Significance
Acute myeloid leukemia (AML) consists of a group of hematopoietic malignancies with considerable diversities in clinical and biological features. Recently, not only genetic abnormalities but also “oncometabolites,” such as 2-hydroxyglutarate (2-HG), have been found to play a role in driving AML pathogenesis and serve as potential disease markers. In this study on a large cohort of AML, we found that the serum 2-HG level was increased in 62 of 367 (17%) cases with distinct hematologic and biological features. Survival analysis performed in 234 patients without prognostic cytogenetic markers showed that increased 2-HG level was a poor predictor, demonstrating the potential of serum 2-HG as an independent marker for outcome evaluation of AML.
Keywords: biomarker, prognosis
Abstract
The 2-hydroxyglutarate (2-HG) has been reported to result from mutations of isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2) genes and to function as an “oncometabolite.” To evaluate the clinical significance of serum 2-HG levels in hematologic malignancies, acute myeloid leukemia (AML) in particular, we analyzed this metabolite in distinct types of human leukemia and lymphoma and established the range of serum 2-HG in appropriate normal control individuals by using gas chromatograph–time-of-flight mass spectrometry. Aberrant serum 2-HG pattern was detected in the multicenter group of AML, with 62 of 367 (17%) patients having 2-HG levels above the cutoff value (2.01, log2-transformed from 4.03 μg/mL). IDH1/2 mutations occurred in 27 of 31 (87%) AML cases with very high 2-HG, but were observed only in 9 of 31 (29%) patients with moderately high 2-HG, suggesting other genetic or biochemical events may exist in causing 2-HG elevation. Indeed, glutamine-related metabolites exhibited a pattern in favor of 2-HG synthesis in the high 2-HG group. In AML patients with cytogenetically normal AML (n = 234), high 2-HG represented a negative prognostic factor in both overall survival and event-free survival. Univariate and multivariate analyses confirmed high serum 2-HG as a strong prognostic predictor independent of other clinical and molecular features. We also demonstrated distinct gene-expression/DNA methylation profiles in AML blasts with high 2-HG compared with those with normal ones, supporting a role that 2-HG plays in leukemogenesis.
Acute myeloid leukemia (AML) represents a group of clonal hematopoietic progenitor malignancies with considerable diversities in clinical and biological features (1). In addition to clinical parameter, such as age and white blood cell counts (WBC), a variety of biomarkers have been shown to be predictive of outcome in AML patients, including cytogenetic characteristics and patterns of recurrent gene mutations in the blasts, as exemplified by nucleophosmin (NPM1), fms-related tyrosine kinase 3 (FLT3), and CCAAT/enhancer binding protein-alpha (CEBPA) (1). Even with the remarkable progress of genomic studies on AML (2, 3), clinically useful biomarkers with prognostic values are still needed in a part of cases for better clinical management, particularly in cytogenetically normal AML (CN-AML) patients. Of note, gene mutations of isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2) were reported recently not only in gliomas but also in AML (4, 5). The prognostic significance of these mutations appears controversial in AML (6–10), although a metaanalysis suggests a poor outcome in cases with IDH1 mutations (11).
IDHs are key enzymes that participate in the tricarboxylic acid metabolic cycle. Three members (IDH1, IDH2, and IDH3) are encoded by the IDH gene family and their activities are NADP(+)/NAD(+)-dependent. IDH1 and IDH2 catalyze the oxidative decarboxylation of isocitrate to α-ketoglutarate (α-KG). Mutant enzymes form a heterodimer, which display reduced catalytic activity to produce α-KG but a newly acquired activity to convert α-KG to 2-hydroxyglutarate (2-HG) (12). Recent studies have suggested that 2-HG may be an “oncometabolite” and play a role in driving malignant phenotype (13–15). Interestingly, 2-HG has been shown to competitively inhibit α-KG–dependent dioxygenases, such as the ten-eleven-translocation 2 (TET2) enzyme, and disturb the epigenetic regulatory network, leading to genome-wide histone and DNA damages with hypermethylation (12–14). Notably, in patients with d-2-hydroxyglutaric aciduria, there is an elevated risk of brain tumors (15). 2-HG can be metabolized by the enzyme 2-HG dehydrogenase (D2HGDH), but until now the possible contribution of genetic variants of this enzyme to the turnover of 2-HG has not yet been addressed.
Although IDH1/2 mutations are biomarkers in gliomas, the serum levels of 2-HG in these mutations are usually normal (16). In AML harboring IDH1/2 mutations, serum 2-HG concentrations were elevated in some cases (17–21). Very recent reports suggested that high 2-HG could predict IDH1/2 mutations in AML and be used as a marker for minimal residual disease during clinical remission (21, 22). However, the significance of serum 2-HG levels in prognosis remains obscure. In this work, we examined the levels of 2-HG as a part of the metabolomic study in a large series of AML cases, as well as other hematologic malignancies and healthy controls, and characterized their relevant clinical and molecular features. In particular, we attempted to address the potential prognostic value of 2-HG in AML.
Results
Serum 2-HG Levels in AML and Healthy Controls.
