Significance
In this report we describe a group of highly complex glycosides active against hepatitis C virus and a separate group of natural products active against established targets for the control of diabetes mellitus. These complex metabolites were found in the rare plant Diplostephium rhododendroides Hieron. from the mountains of Ecuador. This report illustrates the human health significance of protecting rare and endangered plants for the control of new and emerging diseases. The extinction of this particular plant would have taken with it promising opportunities to develop unique treatments for the control of two modern-day disease challenges. The genus Diplostephium is represented by several plant species with a history of use in traditional medicine in Central and South America.
Keywords: endangered plants, hepatitis C virus, diabetes mellitus
Abstract
One in five of the world’s plant species is threatened with extinction according to the 2010 first global analysis of extinction risk. Tilman et al. predicted a massive ecological change to terrestrial plants within the next 50–100 y, accompanied by an increase in the number of global plant species facing extinction [Tilman D, et al. (2001) Proc Natl Acad Sci USA 98(10):5433–5440]. Most of the drug-producing plant families contain endangered species never previously studied for their utility to human health, which strongly validates the need to prioritize protection and assessment of these fragile and endangered groups [Zhu F, et al. (2011) Proc Natl Acad Sci USA 108(31):12943–12948]. With little prior attention given to endangered and rare plant species, this report provides strong justification for conservation of the rare plant Diplostephium rhododendroides Hieron., as well as other potential drug-producing endangered species in this and other groups.
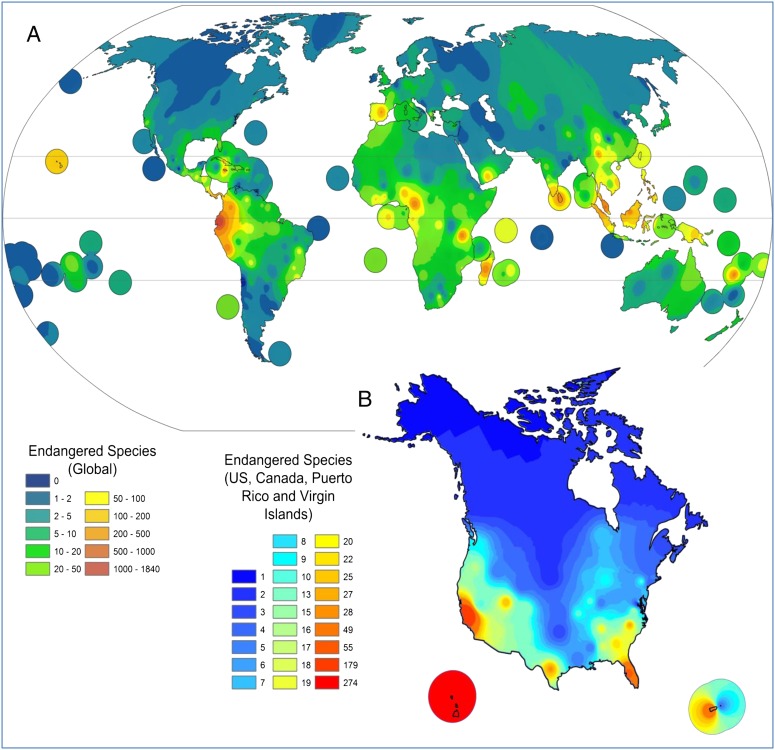
The International Union for Conservation of Nature (IUCN), in its latest Red List (www.iucnredlist.org), detailed the ongoing threats to biodiversity on the planet and assessed a total of 63,837 plant and animal species around the globe. This study revealed that 19,817 are currently threatened with extinction, 3,947 described as critically endangered while more than 10,000 species are listed as vulnerable (Fig. 1). Plants have become more threatened than birds and as threatened as mammals leaving them second only to amphibians or corals in their risk for extinction (1). Habitat loss (2) and climate change are the primary causes of species endangerment; however other biological factors, such as competition, invasion (3), including nonnative fungi (4) or exotic arthropods (5), can also drive plant species to extinction. Climate change has been shown to play a critical role in plant species distribution shifts around the world, and Thuiller et al. projected the distribution of 1,350 European plants species under seven climate change scenarios, where the expected species loss and turnover per pixel across different scenarios over Europe have been shown to be 27–42% and 45–63%, respectively (6). The greatest changes are expected to be in the transition between Mediterranean and Euro-Siberian regions (6). Based on the Palmer Drought Index, the National Oceanic and Atmospheric Administration (NOAA) announced that May 2012 was the second warmest and 27th driest May on record (since 1895) with persistent dryness across the United States (7). In accordance, NOAA showed an increase in the number of wildfires in the United States through June 2012 leading to further stress on endangered groups. The 2005 Global Forest Resource Assessment (8), coordinated by the Food and Agriculture Organization of the United Nations, declared a high rate of deforestation with approximately a 6-million-hectare annual decrease in forest area, which further limits the forests’ role in the conservation of biodiversity. Cochrane and Laurance projected that by the year 2032, more than 70% of the planet’s land surface will have been destroyed or disturbed (9). Interestingly, natural events such as geological and climate change represent a 19% threat to plant life while the greatest threat involves human-induced habitat loss as part of the conversion of natural habitats to agriculture or livestock grazing. Human-induced environmental changes include increased levels of atmospheric CO2, phosphorus, calcium, pH, and other climate changes (10). These changes facilitate plant extinction as plant physiologies are forced to adapt to changing environmental constraints (10). It is estimated that habitat destruction from human activity is the primary cause of risk for 83% of endangered plant species.
