Significance
Gene families significantly influence organismal diversity and adaptation, but how they evolved and are controlled is not fully clear. Using the Arabidopsis F-Box protein superfamily as a model, we show that both genomic and epigenomic forces are consequential with reversible, suppressive chromatin marks potentially helping dampen the adverse effects of altered gene dosage and the emergence of deleterious alleles. Such forces might be relevant to other highly polymorphic gene families impacted by genomic drift evolution.
Keywords: ubiquitylation, population genomics, gene birth-and-death
Abstract
Comparisons within expanding sequence databases have revealed a dynamic interplay among genomic and epigenomic forces in driving plant evolution. Such forces are especially obvious within the F-Box (FBX) superfamily, one of the largest and most polymorphic gene families in land plants, where its frequent lineage-specific expansions and contractions provide an excellent model to assess how genetic variation impacted gene function before and after speciation. Previous phylogenetic comparisons based on orthology, diversity, and expression patterns identified three plant FBX groups—Common, Lineage-Specific, and Pseudo(genized)—whose emergences are consistent with genomic drift evolution. Here, we examined this variance within Arabidopsis thaliana by evaluating SNPs for all 877 FBX loci from 432 naturally occurring accessions and their relationships to variations in natural selection, expression, and DNA/histone methylation. In line with their phenotypic importance, Common FBX loci have low polymorphism but high deleterious mutation rates indicative of stringent functional constraints. In contrast, the Lineage-Specific and Pseudo groups are enriched in genes with basal expression and higher SNP density and more correlated with methylation marks (RNA-directed DNA methylation and histone H3K27 trimethylation) that promote transcriptional silencing. Taken together, we propose that reversible epigenomic modifications helped shape FBX gene evolution by transcriptionally suppressing the adverse effects of gene dosage imbalance and harmful FBX alleles that arise during genomic drift, while simultaneously allowing innovations to emerge through epigenomic reprogramming.
The evolution of gene families is a complex process in which gene duplication events are central (1). These duplications occur globally via whole-genome and segmental duplications and locally by tandem duplications and retrotransposition events. The duplicated loci can then diverge to subfunctionalize or neofunctionalize the encoded protein's functions through changes in expression, location, and/or activity. Extensive phylogenetic studies showed that many gene families are also subject to dynamic birth-and-death processes that retain active loci and eliminate nonfunctional/silenced loci (1).
Because the size of some gene families (e.g., chemo-, olfactory and pathogen receptors, and immunoglobulins) varies extensively even among closely related species, the process of genomic drift has been postulated to explain these variable lineage/species-specific expansions/contractions (2). Here, neutral evolution is proposed to generate such extensive size variation without significantly affecting species fitness. Although many duplicated loci are eventually lost, some provide reservoirs for innovation and become fixed within species if they are adaptive. Although it is presumed that gene family expansions/contractions are largely inconsequential, many genes are sensitive to dosage, with the additions/subtractions detrimentally affecting function by altering the abundance of the encoded protein outside optimal levels [e.g., haploinsufficency and aneuploidy (3, 4)]. How genomic drift accommodates dosage imbalance without fitness cost is unclear, but a role for epigenomic processes is possible (5–9).
To better understand how gene families evolved, we have extensively analyzed the F-Box (FBX) superfamily, which encodes the target recognition subunits of S-phase kinase-associated protein (SKP)-1/Cullin1/FBX (SCF) ubiquitin (Ub) ligases (or E3s) that drive selective protein ubiquitylation (10). SCF E3s are assembled using Cullin1 to scaffold the RING Box (RBX)-1 subunit, which binds Ub-conjugating enzymes, and a SKP1 adaptor, which recruits target-specific FBX proteins with their cognate targets (10). FBX proteins harbor a signature ∼50-amino acid FBX domain that binds SKP1 followed by a diverse assortment of substrate-specific recognition modules (10). In Arabidopsis thaliana alone, almost 900 FBX-domain–encoding loci have been identified (11, 12), ∼5% of which have been connected thus far to numerous essential processes, including cell division, hormone signaling, light perception, self-incompatibility, and abiotic/biotic stress defense (10). Together, the vast number of FBX genes and their striking sequence and functional diversities provide an excellent opportunity to study gene family evolution. In fact, preliminary SNP analyses of 20 accessions revealed that the FBX superfamily is one of the most polymorphic gene groups in Arabidopsis (13).
Our prior phylogenetic studies among a cohort of 18 plant species revealed numerous lineage/species-specific expansion/contraction events within the FBX superfamily consistent with genomic drift evolution (12). Within Brassicales for example, Carica papaya contains as few as 198 predicted FBX loci whereas Arabidopsis lyrata contains as many as 1,350. These numbers differed greatly even among closely related species (11, 12, 14, 15), suggesting that such large size variations were not solely caused by species complexity or evolutionary history.
The plant FBX superfamily could be divided into three groups by either extent of orthology, natural selection (Ka/Ks), or expression: a Common FBX group (previously named Large Taxonomic Scale) enriched in genes with many orthologs, experiencing strong purifying selection, and having diverse expression patterns; a large Lineage-Specific FBX group (previously named Small Taxonomic Scale) enriched in lineage-specific loci with more relaxed selection and typically weak expression; and a poorly expressed FBX Pseudo(gene) group containing members harboring frame-shift or nonsense mutations (12). The Lineage-Specific group closely resembled the Pseudo group by both natural selection and expression criteria, implying that many Lineage-Specific loci are on the path to pseudogenization.
