Abstract
Absorption of UV-B by DNA induces the formation of covalent bonds between adjacent pyrimidines. In maize and arabidopsis, plants deficient in chromatin remodeling show increased DNA damage compared to WT plants after a UV-B treatment. However, the role of enzymes that participate in DNA methylation in DNA repair after UV-B damage was not previously investigated. In this work, we analyzed how chromatin remodeling activities that have an effect on DNA methylation affects the repair of UV-B damaged DNA using plants deficient in the expression of DDM1 and ROS1. First, we analyzed their regulation by UV-B radiation in arabidopsis plants. Then, we demonstrated that ddm1 mutants accumulated more DNA damage after UV-B exposure compared to Col0 plants. Surprisingly, ros1 mutants show less CPDs and 6-4PPs than WT plants after the treatment under light conditions, while the repair under dark conditions is impaired. Transcripts for two photolyases are highly induced by UV-B in ros1 mutants, suggesting that the lower accumulation of photoproducts by UV-B is due to increased photorepair in these mutants. Finally, we demonstrate that oxidative DNA damage does not occur after UV-B exposure in arabidopsis plants; however, ros1 plants accumulate high levels of oxoproducts, while ddm1 mutants have less oxoproducts than Col0 plants, suggesting that both ROS1 and DDM1 have a role in the repair of oxidative DNA damage. Together, our data provide evidence that both DDM1 and ROS1, directly or indirectly, participate in UV-B induced- and oxidative DNA damage repair.
Keywords: UV-B, DNA repair, chromatin remodeling, DNA methylation, arabidopsis
Introduction
Because of their sessile condition, plants are inevitably exposed to ultraviolet-B radiation (UV-B, 290–315 nm); this energetic radiation causes direct damage to DNA, proteins, lipids, and RNA (Britt, 1996; Jansen et al., 1998; Gerhardt et al., 1999; Casati and Walbot, 2004). Absorption of UV-B by DNA induces the formation of covalent bonds between adjacent pyrimidines, giving rise to cyclobutane pyrimidine dimers (CPD) and, to a lesser extent, pyrimidine (6-4) pyrimidone photoproducts (6-4PPs) (Friedberg et al., 1995). These lesions disrupt base pairing and block DNA replication and transcription if photoproducts persist, or result in mutations if photoproducts are bypassed by error-prone DNA polymerases (Britt, 1996). Accumulation of such lesions must be prevented to maintain genome integrity, plant growth and seed viability. Thus, plants have not only developed mechanisms that filter or absorb UV-B to protect them against DNA damage (Mazza et al., 2000; Bieza and Lois, 2001), but also have different DNA repair systems to remove or tolerate DNA lesions (Hays, 2002; Bray and West, 2005; Kimura and Sakaguchi, 2006). At the genome level, the accessibility of DNA is determined by the structure of chromatin, which is subjected to epigenetic regulation. The structure of chromatin can be remodeled by three distinct processes, including covalent modifications of histones, such as phosphorylation, acetylation, methylation, ubiquitylation, sumoylation; ATP-dependent reorganization and positioning of DNA-histones; and methylation of DNA cytosine residues (Verbsky and Richards, 2001; Eberharter and Becker, 2002; Pfluger and Wagner, 2007; Vaillant and Paszkowski, 2007).
In plants, DNA methylation regulates different epigenetic phenomena, including transcriptional silencing of transposons and transgenes, defense against pathogens, regulation of imprinting as well as silencing of genes (Vongs et al., 1993; Jeddeloh et al., 1999; Bender, 2004; Chan et al., 2005; Vanyushin and Ashapkin, 2011; Yaish et al., 2011). DECREASE IN DNA METHYLATION1 (DDM1), is an ATP-dependent SWI2/SNF2 chromatin remodeling factor that is required for normal patterns of genomic DNA methylation in arabidopsis (Vongs et al., 1993; Jeddeloh et al., 1999). Mutations in DDM1 result in a rapid loss of cytosine methylation at heterochromatic repetitive sequences and a gradual depletion of methylation at euchromatic low-copy sequences over successive generations (Kakutani et al., 1996). In ddm1 heterochromatin, DNA methylation is lost and methylation of lysine 9 is largely replaced by methylation of lysine 4 (Gendrel et al., 2002). In addition, DDM1 maintains 5S rDNA methylation patterns while silencing transcription through 5S rDNA intergenic spacers (IGS) (Kurihara et al., 2008). DDM1 also regulates gene imprinting, transposon, gene and transgene silencing, and possibly the occurrence of paramutations (Jeddeloh et al., 1998; Vielle-Calzada et al., 1999; Hirochika et al., 2000). In ddm1 plants, there is a significant DNA decondensation at centromeric and pericentromeric regions rich in repetitive sequences and transposons; and in these mutants, some transposons become transcriptionally active or even undergo transposition (Hirochika et al., 2000; Miura et al., 2001; Singer et al., 2001; Mittelsten Scheid et al., 2002; Soppe et al., 2002; Fransz et al., 2003; Lippman et al., 2003; Probst et al., 2003; Slotkin and Martienssen, 2007; Mirouze et al., 2009; Tsukahara et al., 2009). DDM1 apparently stabilizes the activity of transposons; one of the ddm1-induced abnormalities was shown to be caused by insertion of CAC1, an endogenous CACTA family transposon (Miura et al., 2001). ddm1 plants are also sensitive to NaCl stress and are deficient in DNA repair by methyl methane sulfonate (Yao et al., 2012); DDM1 participates in homologous recombination, and plants deficient in the expression of this gene show sensitivity to γ and UV-C radiation; demonstrating that DDM1 plays a role in response to DNA damage (Shaked et al., 2006).
