Significance
TATA-binding protein (TBP)-associated factor 7l (Taf7l), Taf4b, and TBP-related factor 2 (Trf2) were found to be three important atypical testis-specific core promoter recognition factors. However, whether there was any functional cross-talk between them and at which stages of spermatogenesis remained uncharacterized. Here we report that Taf7l−/Y becomes sterile rather than merely subfertile after backcrossing. Importantly, we found that Taf7l cooperates with Trf2, but not Taf4b, at late stages to direct the transcription of key spermatogenic genes. Our findings thus provide unique insights into the function of cell-type–specific Tafs and how two core promoter recognition factors mediate transcriptional control of spermiogenesis. The testis-specific role of Taf7l identifies this regulator as a potential target for male contraceptive intervention.
Keywords: gene regulation, reproduction, RNA-seq, contraceptive medicine
Abstract
TATA-binding protein (TBP)-associated factor 7l (Taf7l; a paralogue of Taf7) and TBP-related factor 2 (Trf2) are components of the core promoter complex required for gene/tissue-specific transcription of protein-coding genes by RNA polymerase II. Previous studies reported that Taf7l knockout (KO) mice exhibit structurally abnormal sperm, reduced sperm count, weakened motility, and compromised fertility. Here we find that continued backcrossing of Taf7l−/Y mice from N5 to N9 produced KO males that are essentially sterile. Genome-wide expression profiling by mRNA-sequencing analysis of wild-type (WT) and Taf7l−/Y (KO) testes revealed that Taf7l ablation impairs the expression of many postmeiotic spermatogenic-specific as well as metabolic genes. Importantly, histological analysis of testes revealed that Taf7l−/Y mice develop postmeiotic arrest at the first stage of spermiogenesis, phenotypically similar to Trf2−/− mice, but distinct from Taf4b−/− mice. Indeed, we find that Taf7l and Trf2 coregulate postmeiotic genes, but none of Taf4b-regulated germ stem cell genes in testes. Genome-wide ChIP-sequencing studies indicate that TAF7L binds to promoters of activated postmeiotic genes in testis. Moreover, biochemical studies show that TAF7L associates with TRF2 both in vitro and in testis, suggesting that TAF7L likely cooperates directly with TRF2 at promoters of a subset of postmeiotic genes to regulate spermiogenesis. Our findings thus provide a previously undescribed mechanism for cell-type–specific transcriptional control involving an interaction between a ”nonprototypic” core promoter recognition factor (Trf2) and an orphan TAF subunit (Taf7l) in mammalian testis-specific gene transcription.
Spermatogenesis is a cyclic process in which diploid spermatogonia differentiate into mature haploid spermatozoa. This process is mainly driven by two—pre- and postmeiotic—transcription waves that are tightly controlled by testis-specific transcription factors. During the premeiotic transcription phase, individual spermatogonia are committed to differentiating into primary spermatocytes that later undergo two meiotic divisions to generate haploid round spermatids connected by intercellular cytoplasmic bridges (1–3). During the postmeiotic transcription phase of spermiogenesis, haploid round spermatids are sculptured into the elongated shape of mature spermatozoa. These latter stages are accompanied by dramatic biochemical and morphological changes, including major remodeling of chromatin with protamines substituting for somatic histones to tightly pack DNA into the sperm nucleus.
Understanding the intricate mechanisms that control spermatogenesis has important implications for human health and reproduction. A key step in the regulation of spermatogenesis occurs at the level of transcription, starting with the use of distinct promoter elements (4) within uniquely reorganized chromatin (5) and driven by the action of several testis-specific transcription factors including cAMP-responsive element modulator (CREM) (6, 7) and the core promoter recognition factors required for global or gene-specific transcription such as testis-specific transcription factor IIA (TFIIAtau)/ALF (a paralogue of TFIIA) (8), TATA-binding protein (TBP)-associated factor 4b (Taf4b; a homolog of Taf4) (9), TBP-related factor 2 (Trf2) (10, 11), and Taf7l (12, 13). For example, mice bearing mutant or deficient CREM showed decreased postmeiotic gene expression and defective spermiogenesis (14). Mice deficient in Taf4b, a testis-specific homolog of Taf4, are initially normal but undergo progressive germ-cell loss and become infertile by 3 mo of age with seminiferous tubules devoid of germ cells. Taf4b depletion blunted the expression of spermatogonial stem cell genes, indicating a critical role in maintenance of spermatogonial stem cells (9). The core promoter recognition factor Trf2 is highly expressed in a finely regulated pattern in the mouse testis during spermatogenesis, and mice lacking Trf2 are viable but sterile due to a complete arrest of late spermiogenesis with largely normal spermatogonia and spermatocytes (15, 16). Taf7l, originally identified in spermatogonia, is a testis- and adipose-specific X-chromosome gene (17). This orphan Taf is expressed throughout male germ-cell differentiation, while its intracellular localization is dynamically regulated from cytoplasm in spermatogonia to nucleus in late spermatogenesis (13). Early studies of Taf7l-deficient mice showed reduced fertility, abnormal sperm structure, low sperm count, and weakened motility (12). Although the spermatogenic defects of various mouse lines mentioned above have been extensively studied, the molecular mechanisms controlling testis-specific gene expression programs remain poorly understood. It has also remained unclear what, if any, functional relationship may exist between these testis-specific transcription factors, and potential cross-talk between these various regulators has remained elusive (15).