Here, we examined at the population level the involvement of genomic drift in shaping the plant FBX superfamily, using the genomic, epigenomic, and transcriptomic information now emerging for A. thaliana populations (13, 16–18). By combining SNPs for all FBX loci in 432 geographically diverse accessions with DNA methylome, histone modification, and transcriptome data, we provide more support for genomic drift controlling FBX gene evolution along with evidence that cytosine and histone methylation marks were also influential. Collectively, we propose that these epigenomic forces helped limit the adverse effects of gene dosage imbalance and the appearance of harmful alleles, while maintaining a reservoir of FBX loci available for evolutionary innovation.
Results
SNP Analysis of FBX Genes.
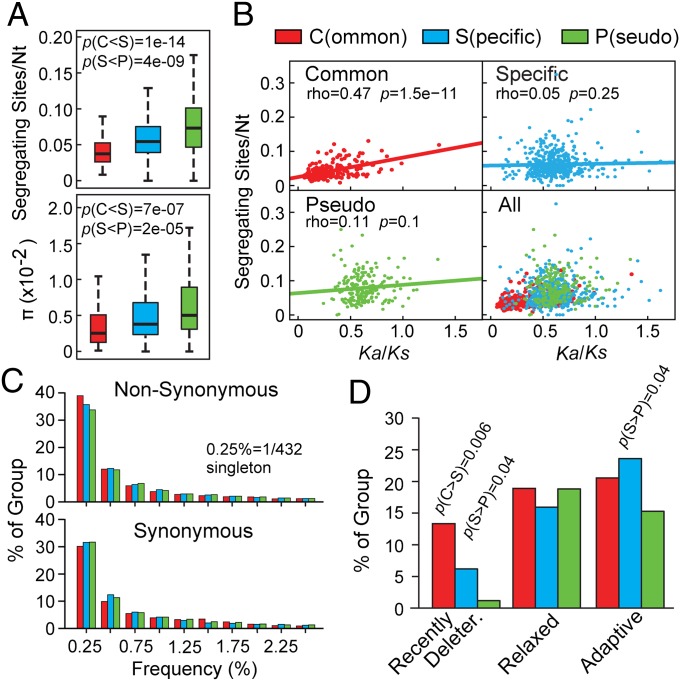
Our and others' phylogenetic analyses of FBX genes among species revealed a dynamic evolution of this superfamily in plants (11, 12, 14, 15). To help understand how this diversity continued within species, we compared sequence polymorphism [segregating sites per nucleotide (nt)] and diversity [average nt differences per site (π)] within the predicted coding sequence of Common, Lineage-Specific, and Pseudo FBX genes (186, 493, and 198 members, respectively) among 432 sequenced Arabidopsis accessions. In agreement with their strong purifying selection among species (12), Common FBX loci displayed the lowest sequence polymorphism within Arabidopsis (Fig. 1A and SI Appendix, Fig. S1).
Fig. 1.
SNP analyses of Common, Specific, and Pseudo FBX genes from 432 Arabidopsis accessions. (A) Comparisons of sequence polymorphism (segregating sites per nt) (Upper) and diversity (average nt differences per site, π) (Lower). Each box plot shows the median (solid line), the 25th and 75th percentiles (boxes), and the fifth and 95th percentiles (dashed lines). (B) Spearman rank correlation test between sequence polymorphism and natural selection (Ka/Ks). Correlation coefficients (rho), P values, and lines of best-fit linear regression are included. (C) Frequency spectrum of rare alleles with nonsynonymous and synonymous mutations. (D) Distribution of recently deleterious (MAF < 5%), relaxed, and adaptive mutations (McDonald–Krietman test). P values in A and D were calculated by Wilcoxon rank and Fisher’s exact tests, respectively.
Although our previous study showed similar relaxed selection on genes within the Lineage-Specific and Pseudo groups among species (12), we found significantly lower genetic variation in the former group within Arabidopsis, which could reflect functional constraints for some Lineage-Specific genes. To test this scenario, we compared by the Spearman rank test the correlation between sequence polymorphism and natural selection [as estimated by the ratio of the number of nonsynonymous substitutions per nonsynonymous site (Ka) to the number of synonymous substitutions per synonymous site (Ks) (12)] for FBX coding sequences. Interestingly, our results showed that whereas the Common group had a significant correlation between sequence polymorphism and natural selection, the Lineage-Specific group, as for the Pseudo group, did not (Fig. 1B), supporting the idea that most lineage-specific FBX genes resemble Pseudo FBX genes in having low functional constraints. The ∼10-fold lower median value for diversity (π) than that for segregating sites/nt (Fig. 1A) suggested that most FBX mutations are rare. Indeed, 34% of the 57,692 total SNP alleles were detected only once among the 432 accessions (Fig. 1C).
The preponderance of rare alleles implied that many FBX mutations are deleterious. Based on the enrichment of nonsynonymous polymorphic mutations (Pn) in alleles with Minor Allele Frequencies (MAF) (<5% MAF), which typically contain harmful mutations (19), the Common group was predicted to be enriched in recently deleterious polymorphisms (13% of loci) compared with the Lineage-Specific (6%) and Pseudo (1%) groups, suggesting that the latter two groups tolerate mutations better (Fig. 1D). After removing the potentially deleterious polymorphic alleles, the remaining “neutral” polymorphic alleles were subjected to the McDonald–Kreitman test (20), which analyzes the effect of Darwinian selection on the fixation of nonsynonymous mutations (Dn). We surprisingly found near equal percentages (∼20%) of FBX genes under either relaxed or adaptive selection in the three groups, implying that the Common and Lineage-Specific groups play similar roles in plant adaptation, and that adaptive mutations might suppress harmful FBX alleles (Fig. 1D).