Fig. 1.
Global (A) and the United States and Canada (B) distribution of endangered plants. (Global and US–Canada data are from The IUCN Red List and US Department of Agriculture, respectively.)
According to Winter et al., human activities mainly affect two fundamental processes: native extinction and alien introduction. These two processes affect taxonomic and phylogenetic characteristics (11). The region of greatest global biodiversity is the Andes, and in particular the rain forest, cloud forest, and paramo, where the DNA sequence in the endemic shrub Cyathostegia mathewsii indicates the region has been isolated for at least 5 My, separated by about 600 km of high cordillera in Peru (12). The IUCN has demonstrated that the paramo areas in both Ecuador and Colombia are hot spots for endangered and threatened plant species.
These natural threats come at a time when emerging infectious diseases are testing our current armamentarium of drugs. Plants, marine organisms, and microbes provide a wide variety of important drug products, such that 26% of the new entities approved by the Food and Drug Administration (FDA) in 2009–2010 were naturally derived in some fashion (13, 14). Endophytic microorganisms with their wide distribution in almost every plant on the earth and various symbiotic to slightly pathogenic relations represent a potential source of producing novel natural products with remarkable applications (15). Naturally derived drugs are mainly generated by specific gene clusters found in certain drug-producing families, where 80% of the approved drugs and 67% of the drugs in clinical trials are concentrated in 17 and 30 drug-prolific families, respectively (13). The US Fish and Wildlife Service declared that 795 plant species are at risk for extinction throughout the United States and according to Newman and Cragg, over 60% of cancer therapeutics were derived from naturally occurring products (14). Examples of highly significant drugs used routinely like digitalis from Digitalis purpurea for the treatment of heart disease, galantamine from Galanthus used for the treatment of Alzheimer’s disease, vincristrine and vinblastine from Catharanthus roseus used for chemotherapy, and paclitaxel (the anticancer drug Taxol) from the bark of the Pacific yew tree (SI Appendix). Gilbert reported in 2010 that more than 17,000 species of fauna and flora were at risk for extinction in 2009, and over half of all medicinal plants in Africa face extinction (16). In 2008, Plantlife reported that roughly 15,000 of the 50,000 wild medicinal plants are at risk and headed for extinction (17). This creates serious concern, as the World Health Organization (WHO) estimates that 80% of the developing countries population relies on medicines derived from traditional plants for their pharmacopeia (18).
Results
Of the emerging new threats to human health, hepatitis C virus (HCV) and diabetes mellitus (DM) are two of the most severely life-threatening. According to WHO, ischemic heart diseases, stroke and other cerebrovascular diseases, HIV/AIDS, and DM have 12.8%, 10.8%, 3.1%, and 2.2% mortality rates worldwide, respectively (19). Hepatitis epidemics were first recorded as early as 2,000 B.C., but it was not until 1989 that HCV also known as “non-A, non-B viral hepatitis” was identified using a novel molecular cloning approach (20). As many as 4 million individuals in the United States and 130–180 million worldwide are infected with HCV (21, 22). According to the Centers for Disease Control and Prevention (CDC), there were up to 18,000 new HCV infections in the United States in 2008, and HCV is responsible for 12,000 deaths annually (23). According to Hepatitis Central, HCV accounts for 20% of acute hepatitis, 70% of chronic hepatitis, 40% of end-stage cirrhosis, 60% of hepatocellular carcinoma, and 30–40% of liver transplants (24). The CDC estimates that in the next two decades, chronic HCV will be a major burden on the health care system. The direct medical costs of chronic HCV infections are estimated to exceed $10.7 billion, to which should be added both the enormous societal cost of premature mortality ($54.2 billion) and the costs of disability due to HCV infections ($21.3 billion). Recently, it has been shown that US mortality rates from HCV now exceed those from HIV-1 (25). HCV has six major genotypes; genotype 1 is the most common in the United States and is associated with more severe liver diseases and a higher risk of hepatocellular carcinoma than the other genotypes (26). According to Murray and Rice (27), HCV replication requires a series of virally coded nonstructural proteins, including two proteases (NS2-3 and NS3-4A), a helicase (NS3), and a polymerase (NS5B). Of these, NS3-4A and NS5B have attracted the most attention as drug targets. In 2010, Gao et al. (28) reported a potent NS5A inhibitor, BMS-790052, that is under evaluation in combination with other drugs, and this has stimulated studies targeting proteins which lack intrinsic enzymatic or receptor functionality. The most frequently used treatment involves a weekly injection of a combination of ribavirin plus pegylated α-IFN [IFN is a family of cytokines, which are expressed as antiviral responses and function by stimulating the cytotoxic T cells, and by inducing a number of intracellular genes that prevent virus replication], which are only 50% effective and have serious side effects including depression and influenza-like symptoms in more than 60% of the patients (29). More recently, Telaprevir and Bocepravir were approved by the FDA (30), but even these potent protease inhibitors are effective at most 80% of the time when combined with IFN and have numerous side effects including rash and anemia.