Here we report that backcrossed Taf7l−/Y males from N5 to N9 leads to infertility and Taf7l−/Y testes show obvious deficiencies during spermiogenesis. The more severe infertility phenotypes observed in this study relative to previous reports suggest that additional backcrossing of Taf7l−/Y mice may have uncovered a fuller range of Taf7l functions in spermatogenesis. Perhaps equally importantly, recent studies of human oligozoospermia patients have found mutations in human Taf7l (18, 19), providing a potential link between the critical role of Taf7l in mouse spermatogenesis to human infertility. These unique findings give further impetus to more fully dissect the underlying molecular mechanisms by which Taf7l regulates spermatogenesis. In this report we explore in greater depth how Taf7l functions to regulate the differentiation of germ cells. We carried out genome-wide expression profiling and direct binding studies with TAF7L and identified many spermiogenesis-specific gene promoters targeted by TAF7L. Interestingly, Taf7l impairs spermatogenesis at a similar stage (spermiogenesis) to Trf2 and is translocated into the nucleus when Trf2 is highly expressed (13, 20). Moreover, we find that Taf7l regulates many known Trf2-targeted testis genes (11, 16), and biochemical studies reveal that TAF7L interacts with TRF2 in vitro and in testis by coimmunoprecipitation (co-IP) analysis. Together these data suggest that Taf7l might cooperate with Trf2 to control transcription in the postmeiotic stage of spermatogenesis, thus providing an important example of functional cross-talk between two atypical core promoter recognition factors operating coordinately to direct tissue-specific gene transcription.
Results
Taf7l Is Essential for Spermatogenesis.
Initially, four Taf7l heterozygous (Taf7l+/−) females and two Taf7l-null (Taf7l−/Y) males (N6) were obtained from the University of Pennsylvania, and mating cages were set up for all six mice with either WT males or females. Litters were obtained from all Taf7l+/− females mating with WT males, but no litter was born from the two Taf7l−/Y males mating with WT females for >1 y. Further matings were carried out with the offspring (Taf7l+/− or Taf7l−/Y), and again litters were only obtained from Taf7l+/− females mating with WT males, but none from Taf7l−/Y males mating with either WT or Taf7l+/− females. These observations indicated that N7–9 Taf7l−/Y males, after two to four additional rounds of backcrossings, might have become sterile.
To more carefully assess the fertility of Taf7l−/Y males, we carried out a comprehensive analysis documented in Fig. 1A. Out of the 15 matings with Taf7l−/Y males (N9) crossed with Taf7l+/− females, only 1 produced progeny (litter of six). Likewise, when Taf7l−/Y males crossed with WT females, only one litter with five progeny was produced. In both these cases, the single successful mating occurred with younger males (age 6–8 wk) with no further pup produced over 7 mo (Fig. 1A). By contrast, WT males mated with Taf7l+/− females were successful 73 times, producing a total of 450 progeny in 5 mo. Genotyping of the offspring by PCR analysis confirmed a distribution (Taf7l+/+, Taf7l+/−, Taf7l+/Y, and Taf7l−/Y) that follows the expected Mendelian ratio. These data confirm that the reproductive potential of Taf7l−/Y males is severely compromised (Fig. 1 B and C) and that Taf7l−/Y males are essentially infertile. This severe male sterility also made Taf7l−/− females effectively unavailable because Taf7l is an X-chromosome–linked gene and heterozygous Taf7l−/Y behave as Taf7l-null males, severely limiting our ability to assess the reproductive capacity of Taf7l−/− females.
Fig. 1.
Taf7l is essential for male reproduction abilities. (A) Progeny produced by 15 mating cages with WT males (Taf7l+/Y) and heterozygous Taf7l−/+ females, Taf7l−/Y males and Taf7l−/+ females, and Taf7l−/Y males and WT females (Taf7l+/+) for 5 mo. (B and C) Progeny numbers and litter times of Taf7l−/Y males (−/Y) relative to WT males (+/Y) of mating analysis in A. (D) Number of sperm in WT and Taf7l−/Y (KO) male testis. (E) Testis weight of WT and KO mice. (F) Serum testosterone levels of WT and KO mice. (G) Body weights were measured for WT and KO mice. D–G show results with 3-mo-old mice. Values in D–G represent the mean ± SEM of mice (n = 7–10). Asterisks denote statistically significant differences of KO compared with WT. *P < 0.05; **P < 0.01 (Student t test).
As part of our phenotyping, we also measured sperm count, testis weight, and serum testosterone levels as reported (Fig. 1 D and E) (12). Taf7l−/Y males also have slightly reduced testosterone levels, which may be related to our recent finding that Taf7l KO animals show defects in the synthesis of white adipocytes and cholesterol (Fig. 1F) (21). Otherwise, Taf7l−/Y males appear normal and healthy; they showed no apparent abnormalities in major organs, no obvious differences in body weight (Fig. 1G), and no detectable overall metabolic changes under normal feeding conditions.
Taf7l Ablation Disrupts Normal Gene Expressions in Testis.
To gain a better understanding of the more severe phenotypes observed, we compared the genome-wide expression profiles of 3-mo-old WT and Taf7l−/Y testes by mRNA sequencing (mRNA-seq). RNAs from six WT and six Taf7l−/Y (KO) testes were extracted and mixed separately to avoid individual differences; RNA-seq libraries were generated for both WT and KO samples and then analyzed by Illumina deep sequencing. The genome-wide expression data identified 726 genes down-regulated (by threefold or more) and 894 genes up-regulated in mouse testes lacking Taf7l (Fig. 2A). In good agreement with a previous study, the six genes identified as potential targets of Taf7l [Cpa6, Adc, Fascin (Fscn1), Sfmbt2, 4732473B16Rik, and D1Ertd622e] by microarray analysis (12) were also found to be reduced (approximately twofold) in Taf7l−/Y testes (Fig. 2D and Fig. S1). Gene Ontology analysis revealed that many genes implicated in spermatogenesis and metabolism were substantially down-regulated in the testis of Taf7l KO males. By contrast, the up-regulated genes include those involved in antimicrobial activity, growth regulation, metabolism, and immune system development, but not spermatogenesis (Fig. S2).