Correlations Between DNA Methylation and FBX Sequence Polymorphisms.
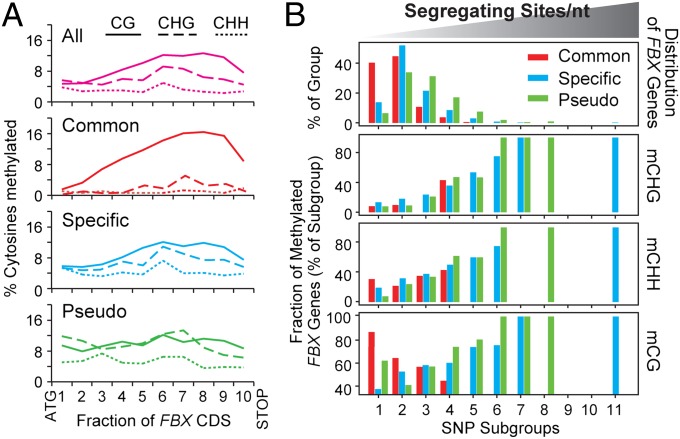
Besides SNPs, cytosine DNA methylation is an influential source of inherited variability through its ability to regulate gene expression (16, 21). Therefore, we examined differential coding region methylation patterns among the three aforementioned FBX groups with respect to modification at symmetric CG and CHG sites and asymmetric CHH sites (H = A, C, or T) within the Col-0 accession (21). We noticed that CHG and CHH methylation, which is often associated with RNA-directed DNA methylation (RdDM) and transcriptional repression (22), increased substantially among the FBX groups as their selective constraint and/or orthology levels decreased [Common/Lineage-Specific/Pseudo = 1.6/5.8/9.8% and 0.9/3.1/5.2% for average methylation levels and 9.1/21.9/24.2 and 27.4/34.2/36.6% for the frequency of CHG and CHH methylated genes, respectively (Fig. 2A and SI Appendix, Table S1)], indicating a positive link between these marks and FBX gene polymorphism. For CG methylation, no clear trend was observed with methylated Common, Lineage-Specific, and Pseudo members having 10.4, 7.8, and 10%, respectively, of their coding-sequence CG cytosines being methylated. However, when the FBX coding sequences were dissected into subregions, a striking difference in CG methylation patterns was detected for the Common group. In agreement with other moderately expressed, slowly evolving Arabidopsis genes (7, 21), members of the Common group were preferentially enriched in CG methylation toward their 3′ ends, which was not evident for the CHG and CHH modifications (Fig. 2A). The Common FBX group was also enriched for loci bearing CG methylation only in the transcribed region, which is prominent in conserved, phenotypically important genes (7, 21). Such “gene-body” methylated genes comprised 17.2, 4.9, and 3.1% of the Common, Lineage-Specific, and Pseudo groups, respectively (SI Appendix, Table S1).
Fig. 2.
Correlation between SNPs and DNA methylation patterns within the Common, Specific, and Pseudo FBX gene groups. (A) Average distribution of methylated CG (solid), CHG (dashed), and CHH (dotted) sites along the coding sequences. “All” represents the average distribution of all 869 FBX loci. (B) Frequency distributions of methylated FBX genes at CHG, CHH, and CG contexts in relation to sequence polymorphism. All FBX loci were binned into 11 groups with equal intervals based on their number of segregating sites per nt. The distributions of the Common, Specific, and Pseudo FBX genes among the intervals are shown in the Top.
To explore the link between cytosine methylation and polymorphism further, the entire FBX gene collection was binned based on SNP frequencies into 11 incremental clusters (0 to >0.325 segregating sites/nt) and then analyzed for CG, CHG, and CHH methylation frequencies in each cluster. The frequencies of all three methylation patterns rose for Lineage-Specific and Pseudo genes in concert with increased SNPs (Fig. 2B). A similar correlation was observed in the Common group for CHG and CHH methylation but, strikingly, CG methylation had the opposite trend, being more frequent among less polymorphic loci. We further compared by the Spearman rank test the correlations between the extent of methylation and either SNP rate or nucleotide diversity of methylated FBX genes. In accordance with the frequency distribution, CHG and CHH, but not CG, methylation levels correlated with both (SI Appendix, Fig. S2). Together, these data show that differential cytosine methylation also might have influenced Arabidopsis FBX gene divergence.
Expression Variance of FBX Genes.