DM is also emerging as a serious health care problem as shown by the annual report that about 18 million people die from cardiovascular disease, for which DM is a significant predisposing factor along with hypertension (31). The global number of patients with DM in 2000 was 171 million and is projected to reach 366 million by 2030 (32). The increasing prevalence of type-2 DM, which accounts for 95% of all DM cases, is attributed to the rise in obesity levels and the associated metabolic syndrome. Such an elevated incidence of type-2 DM and obesity has prompted a search for new drugs leads to control this disease. One of the recently discovered targets for DM is protein tyrosine phosphatase 1B (PTP1B), which blocks insulin signaling through dephosphorylation of the insulin receptor and insulin receptor substrates (33). Previous pioneering studies that used PTP1B knockout mice exhibited reduced circulating insulin, glucose, and triglyceride levels after being injected with insulin and showed enhanced insulin sensitivity (34). Thus, inhibition of PTP1B has been gaining interest as an attractive therapeutic target for the treatment of metabolic diseases such as type-2 DM and obesity. There have been intensive efforts toward the development of PTP1B inhibitors, but the attempts to discover selective PTP1B inhibitors with desirable pharmacological properties have so far failed. One of the challenging problems for the development of potent inhibitors which bind directly to the PTP1B active site is poor bioavailability, originating from their structural similarity with phosphotyrosine, the substrate for the enzyme. Therefore, it is difficult to develop an inhibitor for PTP1B that is selective over other members of the PTP family, unless we consider inhibitors binding to a region outside the active site. Recently, a selective noncompetitive PTP1B inhibitor, trodusquemine, proceeded to phase I clinical trials and demonstrated promising pre- and early clinical results in suppressing appetite and hyperglycemic level, which indicates that allosteric inhibition of PTP1B could be a feasible therapeutic target in the treatment of type-2 DM (35).
Recently, we began to evaluate HCV and PTP1B inhibitory activity of a rare Latin American plant extract from the Natural Products Branch of the NCI. Repository sample N077951 was collected from Sarayo, Ecuador under a specific material transfer agreement that protects the rights of Ecuador. Diplostephium rhododendroides Hieron. (Asteraceae) is found in the mountains of Ecuador and Colombia and because of its rarity, geographical location, and terrain, this species was not easy to reach. The geographic coordinates are 78° W, 1° N to the nearest degree and a voucher sample is held at the National Herbarium in Washington DC, part of the Smithsonian Institution under the code 0CJS1259. The plant was a 180-cm shrub with a purple inflorescence and was collected at an altitude of 3,600 m. Diplostephium is a genus composed of ∼110 species of which eight are endangered. Sixty-three Diplostephium species are found in Colombia, and it is the third most diverse genus of the paramos (36). Several Diplostephium species have reported use in traditional medicine; Diplostephium foliosissimum S. F. Blake is used in the northern Peruvian Andes for the treatment of gastric pain, hypotension, and systemic debilitation whereas Diplostephium gynoxyoides Cuatrec is used to treat common cold and inflammation of the lungs (37, 38). The Asteraceae family represents one of 66 drug-producing families in the Viridiplantae kingdom; many of these families include endangered species. Artemisinin represents an example isolated from Artemisia annua, a member of the Asteraceae family. In April 2009, the FDA approved the first Artemisinin-based combination therapy for malaria, known as “Coartem.” The bioactivity-guided isolation of the active metabolites from D. rhododendroides Hieron. was highly challenging due to the limited amount of extract and the absence of additional fresh plant material. Evaluation of NMR, overlaying 2D-NMR experiments, gas chromatography (GC), and high-resolution electron spray ionization mass spectroscopy (HRESIMS) data permitted the identification and characterization of potent HCV and DM leads from two different structural classes.