Biochemical and genetic evidences have shown that plants possess DNA glycosylases that specifically remove 5-meC from DNA, initiating its replacement by unmethylated cytosine through a base excision repair process (Gehring et al., 2009; Roldan-Arjona and Ariza, 2009; Zhu, 2009). The in vivo functions of plant 5-meC DNA glycosylases are not fully understood, but they seem to contribute to the stability and flexibility of the plant epigenome. Plant 5-meC DNA glycosylases comprise a subfamily of atypical HhH-GPD enzymes, examples of enzymes in this group are the arabidopsis proteins ROS1 (repressor of silencing 1), DME (Demeter), DML2, and DML3 (Demeter-like proteins 2 and 3) (Choi et al., 2002; Gong et al., 2002; Penterman et al., 2007; Ortega-Galisteo et al., 2008). ROS1 was identified in a screen for mutants with increased silencing of the repetitive RD29ALUC transgene (Gong et al., 2002). Together with paralogs DML2 and DML3, ROS1 is needed to regulate the DNA methylation pathway at discrete regions across the plant genome, and probably protect the genome from excess methylation (Penterman et al., 2007; Zhu et al., 2007; Ortega-Galisteo et al., 2008). ROS1 and its homologs are bifunctional DNA glycosylases/lyases that cleave the phosphodiester backbone at the 5-meC removal site by b-elimination, generating a 3′ phospho a,b-unsaturated aldehyde at the strand break (Agius et al., 2006; Gehring et al., 2006; Morales-Ruiz et al., 2006; Penterman et al., 2007; Ortega-Galisteo et al., 2008). The final reaction product generated by ROS1 is a single-nucleotide gap flanked by 3′-phosphate and 5′-phosphate termini. The phosphate group present at the 3′ end of the single-nucleotide gap generated by ROS1 is removed by a DNA 3′phosphatase (Martínez-Macías et al., 2012). Finally, a yet unknown DNA polymerase must fill this gap with an unmethylated cytosine before a DNA ligase can seal the remaining nick. In addition to 5-meC, ROS1 also excise with less efficiency its deamination product thymine (5-methyluracil) from T_G mispairs, but do not show detectable activity on either C_G pairs or U_G mispairs; and ROS1 activity is facilitated at mismatched 5-meC residues (Morales-Ruiz et al., 2006; Ponferrada-Marín et al., 2009). The ros1 mutation increases the telomere length in arabidopsis (Liu et al., 2010b); however, ros1 mutants have not previously shown any differential response in DNA repair when compared to WT plants (Liu et al., 2010a).
We have previously demonstrated that arabidopsis plants deficient in 4 chromatin remodeling proteins NFC4, SDG26, HAM1, and HAM2 show more damaged DNA than WT plants after 4 h of UV-B exposure (Campi et al., 2012). In addition, plants treated with an inhibitor of histone acetyltransferases, curcumin, previous to the UV-B treatment show deficiencies in DNA repair; demonstrating that histone acetylation is important during DNA repair in arabidopsis. These results showed that chromatin remodeling, and histone acetylation in particular, are essential during DNA repair by UV-B; demonstrating that both genetic and epigenetic effects control DNA repair in plants. However, the role of enzymes that participate in DNA methylation in DNA repair after UV-B damage has not been investigated yet. Therefore, the aim of this work was to analyze the role of chromatin remodeling proteins that have a role in DNA methylation in the repair of CPDs and 6-4PPs using plants deficient in the expression of DDM1 and ROS1. First, we analyzed their regulation by UV-B radiation in WT plants. Then, using plants with decreased transcript levels of DDM1 and ROS1, we demonstrated that ddm1 mutants accumulated more DNA damage after UV-B exposure compared to Col0 WT plants. Surprisingly, ros1 mutants show less CPDs and 6-4PPs than Col0 plants after the treatment under light conditions, while the repair under dark conditions is impaired. Transcripts for two photolyases are highly induced by UV-B in ros1 mutants, suggesting that the lower accumulation of photoproducts by UV-B is due to increased photorepair in these mutants. Finally, we here demonstrate that oxidative DNA damage does not occur after UV-B exposure in arabidopsis plants; however, ros1 plants accumulate high levels of oxoproducts, while ddm1 mutants have less oxoproducts than Col0 plants, suggesting that both ROS1 and DDM1 have a role in the repair of oxidative DNA damage. Together, our data provide evidence that both DDM1 and ROS1, directly or indirectly, participate in UV-B induced- and oxidative DNA damage repair.
