent 5hmC content (Fig. 5A).
Fig. 5.
A model for the mechanism based on asymmetric 5hmC content by which immortal DNA chromosomes are specified and maintained during nonrandom sister chromatid segregation in DSCs. Shown is the progression of inheritance of one representative mammalian DSC chromosome composed of DNA strands (thin solid lines) of different ages as a consequence of semiconservative DNA replication. The modeled progression begins with an sDSC undergoing random chromosome segregation in the late G1 phase of the cell cycle (G1L). The pie charts denote the 5hmC content of the adjacent DNA strands. A completely open pie chart denotes the lowest content, and a completely filled pie chart denotes the highest content. Reflecting the different ages of the future DNA template strands, the older (o) strand has less 5hmC than the younger (y) strand. This difference exists because the older strand has had more time for the removal of 5hmC in its role as an intermediate in TET enzyme demethylation of DNA strands after their replication-dependent methylation in S phase. As the DSC progresses by SSR through S phase and G2 phase (S–G2), the amount of 5hmC in these template DNA strands continues to decrease by the same mechanism. Mitosis (M) of the DSC produces two new sDSCs, with new chromosomes that each contain one newly synthesized DNA strand (short dashes; n) that has the maximum content of 5hmC (filled pie charts) before it begins to decline as a result of demethylation. (A) Continued SSR by a sister DSC (dashed rectangle), which maintains normal levels of TET enzyme activity, recapitulates the preceding patterns of symmetric chromosomal 5hmC content on average. (B) The switching of an sDSC sister to an ASR program (aDSC), with coordinated loss of TET enzyme activity (−TET), prevents subsequent loss or production of 5hmC. As a result, in subsequent mitoses of aDSCs, total 5hmC content is predicted to be reduced overall and distributed asymmetrically, being higher on chromosomes that contain the oldest, immortal (i) DNA strands. The model also can explain why pulses of BrdU given during the indicated period in label-retention experiments result in newly selected immortal DNA strands containing BrdU. Dashed rectangles indicate cells followed through next mitosis; long dashed lines indicate newly synthesized DNA strands; bold solid lines indicate newly synthesized DNA strands. DC, differentiating cell or cell lineage. m, mortal DNA strands.
If a symmetrically self-renewing DSC (sDSC) that switched to ASR were also to lose significant TET enzymatic activity, the difference in 5hmC content between sister chromatids would increase dramatically after a second asymmetric cell cycle (Fig. 5B, −TET, ASR, aDSC). Although newly synthesized DNA strands would contain 5mC, as usual, it would not be converted into 5hmC (Fig. 5B, open pie charts). DNA strands with this lowest 5hmC level would be produced as long as TET activity levels remained low. More importantly, for the DNA strands that were synthesized most recently during SSR and had achieved a maximal level of 5hmC, their higher 5hmC content would be stabilized upon switching to ASR (Fig. 5B, filled pie charts). We propose that such stabilized higher 5hmC content in the most recently synthesized DNA strands specifies the chromosomes that contain them for future nonrandom cosegregation. In subsequent ASR divisions with continued reduced TET activity (Fig. 5B, ASR, aDSC), the higher 5hmC mark would be maintained passively. The marked strands would progress from initially being the younger template DNA strands (Fig. 5B, y) to becoming the older template DNA strands and finally the oldest immortal DNA strands in aDSCs (Fig. 5B, ASR, aDSC, o/i).
Our model for DSC immortal DNA strand specification and maintenance based on 5hmC chromosomal asymmetry is, in essence, an elaboration of the inherent information asymmetry of semiconservatively replicated double-stranded DNA. The essential features and predictions from the model are readily accessible to experiment. In fact, already, the model provides an explanation for a long-standing, apparently paradoxical feature of nonrandom segregation analyses. This paper (Fig. 4, Co-Asym) and previous reports (4, 11, 12) show that, when cells are labeled with BrdU before switching from SSR and random segregation to ASR and nonrandom segregation, newly specified immortal DNA strands have incorporated BrdU. This experimental observation cannot be explained by mechanisms that specify the “oldest” DNA strands per se. However, the proposed 5hmC-asymmetry model accounts for this finding, because it predicts that the more recently synthesized BrdU+ DNA strands will be specified for nonrandom segregation because of their higher 5hmC content (Fig. 5, BrdU). Because of the metabolic pathways involved in the regulation of TET enzymatic activity, the model also has the potential to account for the long-unexplained role of guanine ribonucleotide biosynthesis in the regulation of DSC ASR and nonrandom segregation (Fig. S2 and SI Results and Discussion) (4, 11, 12, 32).