Discussion
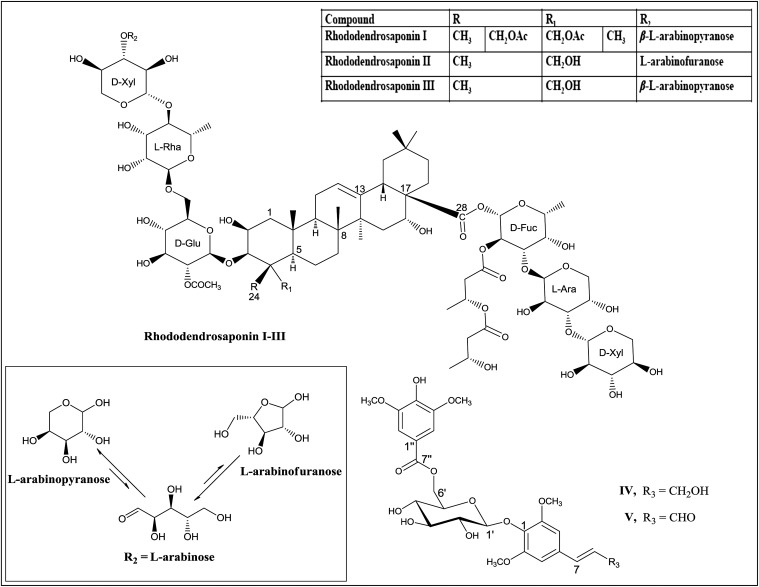
HCV-identified glycosides (Rhododendroglycosides I–III) represent a high degree of complexity and share the core oleanane triterpene skeleton. The total carbohydrate content by weight for the major glycoside, rhododendroglycoside III, has been shown by GC analysis to be 73.8%. The processed data revealed the presence of the following units: a glucosyl, two arabinosyls, a rhamnosyl, a fucosyl, and two xylosyls. Rhododendroglycosides II and III were shown by (+)-HRESIMS to possess the same mass with m/z 1,722.6523, and 1,722.6498 for the sodiated species, [C78H124O40+Na]+ whereas rhododendroglycoside I showed a mass at m/z 1,764.7908 for the sodiated species [C80H126O41+Na]+ and the presence of an extra acetyl group, which was confirmed by superimposed proton and carbon data for glycosides I–III (methanol-d4, 600 MHz). Overlaid heteronuclear single-quantum coherence (HSQC) data for rhododendroglycosides I and III revealed a high level of homology with interchangeable α- and β-configuration at C-24 of rhododendroglycoside I. In a similar manner, stacked HSQC studies for glycosides II and III revealed similar carbohydrate units, which were confirmed using carbohydrate analysis data (SI Appendix). Interestingly, the superimposed data clearly demonstrated the presence of the uncommon Amadori-type pyranose–furanose isomerism of the terminal l-arabinosyl moiety. It was possible to monitor the presence of the acyclic intermediate (formyl; CHO) at 217.6 ppm.
Interestingly, PTP1B inhibitory bioactivity-guided isolation from the same plant led to the isolation and identification of a unique and different class of compounds: phenolic glycosides (IV and V) (Fig. 2). Compound IV showed a pseudomolecular ion peak at m/z 575.1694 [M+Na]+ corresponding to the molecular formula C26H33O13 in the HRESIMS. The 1H and 13C NMR data of IV indicated the presence of two symmetrical 1,3,4,5-tetrasubstituted benzene moieties. The connectivity of a syringoyl moiety, sinapyl alcohol, and glucose were confirmed by analysis of heteronuclear multiple-bond correlation data. On the basis of the coupling constant of the anomeric proton (J = 7.2 Hz), the glycosidic linkage of the glucopyranoside was determined to be β-orientation. GC analysis indicated that the glucosyl moiety in IV belonged to the D-series of hexoses. Consequently, the structure of IV was elucidated as 1-[6-O-(3,5-dimethoxy-4-hydroxybenzoyl)-β-d-glucopyranosyloxy]-2,6-dimethoxyphenylprop-7E-en-9-ol, entitled as diplostephioside A (IV). The molecular formula of compound V was determined to be C26H30O13 by a pseudomolecular ion peak at m/z 551.1745 [M + H]+ observed in the HRESIMS. 1H and 13C NMR data as well as the physical data for V were similar to those of IV. The difference of two mass units and the NMR signals at δH 9.62 (1H, d, J = 7.6 Hz) and δC 194.7 (C-9), characteristic of formyl group, clearly indicated that the oxymethylene at the C-9 position of IV is transformed to an formyl group in V; compound V was identified as 1-[6-O-(3,5-dimethoxy-4-hydroxybenzoyl)-β-d-glucopyranosyloxy]-2,6-dimethoxyphenylprop-7E-en-9-al and named diplostephioside B (V). Comprehensive docking experiments using computational modeling techniques suggested that the isolated compounds may act on the allosteric inhibition site of PTP1B (39).
Fig. 2.
Structures of isolated metabolites I-V from D. rhododendroides and Amadori-type pyranose–furanose isomerism in II and III.
