Significance
More than a million ribonucleotides may be incorporated into the mammalian nuclear genome during each round of DNA replication. When these ribonucleotides are not removed, they persist in the DNA template used for the next round of replication. Here we show that replicases stall when attempting to bypass ribonucleotides in DNA templates, with stalling increasing as the number of consecutive ribonucleotides increases from one to four. Structural analysis reveals that stalling is associated with displacement of a conserved tyrosine residue that is important for template strand interactions and with an unpreferred C2´-endo conformation for the ribose. Replication fork stalling during ribonucleotide bypass is likely to be relevant to both negative and positive consequences of ribonucleotides in DNA.
Keywords: translesion synthesis, replication stalling, DNA replication
Abstract
Ribonucleotides are frequently incorporated into DNA during replication, they are normally removed, and failure to remove them results in replication stress. This stress correlates with DNA polymerase (Pol) stalling during bypass of ribonucleotides in DNA templates. Here we demonstrate that stalling by yeast replicative Pols δ and ε increases as the number of consecutive template ribonucleotides increases from one to four. The homologous bacteriophage RB69 Pol also stalls during ribonucleotide bypass, with a pattern most similar to that of Pol ε. Crystal structures of an exonuclease-deficient variant of RB69 Pol corresponding to multiple steps in single ribonucleotide bypass reveal that increased stalling is associated with displacement of Tyr391 and an unpreferred C2´-endo conformation for the ribose. Even less efficient bypass of two consecutive ribonucleotides in DNA correlates with similar movements of Tyr391 and displacement of one of the ribonucleotides along with the primer-strand DNA backbone. These structure–function studies have implications for cellular signaling by ribonucleotides, and they may be relevant to replication stress in cells defective in ribonucleotide excision repair, including humans suffering from autoimmune disease associated with RNase H2 defects.
Replication of the eukaryotic nuclear genome initiates when RNA primase synthesizes RNA primers of about 10 nucleotides (1). Because this occurs at multiple replication origins and at ∼200-bp intervals on the lagging strand template, about 5% of the genome is initially synthesized as chains of consecutive ribonucleotides. These ribonucleotides are subsequently removed during Okazaki fragment maturation by the combined action of ribonucleases (RNases) H (2) and flap endonucleases (3). Ribonucleotides are also incorporated into DNA by DNA polymerases (Pol) α, δ, and ε, because they discriminate against ribonucleoside triphosphates (rNTPs) efficiently but imperfectly (4) and because cellular rNTP concentrations are much higher than dNTP concentrations (4). As a consequence, large numbers of ribonucleotides are incorporated during replication, and are present in the genomes of cells defective in the repair enzymes that initiate their removal, RNase H2 (5-9) and topoisomerase 1 (10).
Ribonucleotides in DNA are a dual-edged sword, in that they have both beneficial and deleterious consequences. On the beneficial side, two consecutive ribonucleotides in the genome are signals for mating type switching in Schizosaccharomyces pombe (11). In addition, recent evidence suggests that RNase H2-dependent processing of ribonucleotides incorporated into the Saccharomyces cerevisiae genome by Pol ε, the primary leading strand replicase, generates a signal that can direct mismatch repair (MMR) to correct replication errors in the nascent leading strand (9, 12). Other possible beneficial signaling roles for ribonucleotides have also been considered (4, 13).
On the deleterious edge of the sword, the 2′-oxygen on a ribose sugar in DNA can attack the backbone and render DNA chemically unstable. Yeast strains defective in RNase H2-dependent ribonucleotide excision repair (RER) (5, 14) exhibit several characteristics of replicative stress, including strongly elevated rates for deleting 2–5 bp from repetitive DNA sequences (5, 15), events that are initiated by topoisomerase 1 cleavage of a ribonucleotide in DNA (10, 16). Yeast strains defective in RNase H2 and RNase H1 progress slowly through S phase, accumulate ubiquitylated proliferating cell nuclear antigen (PCNA), and are sensitive to treatment with hydroxyurea (17). Moreover, their survival in the presence of hydroxyurea partly depends on MMS2-dependent template switching and on REV3, which encodes the catalytic subunit of the translesion synthesis (TLS) enzyme Pol ζ. When ribonucleotide incorporation during leading strand replication is increased by a M644G substitution in the Pol ε active site, a defect in RNase H2 results in elevated deletion mutagenesis, elevated dNTP pools, slow growth and activation of the S-phase checkpoint (5, 10, 18), and concomitant deletion of the RNH1 gene encoding RNases H1 is lethal (17). In mice, knocking out any of the genes encoding the three subunits of RNase H2 is embryonic lethal (7, 19). RNase H2 null embryos grow slowly due to reduced cell proliferation and exhibit genome instability and a p53-dependent DNA damage response. Fibroblasts from these embryos contain more than a million single and/or di-ribonucleotides in their genomes and elevated numbers of strand breaks, γ-H2A histone family, member X foci, micronuclei, and chromosomal aberrations. In humans, mutations in the genes encoding RNase H2 are associated with Aicardi-Goutières syndrome, a rare neuroinflammatory condition resembling congenital viral infection (20).
These phenotypes of RNase-deficient cells are characteristic of stress that could arise from difficulty in replicating DNA templates containing unrepaired ribonucleotides. This idea is consistent with knowledge that replicases require normal DNA helix geometry to achieve efficient and accurate DNA synthesis, and with crystallographic and NMR studies (21–24) showing that ribonucleotides in DNA alter helix parameters. Recent studies have shown that Pols δ and ε have difficulty bypassing ribonucleotides in DNA templates (4), whereas Pol ζ does not (17). The probability that Pol ε will pause during single ribonucleotide bypass increases after dNTP insertion opposite the ribonucleotide and for several additional insertions opposite deoxynucleotides (5, 25).