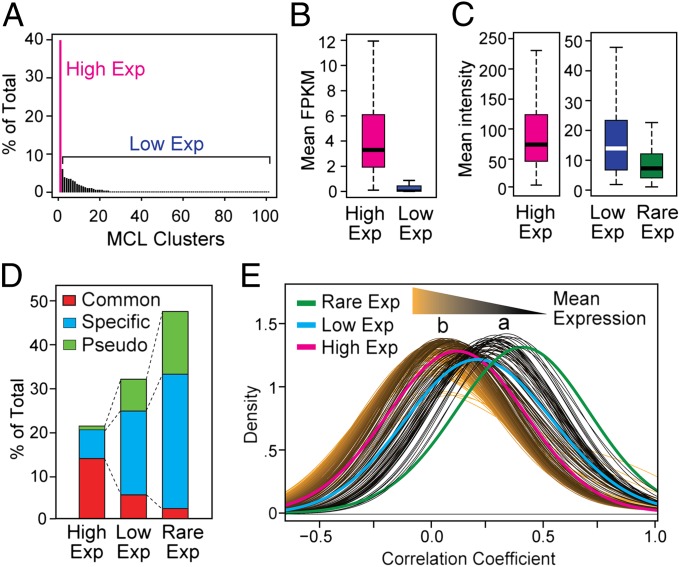
Besides orthology and natural selection comparisons, expression patterns based on the extensive Nottingham Arabidopsis Stock Centre microarray datasets (NASCArrays) for the Col-0 accession allowed the identification of three distinct FBX clusters (12). One cluster enriched for Common loci displayed high and variable expression patterns, a second cluster enriched in Lineage-Specific and Pseudo loci had low and more correlated expression patterns, and a third cluster almost exclusively containing Lineage-Specific and Pseudo loci had no evidence of expression. We also found similar groupings at the population level by Markov Cluster analysis of the leaf RNA-seq expression profiles currently available for 19 Arabidopsis accessions (23). The 877 FBX loci could be split into High, Low, and Rare Exp(ression) clusters with 183, 276, and 418 respective members, which displayed high, low, and no evidence of expression (Fig. 3 A–C). When using the Col-0 NASCArrays datasets, the High Exp cluster had 5.3- and 10.1-fold higher median transcript level than the Low and Rare Exp clusters, respectively (Fig. 3C). (The evidence of expression for some Rare FBX loci likely reflected the accumulation of transcripts in tissues/conditions not analyzed by RNA-seq.) Strikingly, 73% of High Exp FBX genes were from the Common group, whereas the Lineage-Specific and Pseudo FBX genes mostly populated the Low and Rare Exp clusters, reflecting a strong inverse connection between FBX expression and polymorphisms/DNA methylation (Fig. 3D).
Fig. 3.
Clustering of FBX genes by their expression pattern variances. (A) Markov Clustering (MCL) of FBX genes based on the leaf RNA-seq expression profiles of 19 Arabidopsis accessions (Pearson's correlation coefficient cutoff = 0.9). The single largest FBX MCL cluster was designated as the High Exp group (40% of total, 183 loci). All other FBX clusters with evidence of expression were designated as Low Exp (276 loci), and FBX loci with no expression were designated as Rare Exp (418 loci). (B) Mean expression levels for the High and Low Exp FBX groups identified in A based on the RNA-seq analysis of 19 accessions [fragments/kb of exon/106 fragments mapped (FPKM)]. (C) Mean expression of High, Low, and Rare FBX loci based on the Col-0 NASCArrays datasets. See Fig. 1 for description of box plots. (D) Distribution of Common, Specific, and Pseudo FBX genes in the High, Low, and Rare Exp groups. (E) Transcription correlation profiles of High, Low, and Rare FBX genes compared with the entire Arabidopsis Col-0 transcriptome binned into 230 subgroups based on their mean NASCArray expression levels (SI Appendix, Fig. S3). The estimated distribution of pairwise Pearson’s correlation coefficients of expression in each group is displayed using a Gaussian kernel density curve (bandwidth = 0.25).
The poor transcriptional evidence for Low and Rare Exp FBX genes suggested that they, like many pseudogenes, are transcriptionally silent. To support this possibility, we compared their NASCArrays transcription profiles to that for the entire transcriptome. Here, 23,019 analyzed nuclear transcripts from the Col-0 accession were binned based on mean expression level into 230 subgroups with ∼100 transcripts each (SI Appendix, Fig. S3 A and B). Through pairwise comparisons among transcripts in each subgroup, a bimodal distribution emerged: one collection (collection a) encompassing the first 49 subgroups had low-expression and high average-expression correlation coefficients (mean r∼0.28), and a second collection (collection b) containing the remaining 181 subgroups had higher expression levels and lower correlation coefficients (mean r∼0.07) (Fig. 3E and SI Appendix, Fig. S3 C–E). A demarcation was evident around subgroup 50, suggesting that ∼5,000 Arabidopsis genes (21%) share a basal expression pattern akin to transcriptional noise (24). Interestingly, the expression correlation coefficient distributions for the Low and Rare Exp FBX clusters overlapped with that for collection a, whereas the distribution for the High Exp FBX cluster overlapped with that for collection b (Fig. 3E). This distinction could also be seen from the frequency distribution of FBX genes in relation to all pseudogenes within the 230 subgroups (SI Appendix, Fig. S4), confirming that most Low and Rare Exp FBX loci are basally expressed and that most High Exp FBX loci are transcriptionally up-regulated.
A Machine-Learning Approach to Explain the FBX Expression Variance.
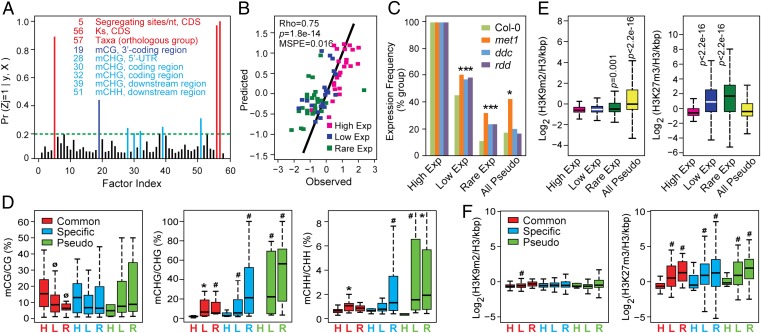
To identify parameters that differentiate the Common, Lineage-Specific, and Pseudo FBX groups, we developed a multivariate linear regression model to quantify how various gene features, including sequence polymorphism, DNA methylation, recombination, and natural selection, might collectively impact expression. Through a Bayesian approach assessing 58 gene features (SI Appendix, Fig. S5 and Table S1), we performed 10,000X Gibbs samplings that differentially combined gene features to mimic the effect of uncertain environmental changes.