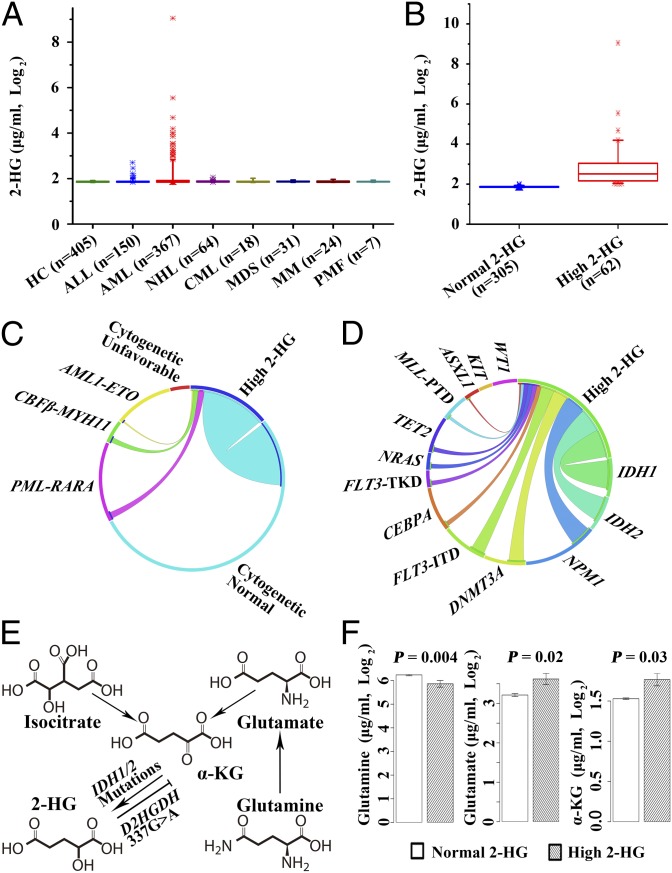
We assayed serum 2-HG in healthy controls and patients with hematologic malignancies. Because the distribution pattern of serum 2-HG levels among 405 healthy controls was rather non-Gaussian, a log2 transformation was performed so that the distribution tended to be close to normal, and the log2-transformed values of 2-HG (µg/mL, log2) were thus used in all of the subsequent steps of analysis. Of note, there was no difference in 2-HG levels between 233 males and 172 females [1.86 ± 0.03 vs. 1.86 ± 0.04 (µg/mL, log2), P = 0.39], and no correlation between age and 2-HG levels was observed (P = 0.83). As shown in Fig. 1A, in all disease groups investigated statistically significant differences of serum 2-HG levels were noted only between AML patients and normal control individuals. We then established a cutoff of 2.01 (log2-transformed from 4.03 μg/mL) (Fig.1A) based on the mean plus 5 SD of 2-HG values, according to a previously reported method for metabolomic investigations (23). Notably, the age groups [median (range), 46 (16–82) y, P = 0.66] and sex ratio (male/female: 233/172, P = 0.93) of the normal controls were comparable to AML patients [age, median (range), 46 (15–82) y; male/female: 210/157]. Among 367 AML cases in this multicenter series, 305 cases fell into the normal 2-HG group with 2-HG values less than the cutoff [median (interquartile range, IQR) serum 2-HG: 1.85 (1.84–1.88) (µg/mL, log2)], whereas the other 62 cases were defined as the high 2-HG group [median (IQR) serum 2-HG: 2.52 (2.14–3.07) (µg/mL, log2)] (Fig. 1B).
Fig. 1.
The distribution of 2-HG in different hematological malignancies and healthy controls and the correlation between a high level of 2-HG and cytogenetic abnormalities, gene mutation status, and metabolic pathway changes. Box plots show the distribution of serum 2-HG levels in healthy controls (HC) and several hematological malignancies (A), and in AML patients with normal or high 2-HG levels (B). ALL, acute lymphoblastic leukemia; CML, chronic myeloid leukemia; MDS, myelodysplastic syndrome, MM, multiple myeloma; NHL, non-Hodgkin lymphoma; PMF, primary myelofibrosis. Note that rare cases with high 2-HG were observed in ALL (5 of 150, 3.3%) and NHL (1 of 64, 1.6%), whereas no case with increased 2-HG was found in other disease types. Circos diagrams representing correlation between high levels of 2-HG and distinct cytogenetic abnormalities in AML (C) or with gene mutation events in CN-AML (D). Cytogenetic subtypes: favorable: t(15;17)/PML-RARA, t(8;21)/AML1-ETO, and inv16/CBFβ-MYH11; unfavorable: t(9;22), inv(3)/t(3;3), −5, −7, del(5q), del(7p), 11q23, and complex translocations. CN-AML indicates cases having no cytogenetically identifiable abnormalities. Note the comparison of concentrations of metabolites directly related to the 2-HG production pathway (E) between high and normal 2-HG AML groups (F).
Clinical Characteristics, Immunophenotype Attributes, and Molecular Hallmarks of AML with High 2-HG.
For the purpose of stratifying the enrolled patients for outcome analysis, we characterized the basic features, including clinical and cytogenetic ones, with regard to 2-HG levels in AML patients. We found that patients with high 2-HG were of significantly older age (P < 0.001), with higher bone marrow (BM) blast percentage (P = 0.008), higher frequency of AML-M0/M1 subtypes (P < 0.01), and less AML with t(8;21) subtype (P = 0.006) than those in the normal 2-HG group (Table 1). However, no significant correlation between 2-HG levels and WBC was observed (P = 0.18).
Table 1.