Rhododendroglycosides I–III were evaluated at six different concentrations for anti-HCV activity in Huh-7 replicon cells, where the compounds I–III showed EC50 of 0.2 μg·mL−1, EC90 of 0.3–0.9 μg·mL−1, and the 50% cytotoxic concentration (CC50) of 5.3–7.0 µg/mL compared with EC50 of 0.6 μg·mL−1, EC90 of 2.3 μg·mL−1, and CC50 of >2.6 μg·mL−1 for the positive control 2′-C-methylcytidine (2′-C-Me-C). We were able to develop two independent resistant HCV replicons to the compound, however they showed the absence of a consistent mutation to a viral target suggesting a cellular or entry target. Pharmacokinetic studies of a similar natural product-derived molecule from the metabolism of a ginseng saponin, 20-O-β-D-glucopyranosyl-20(S)-protopanaxadiol (IH-901), revealed the potential for these high-molecular-weight glycosides to target the liver where rapid disappearance from the plasma at α-phase followed by slow disappearance at β-phase after i.v. administration were shown (40). The liver levels of the IH-901 glycoside exceeded initial plasma concentration by sixfold shortly after i.v. administration. At steady state C (ss, liver) is about 11-fold higher than C (ss, plasma), whereas C (ss, bile) is about 1/2 C (ss, liver) after i.v. infusion. The activity profile clearly revealed significant potency for these metabolites comparable to the recently approved drug Telaprevir, (EC50 0.1–0.2 μg·mL−1) (41) and the therapeutic window warrants further investigations.
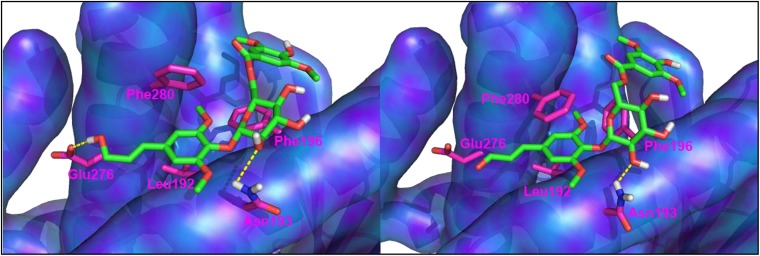
Compounds IV and V were tested for inhibitory activity against PTP1B. The known PTP1B inhibitors, RK-682 (IC50 = 4.5 μM), and ursolic acid (IC50 = 3.2 μM) were used as positive controls in this assay (SI Appendix). Compounds IV and V inhibited PTP1B activity with IC50 values of 81.0 and 25.9 μM, respectively, to afford a starting point for structure–activity relationship investigations. Comprehensive induced-fit docking simulations with the available PTP1B crystal structures revealed that compounds IV and V showed excellent docking scores with allosteric binding sites of PTP1B (1T48, 1T49 and 1T4J). According to the predicted binding modes of compounds IV and V, two benzene cores sit in the hydrophobic binding pocket constructed by the lipophilic side chain of Leu192, Phe196, and Phe280 (Fig. 3) (39). A hydroxy group attached to C-2′ in the glucose residue centered between the two aromatic groups forms a hydrogen bond to the side chain of Asp193, which is thought to be crucial to bind to the allosteric site. The hydrogen bond observed between the hydroxy group attached to C-9 in compound IV and the carboxylate side chain of Glu276 probably accounts for its better docking score than compound V. The predicted binding mode suggests the possibility that glucose flanked with a bis-benzene scaffold could be a reasonable starting point in the design of a potential allosteric site inhibitor of PTP1B. Compounds IV and V found in the stems of D. rhododendroides are distinguished from the constituents in the other species studied so far, which indicates that the compounds could be considered as marker compounds for establishing the chemotaxonomy of D. rhododendroides. The interesting arrangement of two aromatic groups centered with a glucose suggests a unique scaffold of a glucose-based PTP1B inhibitor given the fact that a range of glycolipids were evaluated for their PTP1B inhibition potency, and most of the structures were proven to be ineffective (42).
Fig. 3.
Predicted binding mode of compounds IV (Left) and V (Right). Dotted lines indicate hydrogen bonds. Model rendering in Pymol (version 1.3, Schrödinger).
In conclusion, the opportunity for identifying new drugs for emerging diseases depends on a diversity of drug-producing species. Endangered plants have been shown to cluster with previously established drug-producing families, and if the endangered species becomes extinct, a significant opportunity to identify novel prototypes to emerging diseases will also be lost. The rare plant D. rhododendroides Hieron. represents a vital opportunity in the control of HCV and type-2 DM, which have emerged as major global health threats, and strongly supports the value in protecting endangered and rare plants.