In this study, we quantify stalling by yeast replicative Pols δ and ε as the number of consecutive ribonucleotides in the DNA template increases from one to four. We show that stalling increases as the number of consecutive ribonucleotides in the DNA template increases, with Pol δ being more efficient at ribonucleotide bypass than Pol ε. We then examine the structural basis for difficulty in ribonucleotide bypass using a homologous B family replicase, bacteriophage RB69 DNA polymerase, as a surrogate that is highly amenable to structural studies (26, 27). To promote the structural analysis, we used a variant of RB69 Pol containing a phenylalanine substituted for Leu415 (28). Leu415 is adjacent to invariant Tyr416, which interacts with the sugar of the incoming dNTP and has an important role in preventing rNTP incorporation (29). An initial crystal structure of L415F RB69 Pol with correctly base-paired dTTP opposite template dA (28) revealed that the phenylalanine ring is accommodated within a cavity present in the WT polymerase without steric clash or major change in active site geometry, consistent with retention of high catalytic efficiency for correct incorporation. Moreover, L415F RB69 Pol can also bypass 8-oxo-guanine more efficiently than can WT RB69 Pol (28). These bypass results were encouraging because the yeast replicases bypass single ribonucleotides with efficiencies somewhat similar to those for bypass of 8-oxo-guanine, and L415F RB69 Pol may therefore facilitate crystallization of ternary complexes with ribonucleotides in the DNA template (25). We show that WT and L415F RB69 Pol also stall during ribonucleotide bypass, to a degree most closely resembling stalling by Pol ε. We describe seven unique crystal structures relevant to L415F RB69 Pol bypass of one or two ribonucleotides. The data are discussed in relation to the consequences of ribonucleotides in the genomes of cells defective in their removal.
Results
Measuring Ribonucleotide Bypass Parameters.
All bypass reactions contain excess primer template over polymerase, such that DNA products largely reflect one cycle of DNA synthesis. The primer template excess was empirically demonstrated by the fact that the probability of termination of synthesis at each position remained constant over the time course of the reaction (30). This approach allows direct comparisons of relative bypass efficiencies and site-specific termination probabilities among all substrates and enzymes examined.
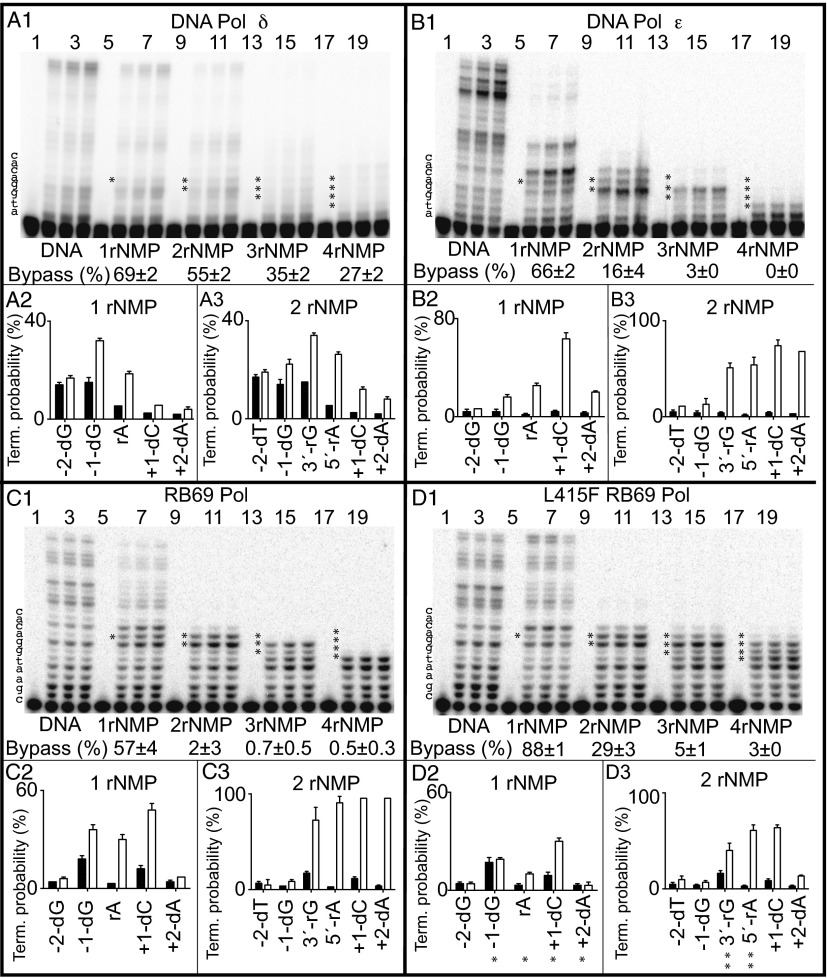
Bypass by Yeast Pol δ.
Relative to the all-DNA template (Fig. 1A1, lanes 2–4), Pol δ bypassed a rA (Fig. 1A1, lanes 6–8) with 69% efficiency. The slight stalling due to the rA reflects increased termination after dNTP insertion opposite the dG preceding the rA, opposite the rA itself, and opposite dC at +1 and dA at +2 (Fig. 1A2). These results recapitulate earlier studies (25) and show that Pol δ recognizes a single ribonucleotide in a DNA template and reacts by slightly stalling. Stalling is also observed for Pol δ bypass of two, three, or four consecutive ribonucleotides, with bypass efficiency decreasing and termination probabilities increasing at multiple positions from –1 through +2 as the number of consecutive ribonucleotides present in the template increases (Fig. 1A1). Despite stalling, some bypass is achieved with all four templates, including bypass of four consecutive ribonucleotides at 27% efficiency (Fig. 1A1).
Fig. 1.
Bypass of ribonucleotides by Pol δ, Pol ε, and WT and L415F RB69 Pol. Bypass reactions were performed as described in Materials and Methods, in each case using a large excess of primer template over polymerase. Results are for primer extension by yeast Pol δ (A), yeast Pol ε (B), RB69 Pol (C), and L415F RB69 Pol (D). A1, B1, C1, and D1 includes PAGE phosphorimages of DNA products when copying each of five different primer templates (Table S1), from reactions incubated for 0, 4, 8, and 12 min. The efficiency of ribonucleotide bypass relative to the all-DNA control was calculated as described previously (30), and the values are shown as percentages below each set of four lanes. Termination probabilities are also shown, again calculated as described previously (30), after incorporation at each of several template positions during copying of the all-DNA template (black bars) or templates containing either one (A2, B2, C2, and D2) or two ribonucleotides (A3, B3, C3, and D3) (white bars). The asterisks in D2 correspond to the structures depicted in Fig. 2, and the double asterisks in D3 correspond to the structures depicted in Figs. 3 and 4.
Bypass by Yeast Pol ε.