Fig. 2.
Taf7l depletion dramatically deregulates testis gene expression by RNA-seq analysis. (A) Global gene expression of testis in Taf7l−/Y (TAF7L KO) (horizontal axis) and WT (vertical axis) mice. Red dots represent genes down-regulated above threefold (726 genes); green dots represent genes up-regulated above threefold (894 genes); blue dots represent genes with low expression and genes with unaltered expression. (B) Relative expression levels of representative spermatogenic activators and markers. (C) Relative expression levels of representative genes important for sperm structure and motility. (D) Relative expression levels of genes involved in metabolism. (B–D) Expression level of genes in WT testis were arbitrarily assigned a value of 1; their corresponding expression levels in KO testis are expressed relative to this value.
Based on Gene Ontology, we have subdivided the genes down-regulated by the loss of Taf7l into three classes (Fig. 2). Note that some genes could just as well be classified within multiple subgroups (i.e., the Tssk3 gene that is involved in metabolism and sperm motility). One prominent group of genes dependent on Taf7l represents well-documented spermatogenic activators and markers such as AR, Zfx, Spz1, and Spem1 (22–26). A second group of Taf7l-regulated genes [Odf1, testis-specific serine kinase 6 (Tssk6), sperm motility kinase 2a/b (Smok2a/b), and Smok3a/b] have been implicated in sperm structure and motility during late stages of spermatogenesis (Fig. 2 B and C) (27–30). Additionally, a group of genes that have generally been classified as ones involved in metabolism are significantly down-regulated in the testis upon loss of Taf7l and likely reflect both metabolic and spermatogenesis functions. These genes include the Tssk3 or Tsks testis-specific kinases (31, 32), as well as Camk4, a protein kinase involved in phosphorylating protamines (33); Hk1 and GCK hexokinases involved in sperm glycolysis linked to motility (34); and Oaz3 (ornithine decarboxylase antizyme) that controls polyamines required for proper sperm formation (35, 36) (Fig. 2D). To confirm the RNA-seq results, we carried out quantitative RT-PCR (RT-qPCR) on a handful of genes selected from these three representative classes (Fig. S3), providing additional evidence that Taf7l regulates a subset of spermatogenic and metabolic genes in testis. These findings also provide some insight into the relationship between metabolism and reproduction that has been reported in the study of various animal models (37–40) linking dysregulated metabolism with sterility. Our genome-wide expression data thus point to the likely involvement of Taf7l in regulating both metabolic and spermatogenesis genes in testis as being at least partly responsible for the observed infertility.
TAF7L Binds to Promoters of Target Genes in Testis.
Given that hundreds of genes are down- or up-regulated by threefold or more in Taf7l−/Y testis, we next set out to determine whether TAF7L directly binds to the promoters and/or enhancers of those spermatogenic and metabolic genes in testis. To this end, we carried out ChIP-sequencing (ChIP-seq) mapping of TAF7L-binding sites in WT testes, using RNA polymerase II (Pol II) binding sites as positive controls to mark the actively transcribed genes and IgG as negative controls. First, mapped ChIP tags from deep sequencing using Bowtie were analyzed by intersecting MACS and Grizzly Peak algorithms to identify binding regions for TAF7L and Pol II (41, 42). This analysis identified 28,979 significant peaks for Pol II, 10,352 significant peaks for TAF7L (Fig. 3C), and no significant peaks for IgG control. Next, we analyzed the distances of the TAF7L binding peaks to transcription start sites (TSS) and found that 95% of TAF7L peaks are within 1 kb of a TSS. Colocalization analysis between TAF7L and Pol II peaks revealed that almost all of the TAF7L peaks overlapped with Pol II peaks (Fig. 3 A and B), suggesting that TAF7L largely associates with the promoters of actively transcribed genes in the testis. In contrast to the Taf7l-dependent genes in testis, our direct binding data revealed that nearly all of the genes up-regulated by the loss of Taf7l in the testis (Fig. S1) likely resulted from indirect secondary effects of Taf7l depletion. We found that many down-regulated spermatogenic and metabolic genes identified by RNA-seq analysis bear direct TAF7L binding sites at or near their promoters. A few representative gene loci such as nuclear receptor subfamily 6, group A, member 1 (Nr6a1), Tssk3, and serine/threonine-protein kinase 1 (Sgk1) clearly show that TAF7L colocalizes with Pol II at promoter sites (Fig. 3 D–F). ChIP-qPCR analysis using WT and Taf7l-null testes confirmed that Pol II binding becomes dramatically diminished in Taf7l-null testes at target promoters (Nr6a1, testis-specific serine kinase 3 (Tssk3), Sgk1, protamine 1/2 (Prm1/2), and Fscn3), but not at a control actin intron (Fig. S4), suggesting that the presence of TAF7L is indeed critical for the proper expression of target genes in testis. Together these data suggest that TAF7L directly binds to gene promoters and that this binding is required for the expression of spermatogenic and metabolic genes in testis.
Fig. 3.
ChIP-seq analysis identifies TAF7L and Pol II binding sites in testis. (A and B) Percentage of TAF7L (A) and Pol II (B) binding peaks vs. the distance of the peaks to transcription start sites (TSS) on genome-wide scale in testis. (C) Overlapping between TAF7L and Pol II binding peaks in testis. (D–F) Read accumulation of Pol II and TAF7L were shown on the Nr6a1 (D), Tssk3 (E), and Sgk1 (F) gene loci. Vertical axis is 0–500 reads for each factor; colocalized peaks are marked with red solid boxes. IgG served as negative control for ChIP-seq analysis.
Taf7l−/Y Resembles Trf2−/− Mice and Targets Similar Postmeiotic Genes.