Results
UV-B regulation of ddm1 and ros1, mutant analysis and physiological effects
Chromatin remodeling has previously been shown to be crucial for UV-B damage repair in plants (Casati et al., 2006; Campi et al., 2012). Different chromatin landscapes control the accessibility of the DNA repair machinery to damaged DNA. In several organisms, a major factor affecting chromatin accessibility is DNA methylation. Therefore, we sought to determine if enzymes that have a role in DNA methylation participate in UV-B damage repair in arabidopsis. Provided that ros1 and ddm1 mutants were previously reported to contain altered levels of DNA methylation in their genomes (Kakutani et al., 1996; Xia et al., 2006), they confer an adequate system to analyze how DNA methylation affects the repair of UV-B induced DNA lesions. A. thaliana mutants defective in DDM1 and ROS1 were identified in the SALK collection. For ros1, two independent T-DNA insertional lines, SALK_135293 and SALK_045303, with insertions in the 3′ UTR and the 16th exon, respectively, were identified by a PCR screen using gene-specific primers and one specific primer for the T-DNA left border (Figures S1, S2 and Table S1 in Supplementary Material). Insertional inactivation of ROS1 in both lines was confirmed by RT-PCR (Figures S1, S2). For the DDM1 gene, two independent T-DNA insertional lines, SALK_000590 and SALK_093009 (ddm1-10, Jordan et al., 2007), with insertions in the 16th exon and the 15th intron, respectively, were identified by a PCR screen using gene-specific primers and one specific primer for the T-DNA left border (Figure S3 and Table S1 in Supplementary Material). Decreased expression of DDM1 in both lines was confirmed by RT-PCR (Figure S4 in Supplementary Material). ddm1 mutants show hypomethylation in several regions of the DNA; in particular, the AtMu1 transposon, which is usually methylated and its transposase is not transcribed in WT plants, it is actively transcribed when it is hypomethylated in ddm1 mutants (Singer et al., 2001). Figure S5 in Supplementary Material shows that AtMu1 is highly transcribed in the SALK_093009 mutant, while is not expressed in the Col0 plants. In addition, because DNA hypomethylation induces the misregulation of the expression of diverse genes, ddm1 mutants show an abnormal phenotype, with small and curved leaves (Kakutani et al., 1996). The SALK_093009 mutant has already been described to show a ddm1 mutant phenotype, showing up-regulation of genes as a consequence of hypomethylated DNA (Jordan et al., 2007). In addition, Figure S5 shows that both the SALK_093009 and the SALK_000590 mutants have a similar phenotype as that described for other ddm1 mutants (Vongs et al., 1993; Jordan et al., 2007). The SALK_000590 mutants also show high expression of AtMu1 and a similar phenotype as that of SALK_093009 plants (not shown), suggesting that both mutants are probably deficient in DNA methylation. It is important to mention that we have not tested the methylation profile of the SALK_093009 and the SALK_000590 mutants, but we are confident, according to the observed phenotypes, transcription activation of AtMu1 transposon and the published data (Vongs et al., 1993; Kakutani et al., 1996; Singer et al., 2001; Jordan et al., 2007) that the two mutants behave as methylation deficient.
We first investigated the effects of UV-B on physiological parameters in ddm1 and ros1 mutants. UV-B induces flavonoid accumulation such as anthocyanins and other UV sunscreens in many plants (Li et al., 1993; Landry et al., 1995; Ormrod et al., 1995). After a 4 h-UV-B treatment, the concentration of these molecules was 1.76-fold higher than under control conditions in Col0 plants. Similar increases were observed for the two ros1 mutants analyzed (1.63- and 1.73-fold, respectively; Figure 1A). On the contrary, plants with decreased levels of DDM1 transcript have altered accumulation of UV sunscreen photoprotectors. ddm1 mutants showed a significantly higher increase in the level of these pigments after the UV-B treatment (2.24 and 2.16-fold increase, respectively; Figure 1A). Moreover, when pigment levels were compared in baseline control conditions in the absence of UV-B, ddm1 mutants showed already elevated flavonoid levels similar to those in Col0 plants after the UV-B treatment. In addition, UV-B sensitivity was analyzed by inhibition of primary root elongation assays (Tong et al., 2008). One day after the end of the UV-B treatment, both Col0 and ros1 seedlings showed a slight although significant decrease in primary root elongation compared to untreated plants (Figure 1B). However, 2 days after the treatment, ros1 plants showed a lower decrease in primary root growth than Col0 plants. In contrast, ddm1 seedlings showed a significant higher inhibition of root elongation by UV-B than Col0 plants (Figure 1C). Together, these results suggest that ddm1 mutants are more sensitive to UV-B radiation than Col0 plants; while ros1 mutants are less responsive to this radiation.