Implications.
The scarcity of DSCs in mammalian tissues and isolated cell fractions has been a major impediment to experimental investigation of the mechanism of nonrandom segregation. In our earliest studies, we circumvented this barrier by investigating immortalized cell lines that were genetically engineered to express ASR with associated nonrandom segregation (11, 32). More recently, we have advanced our studies to ex vivo-expanded natural DSCs, such as those from mouse hair follicles used in the present studies (3, 4). Given the universal role of ASR in DSC tissue function, if the model proposed applies to this specific DSC strain, it should apply to mammalian tissue DSCs in general. This advance opens a door to further elucidation of the molecular mechanisms responsible for nonrandom segregation. Indeed, the advance of our specific hypotheses and predictions make the investigation of these mechanisms more accessible to molecular genetic approaches that do not require DSCs in high quantity or high purity.
To our knowledge, in the 38 y since Cairns (9) first introduced the immortal strands hypothesis, no subsequent hypothesis, based on experimental findings, has been advanced that accounts for how immortal DNA strands might be specified and maintained in DSCs. In 2002, we reported the direct visualization and experimental control of nonrandom chromosome cosegregation in an experimentally accessible cell model (11). Since then we (3, 4, 12) and others (18–30) have confirmed the biological breadth of DSC nonrandom segregation by its demonstration in diverse mammalian tissues and species. This report presents a richly integrated framework to guide future experimental investigation of molecular and biochemical mechanisms responsible for nonrandom segregation by DSCs. Continued interrogation of the proposed mechanisms of nonrandom segregation also may reveal insights to the equally challenging question of the biological significance of DSC nonrandom segregation in mammalian life and evolution.
Materials and Methods
Cells.
Hair follicle DSCs from mouse strains 3C5 and 5B8 were maintained as previously described (4). The properties of these DSCs, including purine-dependent ASR, purine-dependent nonrandom segregation, long-term self-renewal, and production of multiple differentiated cell types of the skin and hair follicles, have been described (4).
5mC and 5hmC ISIF Analyses.
Fixed cells were washed in PBS and blocked with 10% (vol/vol) normal goat serum. Respective mouse monoclonal anti-5mC antibodies (catalog no. NA81; EMD Biosciences, Inc.) or rabbit polyclonal anti-5hmC antibodies (catalog no. 39769; Active Motif) diluted 1:300 in 10% (vol/vol) normal goat serum in PBS were applied for 16–24 h at 4 °C. At the end of this period, slides were washed with a solution of 0.5% BSA in PBS and then were incubated at room temperature for 1 h with respective Alexa Fluor 568-conjugated secondary goat anti-mouse IgG (catalog no. A-11004; Invitrogen, Inc.) or goat anti-rabbit IgG (catalog no. A-11011; Invitrogen) antibodies diluted 1:300 in 2% (vol/vol) goat serum in PBS. Thereafter, slides were washed and mounted with DAPI containing VectaShield mounting medium (Vector Laboratories, Inc.).
Control analyses with omission of anti-5mC or anti-5hmC antibodies were evaluated to ensure that all detected fluorescence was specific. In addition, antigen-blocking experiments were performed with nucleoside and nucleotide forms of 5-methylated cytosines to confirm that the antibodies did not cross-react among different cytosine species. Preincubation of the anti-5mC antibodies at 4 °C for 6 h with 0.3 mM 2′-deoxy-5-methylcytidine (catalog no. D3610; TCI America) prevented subsequent detection of chromosomal 5mC, whereas 0.3 mM 5-hydroxymethyl-2′-deoxycytidine-5′-triphosphate (catalog no. Bio-39046; Bioline USA) or 2′-deoxycytidine (catalog no. D3897; Sigma-Aldrich) did not. Similarly, preincubation of the anti-5hmC antibodies at room temperature for 1 h with 3.0 mM 5-hydroxymethyl-2′-deoxycytidine-5′-triphosphate prevented detection of chromosomal 5hmC, whereas 3.0 mM 2′-deoxy-5-methylcytidine or 2′-deoxycytidine did not (See Fig. S1).