Clinical characteristics of AML patients with high and normal levels of 2-HG
| Variable | Normal 2-HG | High 2-HG | P value |
| Patients, n (%) | 305 (83) | 62 (17) | |
| Age (yrs), median (range) | 43 (15-82) | 54 (16-77) | <0.001 |
| Male, n (%) | 172 (56.39) | 38 (61.29) | 0.48 |
| Blasts (%), median (range) | 66 (18-99) | 78 (28-97) | 0.008 |
| WBC (109/L), median (range) | 6.62 (0.1–290) | 13.32 (0.1–292) | 0.18 |
| FAB subtype, n (%) | <0.001 | ||
| M0 | 5 (1.60) | 6 (9.68) | 0.004 |
| M1 | 4 (1.30) | 8 (12.90) | <0.001 |
| M2 | 73 (24.00) | 6 (9.68) | 0.01 |
| M3 | 55 (18.00) | 5 (8.06) | 0.05 |
| M4 | 70 (23.00) | 14 (22.58) | 0.95 |
| M4eo | 8 (2.60) | 3 (4.84) | 0.41 |
| M5 | 77 (25.20) | 19 (30.65) | 0.38 |
| M6 | 10 (3.30) | 1 (1.61) | 0.70 |
| Not classified* | 3 (1.00) | 0 (0) | 1.00 |
| Cytogenetics, n (%) | <0.001 | ||
| t(15;17)/PML-RARA | 55 (18.03) | 5 (8.06) | 0.05 |
| t(8; 21)/AML1-ETO | 43 (14.10) | 1 (1.61) | 0.006 |
| inv(16)/CBFβ-MYH11 | 12 (3.93) | 4 (6.45) | 0.49 |
| CN-AML† | 182 (59.67) | 52 (83.87) | <0.001 |
| Unfavorable‡ | 13 (4.26) | 0 (0) | 0.48 |
| Gene mutations§ | |||
| IDH1, n (%) | 5 (1.67) | 20 (32.26) | <0.001 |
| IDH2, n (%) | 8 (2.67) | 16 (25.81) | <0.001 |
| FLT3-ITD, n (%) | 29 (9.83) | 10 (16.95) | 0.11 |
| CEBPA, n (%)¶ | 26 (9.09) | 3 (5.66) | 0.59 |
| NPM1, n (%) | 38 (13.15) | 15 (25.86) | 0.01 |
| DNMT3A, n (%) | 21 (7.19) | 11 (19.30) | 0.004 |
| MLL-PTD, n (%) | 10 (3.57) | 3 (5.56) | 0.45 |
| Treatment protocols‖ | 0.06 | ||
| P1 | 23 (12.64) | 4 (7.69) | |
| P2 | 118 (64.84) | 28 (53.85) | |
| P3 | 41 (22.53) | 20 (38.46) |
P values were calculated by means of nonparametric test for continuous variables and a χ2 test for categorical variables, respectively.
Not classified: AML patients without typical morphological characteristics defined in the FAB nomenclature.
Cytogenetically normal cases (CN-AML) and
patients with unfavorable cytogenetic features were defined in the legend to Fig. 1.
Gene mutation data were available in 342 (93%) patients for analysis.
Double-allele CEBPA mutations.
Treatment protocols for CN-AML: P1, homoharringtonin-based treatment; P2, DA regimen 45 mg⋅m2⋅d); P3, elderly CN-AML patients who received individualized treatment.
Now that biological data at disease onset in the majority of AML cases in this series were available, we were interested in those features related to high serum 2-HG levels. Notably, CN-AML cases were seen as more common in the high 2-HG group (52 of 62 cases, 83.87%) than in the normal 2-HG group (182 of 305 cases, 59.67%, P < 0.001) (Fig. 1C and Table 1). With regard to immunophenotype, expression of CD15 (P = 0.02) and CD64 (P = 0.01) was at a lower level in BM blasts in the high 2-HG group (Table S1). These negative correlations were still observed in CN-AML with respect to expression of CD15 (P = 0.001), CD64 (P = 0.02), and HLA-DR (P = 0.003), respectively (Table S1). In univariate analysis, more frequent mutations of IDH1 (P < 0.001), IDH2 (P < 0.001), NPM1 (P = 0.01), and DNMT3A (P = 0.004) were found in the high 2-HG group compared with that with normal 2-HG (Table 1). However, only IDH1 [P < 0.001, odds ratio (OR) = 97.33] and IDH2 (P < 0.001, OR = 43.30) mutations were closely associated with high levels of 2-HG in multivariate analysis (Table S2). In contrast, fusion genes of PML-RARA and AML1-ETO were rare in the high 2-HG group (Fig. 1C). Similar association of high 2-HG and genetic events was also found in CN-AML (Fig. 1D).
When the relationship of 2-HG levels and IDH1/2 hotspot mutations was further examined in all AML (367 cases), 36 of 62 (58.06%) patients with high 2-HG had these mutations, whereas only 13 of 305 (4.26%) patients with normal 2-HG exhibited mutations of IDH1/2 genes. We proceeded to make an in-depth analysis of 10 cases with high 2-HG but negative IDH1/2 hotspot mutations. On the one hand, to exclude the possibility of gene mutations in a subpopulation of leukemic cells, which might be below the sensitivity of routine Sanger sequencing, we used a very sensitive allele-specific PCR for detecting hotspot mutations of IDH1/2 gene. On the other hand, we conducted sequencing of the whole IDH1/2 coding region for possible mutations beyond the hotspot. The results showed that no IDH1/2 mutations were detected in these cases. We then scrutinized the high 2-HG levels and mutation status in two subsets divided by the median of 2-HG levels [2.53 (µg/mL, log2)]. Interestingly, in 31 cases with very high 2-HG [> 2.53 (µg/mL, log2)], 27 of 31 cases (87%) showed IDH1/2 hotspot mutations. In contrast, among 31 cases with moderately increased 2-HG [2.01–2.53 (µg/mL, log2)], only 9 of 31 cases (29%) had these mutations. This result suggested that among the high 2-HG and wild-type IDH1/2 cases, particularly those with moderately increased 2-HG level, effects of gene/protein variations other than IDH1/2 could contribute to the abnormal serum levels of this metabolite (see below and Table S3).
Relationship of High Serum 2-HG Levels to Metabolite Precursors and Variants of D2HGDH.
Because 2-HG is a product of the glutamine metabolic pathway under the neomorphic activity of mutant IDH1/2 (Fig. 1E), we compared the levels of glutamine, glutamate, and α-KG in both high and normal 2-HG groups using nonparametric test. Indeed, the high 2-HG group showed higher serum levels of glutamate (P = 0.02) and α-KG (P = 0.03) and lower level of glutamine (P < 0.01) (Fig. 1F), with higher ratios of 2-HG to α-KG and glutamate to glutamine (Table S4). Multivariable regression analysis indicated that both IDH1/2 mutations and serum α-KG levels were significantly correlated to 2-HG levels (Table S2).