Figure 1.
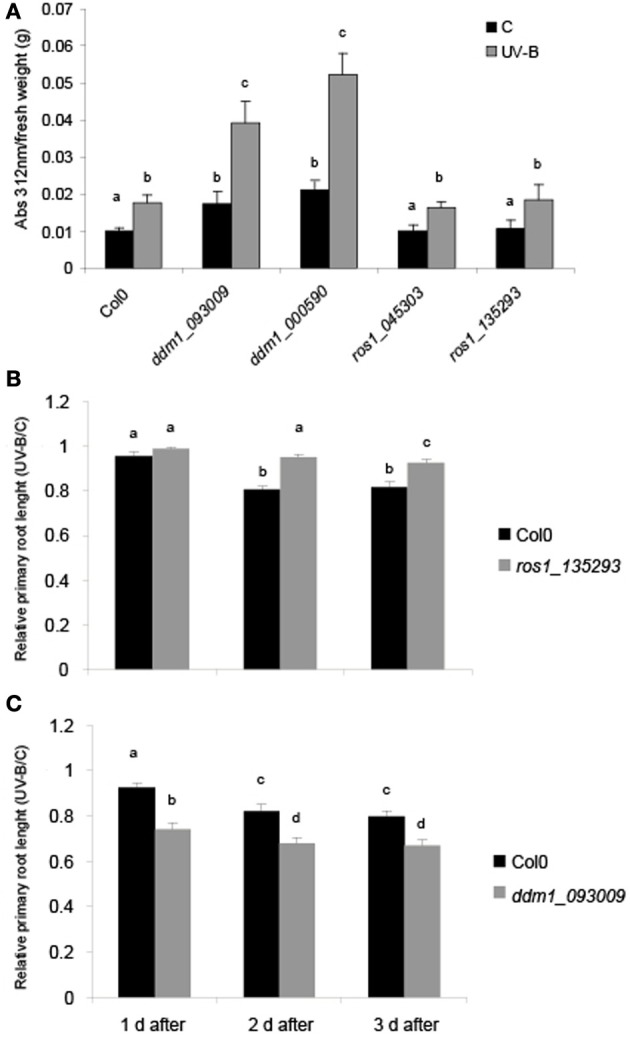
Physiological responses in ddm1 and ros1 mutants after UV-B exposure. (A) Total UV-B absorbing compounds were assayed after 4 h UV-B (UV-B) compared to untreated controls (C) in Col0 plants, and ddm1 and ros1 mutants. Measurements are the average of six adult leaves from six different plants. (B andC) Graph of average root lengths in Col0, ros1 (B) and ddm1 (C) mutants up to 3 days after a UV-B treatment. Error bars represent S.E.M. Statistical significance was analyzed using ANOVA, Tukey test with P < 0.05; differences from the control are marked with different letters.
Previously, four arabidopsis chromatin remodeling genes NFC4, SDG26, HAM1 and HAM2 were reported to be induced by UV-B; and plants deficient in the expression of these genes all showed increased accumulation of CPDs compared to WT plants of the Col0 ecotype when exposed with UV-B light (Campi et al., 2012). Therefore, we investigated if DDM1 and ROS1 were also regulated by this radiation. 4-weeks-old Col0 (WT) plants grown in the absence of UV-B were exposed under UV-B lamps for 4 h in a growth chamber. After the treatment, leaf tissue was collected for RNA extraction and qRT-PCR analysis. Contrary to the up-regulation reported for NFC4, SDG26, HAM1 and HAM2, Figure 2 shows that DDM1 and ROS1 are significantly down regulated by UV-B. The transcript of the arabidopsis CPD photolyase UVR2 (At1g12370), a UV-B inducible gene, was used as a positive control.
Figure 2.