Having found that TAF7L binds to a specific subset of promoter regions of genes involved in spermatogenesis, we next set out to assess the possibility that Taf7l associates with other testis-specific core promoter recognition factors such as Taf4b and/or Trf2 to work combinatorially to target genes directing spermatogenesis. First, we examined the spermatogenic defects of Taf7l−/Y mice relative to Taf4b−/− and Trf2−/− mice. H&E staining of testes from 3-mo-old WT and Taf7l−/Y (KO) mice revealed dramatically decreased elongated spermatids (Fig. 4A). The phenotype was highly reminiscent of Trf2−/− animals, but quite distinct from Taf4b−/− mice that showed defects in the maintenance of germ stem cells (9, 16). Next, we compared the target genes regulated by Taf7l, Taf4b, and Trf2 in testis identified by RNA-seq (9, 16). As shown in Fig. S5, Taf7l and Taf4b appear to regulate largely nonoverlapping sets of genes expressed in the testis. For example, GDNF, stimulated by retinoic acid 8 (Stra8), Stag3, and disrupted meiotic cDNA 1 (Dmc1) are robustly down-regulated by the loss of Taf4b but are slightly increased in Taf7l−/Y testes, suggesting that Taf7l is not required for maintenance of male germ stem cells. Instead, it appears that loss of Taf7l blocks spermatogenesis at a postmeiotic stage and that Taf7l−/Y testes may actually accumulate more stem and meiotic cells (Fig. 4 A and B).
Fig. 4.
Taf7l depletion blocks spermiogenesis and impairs postmeiotic gene expression in a similar way to Trf2−/− in testis. (A) H&E staining on 3-mo-old WT and KO testis at magnification ×200 (I–III) and magnification ×600 (IV–VI). (B) Expression of Trf2-regulated postmeiotic and control genes in Taf7l−/Y testis was compared with WT testis from mRNA-seq analysis. Expression levels of genes in WT testis were arbitrarily assigned a value of 1, and their corresponding expression levels in KO testis are expressed relative to this value. NS, not significant.
We also examined the influence of Taf7l depletion on the expression of TFIID subunits by RNA-seq and RT-qPCR analysis and found that, except for Taf7l itself, all of the other TAFs were largely unaltered or slightly up-regulated (Fig. S6). These findings suggest that Taf7l is a “nonprototypical” testis-specific TAF subunit whose absence does not influence overall levels of TFIID. Strikingly, however, loss of Taf7l—like the loss of Trf2—down-regulates the same subset of genes directing spermatogenesis. For example, Taf7l does not influence the transcription of premeiotic genes such as Proacrosin, H1.2, and Hsp70-2 but dramatically down-regulates Trf2-regulated postmeiotic genes such as Gapdhs, transition proteins 1/2(Tnp1/2), Prm1/2, Fscn1/2, and Smcp (Fig. 4B). This requirement for both Taf7l and Trf2 for proper expression of postmeiotic genes is also consistent with the spermiogenesis defects observed in both Taf7l−/Y and Trf2−/− testes (Fig. 4A). These findings suggest that Taf7l likely operates together with Trf2, but not Taf4b, to control postmeiotic genes, as deduced from our RNA-seq (Fig. 4B and Fig. S5), RT-qPCR (Fig. S7), and ChIP-seq analysis (Fig. 3 and Fig. S4). We found, for instance, that Taf7l binds efficiently to the TSS regions of Trf2-regulated gene Gapdh, but not at the promoter regions of Taf4b-regulated genes such as Stra8 and Dmc1 (Fig. S8).
We also assessed the expression levels and nuclear localization patterns of Taf4b, Trf2, and Taf7l during spermatogenesis to probe potential direct crosstalk between these factors based on previous studies. Taf4b is highly expressed in gonocytes, nuclei of spermatogonia, and spermatids, but not at other stages of spermatogenesis or in somatic cells (9). By contrast, Taf7l is expressed in germ cells, not testis somatic cells, during most stages of spermatogenesis, but it remains cytoplasmically localized in spermatogonia, and its expression becomes silenced at meiotic stages due to sex-chromosome inactivation (MSCI). Taf7l transcription becomes reactivated and translocated into nucleus of spermatids (13, 17) at a stage when Trf2 is also specifically highly expressed (11, 20). Together, losses of Taf7l and Trf2 exhibit a similar spermiogenesis deficiency phenotype; they appear in the nuclei of postmeiotic spermatids at similar times, bind to core promoter elements in an overlapping subset of genes, and regulate their expression in the testis. These findings raise the possibility that Taf7l and Trf2 may actually work together in regulating spermatogenesis.
Taf7l and Trf2 Coregulate a Subset of Postmeiotic Genes.