It has been established that D2HGDH and l-2-hydroxyglutarate dehydrogenase (L2HGDH) are enzymes that convert 2-HG to α-KG. We therefore analyzed serum D2HGDH in AML patients with available samples and found that there was no difference of D2HGDH protein levels between the high and normal 2-HG groups regardless of IDH1/2 mutations (Fig. S1A). Nevertheless, it is worth pointing out that an SNP-related (D2HGDH, 337G > A) amino acid substitution of D2HGDH, R55Q, exhibited significantly higher incidence in patients with high 2-HG but wild-type IDH1/2 (8 of 12 cases tested, 66.7%) than those with normal 2-HG and wild-type IDH1/2 (5 of 15 cases investigated, 25%) (P = 0.03); whereas patients with normal 2-HG but IDH1/2 mutations had no differences in terms of frequency of D2HGDH R55Q compared with those with normal 2-HG and wild-type IDH1/2 (Fig. S1B). Of note, patients with high 2-HG but wild-type IDH1/2 displayed the highest levels of serum α-KG [median (IQR), 1.68 (1.47–2.52), (µg/mL, log2)], whereas cases with normal 2-HG but IDH1/2 mutations tended to have the lowest serum α-KG [median (IQR), 1.45 (1.40–1.59), (µg/mL, log2)] (Fig. S1C). Moreover, statistically higher 2-HG levels were observed in the D2HGDH R55Q SNP group than in the wild-type D2HGDH group (P = 0.04) (Fig. S1D). We did not find mutations in L2HGDH in BM cells from 10 patients with moderately high 2-HG and wild-type IDH1/2 as well as 14 cases with normal 2-HG and wild-type IDH1/2. Meanwhile, there were no significant differences in the expression levels of D2HGDH (P = 0.49) or L2HGDH (P = 0.31) between the two above groups.
High 2-HG Is Associated with Poor Clinical Outcome in CN-AML.
In the analysis of clinical outcome, we focused on the prognostic significance of 2-HG in CN-AML patients (234 cases), in that prognostic impact of both favorable and unfavorable cytogenetic markers were eliminated in this group. Median follow-up time of patients who were still alive was 22 mo. The estimated 2-y overall survival (OS) and event-free survival (EFS) rates were 38% [95% confidence interval (CI), 31–45%] and 32% (95% CI, 25–39%), respectively.
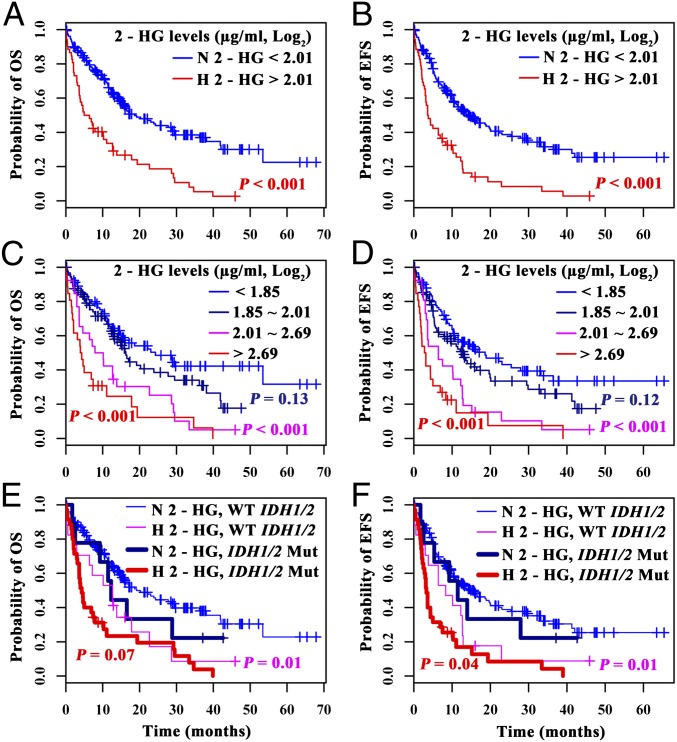
Because of the characteristic of the distribution of 2-HG levels (Fig. 1B), the metabolite was suitable for use as a dichotomous or categorical variable in outcome analysis in CN-AML. As such, when 2-HG levels were used as a dichotomous variable, 22 of 52 patients (42%) with high 2-HG reached complete remission (CR), in contrast to the normal 2-HG group where CR was achieved in 118 of 182 patients (65%) (P < 0.01). In terms of survival data, the median OS of 2-HG high and normal groups were 6 and 19 mo, respectively (P < 0.001) (Fig. 2A), whereas the median EFS of the two groups were 4 and 14 mo, respectively (P < 0.001) (Fig. 2B). It is worth pointing out that there was no statistical difference in terms of therapeutic protocols between the high and normal 2-HG groups (Table 1). When 2-HG levels were used as a four-category variable [<1.85 (µg/mL, log2); 1.85–2.01 (µg/mL, log2); 2.01–2.69 (µg/mL, log2); >2.69 (µg/mL, log2), by using the medians of 2-HG levels in both normal and high groups], the metabolite exerted a statistically significant effect on the outcome with a dose-dependent tendency (Fig. 2 C and D).
Fig. 2.
Kaplan–Meier survival analysis of CN-AML patients. H 2-HG, high 2-HG; N 2-HG, normal 2-HG; WT IDH1/2: wild-type IDH1/2; IDH1/2 Mut: IDH1/2 mutations. (Top row) OS (A) and EFS (B) curves of the patients according to 2-HG levels. (Middle row) OS (C) and EFS (D) curves of patients with four categories of 2-HG levels [<1.85 (µg/mL, log2); 1.85–2.01 (µg/mL, log2); 2.01–2.69 (µg/mL, log2); >2.69 (µg/mL, log2), by using the median of 2-HG levels in normal and high groups in CN-AML]. Note the dose–effect tendency of the four curves depending on 2-HG levels for both OS and EFS, with the two curves of high 2-HG being statistically significantly worse compared with the curve with lowest 2-HG (P values: < 0.001, respectively). (Bottom row) Cases are stratified into four subgroups: N 2-HG with WT IDH1/2, H 2-HG with WT IDH1/2, N 2-HG with IDH1/2 Mut and H 2-HG with IDH1/2 Mut. P values of OS (E) and EFS (F) are shown in subgroup analysis with stratification on IDH1/2 mutation status. P values were calculated by the log-rank test.