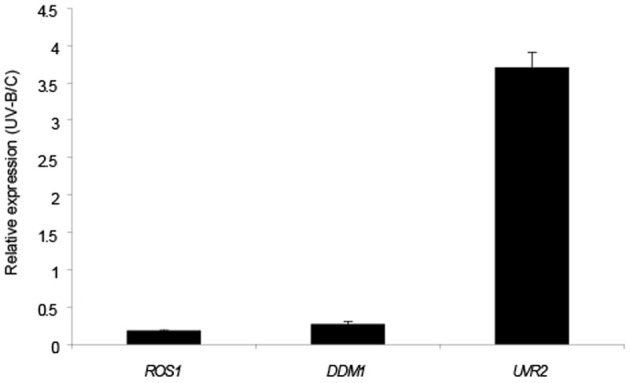
Relative transcript levels of A. thaliana ROS1 and DDM1 genes measured by qRT-PCR. Plants were irradiated with UV-B light for 4 h (UV-B) or kept under control conditions (C) as indicated in Materials and Methods. CPK3 transcript was used for normalization and UVR2 as a control of a UV-B inducible gene. Data show mean values ± S.E.M. of at least three independent experiments.
Opposing impact of ros1 and DDM1 on UV-B DNA damage repair in arabidopsis
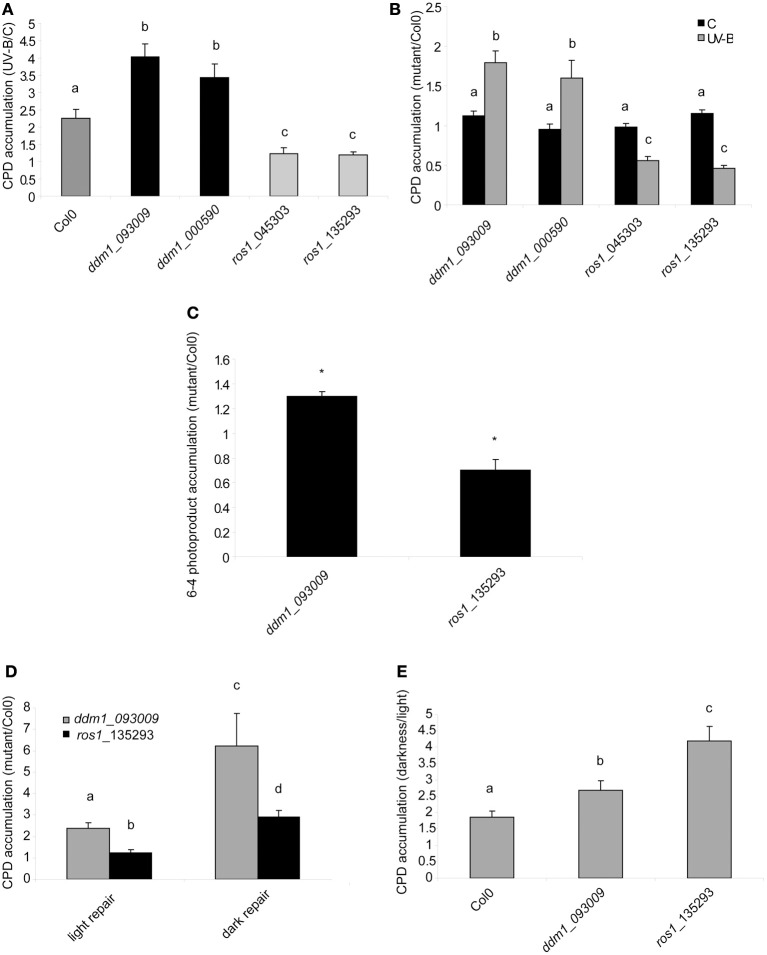
To test the participation of ROS1 and DDM1 in UV-B-damaged DNA repair, 4-weeks-old Col0, ros1 and ddm1 plants were irradiated with UV-B for 4 h. Leaf samples from control and treated plants were collected immediately after the treatment under light conditions that allow photoreactivation. Genomic DNA was extracted to evaluate CPD abundance after UV-B exposure, both in Col0 and mutant plants (Figures 3A,B). CPD levels were measured by an immunological sensitive assay; this assay detects CPDs by monoclonal antibodies specifically raised against them. 1.5 μg of DNA was used for each sample, as that there is a linear relationship of signal values of UV-B treated samples vs. the corresponding amounts of DNA loaded up to 2 μg of DNA (Lario et al., 2011). In the absence of UV-B, the steady state levels of CPDs in Col0 and mutant plants were similar [about 200 intensity of the optical density (IOD) in all samples; Figure 3B]. After 4 h UV-B exposure, unrepaired lesions accumulated in all plants (Figure 3A) CPD levels in ddm1 mutants were significantly higher than in Col0 (Figures 3A,B). Interestingly, ros1 mutants showed only a minor, although still significant increased accumulation of CPDs after the UV-B treatment. Consistent with the lack of UV-B sensitivity observed in the root elongation assay, ros1 plants accumulate lower levels of CPDs than Col0 (Figures 3A,B). These results confirm the participation of ROS1 and DDM1 in UV-B damage repair and also evidence the opposing effects of these two proteins in UV-B response.