Experimental Procedures
The antiviral effectiveness of the compounds was calculated by subtracting the threshold RT-PCR cycle of the test compound from the threshold RT-PCR cycle of the nondrug control (ΔCt HCV). A ΔCt of 3.3 equals a 1-log reduction (equal to 90% less starting material) in replicon RNA levels. The cytotoxicity of the compounds was calculated by using ΔCt rRNA values and 2′-C-Me-C used as a control. To determine EC50 and CC50 values, ΔCt values were first converted into fractions of starting material and then used to calculate the percent inhibition (SI Appendix). For end point determination, Huh-7 B cells containing HCV genotype 1 replicon RNA were seeded in a 96-well plate at 3,000 cells per well, and the compounds were added at concentrations of 10, 3, 1, 0.3, 0.1, and 0.03 μg·mL−1 in triplicate immediately after seeding. Following a 5-d incubation [37 °C, 5% (vol/vol) CO2], total cellular RNA was isolated using the Manual Perfect Pure RNA 96 Cell Vac kit (5 Prime). Replicon RNA and internal control (TaqMan rRNA control reagent, Applied Biosystems) were amplified in a single-step multiple real-time RT-PCR assay. The homology in the structures of the isolated metabolites was echoed in a similar anti-HCV activity pattern.
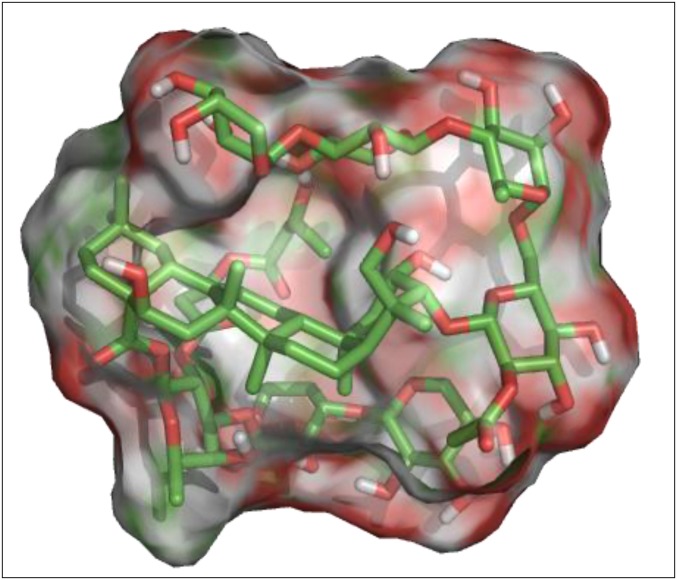
The 3D structure of the major glycoside III was minimized in Macromodel from Schrödinger (Schrödinger, LLC., New York) using the optimized potential for liquid simulations (OPLS_2005) force field with extended Van der Waals, electrostatic, and H-bond cutoffs of 8, 20, and 4 Å, respectively. All stereogenic centers were fixed during the calculation. The Polak–Ribière-type conjugate gradient (PRCG) method was used with 0.05 kJ−1·mol−1·Å−1 as the rms gradient convergence threshold. Mixed torsional/low-mode sampling was used for conformational analysis with 1,000 steps. PyMol 1.4 was used to generate the 3D surface with element color scheme as shown in Fig. 4.
Fig. 4.
The minimized 3D structure surface generated by PyMol 1.4 for the major metabolite, rhododendroglycoside III, where red represents oxygen and green and white represent carbon and hydrogen.
Before the docking simulations of compounds IV and V, the docking protocols were validated by predicting the binding mode of the cocrystalized compounds. The Induced Fit protocol successfully predicted their binding modes with Root mean square (rms) deviations of less than 1.5 Å, and the generated docking scores are almost in accordance with the biological potency. Schrödinger was used for comprehensive PTP1B docking studies with the isolated compounds. Protein structures were imported from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (www.rcsb.org/) with relevant protein accession codes. Imported protein crystal structures were prepared using the Protein Preparation Wizard (Schrödinger LLC.) under default settings (SI Appendix) with exhaustive sampling selected. Docking simulation was performed using Induced Fit (Schrödinger LLC.) (SI Appendix) with default settings except for the change on the energy window from 2.5 to 20 Kcal·mol−1 in conformational sampling. After docking simulations with the isolated compounds to reported PTP1B crystal structures, the crystal structures which yielded a docking score lower than −8.5 (1T48, 1T49, and 1T4J) were selected to simulate a plausible binding pose of the isolated compounds. PTP1B (human, recombinant) was purchased from BIOMOL International LP. The enzyme activity was evaluated using p−nitrophenyl phosphate, as described in SI Appendix.