Next we examined the ability of WT, four-subunit yeast Pol ε to bypass ribonucleotides in the same templates. Relative to the all-DNA template (Fig. 1B1, lanes 2–4), Pol ε bypassed a rA (Fig. 1B1, lanes 6–8) with 66% efficiency, a value similar to that for Pol δ. This result recapitulates our initial study (4), showing that Pol ε stalls during bypass of a single ribonucleotide in a DNA template, with increased termination after dNTP insertion opposite positions –1 through +4 (Fig. 1B2). Once the rA-containing base pair is located 5 bp upstream of the active site, more processive synthesis is observed (Fig. 1B1, upper bands in lanes 6–8). Compared with Pol δ, Pol ε is about threefold less efficient at bypassing two ribonucleotides (Fig. 1 A1 and B1), a difference that reflects strong increases in termination at several positions (Fig. 1B3). Pol ε is about 10-fold less efficient than Pol δ at bypassing three ribonucleotides, and it is unable to bypass four consecutive ribonucleotides during one cycle of processive synthesis (Fig. 1 A1 and B1).
Ribonucleotide Bypass by RB69 Pols.
To set the stage for structural studies, we compared the ribonucleotide bypass ability of RB69 Pol to that of Pol δ and Pol ε. The results (Fig. 1C) reveal that bypass by RB69 Pol most closely approximates that of Pol ε with respect to efficiency and termination probability at multiple positions. Initial attempts to obtain crystals relevant to RB69 Pol bypass of a single ribonucleotide failed. We therefore measured bypass by L415F RB69 Pol. L415F RB69 Pol was previously found to be more efficient than its WT parent at bypassing 8-oxo-dG and at inserting a dNTP opposite an abasic site (28). These bypass data suggest that its ribonucleotide bypass efficiency might also be higher, perhaps increasing the probability of obtaining crystals. Both expectations were met. L415F RB69 Pol was more efficient at bypassing ribonucleotides, especially two consecutive ribonucleotides, where relative bypass by the L415F variant was 14-fold higher than its WT parent (Fig. 1 C1 and D1). The variant Pol also exhibited less (but measurable) termination opposite each of several positions (Fig. 1 C2, D2, C3, and D3).
Structural Analysis of L415F RB69 Pol.
We obtained seven crystal structures of L415F RB69 Pol that are relevant to ribonucleotide bypass (Table S2). These structures all contain a 14-mer primer hybridized to an 18-mer template of similar sequence to that in the 1.8-Å structure of RB69 Pol (27). Similar to most published structures of RB69 Pol, all seven unique structures contain Ca2+ at the polymerase active site. These structures allow metal-independent comparisons between our all-DNA structure and the ribonucleotide-containing structures, such that the structural changes resulting from ribonucleotides in the template are more likely to be independent of the metal.
The all-DNA structure (2.2 Å) contains an incoming nonhydrolyzable nucleotide analog (dUMPNPP) correctly paired opposite template dA. This structure overlays well with the recently described (27) 1.8-Å structure of WT RB69 Pol (RMSD for Cα, 0.347 Å) with no significant structural changes other than the L415F mutation.
Structures Relevant to Bypassing a Single Ribonucleotide.
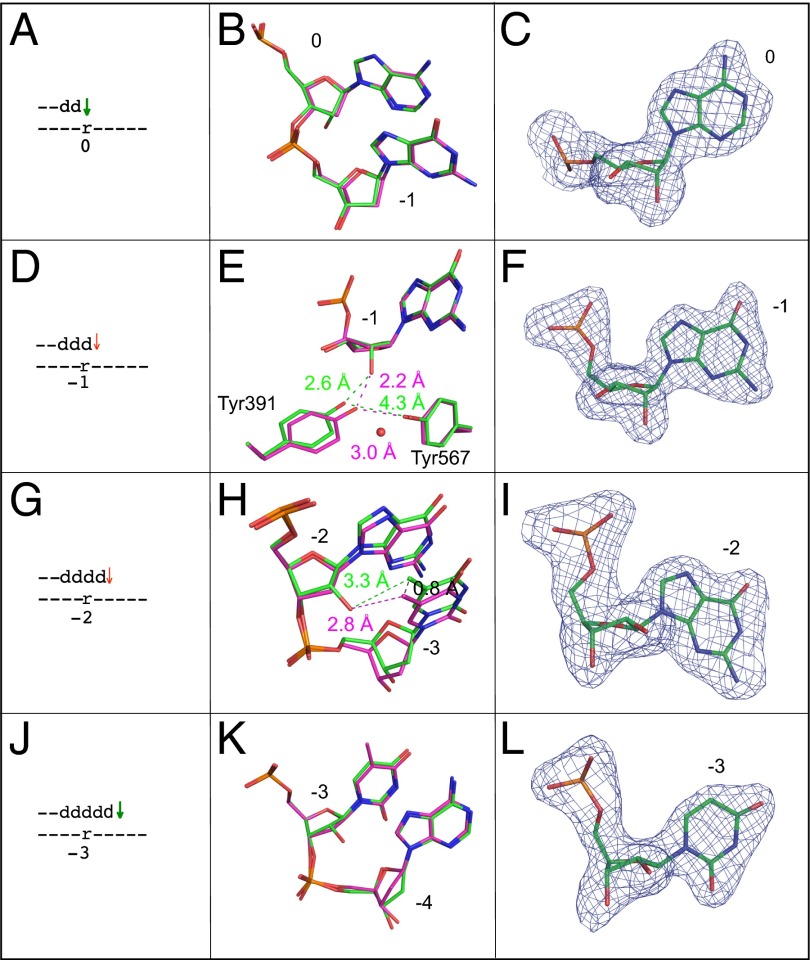
Four structures with a single ribonucleotide in the template were obtained (Fig. 2). Each contains a ddC primer terminus and dTTP paired with adenine, but differs with respect to the location (position 0, −1, −2, and −3) of the ribonucleotide (Fig. 2 A, D, G, and J, respectively), to model four consecutive insertions during bypass.
Fig. 2.