Using 255 randomly chosen FBX genes within the NASCArrays expression datasets for training [85 representatives from the High, Low, and Rare Exp categories (SI Appendix, Fig. S6)], our Bayesian analysis revealed that segregating sites per nt, the Ks value for the FBX coding sequence, and the scale of the corresponding orthologous group within plants were dominant in estimating the expression of an FBX gene [posterior probability (PP) > 80%]. In addition, the extent of CG methylation at the 3′-coding region acted at a second level (PP > 40%) in promoting expression. Our formula predicted that the extent of CHG methylation at upstream, coding, and downstream regions of an FBX gene and the extent of downstream CHH methylation were also influential (PP > 20%) (Fig. 4A and SI Appendix, Fig. S5). Testing with 75 additional randomly selected FBX genes (25 from each group) confirmed the accuracy of the model based on group clustering, tight correlation, and low mean-squared predictive error (MSPE) between the observed expression values and those predicted by the model (Fig. 4B). Such differential genomic and epigenomic impacts could also be visualized from heat maps of all FBX loci, which showed that High Exp FBX genes in general have higher Ks, taxa, and gene-body CG methylation values but lower sequence polymorphism and suppressive CHG and CHH methylation values (SI Appendix, Fig. S7).
Fig. 4.
Quantitative analyses of the impact of genomic and epigenomic variations on FBX gene expression. (A) PP of a nonzero effect of 58 gene parameters on FBX gene expression. See SI Appendix, Fig. S5 for the parameter descriptions. The parameters highlighted with red, dark blue, and light blue have a PP > 0.2. (B) Spearman rank correlation between the predicted mean expression values of FBX genes from the test sample containing 25 representatives from the High, Low, and Rare Exp groups with those observed within NASCArrays. Logarithmic transformations fit the data to an approximately normal distribution for Bayesian analysis. (C) Expression frequency of High, Low, and Rare FBX genes, as well as the full set of Arabidopsis pseudogenes in three methylation defective mutants (met1, ddc, rdd) compared with Col-0. (D) Enrichment of coding sequence DNA methylation at CG, CHG, and CHH contexts within the Common, Lineage-Specific, and Pseudo FBX groups further subdivided based on their expression levels (High, Low, and Rare Exp). (E) Occupancy of H3K9m2 (Left) and H3K27m3 (Right) in the coding regions of High, Low, and Rare FBX genes, and all Arabidopsis pseudogenes in the Col-0 accession. (F) Occupancy of H3K9m2 and H3K27m3 within the coding regions of Common, Specific, and Pseudo FBX genes further subdivided based on their expression levels (High, Low, and Rare Exp). See Fig. 1 for description of box plots. P values in C were calculated by Fisher’s exact test, and P values in D–F were calculated by Wilcoxon rank test. ØP(Low/Rare < High) < 0.05. *P(Low/Rare > High) < 0.05. #P(Low/Rare > High) < 0.01.
Connections between DNA/Histone H3 Methylation and FBX Expression.
To further confirm that cytosine methylation influences FBX gene transcription, we analyzed FBX transcript levels within the RNA-seq datasets of ref. 21, which compared the floral transcriptomes from Arabidopsis Col-0 mutants abrogated in key cytosine methyl transferase and demethylase activities. The mutants included methytransferase (met)1-3 that is defective in CG maintenance methylation, the drm1-2 drm2-2 cmt3-11 triple mutant (ddc) that blocks most non-CG and de novo DNA methylation, and the ros1-3 dml2-1 dml3-1 triple mutant (rdd) that inhibits demethylation. None of these mutants influenced the expression of the High Exp FBX group, based on either expression frequency or strength (Fig. 4C and SI Appendix, Fig. S8). In contrast, the expression of some loci within the Low and Rare Exp FBX groups was significantly derepressed in all three backgrounds, thus empirically supporting a role for DNA methylation in controlling the transcription of these loci. Interestingly, like the complete collection of Arabidopsis pseudogenes, members of both Low and Rare Exp FBX groups were most strongly derepressed by loss of MET1 (Fig. 4C and SI Appendix, Fig. S8). To assess whether RdDM also contributed, we examined the FBX expression patterns in Col-0 floral tissue abrogated in key RdDM components, AGO4, DRM2, and NRPE1 (22, 25). A slight but significant increase in Low/Rare Exp FBX transcripts was observed that was not seen for High Exp transcripts, suggesting that RdDM helps transcriptionally suppress Low and Rare FBX genes (SI Appendix, Fig. S9).
To better connect DNA methylation with functional divergence, we partitioned the Common, Lineage-Specific, and Pseudo FBX loci into the High, Low, and Rare Exp groups and measured enrichment for CG, CHG, and CHH methylation separately. CG methylation, but not CHG and CHH methylation, rose significantly among Common FBX genes as their expression strength increased (High > Low > Rare), further linking CG methylation to up-regulated expression (Fig. 4D). Conversely, CHG and CHH methylation increased significantly for all three groups (Common, Lineage-Specific, and Pseudo) as their expression dropped, implicating these suppressive marks, likely through RdDM (12, 19), in dampening transcription of a subset of loci within each group (Fig. 4D).