Supplementary Material
Acknowledgments
The authors thank Ms. Christina Marrongelli and Dr. John S. Williamson for help with purification of the metabolites; Mr. Khaled Elokely for help with generation of the 3D minimized structure; and the Complex Carbohydrate Research Center (The University of Georgia) for carbohydrate analysis. The authors also acknowledge National Institutes of Health (NIH) , National Center for Complementary and Alternative Medicine (NCCAM) Grant R01-AT-007318, National Center for Research Resources (NCRR) Award CO6-RR-145503 and a grant from Kraft Foods Group, Inc.; fellowship support provided through the Ministry of Higher Education and Scientific Research–Egypt (to M.A.I.); the Department of Veterans Affairs and NIH Centers for AIDS Research (CFAR) Grant 2P30-AI-050409 (to R.F.S.); and National Research Foundation of Korea (NRF) NRF-C1ABA001-2010-0020484 (to M.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: A Diplostephium rhododendroides Hieron. (Asteraceae) voucher sample was deposited in the National Herbarium at the Smithsonian Institution (code 0CJS1259).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1311528110/-/DCSupplemental.
References
- 1. Gilbert N (2010) Threats to the world’s plants assessed. Nature, 10.1038/news.2010.499.
- 2.Feeley KJ, Silman MR. Extinction risks of Amazonian plant species. Proc Natl Acad Sci USA. 2009;106(30):12382–12387. doi: 10.1073/pnas.0900698106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klironomos JN. Feedback with soil biota contributes to plant rarity and invasiveness in communities. Nature. 2002;417(6884):67–70. doi: 10.1038/417067a. [DOI] [PubMed] [Google Scholar]
- 4.Gundale MJ. Influence of exotic earthworms on the soil organic horizon and the rare fern. Botrychium mormo. Conserv Biol. 2002;16:1555–1561. [Google Scholar]
- 5.Pimentel D, Zuniga R, Morrison D. Update on the environmental and economic costs associated with alien-invasive species in the United States. Ecol Econ. 2005;52:273–288. [Google Scholar]
- 6.Thuiller W, Lavorel S, Araújo MB, Sykes MT, Prentice IC. Climate change threats to plant diversity in Europe. Proc Natl Acad Sci USA. 2005;102(23):8245–8250. doi: 10.1073/pnas.0409902102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. National Climatic Data Center (NCDC), National Oceanic and Atmospheric Administration (NOAA) (May, 2012) NCDC Releases May 2012 U.S. Monthly Climate Report: US experiences second warmest May and warmest Spring on record. Available at http://www.ncdc.noaa.gov. Accessed February 15, 2013.
- 8. Global Forest Resources Assessment (2005) 15 Key findings. Available at http://foris.fao.org/static/data. Accessed February 15, 2013.
- 9.Cochrane M, Laurance W. Fire as a large-scale edge effect in Amazonian forests. J Trop Ecol. 2002;18:311–325. [Google Scholar]
- 10.Tilman D, Lehman C. Human-caused environmental change: Impacts on plant diversity and evolution. Proc Natl Acad Sci USA. 2001;98(10):5433–5440. doi: 10.1073/pnas.091093198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winter M, et al. Plant extinctions and introductions lead to phylogenetic and taxonomic homogenization of the European flora. Proc Natl Acad Sci USA. 2009;106(51):21721–21725. doi: 10.1073/pnas.0907088106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pennington RT, et al. Contrasting plant diversification histories within the Andean biodiversity hotspot. Proc Natl Acad Sci USA. 2010;107(31):13783–13787. doi: 10.1073/pnas.1001317107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu F, et al. Clustered patterns of species origins of nature-derived drugs and clues for future bioprospecting. Proc Natl Acad Sci USA. 2011;108(31):12943–12948. doi: 10.1073/pnas.1107336108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newman DJ, Cragg GM. Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007;70(3):461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 15.Strobel G, Daisy B. Bioprospecting for microbial endophytes and their natural products. Microbiol Mol Biol Rev. 2003;67(4):491–502. doi: 10.1128/MMBR.67.4.491-502.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilbert N. Biodiversity hope faces extinction. Nature. 2010;467(7317):764. doi: 10.1038/467764a. [DOI] [PubMed] [Google Scholar]
- 17. Hamilton A (2008) Medicinal plants in conservation and development: Case studies and lessons learnt. (Plantlife, Salisbury, United Kingdom)
- 18. Kasilo OMJ, Trapsida JM, Mwikisa CN, Lusamba-Dikassa PS (2010) An overview of the traditional medicine situation in the African region. (World Health Organization), 14:1–15. Available at www.aho.afro.who. Accessed July 28, 2013.
- 19. Wittenauer R, Smith L (2012) Update on 2004 background paper, BP 6.6 Stroke (World Health Organization), pp 1--46. Available at www.who.int/medicines. Accessed February 15, 2013.