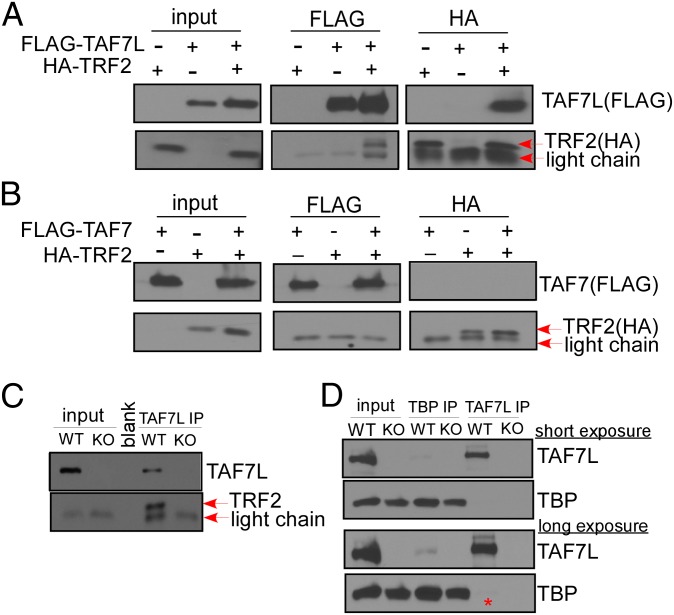
To further explore the possibility of interaction between TAF7L and TRF2 in regulating postmeiotic gene expression during spermatogenesis, we used FLAG-tagged TAF7L or TAF7 to co-IP with HA-tagged TRF2 coexpressed in 293T cells. These co-IP experiments showed that TAF7L could efficiently pull down TRF2 and vice versa (Fig. 5A); by contrast, TAF7 (a paralogue of TAF7L) was unable to coimmunoprecipitate TRF2 (Fig. 5B). To confirm that the association between TAF7L and TRF2 is directed by protein:protein interactions and not mediated via indirect DNA/chromatin interactions, we included benzonase treatment in our co-IP assays. Eliminating DNA in these co-IP experiments did not alter the binding interactions we observed between TAF7L and TRF2. Next, we used affinity-purified TAF7L antibody to coimmunoprecipitate endogenous TRF2 in WT and Taf7l−/Y testis. The results showed that TAF7L could pull down endogenous TRF2 from WT testes, but not from Taf7l−/Y testes (Fig. 5C), suggesting that TAF7L is associated with TRF2 in testes. The association between TAF7L and TRF2 in vitro and in vivo prompted us to explore further the relationship between TAF7L and TRF2 or TBP in testes. A previous study reported that TBP could be an interacting partner of TAF7L in testes (12). For these experiments, we used TAF7L and TBP antibodies to coimmunoprecipitate each other in WT and Taf7l−/Y testes and found that only a small portion of TAF7L weakly associated with TBP in mouse testes (Fig. 5D). A direct comparison of TRF2 and TBP signals coimmunoprecipitated by TAF7L (Fig. 5 C and D) confirmed that TAF7L associates more efficiently with TRF2 than with TBP in mouse testes when all three proteins are present. These studies suggest that two atypical testis-specific core promoter recognition factors, TAF7L and TRF2, likely work in concert to regulate a subset of postmeiotic genes required for spermiogenesis. Ablation of either factor results in a similar blockade of spermiogenesis, leading to male infertility.
Fig. 5.
TAF7L associates with TRF2 both in vitro and in vivo. (A) FLAG–TAF7L and HA–TRF2 were overexpressed in 293T cells, and IPs were performed on both FLAG and HA antibodies, followed by Western blotting analysis with FLAG and HA antibodies. (B) The same procedures were performed on FLAG–TAF7 and HA–TRF2. (C) IPs were performed on WT and Taf7l−/Y testis lysates with TAF7L antibody and followed by Western blotting with TAF7L and TRF2 antibodies. (D) IPs were performed on WT and Taf7l−/Y testis lysates with either TAF7L or TBP antibody, followed by Western blotting with TAF7L and TBP antibodies. Both images are from the same Western blots with short (Upper) and long (Lower) exposure time. Red star marks the TBP signal from TAF7L IP.
Discussion
Cell-type–specific transcription is a key driver of tissue and organ formation during embryonic development. These complex expression networks are controlled by tissue-specific enhancer/promoter binding factors as well as core promoter recognition factors including Tafs, mediators, and TRFs. In several previous studies, Taf7l and Trf2 have separately been found to function as testis-specific transcription factors. In this study, we provide unique evidence suggesting that these two atypical core promoter recognition factors work together to drive testis-specific gene expression.
Our model proposes that during the postmeiotic wave of transcription that occurs at the spermatogenesis–spermiogenesis transition, Taf7l is reactivated from MSCI and translocated into the nuclei of pachytene/round spermatids where Trf2 is highly expressed. We postulate that a testis-specific transcription preinitiation complex containing both TAF7L and TRF2 is formed and targeted to a subset of “TATA-less” promoters to regulate postmeiotic Trf2-dependent gene expression (43–45) (Fig. S9). At the same time, prototypic TBP-containing core promoter recognition complexes can operate to direct TBP-dependent housekeeping genes and possibly some testes-specific genes. TAF7L, TRF2, and TBP all seem to be required for spermatogenesis, and each may provide some nonredundant testis-selective transcription function. Previous studies found that TRF2 can associate with multiple proteins to form a complex of >500 kDa (10). It will be interesting to see whether there is a TRF2–TAF complex of similar size in testes and eventually identify the other associated factors to more fully dissect the molecular mechanisms of Taf7l- and Trf2-driven testis-specific transcription. It will also be interesting to test whether, beyond its roles in testis-specific transcription control, Taf7l is required for maintaining chromatin architecture in round spermatids.
Given that Taf7l is highly expressed in germ cells, but not somatic cells in testis, we analyzed the global influence of Taf7l depletion in the whole testis. As a result, both up- and down-regulated genes found in this study by RNA-seq or RT-qPCR were probably the direct result of Taf7l KO. It is also likely that diminished elongated spermatids observed are due to blockade of spermiogenesis upon loss of Taf7l. Although our ChIP-seq data supported a role for TAF7L in regulating testis-specific genes by direct binding to their promoters, future studies with stage-specific fractionated germ cells will be helpful to elucidate a more detailed molecular mechanism.
We recently reported that TAF7L associates with the adipocyte-specific transcription factor PPARγ during adipogenesis (21). Here in the context of testis-specific transcription, TAF7L teams up with TRF2, another testis-specific core promoter recognition factor, to direct male sperm formation. The association of an orphan TAF and a TRF is reminiscent of the TRF3/TAF3 scenario reported for myogenesis (46). These studies, taken in aggregate, suggest that various cell lineages take advantage of diversified core promoter factors in a combinatorial fashion to regulate tissue-specific programs of transcription.