Superposition of all-DNA structure (magenta) with four different single ribonucleotide structures (green). (A) Schematic depicting the ribonucleotide in the nascent base pair binding pocket (position 0). A green arrow implies that dNTP insertion is not strongly reduced, as inferred from the relatively normal termination after the preceding incorporation (Fig. 1 D1 and D2, −1-dG). (B) Overlay of the ribonucleotide-containing structure depicted in A with the all-DNA structure, showing template positions 0 and −1. (C) Simulated annealing Fo-Fc omit map contoured at 3 σ for the ribonucleotide in the nascent base pair binding pocket. (D) Stick diagram depicting the location of ribonucleotide in the primer-terminal base pair (−1 position). The red arrow implies that dNTP insertion at this position (or possibly translocation before insertion) is problematic, as indicated by the increased termination following insertion opposite rA (Fig. 1 D1 and D2, rA). (E) Overlay of the ribonucleotide containing structure depicted in D with the all-DNA structure, showing template position −1, Tyr391, and Tyr567. The red sphere indicates the position of an additional water molecule in the ribonucleotide-containing structure. (F) Simulated annealing Fo-Fc omit map contoured at 3 σ for the ribonucleotide in the −1 position. (G) Stick diagram of the position of the ribonucleotide located 2 bp upstream of the active site (−2 position). As above, the red arrow implies that dNTP insertion at this position is problematic (Fig. 1 D1 and D2, +1-dC). (H) Overlay of the ribonucleotide containing structure with the all-DNA structure at template positions −2 and −3. (I) Simulated annealing Fo-Fc omit map contoured at 3 σ for the ribonucleotide at position −2. (J) Stick diagram of the position of ribonucleotide located 3 bp upstream of the active site (−3 position). As explained above, the green arrow indicates that insertion is normal (Fig. 1 D1 and D2, +2dA). (K) Overlay of the ribonucleotide-containing structure with the all-DNA structure at template positions −3 and −4. (L) Simulated annealing Fo-Fc omit map contoured at 3 σ for the ribonucleotide at position −3.
Ribonucleotide in the nascent base pair binding pocket (position 0).
This 2.1-Å structure overlays well with the all-DNA ternary complex and shows no obvious structural difference (Fig. 2 A and B). In both structures, the sugar pucker for the templating base is in a C3′-endo conformation (Fig. 2C), the preferred conformation for a ribonucleotide.
Ribonucleotide in the primer-terminal base pair (position −1).
This 2.1-Å structure also overlays well with the all-DNA structure (Fig. 2 D and E). The sugar pucker for the relevant nucleotide at this position, designated as –1 because it is 1 bp upstream of the nascent base pair binding pocket, is again found in the C3′-endo conformation in both structures (Fig. 2F). However, the presence of the ribonucleotide displaces Tyr391 from its location in the all-DNA structure (Fig. 2E). This displacement increases the distance between the phenol oxygen atoms of Tyr391 and Tyr567 from 3.0 to 4.3 Å, thus disrupting an H-bond between these residues and placing the phenol oxygen of Tyr391 2.6 Å from the 2′O atom on the ribose. In addition to the five previously observed, well-ordered waters interacting in the DNA minor groove (27), an additional water is observed 2.7 Å from the phenol oxygen of Tyr391 and adjacent to Tyr567.
Ribonucleotide located 2 bp upstream of the active site (position −2).
This 2.4-Å structure also overlays well with the all-DNA structure (Fig. 2 G and H), but now the sugar pucker of the ribonucleotide is in the unpreferred C2′-endo conformation, similar to the all-DNA sugar pucker (Fig. 2I). This conformation places the 2′O atom within 3.3 Å of the methyl group of the adjacent upstream template T (Fig. 2H), slightly altering its position (0.8 Å) with respect to the all-DNA structure.
Ribonucleotide located 3 bp upstream of the active site (position −3).
This 2.2-Å structure overlays well with the all-DNA structure (Fig. 2 J and K). The sugar pucker is again in the unpreferred C2′-endo conformation, similar to the all-DNA sugar pucker (Fig. 2L).
Structures Relevant to Bypassing Two Consecutive Ribonucleotides.
We obtained two crystal structures of L415F RB69 Pol relevant to bypassing two consecutive ribonucleotides. These structures correspond to the positions of the two ribonucleotides that are associated with the termination of synthesis after dNTP insertion opposite the 3′-ribonucleotide and opposite the 5′-ribonucleotide.
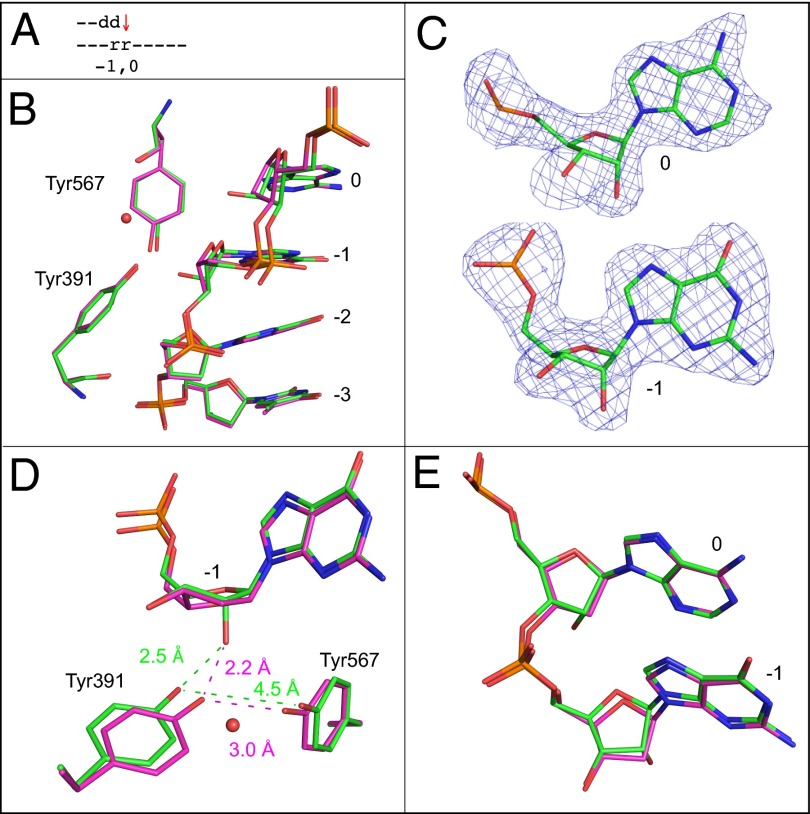
Ribonucleotides in the binding pocket and in the primer terminal base pair (positions 0 and −1).
This 2.4-Å structure aligns well with the all-DNA structure, but the backbone between the two ribonucleotides is distorted (0.5 Å) (Fig. 3 A, B, and E). The sugar pucker conformations of the ribonucleotides are similar to the all-DNA structure and are C3′-endo (Fig. 3C). The distance between Tyr391 and Tyr567 is increased from 3.0 to 4.5 Å, again disrupting the hydrogen bond found in the all-DNA structure. The additional water molecule seen in the structure with a single ribonucleotide mentioned above is again observed (Fig. 3D).