In concert with DNA methylation, histone methylation substantially impacts gene activity with the appearance of histone H3 dimethylation at lysine-9 (H3K9m2) or trimethylation at lysine-27 (H3K27m3) promoting transcriptional silencing (22, 26). To test whether the FBX superfamily was influenced by these suppressive marks, we examined the genome-wide H3K9m2 and H3K27m3 maps from ref. 26 for enrichment within FBX loci. Based on occupancy (H3K9m2 or H3K27m3/H3/kbp), none of the three FBX groups (High, Low, or Rare Exp) were enriched in H3K9m2 in contrast to the complete collection of Arabidopsis pseudogenes which showed a significant enrichment (1.5-fold higher than High Exp loci) (Fig. 4E). H3K27m3 is common in silent protein-coding regions (26). In agreement, we found that the High Exp FBX loci were not enriched in this modification, and, in fact, these loci were modified to a similar extent as all Arabidopsis pseudogenes (Fig. 4E). Strikingly, the Low and Rare Exp FBX loci were significantly more impacted by H3K27m3; they had 2.8- and 4.9-fold more of this mark on average relative to the High Exp loci, respectively (Fig. 4E). For further support, we used the same expression partitions used for cytosine methylation (Fig. 4D) and found that H3K9m2 was not preferentially enriched in eight of the nine categories, whereas all six Low and Rare Exp FBX categories had substantially more H3K27m3 than all three High Exp categories (Fig. 4F). These strong correlations imply that H3K27m3 and its associated Polycomb repression machinery (26) actively suppress FBX expression.
Because chromosomal origin can significantly affect gene function, we compared the distribution of FBX genes in the syntenic blocks shared between A. thaliana and A. lyrata, as well as the differences in DNA methylation, gene expression, and occupancy of H3K9m2 and H3K27m3 between syntenic and nonsyntenic FBX loci (SI Appendix, Fig. S10). Consistent with our previous study (12), syntenic blocks were enriched for Common loci, whereas nonsyntenic blocks had more Lineage-Specific and Pseudo loci. Not surprisingly, nonsyntenic FBX genes also had higher DNA and H3K27m3 methylation and lower expression levels, implying a genome-wide epigenomic-genomic coinheritance (27). To test whether epigenomic programming is common among large Arabidopsis gene families, we examined 40 other families in the Col-0 accession (16, 21) before or after subdividing them into syntenic and nonsyntenic clusters (SI Appendix, Figs. S11–S13). All comparisons revealed that the FBX superfamily is one of a small group (others include MYB and MADS) enriched for suppressive CHG/CHH DNA and H3K27m3 methylation marks but with medium CG methylation and low H3K9m2 marks.
Discussion
After the discovery that land plant genomes encode a myriad of Ub E3s [>1,500 possible in Arabidopsis (10, 28)], the question emerged as to why so many. An obvious answer in line with the multitude of ubiquitylated proteins (29) was that each E3 has a dedicated target. However, the recent realizations that the FBX superfamily is one of the more rapidly evolving families in plants (12, 14) and that strong differences in FBX gene numbers (but not likely targets) exist even among closely related species offer an alternative hypothesis for this E3 subtype; it has evolved via genomic drift, and much fewer FBX loci actively direct ubiquitylation (12). Our polymorphism and transcriptome analyses of this superfamily within Arabidopsis further support this scenario with strong evidence that only a subset (∼21%) of FBX loci (mostly Common/High Exp) are under more stringent functional constraints and actively expressed. Notably, the predicted active FBX loci include those known to control important phenotypic responses in Arabidopsis and other species (12) and/or to bind SKP1 (11, 30). Most of the remaining FBX loci are under more relaxed selection and, as for pseudogenes, are often expressed at basal levels. A number of these poorly expressed loci are transcribed in pollen (12); we presume that this expression results from epigenetic reprogramming active in pollen (31) and not their functional relevance in this tissue. If collectively true, then the number of influential SCF E3s would collapse dramatically and be mostly limited to those assembled with the 100–200 well-expressed FBX proteins with extensive orthology among plants (12).
With respect to genomic drift driving gene family evolution (2), two heretofore unsolved complications arise. One is the need to accommodate gene dosage imbalances as families randomly expand or contract. Although dosage changes in some families might be neutral to fitness initially (e.g., chemo-, olfactory, and pathogen receptors), changes in others could have immediate fitness costs if the levels of the corresponding proteins are crucial (3, 4). Such penalties might be particularly substantial for Ub-26S proteasome system (UPS) E3s, which fine-tune through directed proteolysis the abundance of rate-limiting metabolic enzymes, signaling components, and numerous transcriptional activators/repressors (10, 28). For example, in addition to the severe consequences of knockdown FBX mutations, several studies have shown that overexpression of individual FBX genes can significantly perturb plant growth by influencing hormone responses, histone methylation, and RNA-directed posttranscriptional gene silencing. Large-scale expansions/contractions could also profoundly affect the entire superfamily by impacting competition among FBX proteins for the common core SKP1/Cullin1/RBX1 machinery (10). Another complication to genomic drift is the need to suppress the expression of detrimental alleles that arise spontaneously and to reactivate suppressed alleles that improve fitness. Nascent E3 mutants that recognize unintended targets could be particularly harmful by acting in effect as target knockdown alleles.