Nonrandom Segregation and Chromosomal 5hmC Analyses.
Cells were labeled for 24 h with 5 μM BrdU and chased for 96 h in BrdU-free medium supplemented with 50 μM thymidine either with or without Xn supplementation. For CD treatment, culture was continued for 14 h in respective media supplemented to 2 μM CD. Thereafter, the cells were fixed with ice-cold 70% ethanol in distilled water for 30 min. All subsequent incubations for BrdU detection were performed at room temperature. After washing with PBS, the cells were treated with 2N HCl for 10 min. After washing again with PBS, they were incubated in blocking solution (PBS, 0.5% BSA, 0.05% Tween-20) for 10 min. The blocked cells were incubated for 3 h with mouse monoclonal anti-BrdU antibodies (catalog no. MAB3424; Millipore Corp.) diluted 1:100 in blocking solution. After washing with blocking solution, incubation with Alexa Fluor 488-conjugated secondary goat anti-mouse IgG (catalog no.11001; Invitrogen, Inc.) diluted 1:300 in blocking solution was maintained for 1 h. Then the cells were washed in PBS and blocked with 10% normal goat serum. Rabbit polyclonal anti-5hmC antibodies (catalog no. 39769; Active Motif), diluted 1:300 in 10% (vol/vol) normal goat serum in PBS, were applied for 16–24 h at 4 °C. At the end of this period, slides were washed with a solution of 0.5% BSA in PBS and then were incubated at room temperature for 1 h with Alexa Fluor 568-conjugated secondary goat anti-rabbit IgG (catalog no. A-11011; Invitrogen, Inc.) antibodies diluted 1:300 in 2% (vol/vol) goat serum in PBS. Thereafter, slides were washed sequentially with 0.5% BSA in PBS and in PBS alone and then were mounted with DAPI-containing VectaShield mounting medium. To ensure the specificity of the respective anti-BrdU and anti-5hmC fluorescence signals, control analyses were performed that omitted each primary antibody individually or both together.
Digital Imaging Quantification.
Epifluorescence images were captured with a Leica DMR microscope and Leica DC300F digital camera system. The mean pixel intensity of fluorescent nuclei or chromosome sets was quantified using National Institutes of Health Image J software as previously described (2, 4). The percent difference in mean fluorescence intensity between sister nuclei or sister chromosome sets was calculated as [(A−B)/A] × 100%, where A ≥ B. Total nuclear or chromosomal set fluorescence equals A+B.
Statistical Analyses.
The statistical significance of observed differences in categorical data was evaluated in 2 × 2 contingency tables using Fisher’s two-tailed exact test. The statistical significance of observed differences in data distributions was evaluated using the Kolmogorov–Smirnov test. Student t test was used to evaluate the statistical significance of differences in the mean fluorescence intensity of compared groups of nuclei or chromosome sets corresponding to the members of pairs that had either the higher or lower fluorescence intensity. For the t test analyses, the cutoff for a symmetric versus an asymmetric 5hmC chromosomal pattern was determined to be two SDs from the mean of the distribution of percent differences for the DAPI fluorescence of the corresponding sister nuclei or sister chromosome sets. These differences were 20%, 23%, and 18% for the data from Xn-free SPr, CD, and mitotic cell assays, respectively (Fig. 2 D–F).
Supplementary Material
Acknowledgments
We thank Dr. H. Paulus for review of the manuscript and suggestions for its improvement; Drs. R. Taghizadeh and J. A. Lansita for critical reading of the manuscript; and Dr. J. Garlick for critical discussion of the manuscript’s implications for future DSC research. This research was supported by National Institutes of Health–National Institute of General Medical Sciences Director’s Pioneer Award 5DP1OD000805 and by a grant from the Lee Iacocca Family Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310323110/-/DCSupplemental.