Because of adverse side effects of female contraceptive medicines and the lack of male ones, an effective and safe therapeutic target has become a focus in the male germ-cell research field. Thus far, hundreds of genes have been found to influence spermatogenesis when mutated or depleted, including core promoter recognition factors Trf2 and Taf4b, leptin ob and leptin receptor db genes, and other genes such as Daz and Ddx4 (38, 47–49). However, most of these genes have relatively high expression levels and function in tissue or organs other than testes. Therefore, their depletion or inhibition often results in side effects and unacceptable complications that can range from minor to severe. For example, depletion of the ob/db gene results in obesity and diabetes (38). By contrast, Taf7l is more highly expressed in testis than all other tissues that were sampled, including adult white adipose tissue, and, not surprisingly, depletion of Taf7l causes >98% male infertility in mice. Most importantly, ablation of Taf7l leaves most of the male germ stem cells intact, suggesting that the effect of contraceptives targeting Taf7l will likely be reversible upon removal of the treatment. It is also interesting to note that Taf7l is an X-chromosome–linked gene; males carry only a single copy, and therefore the chances of introducing random mutations in Taf7l is twice as high as autosomal regulatory genes such as Taf4b and Trf2. These characteristics together may explain why multiple Taf7l mutations have been found in human infertile patients with oligozoospermia (low concentration of sperm). Our mouse studies thus may also help identify a potential candidate target gene affecting human male infertility.
Materials and Methods
Vectors and Plasmids.
Taf7l, Taf7, and Trf2 full-length cDNAs were cloned into the pCS2+ vector with either HA or FLAG tag at their N terminus.
RNA Isolation and Real-Time PCR Analysis.
A complete description of RNA isolation and real-time PCR analysis is provided in SI Materials and Methods.
Western Blot Analysis and IP.
A complete description of Western blot analysis and IP is provided in SI Materials and Methods.
Animal Care and Permission.
Taf7l-KO mice were generated previously (12). All animal experiments were performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals from the National Institutes of Health (50). All of the animals were handled according to approved animal use protocols (no. R007) by the Animal Care and Use Committee of the University of California, Berkeley.
Animals Reproduction Capability Analysis.
Fifteen mating cages of three mating conditions were used as follows: WT males (Taf7l+/Y) with heterozygous Taf7l+/− females and Taf7l-null (Taf7l−/Y) males with either Taf7l+/− or WT females were set up for over 7 mo. Males were started at 4 wk of age, and females were started at 6 wk of age in all of the mating cages, and mating cage numbers, total progeny produced, and total litter production times were recorded for 5 mo.
ChIP, ChIP Library Preparation, and Deep Sequencing (ChIP-seq).
WT and Taf7l KO testis tissue from 10 mice at 3 mo of age was homogenized, followed by fixation with 1% (vol/vol) formaldehyde for 15 min at room temperature, 0.125 M glycine to stop the cross-linking for an additional 5 min, and collection of tissue by spinning down. The following steps are described in SI Materials and Methods.
Mapping Sequencing Reads and Peak Calling Methods.
A complete description of sequencing reads mapping and peak calling methods is provided in SI Materials and Methods.
mRNA-seq Library Preparation, Deep Sequencing, and Data Analysis.
A complete description of mRNA-seq library preparation, deep sequencing, and data analysis is provided in SI Materials and Methods.
Data Availability.
Raw sequencing reads are available from the National Center for Biotechnology Information's GEO database (www.ncbi.nlm.nih.gov/geo/) under accession no. GSE50807.
Additional materials and methods are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank R. Steven and D. Schichness for help with H&E staining techniques; L. P. Jennings and A. Howell for help with animal care and blood sample extraction; S. Gerald and M. Jurczak for help with metabolic studies; and all R.T. laboratory members for valuable scientific discussions and suggestions. H.Z. is a research associate of the Howard Hughes Medical Institute. T.K. is a member of the Israeli Center of Excellence (I-CORE) for Gene Regulation in Complex Human Diseases (no. 41/11). R.T. is an investigator of the Howard Hughes Medical Institute. This work was supported by the Howard Hughes Medical Institute and the National Institutes of Health (R.T.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE50807).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1317034110/-/DCSupplemental.