Figure 3.
CPD and 6-4PPs levels in the DNA of Col0, ddm1 and ros1 arabidopsis plants. (A) CPD levels in DNA of UV-B treated Col0, ddm1, and ros1 plants for 4 h, relative to levels under control conditions without UV-B (C). (B) CPD levels in DNA of ddm1 and ros1 plants relative to Col0 plants under control conditions without UV-B (C) and after a 4 h-UV-B treatment (UV-B). (C) 6-4PPs levels in DNA of ddm1 and ros1 plants relative to Col0 plants after a 4 h-UV-B treatment. (D) CPD levels in DNA of ddm1 and ros1 plants relative to Col0 plants after a recovery period in the absence of UV-B for 2 h. Experiments were done under conditions that allowed photorepair in the light (light) or under dark conditions (dark). (E) CPD levels in DNA of Col0, ddm1 and ros1 plants after a recovery period in the absence of UV-B for 2 h in the light relative to levels after recovery under dark conditions. Results represent the average ± S.E.M. of six independent biological replicates. Different letters denote statistical differences applying ANOVA tests using Sigma Stat 3.1. Asterisks denote statistical differences applying Student's t test (P < 0.05).
6-4PPs constitute around 25% of the DNA damage induced by UV-B radiation (Britt, 1996). We investigated how 6-4 photoproducts were accumulated in ddm1 and ros1 mutants. As observed for CPD accumulation, ddm1 plants accumulated significant higher levels of 6-4PPs that Col0 plants after a 4 h-UV-B treatment, while ros1 mutants showed lower accumulation of these products under the same conditions (Figure 3C).
ddm1 and ros1 mutants have altered levels of DNA repair transcripts
The evidence of a role of DDM1 and ROS1 in UV-B damage repair prompted us to investigate their involvement in the regulation of the expression of DNA repair genes. UV-B-induced DNA damage repair is accomplished by two main pathways: nucleotide excision repair (NER) and photoreactivation (PR). Therefore, we measured the transcript levels of some selected NER and PR genes before and after UV-B exposure. First, we evaluated the expression of 2 photolyase genes: UVR2, encoding a CPD photolyase, and UVR3, encoding a 6-4 photoproduct photolyase (At3g15620). Figure 4 and Figure S6 in Supplementary Material show that both genes were up-regulated by UV-B radiation in Col0 plants after the treatment; however, ddm1 mutants constitutively expressed high levels of both photolyases. In previous studies using different mutants that are deficient in homologous recombination and repair of damaged DNA with methylmetane sulphonate, such us abo4 (a mutant in the DNA pol ε, Yin et al., 2009), rfc1 (a mutant in the DNA replication factor C1; Liu et al., 2010a), and polα (a mutant in the DNA pol α, Liu et al., 2010b), DNA repair transcripts were highly and constitutively expressed, suggesting that in these mutants DNA repair-related genes were spontaneously induced. We hypothesize that a similar situation occurs in ddm1 plants. In contrast, ros1 mutants contained wild type amounts of UVR2 and UVR3 transcripts in the absence of UV-B and showed higher levels of both transcripts after the UV-B treatment compared to Col0 (Figure 4 and Figure S6). Thus, the lower accumulation of CPDs in ros1 mutants after the UV-B treatment may be a result of increased photolyases activity.
Figure 4.
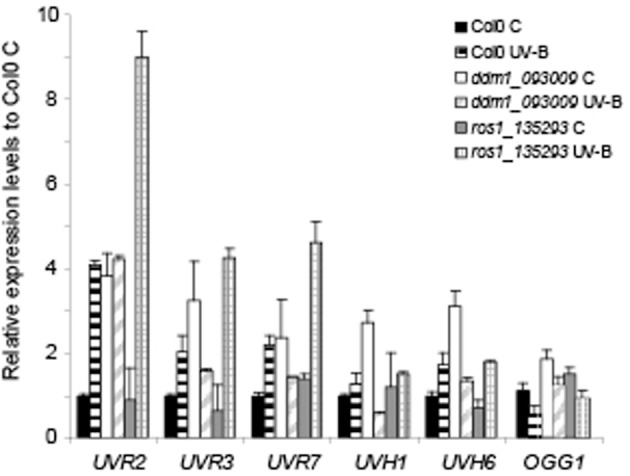
Relative expression of DNA repair transcripts by RT-qPCR in Col0, ddm1 (ddm1_093009 line) and ros1 (ros1_135293 line). Levels of UVR2, UVR3, UVR7, UVH1, UVH6, and OGG1 were assayed in arabidopsis plants that were irradiated with UV-B for 4 h (UV-B) or were kept under control conditions without UV-B (control, C). Expression values are relative to the values in Col0 plants under control conditions in the absence of UV-B. The CPK3 transcript was used as a control. Data show mean values ± S.E.M. of at least three independent experiments.