References
- 1.Sherley JL. A new mechanism for aging: Chemical “age spots” in immortal DNA strands in distributed stem cells. Breast Dis. 2008;29:37–46. doi: 10.3233/bd-2008-29105. [DOI] [PubMed] [Google Scholar]
- 2.Noh M, Smith JL, Huh YH, Sherley JL. A resource for discovering specific and universal biomarkers for distributed stem cells. PLoS ONE. 2011;6(7):e22077. doi: 10.1371/journal.pone.0022077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huh YH, Sherley JL. Molecular cloaking of H2A.Z on mortal DNA chromosomes during nonrandom segregation. Stem Cells. 2011;29(10):1620–1627. doi: 10.1002/stem.707. [DOI] [PubMed] [Google Scholar]
- 4.Huh YH, King J, Cohen J, Sherley JL. SACK-expanded hair follicle stem cells display asymmetric nuclear Lgr5 expression with non-random sister chromatid segregation. Sci Rep. 2011;1:176. doi: 10.1038/srep00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sherley JL. Advancing renewable normal human cell assays for drug discovery. Drug Dev Res. 2013;74(2):127–137. [Google Scholar]
- 6.Loeffler M, Potten CS. In: Stem Cells. Potten CS, editor. San Diego: Harcourt Brace & Co.; 1997. pp. 1–28. [Google Scholar]
- 7.Sherley JL. Asymmetric cell kinetics genes: The key to expansion of adult stem cells in culture. Stem Cells. 2002;20(6):561–572. doi: 10.1634/stemcells.20-6-561. [DOI] [PubMed] [Google Scholar]
- 8.Sherley JL. In: Stem Cell Repair and Regeneration. Habib NA, Gordon MY, Levicar N, Jiao L, Thomas-Black G, editors. London: Imperial College Press; 2005. pp. 21–28. [Google Scholar]
- 9.Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255(5505):197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- 10.Knudson AG. Stem cell regulation, tissue ontogeny, and oncogenic events. Semin Cancer Biol. 1992;3(3):99–106. [PubMed] [Google Scholar]
- 11.Merok JR, Lansita JA, Tunstead JR, Sherley JL. Cosegregation of chromosomes containing immortal DNA strands in cells that cycle with asymmetric stem cell kinetics. Cancer Res. 2002;62(23):6791–6795. [PubMed] [Google Scholar]
- 12.Rambhatla L, Ram-Mohan S, Cheng JJ, Sherley JL. Immortal DNA strand cosegregation requires p53/IMPDH-dependent asymmetric self-renewal associated with adult stem cells. Cancer Res. 2005;65(8):3155–3161. doi: 10.1158/0008-5472.CAN-04-3161. [DOI] [PubMed] [Google Scholar]
- 13.Sherley JL. In: Tissue Stem Cells. Potten CS, Clarke RB, Wilson J, Renehan AG, editors. New York: Taylor Francis; 2006. pp. 37–54. [Google Scholar]
- 14.Lark KG, Consigli RA, Minocha HC. Segregation of sister chromatids in mammalian cells. Science. 1966;154(3753):1202–1205. doi: 10.1126/science.154.3753.1202. [DOI] [PubMed] [Google Scholar]
- 15.Lark KG. Nonrandom segregation of sister chromatids in Vicia faba and Triticum boeoticum. Proc Natl Acad Sci USA. 1967;58(1):352–359. doi: 10.1073/pnas.58.1.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lark KG. Discovering non-random segregation of sister chromatids: The naïve treatment of a premature discovery. Front Oncol. 2013;2:211. doi: 10.3389/fonc.2012.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Potten CS, Hume WJ, Reid P, Cairns J. The segregation of DNA in epithelial stem cells. Cell. 1978;15(3):899–906. doi: 10.1016/0092-8674(78)90274-x. [DOI] [PubMed] [Google Scholar]
- 18.Potten CS, Owen G, Booth D. Intestinal stem cells protect their genome by selective segregation of template DNA strands. J Cell Sci. 2002;115(Pt 11):2381–2388. doi: 10.1242/jcs.115.11.2381. [DOI] [PubMed] [Google Scholar]
- 19.Karpowicz P, et al. Support for the immortal strand hypothesis: Neural stem cells partition DNA asymmetrically in vitro. J Cell Biol. 2005;170(5):721–732. doi: 10.1083/jcb.200502073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith GH. Label-retaining epithelial cells in mouse mammary gland divide asymmetrically and retain their template DNA strands. Development. 2005;132(4):681–687. doi: 10.1242/dev.01609. [DOI] [PubMed] [Google Scholar]