Fig. 3.
Superposition of the all-DNA structure (magenta) with the structure containing ribonucleotides at 0 and −1 position (green). (A) Schematic indicating the positions of the two ribonucleotides. The red arrow indicates that incorporation is problematic here (Fig. 1D3, 3′-rG). (B) Superposition of template nucleotides at positions 0 to −3, Tyr391, and Tyr567. A red sphere indicates the position of an additional water molecule. (C) Simulated annealing Fo-Fc maps contoured at 3 σ for the ribonucleotides at template positions 0 and −1. (D) Overlay with the all-DNA structure, showing the ribonucleotide at the −1 position, as well as Tyr391, Tyr567, and the additional water molecule. (E) Overlay of the ribonucleotides at positions 0 and −1 with the all-DNA structure.
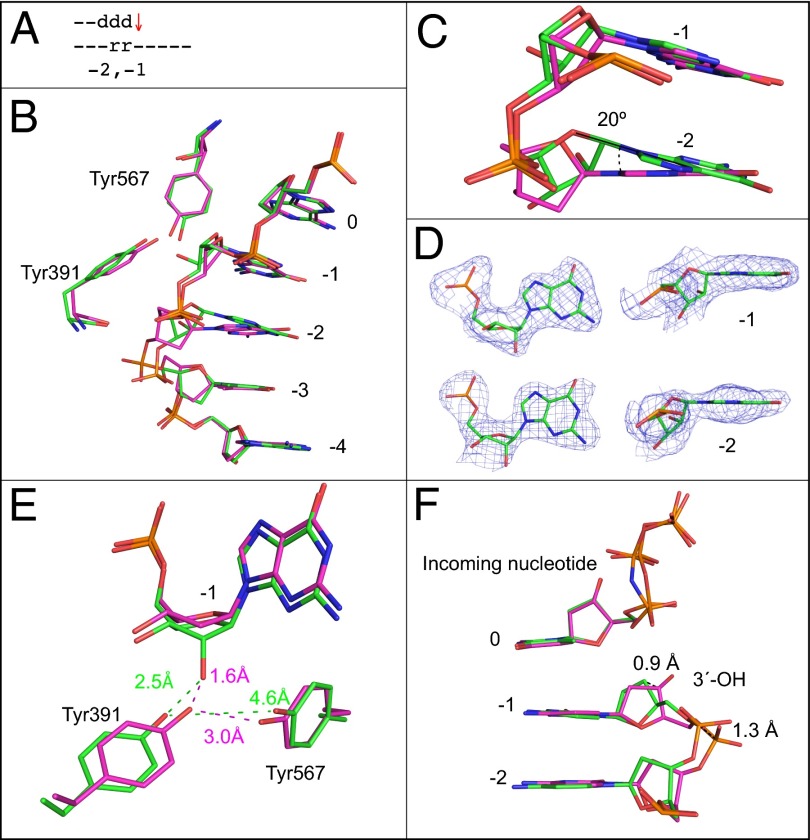
Ribonucleotides 1 and 2 bp upstream of the active site (positions −1 and −2).
This 2.6-Å structure aligns well with the all-DNA structure (Fig. 4 A and B), but the base of the ribonucleotide at the −2 position is 20° out of plane with respect to the all-DNA structure (Fig. 4C). The ribonucleotides maintain the C3′-endo conformation (Fig. 4D). The position of the Tyr391 is once again altered, but this time, the additional water molecule is not observed (Fig. 4E). Unique to this structure, the DNA backbone in the primer strand opposite the two ribonucleotides is displaced by 1.3 Å, and the C3′ atom of the primer terminus is displaced by 0.9 Å (Fig. 4F).
Fig. 4.
Superposition of the all-DNA structure (magenta) with the structure containing ribonucleotides at −1 and −2 positions (green). (A) Schematic indicating the positions of ribonucleotides at −1 and −2. The red arrow indicates that termination frequency is increased ∼10-fold compared with bypass of an all-DNA template, (Fig. 1D3, 5′-rA). (B) Superposition of template bases at 0 to −4 positions, Tyr391, and Tyr567. (C) Overlay of ribonucleotides at the −1 and −2 position with the all-DNA structure. (D) Simulated annealing Fo-Fc omit maps contoured at 3 σ are shown in blue for the two ribonucleotides at the −1 and −2 positions. (E) Overlay with the all-DNA structure showing Tyr391, Tyr567, and the nucleotide at the −1 position. (F) Overlay of primer terminus and incoming nucleotide with the all-DNA structure.
Discussion
These results provide insights into stalling by replicative DNA polymerases during bypass of ribonucleotides and may be relevant to phenotypes associated with ribonucleotides in DNA genomes. All four replicases examined here bypass a single ribonucleotide. Bypass efficiencies for copying a single ribonucleotide are less than twofold lower than for copying a deoxyribonucleotide (Fig. 1). This relatively efficient bypass is consistent with the small structural changes in L415F RB69 Pol ternary complexes bound to ribonucleotide-containing template primers (Fig. 2), as well as with the ability of ribonucleotide repair-deficient yeast (5, 6, 10, 17) and mice (7, 19) to survive and replicate genomes that contain large numbers of ribonucleotides. The fact that ribonucleotides incorporated into the genome by DNA polymerases can be tolerated is consistent with the idea that they can have beneficial signaling functions (4, 9, 11, 12).