On the other hand, we analyzed the expression of the NER genes UVR7 (encoding ERCC1, a DNA excision repair protein, At3g05210), UVH1 (encoding the RAD1 endonuclease, At5g41150), and UVH6 (encoding a protein similar to the human helicase XPD, At1g03190). All these transcripts were induced by UV-B in the Col0 background, and this was also true for ros1 mutants. However, after UV-B exposure the induction of UVR7 was 3-fold higher in ros1 plants compared to Col0 (Figure 4 and Figure S6). In ddm1 mutants, high basal expression of these genes was detected under control conditions, as previously observed for the photolyases. Finally, we analyzed the expression of the 8-oxoguanine DNA glycosylase gene OGG1 (At1g21710), a member of the arabidopsis base excision repair (BER) system. Although the expression of this gene was similar in Col0 and the mutants under control conditions, Col0 showed decreased levels of OGG1 after 4 h UV-B treatment, not measured in either mutant (Figure 4).
Lower accumulation of CPDS in ros1 mutants are probably a consequence of increased levels of photolyases after UV-B exposure
To analyze that the decreased UV-B sensitivity of ros1 mutants is due to increased photolyases activity, we tested the repair of CPDs in the dark and in the light after 2 h of recovery in the absence of UV-B. As expected, all plants repaired CPD damage better in the light, when photoreactivation occurs, than in the dark, when photoreactivation is absent (Figure 3E). After 2 h recovery in the light, ros1 plants showed similar levels of CPDs as Col0 plants as a result of photoreactivation (Figure 3D). However, recovery in the dark was significantly compromised (Figures 3D,E). This result demonstrates that the low levels of CPDs accumulated in the light are probably a consequence of the higher expression of photolyases after UV-B exposure.
On the other hand, ddm1 mutants still showed higher CPD accumulation than Col0 plants after 2 h recovery under both conditions, demonstrating that these mutants have a defect in DNA repair, probably due to a deficiency in chromatin remodeling, as already reported for other types of DNA damage (Shaked et al., 2006; Yao et al., 2012; Figure 3D). It is interesting to note that ddm1 plants were more affected in the dark than in the light repair (Figure 3D), the reason for this may be probably because different proteins participate in the NER repair machinery (the main dark CPD repair system), while photoreactivation requires the action of only one protein, the photolyase. Therefore, chromatin remodeling activities may be more important in the dark repair, which replace the damaged DNA with new, undamaged nucleotides, to allow to spatially accomodate the different proteins that participate in this process.
Together, our results suggest that chromatin remodeling deficient ddm1 plants have increased CPD accumulation by UV-B because DNA repair mechanisms, in particular NER proteins, may require chromatin remodeling by this enzyme for their activities. On the contrary, ros1 mutants are also deficient in CPD dark repair, but have high photoreactivation probably as a result of increased expression of UVR2 and UVR3.
UV-B does not induce the accumulation of oxidized bases in the DNA of arabidopsis plants, but ddm1 and ros1 mutants are affected in oxidative damage repair
The results presented in Figure 4 suggest that both ddm1 and ros1 mutants are deficient in CPD dark repair. In plants, dark pathways fall into two major categories: NER and BER (Britt, 1996). The BER involves the removal of a single damaged base through the action of one of many lesion-specific glycosylases, which leaves the DNA sugar-phosphate backbone intact. Glycosylases and endonucleases specific for cyclobutane dimers have been observed in bacteria and bacteriophages and have been useful as diagnostic agents for the assay of UV-induced damage (Friedberg et al., 1995). On the other hand, UV-B radiation has been described to alter reactive oxygen species metabolism (Hideg et al., 2013). A wide variety of oxidative damage products are induced in DNA by hydroxyl radicals, superoxide, and nitric oxide (Britt, 1996). The most significant oxidized base is 8-hydroxyguanine (8-oxodG); thus, we investigated if UV-B produces base oxidation in arabidopsis. For this aim, we analyzed the accumulation of 8-oxodG after a 4 h UV-B treatment in Col0, ddm1 and ros1 mutants. Interestingly, Col0 plants did not accumulate 8-oxodG after the UV-B treatment (Figure 5). Moreover, the accumulation of 8-oxodG was neither changed in ddm1 nor in ros1 mutants after UV-B. However, both mutants showed significantly different accumulation of this DNA oxidation product compared to Col0 plants (Figure 5). For ros1, 8-oxodG accumulation was higher than Col0 plants (Figure 5). Despite that ROS1 is a glycosyltranferase of the BER repair system that has been described to remove 5-meC and T mismatched to G (Morales-Ruiz et al., 2006), its activity using oxidized bases as substrates has not been previously determined. On the other hand, ddm1 mutants showed significantly lower levels of 8-oxodG than Col0 plants (Figure 5). This is in contrast to which was previously reported for other types of DNA damage, such as treatment with UV-C, γ-radiation and methyl methane sulfonate (Shaked et al., 2006; Yao et al., 2012), and our results in the repair of photoproducts by UV-B (Figure 3), where these mutants show higher levels of DNA damage than WT plants. In particular, ros1 mutants show altered levels of the other 5-meC glycosylases DML2, DML3 and DME1 (Figure S7 in Supplementary Material). Therefore, it is possible that this increase in the accumulation of 8-oxodG may be due to altered expression of different glycosylases in these mutants. Together, our data provide evidence that both DDM1 and ROS1, directly or indirectly, participate in oxidative DNA damage repair in arabidopsis.