- 20.Choo QL, et al. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244(4902):359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 21.Houghton M. Discovery of the hepatitis C virus. Liver Int. 2009;29(Suppl 1):82–88. doi: 10.1111/j.1478-3231.2008.01925.x. [DOI] [PubMed] [Google Scholar]
- 22.Fan H, Starks CM, Hughey H, Eldridge GR, Hu JF. Natural anti-HCV agents. Drugs Future. 2009;34(3):223–236. [Google Scholar]
- 23. National Center for HIV/AIDS, Viral Hepatitis, STD, TB Prevention, Division of Viral Hepatitis (2009), Disease Burden from Viral Hepatitis A, B, and C in the United States (Centers for Disease Control and Prevention), pp 1–5. Available at www.cdc.gov/hepatitis. Accessed July 13, 2012.
- 24. Hepatitis Central (July 2012). Available at www.hepatitis-central.com/hcv/hepatitis. Accessed August 2, 2012.
- 25.Gravitz L. Introduction: a smouldering public-health crisis. Nature. 2011;474(7350):S2–S4. doi: 10.1038/474S2a. [DOI] [PubMed] [Google Scholar]
- 26.Alexopoulou A, Dourakis SP. Genetic heterogeneity of hepatitis viruses and its clinical significance. Curr Drug Targets Inflamm Allergy. 2005;4(1):47–55. doi: 10.2174/1568010053622867. [DOI] [PubMed] [Google Scholar]
- 27.Murray CL, Rice CM. Hepatitis C: An unsuspected drug target. Nature. 2010;465(7294):42–44. doi: 10.1038/465042a. [DOI] [PubMed] [Google Scholar]
- 28.Gao M, et al. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature. 2010;465(7294):96–100. doi: 10.1038/nature08960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Azzam HS, Goertz C, Fritts M, Jonas WB. Natural products and chronic hepatitis C virus. Liver Int. 2007;27(1):17–25. doi: 10.1111/j.1478-3231.2006.01408.x. [DOI] [PubMed] [Google Scholar]
- 30. U.S. Food and Drug Administration (FDA), (May 2011) FDA briefing on boceprevir and telaprevir. Available at www.fda.gov. Accessed July 22, 2012.
- 31.Hossain P, Kawar B, El Nahas M. Obesity and diabetes in the developing world—a growing challenge. N Engl J Med. 2007;356(3):213–215. doi: 10.1056/NEJMp068177. [DOI] [PubMed] [Google Scholar]
- 32.Wild S, Roglic G, Green A, Sicree R, King H. Global prevalence of diabetes: Estimates for the year 2000 and projections for 2030. Diabetes Care. 2004;27(5):1047–1053. doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed] [Google Scholar]
- 33.Popov D. Novel protein tyrosine phosphatase 1B inhibitors: Interaction requirements for improved intracellular efficacy in type 2 diabetes mellitus and obesity control. Biochem Biophys Res Commun. 2011;410(3):377–381. doi: 10.1016/j.bbrc.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 34.Klaman LD, et al. Increased energy expenditure, decreased adiposity, and tissue-specific insulin sensitivity in protein-tyrosine phosphatase 1B-deficient mice. Mol Cell Biol. 2000;20(15):5479–5489. doi: 10.1128/mcb.20.15.5479-5489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lantz KA, et al. Inhibition of PTP1B by trodusquemine (MSI-1436) causes fat-specific weight loss in diet-induced obese mice. Obesity (Silver Spring) 2010;18(8):1516–1523. doi: 10.1038/oby.2009.444. [DOI] [PubMed] [Google Scholar]
- 36.Luteyn JL. A checklist of plant diversity, geographical distribution, and botanical literature. New York Bot Gard. 1999;84:1–278. [Google Scholar]
- 37.De Feo V. Ethnomedical field study in northern Peruvian Andes with particular reference to divination practices. J Ethnopharmacol. 2003;85(2-3):243–256. doi: 10.1016/s0378-8741(03)00017-5. [DOI] [PubMed] [Google Scholar]
- 38.Bussmann RW, Ashley Glenn A. Medicinal plants used in Peru for the treatment of respiratory disorders. Rev Peruana Biol. 2010;17(2):331–346. [Google Scholar]
- 39.Wiesmann C, et al. Allosteric inhibition of protein tyrosine phosphatase 1B. Nat Struct Mol Biol. 2004;11(8):730–737. doi: 10.1038/nsmb803. [DOI] [PubMed] [Google Scholar]
- 40.Lee PS, et al. Pharmacokinetic characteristics and hepatic distribution of IH-901, a novel intestinal metabolite of ginseng saponin, in rats. Planta Med. 2006;72(3):204–210. doi: 10.1055/s-2005-916201. [DOI] [PubMed] [Google Scholar]
- 41.Revill P, Serradell N, Bolos J, Rosa E. Telaprevir. Drugs Future. 2007;32:788–798. [Google Scholar]
- 42.Fürstner A, Ruiz-Caro J, Prinz H, Waldmann H. Structure assignment, total synthesis, and evaluation of the phosphatase modulating activity of glucolipsin A. J Org Chem. 2004;69(2):459–467. doi: 10.1021/jo035079f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.