On the other hand, single ribonucleotides in DNA templates at least partially impede synthesis by RB69 Pol, Pol δ, and Pol ε (Fig. 1). This impediment includes increased termination of synthesis at multiple positions as the polymerases progressively traverse the ribonucleotide (Fig. 1). The greatest increases in termination by L415F RB69 Pol occur following dNTP insertion opposite the ribonucleotide and the next template base (Fig. 1D2, rA and +1-dC). These increases could reflect impairment of any of the steps needed after dNTP insertion at one position through insertion chemistry at the next position. The increase in termination after dNTP insertion opposite the ribonucleotide itself is modeled by the structure in Fig. 2 D–F. Termination correlates with displacement of Tyr391 resulting from the presence of the 2′-oxygen of the ribose (Fig. 2E). This displacement is accompanied by the appearance of a water molecule. Both changes are interesting in light of previous studies of Tyr391 and its interaction with Tyr567. Both tyrosines are invariant in human and yeast Pols α, δ, and ε, implying that the present results with RB69 Pol may be relevant to the eukaryotic replicases. Tyr391 is in the palm subdomain where it interacts with the backbone of the template strand between the –1 and –2 positions (26), with the template-strand sugar of the primer-terminal base pair and with Tyr567 (31). Tyr567 is located in the fingers subdomain, where it forms a hydrogen bond with the template base of the primer-terminal base pair (26, 27), via one of five ordered water molecules that interact with the DNA minor groove (27). Substituting other amino acids for Tyr391 and Tyr567 reduces the fidelity of RB69 Pol and its ability to polymerize using all-DNA primer templates (27, 31–36). These facts suggest that the changes in the positions of Tyr391 and Tyr567 reported here are likely be relevant to the increased termination associated with bypass of one or two template-strand ribonucleotides by RB69 Pol, and by extrapolation, the eukaryotic replicases.
The increased termination observed at the next position is accompanied by a C2′-endo sugar conformation for the ribonucleotide (Fig. 2I). Because C2′-endo is an unfavorable conformation for a ribose in duplex DNA, it seems possible that the energetic cost of achieving and/or maintaining the C2′-endo conformation with a ribose (beyond that needed when a deoxyribose is present, as in the all-DNA substrate) underlies the increased termination at this position. This idea is consistent with a recent study suggesting that slow conformational dynamics at C2′-endo nucleotides have the potential to function as rate-determining molecular switches during catalysis (37). That sugar pucker may be relevant to switching during replication that involves ribonucleotides in DNA is also suggested by a study of Pol α. After RNA primase synthesizes RNA primers to initiate nuclear DNA replication at origins and of Okazaki fragments, Pol α uses these RNA primers to synthesize a DNA primer, followed by a switch that allows Pol δ to synthesize the majority of each Okazaki fragment. Recent data (38) indicate that Pol α recognizes an A-form RNA/DNA helix and that the ensuing synthesis of B-form DNA terminates primer synthesis to allow the switch to Pol δ.
Ribonucleotides in DNA can be considered as lesions because they are noncanonical substrates for DNA polymerases. During translesion synthesis to bypass other lesions, especially those that strongly stall replication, synthesis by one polymerase terminates to allow another polymerase to access the primer template. Our results demonstrate that ribonucleotides become increasingly problematic for replicases as their number increases from one to four (Fig. 1). Pol ε (Fig. 1B) and RB69 Pol (Fig. 1C) have the greatest difficulty in bypassing multiple consecutive ribonucleotides. When two ribonucleotides are present, this difficulty is modeled by the L415F RB69 Pol structures corresponding to the positions of the two ribonucleotides, each associated with increased termination of synthesis (double asterisks in Fig. 1D). For the first of these events (Fig. 1D3, 3′-rG), the position of the DNA backbone between the ribonucleotides at position 0 and −1 changes slightly (Fig. 3 B and E), and similar to the result with a single ribonucleotide (Fig. 2E), the 2′-oxygen of the ribose in the primer-terminal base pair displaces Tyr391 disrupting a hydrogen bond with Tyr567 (Fig. 3D). The latter feature is shared in the structure (Fig. 4E) corresponding to strong termination after the next incorporation opposite rA (Fig. 1D3, 5′-rA). Additionally, in that structure, the base of the ribonucleotide at the −2 position is displaced by 20° (Fig. 4C), and the DNA backbone in the primer strand opposite the two ribonucleotides is displaced by 1.3 Å (Fig. 4F). We suggest that these structural perturbations reduce catalysis and may account for the 29% relative bypass efficiency of L415F RB69 with two consecutive ribonucleotides. Yeast Pol δ is more efficient than Pol ε at bypassing two, three, or four consecutive ribonucleotides by factors of ∼3-, 12-, and ≥20-fold, respectively (Fig. 1 A1 and B1). These differences are interesting in light of evidence that Pol δ is the major lagging strand replicase and Pol ε is the major leading strand replicase (6, 8, 9, 39, 40). The opportunity for Pol δ and Pol ε to replicate templates containing two or more consecutive ribonucleotides could result from (at least) three transactions. Foremost is the 5% of lagging strand replication initially synthesized by primase as stretches of approximately 10 ribonucleotides. Although these primers are normally removed during Okazaki fragment maturation, Okazaki fragment maturation is a very common nuclear DNA transaction, such that even rare persistence of consecutive ribonucleotides could have consequences due to polymerase stalling, perhaps most especially Pol ε stalling. This idea is strongly supported by evidence in S. pombe that a persistent di-ribonucleotide imprint made during lagging strand replication stalls the next round of leading strand replication by Pol ε, thereby initiating mating type switching (13).
A second possibility for consecutive ribonucleotides in DNA that cannot be excluded by current studies in yeast (5, 6, 10, 17) or mice (7, 19) is incorporation of consecutive ribonucleotides during synthesis by DNA replicases, or perhaps by less faithful DNA polymerases. However, a third possibility for bypass of consecutive ribonucleotides was suggested by a study demonstrating that when DNA oligonucleotides containing consecutive ribonucleotides are introduced into yeast, they can direct the template-dependent repair of double strand DNA breaks (41). This observation led to the novel idea that under certain circumstances, RNA may be used as a template for DNA synthesis in vivo. Initial evidence in that study suggested that Pol δ was more likely than Pol ε to copy these ribonucleotide-containing templates in yeast. That interpretation is supported by the results in Fig. 1 showing that Pol δ is substantially more efficient than Pol ε at bypassing multiple consecutive ribonucleotides.
A yeast strain defective in both RNase H2 and RNase H1 progresses slowly through S phase, accumulates ubiquitylated PCNA that is known to be involved in TLS and is sensitive to treatment with hydroxyurea. In the presence of hydroxyurea, survival of the double RNase H-defective strain depends on either MMS2-dependent template switching or Rev3-dependent TLS (17). Dependence on these pathways is not observed for strains defective in either RNase H1 or RNase H2 alone. Because only RNase H2 can repair single ribonucleotides in DNA, but RNases H1 and H2 can both digest substrates containing multiple consecutive ribonucleotides in DNA (2), the collective results suggest that replication fork stalling in the RNases H1/H2 double mutant strain may be due to inefficient bypass of multiple consecutive unrepaired ribonucleotides in DNA, with either template switching or TLS by Pol ζ required to alleviate the stalled fork. Our data showing that Pol δ is more efficient than Pol ε in bypassing consecutive ribonucleotides in DNA templates (Fig. 1 A1 and B1) predicts that the cellular consequences of defects in both RNases H1 and H2 will be less severe for ribonucleotides present in the DNA template used for lagging strand replication than in the DNA template used for leading strand replication.