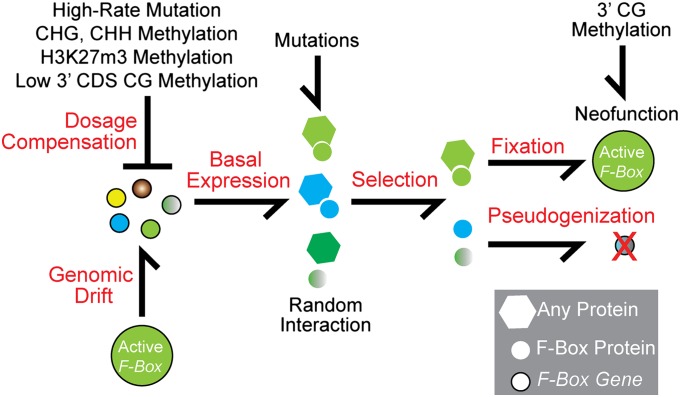
As a consequence, genomic drift likely requires additional layer(s) of control to ameliorate gene dosage imbalances, suppress harmful nascent alleles, and activate innovative alleles. Based on our analyses of the plant FBX superfamily, we propose that evolutionary stable but reversible epigenomic modifications provide such controls (Fig. 5). Through their reprogramming, the expression of recently duplicated FBX loci arising from genomic drift could be selectively suppressed by increased cytosine methylation at CHG and CHH sites or increased histone H3K27 trimethylation, both of which are strong marks for gene silencing. Changes in other suppressive or activating histone modifications are also possible (32). These marks could originate from: (i) the prior chromatin characteristics of the duplicate's insertion site (e.g., duplicates landing in transcriptionally silenced chromatin might be better tolerated by natural selection), (ii) epigenomic RdDM-mediated silencing induced by elevated gene dosage, and/or (iii) random occurrence within the duplicated loci. Presumably, functionally important FBX loci (Common/High Exp genes) would be immune to this negative regulation given the fitness penalties associated with their suppression.
Fig. 5.
Diagram of the random-birth selective-action evolutionary model depicting the innovation of recently duplicated FBX genes. High mutation rates and differential methylation patterns result in basal expression of nascent FBX genes to compensate for increased dosage and to repress harmful FBX alleles. New interactions that emerge via natural selection become fixed and are reactivated by CG methylation to generate neofunctionalized FBX loci. In most other cases, transcriptionally suppressed harmful and poorly functional FBX loci become pseudogenized and eventually eliminated.
Once silenced, these basally expressed, functionally irrelevant FBX genes can innovate without fitness cost. Improvements include subfunctionalization as well as neofunctionalization to recognize and thus control the abundance of new targets (Fig. 5). Although many new alleles would randomly disappear from the population, those that improve fitness would be retained. The expression of new beneficial genes eventually could be reactivated through loss of suppressive CHG/CHH methylation or H3K27m3 marks, along with redistribution of the activating gene-body CG methylation mark toward the 3′ end. In fact, we could replicate this reactivation using Arabidopsis mutants altered in cytosine methylation, which selectively derepressed the expression of some poorly expressed Low and Rare Exp FBX loci (Fig. 4C and SI Appendix, Fig. S8). It is possible that H3K27m3 and DNA methylation (e.g., RdDM or CG gene body) influence in concert FBX gene expression (22, 32), but for the datasets analyzed here such connections were not significant, suggesting that some genes are more affected by their DNA methylation state, whereas others are more affected by H3K27m3 (SI Appendix, Fig. S14).
In summary, epigenomic forces such as DNA and histone methylation have been well connected to the repression of transposable elements, pseudogenes, and pericentromeric regions and as a source of epigenetic variation (22, 32). Here, we propose that these forces were also critical to the genomic drift diversification of the FBX superfamily by reversibly regulating the expression of selected members to mitigate the negative effects of gene dosage imbalance while simultaneously allowing innovations to emerge. It will now be interesting to see how many other gene families within the UPS (RING and BTB E3 families) and elsewhere (MYB, MADS, and NBS-LRR families) show these genomic and epigenomic signatures.
Materials and Methods
SNP Analyses.
SNP datasets for 431 A. thaliana genomes were retrieved from the Arabidopsis 1001 Genomes Project (www.1001genomes.org). SNPs and MAFs were calculated based on single-nucleotide changes (Phred quality score ≥ 25) compared with the Col-0 reference genome. The deleterious mutation and generalized McDonald–Kreitman tests were performed as described in ref. 20, with some modifications (SI Appendix, SI Materials and Methods).
Cytosine and Histone H3 Methylation Analyses.
The number of methylation sites in CG, CHG, and CHH contexts in each FBX gene was counted and normalized by the total number of each context or cytosine sites according to the single base-resolution methylation profile generated by bisulfite sequencing (21). The ChIP-Chip mapping of histone H3 methylation was retrieved from ref. 26.
Expression Analyses.
A Markov Cluster (MCL) Algorithm (33) was applied to identify the High, Low, and Rare Exp FBX groups based on their expression variance in 19 Arabidopsis accessions (23). The Illumina HiSeq expression data from floral tissue in Col-0 and its methylation mutants were retrieved from refs. 21 and 25. The PP of gene factors affecting the FBX gene expression were predicted using a Bayesian multivariate linear regression-modeling approach (SI Appendix, SI Materials and Methods).
Supplementary Material
Acknowledgments
We thank Dr. Ryan Lister for providing the MethylC-seq datasets. This work was supported by US National Science Foundation Grants MCB-0115870 (to R.D.V.) and MCB-0929402 and MCB1122246 (to J.R.E.) and by Howard Hughes Medical Institute (HHMI) and Gordon and Betty Moore Foundation (GBMF) Grant GBMF3034 (to J.R.E.). J.R.E. is an HHMI–GBMF Investigator.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1316009110/-/DCSupplemental.