- 21.Armakolas A, Klar AJS. Cell type regulates selective segregation of mouse chromosome 7 DNA strands in mitosis. Science. 2006;311(5764):1146–1149. doi: 10.1126/science.1120519. [DOI] [PubMed] [Google Scholar]
- 22.Shinin V, Gayraud-Morel B, Gomès D, Tajbakhsh S. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nat Cell Biol. 2006;8(7):677–687. doi: 10.1038/ncb1425. [DOI] [PubMed] [Google Scholar]
- 23.Conboy MJ, Karasov AO, Rando TA. High incidence of non-random template strand segregation and asymmetric fate determination in dividing stem cells and their progeny. PLoS Biol. 2007;5(1):e102. doi: 10.1371/journal.pbio.0050102. 1120–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Capuco AV. Identification of putative bovine mammary epithelial stem cells by their retention of labeled DNA strands. Exp Biol Med (Maywood) 2007;232(10):1381–1390. doi: 10.3181/0703-RM-58. [DOI] [PubMed] [Google Scholar]
- 25.Armakolas A, Klar AJ. Left-right dynein motor implicated in selective chromatid segregation in mouse cells. Science. 2007;315(5808):100–101. doi: 10.1126/science.1129429. [DOI] [PubMed] [Google Scholar]
- 26.Sundararaman B, et al. Asymmetric chromatid segregation in cardiac progenitor cells is enhanced by Pim-1 kinase. Circ Res. 2012;110(9):1169–1173. doi: 10.1161/CIRCRESAHA.112.267716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kajstura J, et al. Tracking chromatid segregation to identify human cardiac stem cells that regenerate extensively the infarcted myocardium. Circ Res. 2012;111(7):894–906. doi: 10.1161/CIRCRESAHA.112.273649. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Hari D, et al. Isolation of live label-retaining cells and cells undergoing asymmetric cell division via nonrandom chromosomal cosegregation from human cancers. Stem Cells Dev. 2011;20(10):1649–1658. doi: 10.1089/scd.2010.0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pine SR, Ryan BM, Varticovski L, Robles AI, Harris CC. Microenvironmental modulation of asymmetric cell division in human lung cancer cells. Proc Natl Acad Sci USA. 2010;107(5):2195–2200. doi: 10.1073/pnas.0909390107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xin H-W, et al. Tumor-initiating label-retaining cancer cells in human gastrointestinal cancers undergo asymmetric cell division. Stem Cells. 2012;30(4):591–598. doi: 10.1002/stem.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rambhatla L, et al. Cellular senescence: Ex vivo p53-dependent asymmetric cell kinetics. J Biomed Biotechnol. 2001;1(1):28–37. doi: 10.1155/S1110724301000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherley JL, Stadler PB, Johnson DR. Expression of the wild-type p53 antioncogene induces guanine nucleotide-dependent stem cell division kinetics. Proc Natl Acad Sci USA. 1995;92(1):136–140. doi: 10.1073/pnas.92.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conerly ML, et al. Changes in H2A.Z occupancy and DNA methylation during B-cell lymphomagenesis. Genome Res. 2010;20(10):1383–1390. doi: 10.1101/gr.106542.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ficz G, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473(7347):398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 35.Koh KP, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8(2):200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pastor WA, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473(7347):394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cortellino S, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146(1):67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo JU, Su Y, Zhong C, Ming G-L, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145(3):423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He Y-F, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333(6047):1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.