As mentioned in the Introduction, unrepaired ribonucleotides also have deleterious consequences in mice. These phenotypes could reflect difficulty in resolving transcriptional R-loops, they could result from endonucleolytic cleavage of ribonucleotides in DNA by topoisomerase 1 to create initially unligatable DNA ends (10), or based on this study, they could reflect difficulty in bypassing unrepaired ribonucleotides during DNA replication that leads to fork stalling. These possibilities are not mutually exclusive. A better understanding of these transactions is motivated by the fact that, in humans, mutations in the RNH201A/B/C genes encoding the three subunits of RNase H2 are associated with Aicardi-Goutières syndrome (20).
Materials and Methods
DNA Polymerases.
WT yeast Pol δ and Pol ε were purified as described previously (42, 43). RB69 Pol and its L415F variant were expressed and purified as described previously (28). In these studies, the RB69 Pol used were exonuclease deficient (D222A/D327A).
Bypass Assays.
All components except the polymerase were mixed on ice and incubated at 37 °C for 2 min. The polymerase was added to initiate reactions, which were terminated after 0, 4, 8, and 12 min. These mixtures were heated at 95 °C for 3 min, and the DNA products were separated by electrophoresis through a 12% (wt/vol) denaturing polyacrylamide gel. A PhosphorImager was used to visualize the DNA products, which were quantified using Image Quant software from Molecular Dynamics. Termination probabilities were calculated as described previously (30).
Protein Crystallization.
Crystals of ternary complexes were formed using the vapor diffusion sitting drop method. The crystals were formed by mixing protein solution, primer template, and nucleotide with the reservoir solution. For data collection, crystals were transferred into a cryo-solution. All crystals were frozen in liquid nitrogen and then mounted on a goniometer in a cold stream of nitrogen gas at 95 K.
Data Collection and Processing.
Data for structures were collected on a Saturn 92 charge-coupled device (CCD) area detector system mounted on a 007HF rotating anode generator equipped with VarimaxHF mirrors or at the Advanced Photon Source, Argonne National Laboratory on the Southeast Regional Collaborative Access Team BM beam line, on a MAR225 CCD area detector. All data were processed using the HKL2000 data processing software (44). Model building was performed using iterative cycles of manual model building using the program COOT (45) and refinement with Phenix (46) with dihedral restraints turned off for the DNA and incoming nucleotide. The quality of the models was assessed using Molprobity (47). A complete description of materials and methods used can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Katarzyna Bebenek and William Beard for thoughtful comments on the manuscript. This work was supported by Project ES065070 (T.A.K.) and Project ES102645 (L.C.P.), both from the Division of Intramural Research of the National Institute of Environmental Health Sciences, National Institutes of Health (NIH). The NIH provided funding for the open access charge. Crystallographic data were collected at the Southeast Regional Collaborative Access Team 22-ID beam line at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at www.ser-cat.org/members.html. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract W-31-109-Eng-38.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4KHQ, 4KHS, 4KHU, 4KHW, 4KHY, 4KI4, and 4KI6).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1309119110/-/DCSupplemental.
References
- 1.Burgers PM. Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem. 2009;284(7):4041–4045. doi: 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cerritelli SM, Crouch RJ. Ribonuclease H: The enzymes in eukaryotes. FEBS J. 2009;276(6):1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balakrishnan L, Bambara RA. Eukaryotic lagging strand DNA replication employs a multi-pathway mechanism that protects genome integrity. J Biol Chem. 2011;286(9):6865–6870. doi: 10.1074/jbc.R110.209502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nick McElhinny SA, et al. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc Natl Acad Sci USA. 2010;107(11):4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nick McElhinny SA, et al. Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol. 2010;6(10):774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyabe I, Kunkel TA, Carr AM. The major roles of DNA polymerases epsilon and delta at the eukaryotic replication fork are evolutionarily conserved. PLoS Genet. 2011;7(12):e1002407. doi: 10.1371/journal.pgen.1002407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reijns MAM, et al. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell. 2012;149(5):1008–1022. doi: 10.1016/j.cell.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lujan SA, et al. Mismatch repair balances leading and lagging strand DNA replication fidelity. PLoS Genet. 2012;8(10) e1003016:e1003016. doi: 10.1371/journal.pgen.1003016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lujan SA, Williams JS, Clausen AR, Clark AB, Kunkel TA. Ribonucleotides are signals for mismatch repair of leading-strand replication errors. Mol Cell. 2013;50(3):437–443. doi: 10.1016/j.molcel.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams JS, et al. Topoisomerase 1-mediated removal of ribonucleotides from nascent leading-strand DNA. Mol Cell. 2013;49(5):1010–1015. doi: 10.1016/j.molcel.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vengrova S, Dalgaard JZ. The wild-type Schizosaccharomyces pombe mat1 imprint consists of two ribonucleotides. EMBO Rep. 2006;7(1):59–65. doi: 10.1038/sj.embor.7400576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghodgaonkar MM, et al. Ribonucleotides misincorporated into DNA act as strand-discrimination signals in eukaryotic mismatch repair. Mol Cell. 2013;50(3):323–332. doi: 10.1016/j.molcel.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalgaard JZ. Causes and consequences of ribonucleotide incorporation into nuclear DNA. Trends Genet. 2012;28(12):592–597. doi: 10.1016/j.tig.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 14.Sparks JL, et al. RNase H2-initiated ribonucleotide excision repair. Mol Cell. 2012;47(6):980–986. doi: 10.1016/j.molcel.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark AB, Lujan SA, Kissling GE, Kunkel TA. Mismatch repair-independent tandem repeat sequence instability resulting from ribonucleotide incorporation by DNA polymerase ε. DNA Repair (Amst) 2011;10(5):476–482. doi: 10.1016/j.dnarep.