Because high serum 2-HG levels were associated with age, BM blasts, and gene mutations, we checked whether the relationship between the poor outcome and high 2-HG was confounded or modified by these factors. The univariate analysis showed age, WBC, and genetic markers, such as FLT3-ITD, IDH1/2, and DNMT3A mutations as risk factors, and biallelic CEBPA mutation as favorable one for OS or EFS (Table S5). By using stratified analysis and testing the interaction effect, we found that high 2-HG was a poor indicator after stratification on IDH1/2 mutation status (Fig. 2 E and F). In the stratified analysis, there was no evidence that the prognostic impact of high 2-HG would be severely confounded or modified by clinical variables or gene mutations (Fig. S2).
We then set out to adjust the prognostic value of 2-HG by using multivariate analysis with the assumption that these factors were the confounders. Importantly, prognostic effect of 2-HG did not depend on IDH1/2 mutations and the other well-known confounding factors, such as age, WBC counts, percentage of BM blasts, treatment protocols, and prognosis-related gene mutation markers, such as FLT3-ITD, NPM1, CEBPA, DNMT3A, and MLL-PTD (MLL-partial tandem duplication) (Table 2). The impact of several genetic markers including IDH1/2 mutations disappeared, whereas CEBPA biallelic mutations remained as favorable maker (Table 2). In the multivariate models, when results from patients older than 60 y or those treated with less-intensive chemotherapy were excluded, high 2-HG was still a poor predictor (Table S6).
Table 2.
Multivariate analysis for OS and EFS in CN-AML
| Variables | Overall survival |
Event free survival |
||
| P value | HR (95% CI) | P value | HR (95% CI) | |
| 2-HG* | 0.03 | 1.89 (1.05, 3.38) | 0.01 | 2.11 (1.20, 3.70) |
| WBC† | 0.001 | 1.006 (1.003,1.01) | 0.002 | 1.006 (1.002,1.01) |
| Age† | 0.04 | 1.02 (1.00, 1.04) | 0.26 | 1.01 (0.99, 1.03) |
| BM blasts (%)† | 0.19 | 1.01 (0.99, 1.02) | 0.14 | 1.01 (0.99, 1.02) |
| P1 vs. P2‡ | 0.24 | 0.71 (0.39, 1.27) | 0.07 | 0.58 (0.32, 1.05) |
| P3 vs. P2‡ | 0.72 | 0.89 (0.48, 1.65) | 0.80 | 0.92 (0.50, 1.71) |
| IDH1/2§ | 0.75 | 1.10 (0.61, 2.00) | 0.54 | 1.19 (0.67, 2.11) |
| FLT3-ITD§ | 0.94 | 1.02 (0.58, 1.80) | 0.60 | 1.16 (0.67, 2.01) |
| CEBPA§ | 0.03 | 0.45 (0.22, 0.91) | 0.04 | 0.51 (0.27, 0.97) |
| NPM1§ | 0.72 | 0.92 (0.57, 1.48) | 0.35 | 0.80 (0.50, 1.27) |
| DNMT3A§ | 0.31 | 1.35 (0.75, 2.41) | 0.34 | 1.32 (0.75, 2.31) |
| MLL-PTD§ | 0.83 | 1.09 (0.50, 2.40) | 0.25 | 1.52 (0.75, 3.09) |
2-HG was used as a dichotomous variable.
Age, WBC, and BM blasts (%) were analyzed as continuous variables.
Treatment protocols: P1, homoharringtonin-based treatment; P2, DA regimen (45 mg⋅m2⋅d); P3, elderly CN-AML patients who received individualized treatment.
Mutation vs. wild-type. Hazard risk (HR) greater than 1 indicates that patients were more likely to bear poorer prognosis than those in other groups. CEBPA: biallelic CEBPA mutations.
Analysis of Gene-Expression Profile and DNA Methylation Patterns in AML Blasts with High 2-HG.
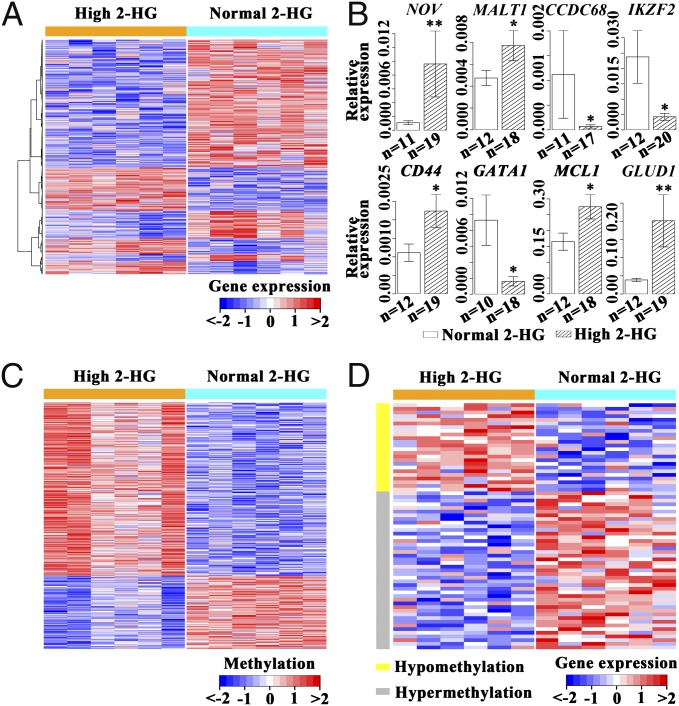
We compared transcriptome patterns of leukemic blasts from six patients with high 2-HG and six patients with normal 2-HG. From these patterns, 846 genes were down-regulated and 378 were overexpressed in the high 2-HG group (Fig. 3A). Quantitative RT-PCR confirmed the RNA microarrays results when 2-HG was taken as a dichotomous variable (Fig. 3B). Kyoto Encyclopedia of Genes and Genomes pathway analysis revealed potential involvement of genes in 78 pathways (Table S7). Notably, in the cancer-related pathway, the oncogenes NOV and MALT-1 showed an overexpression, whereas expression of tumor-suppressors CCDC68 and IKZF2 was repressed. Moreover, antiapoptotic genes MCL1 and NFκB, as well as growth factors FGF14 and FGFR2, were overexpressed. Examination of genes involved in hematopoietic lineages revealed high expression of CD44 (24) and reduced expression of GATA1, CSF1, EPO, and CR1. Among metabolism-related genes, it was noteworthy that GLUD1, the enzyme that converts glutamate to α-KG, was overexpressed. A number of genes involved in the tricarboxylic acid cycle, such as OGDH, and the electron transmission chain, such as COQ3, were down-regulated (Table S7). We also analyzed the DNA methylation patterns in high and normal 2-HG groups and found hypermethylation in 143 genes and hypomethylation in 60 genes (Fig. 3C). However, only 67 genes displayed correlation between DNA methylation status and gene-expression patterns (Fig. 3D).