References
- 1.Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nei M, Niimura Y, Nozawa M. The evolution of animal chemosensory receptor gene repertoires: Roles of chance and necessity. Nat Rev Genet. 2008;9(12):951–963. doi: 10.1038/nrg2480. [DOI] [PubMed] [Google Scholar]
- 3.Conrad B, Antonarakis SE. Gene duplication: A drive for phenotypic diversity and cause of human disease. Annu Rev Genomics Hum Genet. 2007;8:17–35. doi: 10.1146/annurev.genom.8.021307.110233. [DOI] [PubMed] [Google Scholar]
- 4.Veitia RA, Bottani S, Birchler JA. Cellular reactions to gene dosage imbalance: Genomic, transcriptomic and proteomic effects. Trends Genet. 2008;24(8):390–397. doi: 10.1016/j.tig.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Adams KL, Cronn R, Percifield R, Wendel JF. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc Natl Acad Sci USA. 2003;100(8):4649–4654. doi: 10.1073/pnas.0630618100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Madlung A, et al. Remodeling of DNA methylation and phenotypic and transcriptional changes in synthetic Arabidopsis allotetraploids. Plant Physiol. 2002;129(2):733–746. doi: 10.1104/pp.003095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takuno S, Gaut BS. Body-methylated genes in Arabidopsis thaliana are functionally important and evolve slowly. Mol Biol Evol. 2012;29(1):219–227. doi: 10.1093/molbev/msr188. [DOI] [PubMed] [Google Scholar]
- 8.Schmitz RJ, et al. Transgenerational epigenetic instability is a source of novel methylation variants. Science. 2011;334(6054):369–373. doi: 10.1126/science.1212959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becker C, et al. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature. 2011;480(7376):245–249. doi: 10.1038/nature10555. [DOI] [PubMed] [Google Scholar]
- 10.Hua Z, Vierstra RD. The Cullin-RING ubiquitin-protein ligases. Annu Rev Plant Biol. 2011;62:299–334. doi: 10.1146/annurev-arplant-042809-112256. [DOI] [PubMed] [Google Scholar]
- 11.Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD. The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA. 2002;99(17):11519–11524. doi: 10.1073/pnas.162339999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hua Z, Zou C, Shiu SH, Vierstra RD. Phylogenetic comparison of F-Box (FBX) gene superfamily within the plant kingdom reveals divergent evolutionary histories indicative of genomic drift. PLoS ONE. 2011;6(1):e16219. doi: 10.1371/journal.pone.0016219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark RM, et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;317(5836):338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- 14.Xu G, Ma H, Nei M, Kong H. Evolution of F-box genes in plants: Different modes of sequence divergence and their relationships with functional diversification. Proc Natl Acad Sci USA. 2009;106(3):835–840. doi: 10.1073/pnas.0812043106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang X, et al. The F-box gene family is expanded in herbaceous annual plants relative to woody perennial plants. Plant Physiol. 2008;148(3):1189–1200. doi: 10.1104/pp.108.121921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmitz RJ, et al. Patterns of population epigenomic diversity. Nature. 2013;495(7440):193–198. doi: 10.1038/nature11968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao J, et al. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet. 2011;43(10):956–963. doi: 10.1038/ng.911. [DOI] [PubMed] [Google Scholar]
- 18.Long Q, et al. Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat Genet. 2013;45(8):884–890. doi: 10.1038/ng.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fay JC, Wyckoff GJ, Wu CI. Positive and negative selection on the human genome. Genetics. 2001;158(3):1227–1234. doi: 10.1093/genetics/158.3.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mackay TF, et al. The Drosophila melanogaster genetic reference panel. Nature. 2012;482(7384):173–178. doi: 10.1038/nature10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133(3):523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11(3):204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gan X, et al. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature. 2011;477(7365):419–423. doi: 10.1038/nature10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. 2007;14(2):103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 25.Ausin I, et al. INVOLVED IN DE NOVO 2-containing complex involved in RNA-directed DNA methylation in Arabidopsis. Proc Natl Acad Sci USA. 2012;109(22):8374–8381. doi: 10.1073/pnas.1206638109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deleris A, et al. Loss of the DNA methyltransferase MET1 induces H3K9 hypermethylation at PcG target genes and redistribution of H3K27 trimethylation to transposons in Arabidopsis thaliana. PLoS Genet. 2012;8(11):e1003062. doi: 10.1371/journal.pgen.1003062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eichten SR, et al. Heritable epigenetic variation among maize inbreds. PLoS Genet. 2011;7(11):e1002372. doi: 10.1371/journal.pgen.1002372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vierstra RD. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol. 2009;10(6):385–397. doi: 10.1038/nrm2688. [DOI] [PubMed] [Google Scholar]
- 29.Kim DY, Scalf M, Smith LM, Vierstra RD. Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell. 2013;25(5):1523–1540. doi: 10.1105/tpc.112.108613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuroda H, Yanagawa Y, Takahashi N, Horii Y, Matsui M. A comprehensive analysis of interaction and localization of Arabidopsis SKP1-like (ASK) and F-box (FBX) proteins. PLoS ONE. 2012;7(11):e50009. doi: 10.1371/journal.pone.0050009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calarco JP, et al. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell. 2012;151(1):194–205. doi: 10.1016/j.cell.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karlić R, Chung HR, Lasserre J, Vlahovicek K, Vingron M. Histone modification levels are predictive for gene expression. Proc Natl Acad Sci USA. 2010;107(7):2926–2931. doi: 10.1073/pnas.0909344107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Van Dongen SM (2000) Graph clustering by flow simulation. PhD thesis (Utrecht Univ, Utrecht, Netherlands)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.