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim N, et al. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 2011;332(6037):1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lazzaro F, et al. RNase H and postreplication repair protect cells from ribonucleotides incorporated in DNA. Mol Cell. 2012;45(1):99–110. doi: 10.1016/j.molcel.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams JS, et al. Proofreading of ribonucleotides inserted into DNA by yeast DNA polymerase ϵ. DNA Repair (Amst) 2012;11(8):649–656. doi: 10.1016/j.dnarep.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiller B, et al. Mammalian RNase H2 removes ribonucleotides from DNA to maintain genome integrity. J Exp Med. 2012;209(8):1419–1426. doi: 10.1084/jem.20120876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crow YJ, et al. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutières syndrome and mimic congenital viral brain infection. Nat Genet. 2006;38(8):910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- 21.Egli M, Usman N, Rich A. Conformational influence of the ribose 2′-hydroxyl group: Crystal structures of DNA-RNA chimeric duplexes. Biochemistry. 1993;32(13):3221–3237. [PubMed] [Google Scholar]
- 22.Jaishree TN, van der Marel GA, van Boom JH, Wang AH. Structural influence of RNA incorporation in DNA: Quantitative nuclear magnetic resonance refinement of d(CG)r(CG)d(CG) and d(CG)r(C)d(TAGCG) Biochemistry. 1993;32(18):4903–4911. doi: 10.1021/bi00069a027. [DOI] [PubMed] [Google Scholar]
- 23.Ban C, Ramakrishnan B, Sundaralingam M. A single 2′-hydroxyl group converts B-DNA to A-DNA. Crystal structure of the DNA-RNA chimeric decamer duplex d(CCGGC)r(G)d(CCGG) with a novel intermolecular G-C base-paired quadruplet. J Mol Biol. 1994;236(1):275–285. doi: 10.1006/jmbi.1994.1134. [DOI] [PubMed] [Google Scholar]
- 24.DeRose EF, Perera L, Murray MS, Kunkel TA, London RE. Solution structure of the Dickerson DNA dodecamer containing a single ribonucleotide. Biochemistry. 2012;51(12):2407–2416. doi: 10.1021/bi201710q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watt DL, Johansson E, Burgers PM, Kunkel TA. Replication of ribonucleotide-containing DNA templates by yeast replicative polymerases. DNA Repair (Amst) 2011;10(8):897–902. doi: 10.1016/j.dnarep.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franklin MC, Wang JM, Steitz TA. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell. 2001;105(5):657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 27.Wang MN, et al. Insights into base selectivity from the 1.8 Å resolution structure of an RB69 DNA polymerase ternary complex. Biochemistry. 2011;50(4):581–590. doi: 10.1021/bi101192f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong X, Pedersen LC, Kunkel TA. Characterization of a replicative DNA polymerase mutant with reduced fidelity and increased translesion synthesis capacity. Nucleic Acids Res. 2008;36(12):3892–3904. doi: 10.1093/nar/gkn312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang GW, Franklin M, Li J, Lin TC, Konigsberg W. A conserved Tyr residue is required for sugar selectivity in a Pol alpha DNA polymerase. Biochemistry. 2002;41(32):10256–10261. doi: 10.1021/bi0202171. [DOI] [PubMed] [Google Scholar]
- 30.Kokoska RJ, McCulloch SD, Kunkel TA. The efficiency and specificity of apurinic/apyrimidinic site bypass by human DNA polymerase eta and Sulfolobus solfataricus Dpo4. J Biol Chem. 2003;278(50):50537–50545. doi: 10.1074/jbc.M308515200. [DOI] [PubMed] [Google Scholar]
- 31.Yang G, Wang J, Konigsberg W. Base selectivity is impaired by mutants that perturb hydrogen bonding networks in the RB69 DNA polymerase active site. Biochemistry. 2005;44(9):3338–3346. doi: 10.1021/bi047921x. [DOI] [PubMed] [Google Scholar]
- 32.Bebenek A, et al. Interacting fidelity defects in the replicative DNA polymerase of bacteriophage RB69. J Biol Chem. 2001;276(13):10387–10397. doi: 10.1074/jbc.M007707200. [DOI] [PubMed] [Google Scholar]
- 33.Jacewicz A, Makiela K, Kierzek A, Drake JW, Bebenek A. The roles of Tyr391 and Tyr619 in RB69 DNA polymerase replication fidelity. J Mol Biol. 2007;368(1):18–29. doi: 10.1016/j.jmb.2007.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xia S, Beckman J, Wang J, Konigsberg WH. Using a fluorescent cytosine analogue tC(o) to probe the effect of the Y567 to Ala substitution on the preinsertion steps of dNMP incorporation by RB69 DNA polymerase. Biochemistry. 2012;51(22):4609–4617. doi: 10.1021/bi300241m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xia S, Eom SH, Konigsberg WH, Wang J. Structural basis for differential insertion kinetics of dNMPs opposite a difluorotoluene nucleotide residue. Biochemistry. 2012;51(7):1476–1485. doi: 10.1021/bi2016487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang H, Beckman J, Wang J, Konigsberg W. RB69 DNA polymerase mutants with expanded nascent base-pair-binding pockets are highly efficient but have reduced base selectivity. Biochemistry. 2009;48(29):6940–6950. doi: 10.1021/bi900422b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gherghe CM, Mortimer SA, Krahn JM, Thompson NL, Weeks KM. Slow conformational dynamics at C2′-endo nucleotides in RNA. J Am Chem Soc. 2008;130(28):8884–8885. doi: 10.1021/ja802691e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perera R, et al. Mechanism for priming DNA synthesis by yeast DNA polymerase alpha. eLife. 2013;2:e00482. doi: 10.7554/eLife.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pursell ZF, Isoz I, Lundström EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317(5834):127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30(2):137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Storici F, Bebenek K, Kunkel TA, Gordenin DA, Resnick MA. RNA-templated DNA repair. Nature. 2007;447(7142):338–341. doi: 10.1038/nature05720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burgers PM, Gerik KJ. Structure and processivity of two forms of Saccharomyces cerevisiae DNA polymerase delta. J Biol Chem. 1998;273(31):19756–19762. doi: 10.1074/jbc.273.31.19756. [DOI] [PubMed] [Google Scholar]
- 43.Asturias FJ, et al. Structure of Saccharomyces cerevisiae DNA polymerase epsilon by cryo-electron microscopy. Nat Struct Mol Biol. 2006;13(1):35–43. doi: 10.1038/nsmb1040. [DOI] [PubMed] [Google Scholar]
- 44.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolec Crystallogr. Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.