Fig. 3.
Gene-expression/DNA methylation patterns in leukemia blasts of AML patients with high 2-HG compared with those with normal 2-HG. (A) Display of 1,224 genes with significant differences of expression levels between AML cases with high and normal 2-HG. (B) Quantitative RT-PCR results for eight genes showing different expression levels between AML cases with high and normal 2-HG. *P < 0.05, **P < 0.01. (C) DNA segments of 203 genes with significant differences of methylation levels between AML cases with high or normal 2-HG. (D) Display of 67 genes with correlation between modification of DNA methylation patterns and changes in expression levels in the high 2-HG group compared with the normal 2-HG group.
Discussion
It has been well accepted that cytogenetic and molecular factors are independent predictors for the prognosis of AML patients. However, according to the widely used guidelines of molecular markers for the stratification of AML, only half of the patients can be categorized (1). Identification of other markers, including metabolites, may greatly widen the AML stratification toolkit. The 2-HG detected in tumor by using magnetic resonance spectroscopy were recently described as a biomarker in gliomas (25), whereas very recent reports were in support of the value of serum 2-HG as a biomarker of AML (21). Moreover, efforts are underway to explore drugs inhibiting the aberrant activities of IDH1/2, and identification of cases in both hematological malignancies and solid tumors with high 2-HG levels could be eventually helpful for the clinical trials of these drugs. In this context, we carried out a metabolomic study in a large cohort of hematological malignancies from different centers in China.
In a series of metabolomic markers, significantly higher serum 2-HG was detected in AML but not in other hematological malignancies. In AML patients with high 2-HG levels, there were fewer cases with the t(8;21) subtype but significantly more CN-AML. Increased 2-HG tended to be more related to an undifferentiated status of AML cells, because the percentage of BM blasts were much higher and AML-M0/M1 subtypes were more frequent but the level of CD15 was much lower in these cases. Particularly, we found that high 2-HG was related to serum α-KG level and poor molecular markers, such as IDH1/2 and DNMT3A (Table S2). It was revealed that a very high serum level of 2-HG (above the median level in high 2-HG group) could predict IDH1/2 mutations because the mutation rates reached to 87%, similar to a recent report in E1900 trial group (21). However, in cases with moderately increased serum 2-HG, in addition to IDH1/2 mutations (29%), other factors could contribute to the aberrantly increased 2-HG. To this end, the R55Q variant of the D2HGDH gene was more frequently detected in cases with high 2-HG and wild-type IDH1/2 (Fig. S1B), which reminds us of the fact that the genetic disease caused by the mutation of D2HGDH was associated with a high serum 2-HG level (26). Given that accumulation of high 2-HG might require not only a neoenzymatic activity of the IDH1/2 mutant but also the availability of α-KG as a substrate, it was interesting to note that the serum level of α-KG was increased in AML cases with high 2-HG, especially in those with wild-type IDH1/2. Moreover, a significantly enhanced IDH1 expression was detected in AML cells with DNMT3A mutation (6), which could lead to an increased production of α-KG as the precursor of 2-HG. In fact, we found DNMT3A mutations in several AML patients with high 2-HG but wild-type IDH1/2. Hence, we speculate that still other unknown genes/proteins involved in the catabolism of 2-HG, or individual differences with regard to diet cultures or renal excretion of the metabolite, could contribute to the high 2-HG levels. In another aspect, serum 2-HG levels were normal in a small subset of patients with IDH1/2 mutations, and α-KG levels in this group tended to be the lowest we found. Because there are two different 2-HG enantiomers, d-2HG and l-2HG, and previous experiments showed only d-2HG was increased in IDH1/2 mutations (12, 19), the determination of the d-2HG/l-2HG ratio as a more sensitive and specific clinical test may be developed in the future. Taking the above analyses as a whole, we believe that IDH1/2 mutations should be considered primarily responsible for the very highly increased serum 2-HG, whereas variation of genes/proteins leading to elevated expression of IDH1/2 or changed clearance of 2-HG could also be a contributing factor. It is also possible that enhanced activity of the glutamine pathway generating increased α-KG supply could direct excessive synthesis of 2-HG.
In our series, high 2-HG was associated with poor outcome (Fig. 2 and Table 2). Given that many of the well-established AML prognosis parameters were also correlated with high 2-HG in our study (Table 1), it was important to test whether 2-HG is an independent risk factor. Several issues in this regard drew our attention. First, the distribution of 2-HG level was not normal in AML (Fig. 1B), justifying the use of 2-HG as a dichotomous or categorical variable. Second, to reduce the heterogeneity of AML patient populations and to evaluate the prognostic value of 2-HG in a strict way, CN-AML should be addressed instead of the whole AML series. Third, stratification on IDH1/2 mutations or other prognosis-related factors and adjustment in the context of potential confounders were required for evaluating the prognostic value of 2-HG levels. By doing so, we were able to provide evidence to suggest that 2-HG is an independent factor for the prognosis and, hence, clinical stratification of AML cases in China. Of importance, there was no statistically significant confounding between serum 2-HG and potential confounders in stratified survival analysis (Fig. 2 and Fig. S2), indicating that the prognostic impact of high 2-HG should be independent of the other predictors. This independent effect of high 2-HG on poor outcome could also be evidenced in adjustment analysis by IDH1/2 mutation status and other confounders. The fact that IDH1/2 or DNMT3A mutations had no effect on OS and EFS in multivariate analysis implies that 2-HG levels could submerge their significance because of potential correlations (Table 2).