References
- 1.Dym M, Fawcett DW. Further observations on the numbers of spermatogonia, spermatocytes, and spermatids connected by intercellular bridges in the mammalian testis. Biol Reprod. 1971;4(2):195–215. doi: 10.1093/biolreprod/4.2.195. [DOI] [PubMed] [Google Scholar]
- 2.Fawcett DW. A comparative view of sperm ultrastructure. Biol Reprod. 1970;2(Suppl 2) 2, 90–127. [PubMed] [Google Scholar]
- 3.Greenbaum MP, Iwamori T, Buchold GM, Matzuk MM. Germ cell intercellular bridges. Cold Spring Harb Perspect Biol. 2011;3(8):a005850. doi: 10.1101/cshperspect.a005850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kimmins S, Kotaja N, Davidson I, Sassone-Corsi P. Testis-specific transcription mechanisms promoting male germ-cell differentiation. Reproduction. 2004;128(1):5–12. doi: 10.1530/rep.1.00170. [DOI] [PubMed] [Google Scholar]
- 5.Sassone-Corsi P. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science. 2002;296(5576):2176–2178. doi: 10.1126/science.1070963. [DOI] [PubMed] [Google Scholar]
- 6.Lamas M, et al. CREM: A master-switch in the transcriptional response to cAMP. Philos Trans R Soc Lond B Biol Sci. 1996;351(1339):561–567. doi: 10.1098/rstb.1996.0055. [DOI] [PubMed] [Google Scholar]
- 7.Nantel F, Sassone-Corsi P. CREM: A transcriptional master switch during the spermatogenesis differentiation program. Front Biosci. 1996;1:d266–d269. doi: 10.2741/a131. [DOI] [PubMed] [Google Scholar]
- 8.Wang PJ, Page DC. Functional substitution for TAF(II)250 by a retroposed homolog that is expressed in human spermatogenesis. Hum Mol Genet. 2002;11(19):2341–2346. doi: 10.1093/hmg/11.19.2341. [DOI] [PubMed] [Google Scholar]
- 9.Falender AE, et al. Maintenance of spermatogenesis requires TAF4b, a gonad-specific subunit of TFIID. Genes Dev. 2005;19(7):794–803. doi: 10.1101/gad.1290105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rabenstein MD, Zhou S, Lis JT, Tjian R. TATA box-binding protein (TBP)-related factor 2 (TRF2), a third member of the TBP family. Proc Natl Acad Sci USA. 1999;96(9):4791–4796. doi: 10.1073/pnas.96.9.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang D, Penttila TL, Morris PL, Roeder RG. Cell- and stage-specific high-level expression of TBP-related factor 2 (TRF2) during mouse spermatogenesis. Mech Dev. 2001;106(1-2):203–205. doi: 10.1016/s0925-4773(01)00439-7. [DOI] [PubMed] [Google Scholar]
- 12.Cheng Y, et al. Abnormal sperm in mice lacking the Taf7l gene. Mol Cell Biol. 2007;27(7):2582–2589. doi: 10.1128/MCB.01722-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pointud JC, et al. The intracellular localisation of TAF7L, a paralogue of transcription factor TFIID subunit TAF7, is developmentally regulated during male germ-cell differentiation. J Cell Sci. 2003;116(Pt 9):1847–1858. doi: 10.1242/jcs.00391. [DOI] [PubMed] [Google Scholar]
- 14.Nantel F, et al. Spermiogenesis deficiency and germ-cell apoptosis in CREM-mutant mice. Nature. 1996;380(6570):159–162. doi: 10.1038/380159a0. [DOI] [PubMed] [Google Scholar]
- 15.Goodrich JA, Tjian R. Unexpected roles for core promoter recognition factors in cell-type-specific transcription and gene regulation. Nat Rev Genet. 2010;11(8):549–558. doi: 10.1038/nrg2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang D, Penttila TL, Morris PL, Teichmann M, Roeder RG. Spermiogenesis deficiency in mice lacking the Trf2 gene. Science. 2001;292(5519):1153–1155. doi: 10.1126/science.1059188. [DOI] [PubMed] [Google Scholar]
- 17.Wang PJ, Page DC, McCarrey JR. Differential expression of sex-linked and autosomal germ-cell-specific genes during spermatogenesis in the mouse. Hum Mol Genet. 2005;14(19):2911–2918. doi: 10.1093/hmg/ddi322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akinloye O, Gromoll J, Callies C, Nieschlag E, Simoni M. Mutation analysis of the X-chromosome linked, testis-specific TAF7L gene in spermatogenic failure. Andrologia. 2007;39(5):190–195. doi: 10.1111/j.1439-0272.2007.00789.x. [DOI] [PubMed] [Google Scholar]
- 19.Sedivá A, et al. Contiguous X-chromosome deletion syndrome encompassing the BTK, TIMM8A, TAF7L, and DRP2 genes. J Clin Immunol. 2007;27(6):640–646. doi: 10.1007/s10875-007-9123-x. [DOI] [PubMed] [Google Scholar]
- 20.Martianov I, et al. Distinct functions of TBP and TLF/TRF2 during spermatogenesis: Requirement of TLF for heterochromatic chromocenter formation in haploid round spermatids. Development. 2002;129(4):945–955. doi: 10.1242/dev.129.4.945. [DOI] [PubMed] [Google Scholar]
- 21.Zhou H, et al. Dual functions of TAF7L in adipocyte differentiation. Elife. 2013;2:e00170. doi: 10.7554/eLife.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bao J, Zhang J, Zheng H, Xu C, Yan W. UBQLN1 interacts with SPEM1 and participates in spermiogenesis. Mol Cell Endocrinol. 2010;327(1-2):89–97. doi: 10.1016/j.mce.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu SH, Hsieh-Li HM, Li H. Dysfunctional spermatogenesis in transgenic mice overexpressing bHLH-Zip transcription factor, Spz1. Exp Cell Res. 2004;294(1):185–198. doi: 10.1016/j.yexcr.2003.10.029. [DOI] [PubMed] [Google Scholar]
- 24.Luoh SW, et al. Zfx mutation results in small animal size and reduced germ cell number in male and female mice. Development. 1997;124(11):2275–2284. doi: 10.1242/dev.124.11.2275. [DOI] [PubMed] [Google Scholar]
- 25.Wikström AM, Hoei-Hansen CE, Dunkel L, Rajpert-De Meyts E. Immunoexpression of androgen receptor and nine markers of maturation in the testes of adolescent boys with Klinefelter syndrome: Evidence for degeneration of germ cells at the onset of meiosis. J Clin Endocrinol Metab. 2007;92(2):714–719. doi: 10.1210/jc.2006-1892. [DOI] [PubMed] [Google Scholar]