A major difference between this study and a recent report by DiNardo et al. (21) was that in the latter study on AML patients from the United States (E1900 trial), no negative prognostic impact of high serum 2-HG was observed. At least two factors could underlie this difference. In our series, the major treatment for remission induction and consolidation was a DA protocol (daunorubicin; 146 of 234 CN-AML cases, 62%) with a relatively low dose of daunorubicin (45 mg⋅m2⋅d) compared with a very high DA dose used the test group of the E1900 trial (90 mg⋅m2⋅d) (27). This difference in the dosage of anthracycline could also be translated into a difference of outcome, in that the CR rate (60%) and 3-y OS (28%) in our series were close to the control group of E1900 trial (CR rate: 57.3%; 3-y OS: 35%, estimated from survival curve) but inferior to the test group of that trial (CR rate: 70.6%; 3-y OS: 42%, estimated from survival curve). Of note, the previously described negative outcome predictors, such as DNMT3A and IDH1 mutations, were even reported as a positive factor in the test group of E1900 trial (10). Hence, the impact of high 2-HG on the outcome could be abrogated in part because of a much more intensive chemotherapy. Another possible explanation could be that the difference results from the distinct genetic backgrounds of the AML populations investigated in the two studies. Indeed, distinct frequencies of gene mutational events, as well as enzyme variants involved in the tolerance to chemotherapeutic agents, have been reported in Chinese AML patients compared with AML patients of different ancestries (28). It is therefore necessary to study larger patient series in different populations before making a definite conclusion about the prognostic significance of 2-HG in AML.
Because increased 2-HG could be involved in disease mechanism, the pathologic consequence of elevated 2-HG should be investigated in the fresh samples from AML patients. It has been well established that in enzymatic reaction, the ratio between substrate and product should point to its preferential direction. In other words, abnormal metabolites might enter into the serum pool as a result of adaptation of upstream events in the pathway to the downstream abnormalities of enzymatic dynamics. As shown in the present work, higher ratios of 2-HG/α-KG and glutamate/glutamine in high 2-HG AML should reflect an abnormality of the glutamine metabolism pathway. It has been shown that the ultimate effect of 2-HG is to alter DNA and histone methylation, causing aberrant gene expression. A recent study demonstrated that 2-HG could block cell differentiation through inhibiting the H3K9 demethylase KDM4C in the absence of major changes of DNA methylation (29), whereas the prolylhydroxylases, which regulates the stability of the transcription factor HIF1α, could be directly modified by 2-HG (30). Indeed, data from this study are in support of these findings, in that increased 2-HG was correlated to an undifferentiated status of leukemia cells, and a major change of gene-expression patterns (1,224 genes) was observed in fresh blasts from the high 2-HG AML group, whereas only 203 genes showed altered DNA methylation. The number of genes having correlation with altered expression was even smaller. Notably, expression of some oncogenes and antiapoptotic genes was enhanced, whereas expression of several tumor suppressors was down-regulated. Interestingly, there was an overexpression of GLUD1, which encodes a gene for the enzyme catalyzing the conversion of glutamate to α-KG. Our data thus provide unique evidence that 2-HG may contribute to the pathogenesis of AML.
Methods
Patients and Study Design.
From 2007 to 2012, 661 adult patients with hematologic malignancies, including 367 AML and 405 healthy individuals, were entered into this study. The 367 cases included all subtypes of AML in seven hematology centers in Shanghai, Beijing, Suzhou, Hangzhou, Dalian, Nanjing, and Shenyang, China, with their clinical and hematological data at the diagnosis available (Table 1). All AML patients were diagnosed according to the World Health Organization classification system (1). Morphologic/immunophenotypic/cytogenetic analysis was performed in each of the hematology centers and assays of serum metabolites and molecular markers were conducted at the Shanghai Institute of Hematology. Informed consent was obtained from all patients in the seven hematology centers according to the regulation of the Institutional Review Boards of the related Universities/Hospitals in agreement with Declaration of Helsinki. Details of treatment protocols are available in SI Methods.
Sample Preparation and Gas Chromatograph–Time-of-Flight MS Analysis.
Serum samples collected from fasting subjects were stored at −80 °C until use. Samples were pretreated and analyzed by gas chromatograph–time-of-flight MS, as described in SI Methods.
Cytogenetic/Molecular Marker Assays, Microarray Expression Profiling, and Methylation Analysis.
Cytogenetic analysis of BM samples was conducted as previously described (6). Gene mutations/fusions were detected as previously reported (6). Affymetrix Human Genome U133 Plus 2.0 Array GeneChip microarrays were used to assess the total RNA samples. DNA samples were extracted for the HG18 Methylation 2.1 M Deluxe Promoter Array to identify the methylated DNA regions (details are in SI Methods).
Statistical Analysis.
The relation between 2-HG levels and various patient characteristics was determined by nonparametric test for continuous variables and χ2 test for categorical variables. Both OS (with failure defined as death as a result of any cause, live censored at the time of last follow-up) and EFS (with failure defined as relapse or death after CR or induction failure) were considered as endpoints for survival analysis. The Kaplan–Meier method and Cox proportional hazard regression models used in univariate or multivariate analysis were performed to determine the prognostic value of the 2-HG level. Other detailed statistical methods are in SI Methods. All statistical analyses were conducted with R statistical packages, v2.15.0 (www.r-project.org) and SAS software, v9.3 (www.sas.com). The two-sided level of significance was set at P < 0.05.
Supplementary Material
Acknowledgments
We thank Keqin Kathy Li for valuable discussion; Shu-Min Xiong, Bing Chen, Xiang-Qin Weng, Yue-Ying Wang, Fei-Fei Chen, and Wan-Mao Ni for hematologic morphological and cytogenetic analyses; Bing-Hua Su (Shanghai Jiao Tong University School of Medicine), Bai-Rong Shen (Center for Systems Biology, Soochow University), and Ping Liu (University of Western Australia) for statistical assistance; and all patients and investigators from the participating institutions. This work was supported by China 973 Program (2013CB966800, 2010CB529200); a Special Grant of Ministry of Health (201202003); Mega-Projects of Scientific Research for the 12th Five-Year Plan (2013ZX09303302) and National 863 Program (2012AA02A505); the Shanghai Municipal Natural Science Foundation (12ZR1418400); the National Natural Science Foundation of China (81123005); and Samuel Waxman Cancer Research Foundation Co-PI Program.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1315558110/-/DCSupplemental.
References
- 1. Liesveld JL, et al. (2010) Acute myeloid leukemia. Williams Hematology, eds Kaushansky K, et al. (McGraw-Hill, New York), 8th Ed, pp 1277–1330.
- 2.Cancer Genome Atlas Research Network Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen SJ, Shen Y, Chen Z. A panoramic view of acute myeloid leukemia. Nat Genet. 2013;45(6):586–587. doi: 10.1038/ng.2651. [DOI] [PubMed] [Google Scholar]
- 4.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mardis ER, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361(11):1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen Y, et al. Gene mutation patterns and their prognostic impact in a cohort of 1185 patients with acute myeloid leukemia. Blood. 2011;118(20):5593–5603. doi: 10.1182/blood-2011-03-343988. [DOI] [PubMed] [Google Scholar]
- 7.Thol F, et al. Prognostic impact of IDH2 mutations in cytogenetically normal acute myeloid leukemia. Blood. 2010;116(4):614–616. doi: 10.1182/blood-2010-03-272146. [DOI] [PubMed] [Google Scholar]
- 8.Ravandi F, et al. Prognostic significance of alterations in IDH enzyme isoforms in patients with AML treated with high-dose cytarabine and idarubicin. Cancer. 2012;118(10):2665–2673. doi: 10.1002/cncr.26580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boissel N, et al. Prognostic impact of isocitrate dehydrogenase enzyme isoforms 1 and 2 mutations in acute myeloid leukemia: A study by the Acute Leukemia French Association group. J Clin Oncol. 2010;28(23):3717–3723. doi: 10.1200/JCO.2010.28.2285. [DOI] [PubMed] [Google Scholar]
- 10.Patel JP, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366(12):1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou KG, et al. Potential application of IDH1 and IDH2 mutations as prognostic indicators in non-promyelocytic acute myeloid leukemia: A meta-analysis. Leuk Lymphoma. 2012;53(12):2423–2429. doi: 10.3109/10428194.2012.695359. [DOI] [PubMed] [Google Scholar]
- 12.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chowdhury R, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12(5):463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu W, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19(1):17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sonoda Y, Tominaga T. 2-hydroxyglutarate accumulation caused by IDH mutation is involved in the formation of malignant gliomas. Expert Rev Neurother. 2010;10(4):487–489. doi: 10.1586/ern.10.19. [DOI] [PubMed] [Google Scholar]
- 16.Capper D, et al. German Glioma Network 2-Hydroxyglutarate concentration in serum from patients with gliomas does not correlate with IDH1/2 mutation status or tumor size. Int J Cancer. 2012;131(3):766–768. doi: 10.1002/ijc.26425. [DOI] [PubMed] [Google Scholar]
- 17.Sellner L, et al. Increased levels of 2-hydroxyglutarate in AML patients with IDH1-R132H and IDH2-R140Q mutations. Eur J Haematol. 2010;85(5):457–459. doi: 10.1111/j.1600-0609.2010.01505.x. [DOI] [PubMed] [Google Scholar]
- 18.Pollyea DA, et al. 2-Hydroxyglutarate in IDH mutant acute myeloid leukemia: Predicting patient responses, minimal residual disease and correlations with methylcytosine and hydroxymethylcytosine levels. Leuk Lymphoma. 2013;54(2):408–410. doi: 10.3109/10428194.2012.701009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward PS, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17(3):225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gross S, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med. 2010;207(2):339–344. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DiNardo CD, et al. Serum 2-hydroxyglutarate levels predict isocitrate dehydrogenase mutations and clinical outcome in acute myeloid leukemia. Blood. 2013;121(24):4917–4924. doi: 10.1182/blood-2013-03-493197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fathi AT, et al. Prospective serial evaluation of 2-hydroxyglutarate, during treatment of newly diagnosed acute myeloid leukemia, to assess disease activity and therapeutic response. Blood. 2012;120(23):4649–4652. doi: 10.1182/blood-2012-06-438267. [DOI] [PubMed] [Google Scholar]
- 23.Millington D, Koeberl D. Metabolic screening in the newborn. Growth Genetics and Hormones. 2003;19(3):33–38. [Google Scholar]
- 24.Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med. 2006;12(10):1167–1174. doi: 10.1038/nm1483. [DOI] [PubMed] [Google Scholar]
- 25.Kalinina J, et al. Detection of “oncometabolite” 2-hydroxyglutarate by magnetic resonance analysis as a biomarker of IDH1/2 mutations in glioma. J Mol Med (Berl) 2012;90(10):1161–1171. doi: 10.1007/s00109-012-0888-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kranendijk M, et al. Evidence for genetic heterogeneity in D-2-hydroxyglutaric aciduria. Hum Mutat. 2010;31(3):279–283. doi: 10.1002/humu.21186. [DOI] [PubMed] [Google Scholar]
- 27.Fernandez HF, et al. Anthracycline dose intensification in acute myeloid leukemia. N Engl J Med. 2009;361(13):1249–1259. doi: 10.1056/NEJMoa0904544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wheeler HE, et al. Genome-wide local ancestry approach identifies genes and variants associated with chemotherapeutic susceptibility in African Americans. PLoS ONE. 2011;6(7):e21920. doi: 10.1371/journal.pone.0021920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu C, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao S, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324(5924):261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.