- 26.Zhou X, Kudo A, Kawakami H, Hirano H. Immunohistochemical localization of androgen receptor in mouse testicular germ cells during fetal and postnatal development. Anat Rec. 1996;245(3):509–518. doi: 10.1002/(SICI)1097-0185(199607)245:3<509::AID-AR7>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 27.Yang K, et al. The small heat shock protein ODF1/HSPB10 is essential for tight linkage of sperm head to tail and male fertility in mice. Mol Cell Biol. 2012;32(1):216–225. doi: 10.1128/MCB.06158-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, et al. Expression and localization of five members of the testis-specific serine kinase (Tssk) family in mouse and human sperm and testis. Mol Hum Reprod. 2011;17(1):42–56. doi: 10.1093/molehr/gaq071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaudhry PS, Creagh S, Yu N, Brokaw CJ. Multiple protein kinase activities required for activation of sperm flagellar motility. Cell Motil Cytoskeleton. 1995;32(1):65–79. doi: 10.1002/cm.970320108. [DOI] [PubMed] [Google Scholar]
- 30.Chen J, et al. [Differential expression of ODF1 in human ejaculated spermatozoa and its clinical significance] Zhonghua Nan Ke Xue. 2009;15(10):891–894. [PubMed] [Google Scholar]
- 31.Hao Z, et al. Expression analysis of the human testis-specific serine/threonine kinase (TSSK) homologues. A TSSK member is present in the equatorial segment of human sperm. Mol Hum Reprod. 2004;10(6):433–444. doi: 10.1093/molehr/gah052. [DOI] [PubMed] [Google Scholar]
- 32.Xu B, et al. Validation of a testis specific serine/threonine kinase [TSSK] family and the substrate of TSSK1 & 2, TSKS, as contraceptive targets. Soc Reprod Fertil Suppl. 2007;63:87–101. [PubMed] [Google Scholar]
- 33.Wu JY, et al. Spermiogenesis and exchange of basic nuclear proteins are impaired in male germ cells lacking Camk4. Nat Genet. 2000;25(4):448–452. doi: 10.1038/78153. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura N, Shibata H, O’Brien DA, Mori C, Eddy EM. Spermatogenic cell-specific type 1 hexokinase is the predominant hexokinase in sperm. Mol Reprod Dev. 2008;75(4):632–640. doi: 10.1002/mrd.20791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ike A, Yamada S, Tanaka H, Nishimune Y, Nozaki M. Structure and promoter activity of the gene encoding ornithine decarboxylase antizyme expressed exclusively in haploid germ cells in testis (OAZt/Oaz3) Gene. 2002;298(2):183–193. doi: 10.1016/s0378-1119(02)00978-2. [DOI] [PubMed] [Google Scholar]
- 36.Tokuhiro K, et al. OAZ-t/OAZ3 is essential for rigid connection of sperm tails to heads in mouse. PLoS Genet. 2009;5(11):e1000712. doi: 10.1371/journal.pgen.1000712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Luca C, et al. Complete rescue of obesity, diabetes, and infertility in db/db mice by neuron-specific LEPR-B transgenes. J Clin Invest. 2005;115(12):3484–3493. doi: 10.1172/JCI24059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mounzih K, Lu R, Chehab FF. Leptin treatment rescues the sterility of genetically obese ob/ob males. Endocrinology. 1997;138(3):1190–1193. doi: 10.1210/endo.138.3.5024. [DOI] [PubMed] [Google Scholar]
- 39.Hashmi S, et al. Partner in fat metabolism: Role of KLFs in fat burning and reproductive behavior. J Biotechnol. 2011;1(2):59–72. doi: 10.1007/s13205-011-0016-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vijayaraghavan S, et al. Sperm motility development in the epididymis is associated with decreased glycogen synthase kinase-3 and protein phosphatase 1 activity. Biol Reprod. 1996;54(3):709–718. doi: 10.1095/biolreprod54.3.709. [DOI] [PubMed] [Google Scholar]
- 41.Harrison MM, Li XY, Kaplan T, Botchan MR, Eisen MB. Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet. 2011;7(10):e1002266. doi: 10.1371/journal.pgen.1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohbayashi T, et al. Vertebrate TBP-like protein (TLP/TRF2/TLF) stimulates TATA-less terminal deoxynucleotidyl transferase promoters in a transient reporter assay, and TFIIA-binding capacity of TLP is required for this function. Nucleic Acids Res. 2003;31(8):2127–2133. doi: 10.1093/nar/gkg315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Catena R, et al. Proteolytic cleavage of ALF into alpha- and beta-subunits that form homologous and heterologous complexes with somatic TFIIA and TRF2 in male germ cells. FEBS Lett. 2005;579(16):3401–3410. doi: 10.1016/j.febslet.2005.04.083. [DOI] [PubMed] [Google Scholar]
- 45.Teichmann M, et al. Human TATA-binding protein-related factor-2 (hTRF2) stably associates with hTFIIA in HeLa cells. Proc Natl Acad Sci USA. 1999;96(24):13720–13725. doi: 10.1073/pnas.96.24.13720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deato MD, Tjian R. Switching of the core transcription machinery during myogenesis. Genes Dev. 2007;21(17):2137–2149. doi: 10.1101/gad.1583407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matzuk MM, Lamb DJ. The biology of infertility: research advances and clinical challenges. Nat Med. 2008;14(11):1197–1213. doi: 10.1038/nm.f.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Menke DB, Mutter GL, Page DC. Expression of DAZ, an azoospermia factor candidate, in human spermatogonia. Am J Hum Genet. 1997;60(1):237–241. [PMC free article] [PubMed] [Google Scholar]
- 49.Hickford DE, Frankenberg S, Pask AJ, Shaw G, Renfree MB. DDX4 (VASA) is conserved in germ cell development in marsupials and monotremes. Biol Reprod. 2011;85(4):733–743. doi: 10.1095/biolreprod.111.091629. [DOI] [PubMed] [Google Scholar]
- 50. Committee on Care and Use of Laboratory Animals (1985) Guide for the Care and Use of Laboratory Animals (Natl Inst Health, Bethesda), DHHS Publ No (NIH) 85-23.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing reads are available from the National Center for Biotechnology Information's GEO database (www.ncbi.nlm.nih.gov/geo/) under accession no. GSE50807.
Additional materials and methods are described in SI Materials and Methods.