Figure 5.
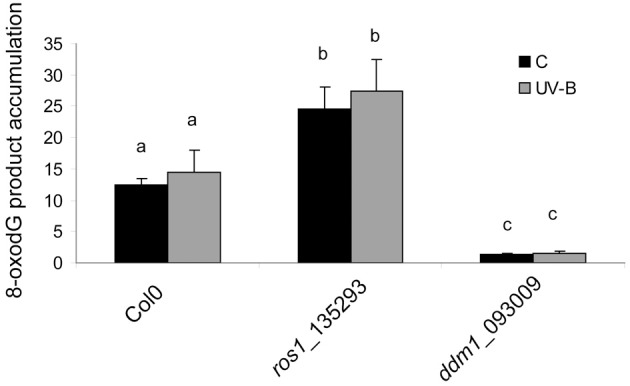
8-oxodG levels in the DNA of Col0, ros1 and ddm1 arabidopsis plants. Plants were assayed under control conditions (C) and after a 4 h UV-B treatment (UV-B). Results represent the average ± S.E.M. of six independent biological replicates. Different letters denote statistical differences applying ANOVA tests using Sigma Stat 3.1.
Discussion
Absorption of UV-B by DNA induces the formation of covalent bonds between adjacent pyrimidines with the formation of CPDs and 6-4PPs (Friedberg et al., 1995); overaccumulation of these lesions must be prevented to maintain genome integrity, plant growth and seed viability. Plants have evolved mechanisms that filter or absorb UV-B to protect against DNA damage (Mazza et al., 2000; Bieza and Lois, 2001), and also have DNA repair systems to remove DNA lesions (Hays, 2002; Bray and West, 2005; Kimura and Sakaguchi, 2006). The genome of plants is organized into chromatin, which limits the accessibility of DNA, affecting the rates of processes such as DNA recombination and repair. The disruption of the interactions of nucleosome–DNA or the remodeling of chromatin can stimulate or repress DNA repair. In yeast, RAD54, RAD26 and RDH54, which all belong to the switch2/sucrose non-fermenting2 (Swi2/Snf2) superfamily, participate in meiosis and also in various aspects of DNA repair, for example in homologous recombination and in nucleotide excision and transcription-coupled repair (Eisen et al., 1995; Klein, 1997; Shinohara et al., 1997). In arabidopsis, the Swi2/Snf2-related SWR1 complex, which deposits histone H2A.Z, is important for DNA repair (Rosa et al., 2013). Mutations in genes for different subunits of the SWR1 complex cause hypersensitivity to various DNA damaging agents; and even without additional genotoxic stress, these mutants show symptoms of DNA damage accumulation (Rosa et al., 2013). In maize, chromatin remodeling has been implicated in UV-B responses. Transgenic maize plants knockdown for chromatin remodeling genes were found to be acutely sensitive to UV-B at doses that do not cause visible damage to maize lacking flavonoid sunscreens (Casati et al., 2006). In maize and arabidopsis, plants deficient in chromatin remodeling show increased DNA damage compared to WT plants after a UV-B treatment (Campi et al., 2012). However, the role of enzymes that participate in DNA methylation in DNA repair after UV-B damage was not previously investigated yet. Therefore, in this work, we analyzed the role of enzymes that participate in DNA methylation in the repair of CPDs and 6-4PPs using mutant plants in DDM1 and ROS1.
First, we analyzed the expression of both DDM1 and ROS1 by UV-B radiation in arabidopsis. Interestingly, both genes are repressed after the treatment, suggesting that DDM1 and ROS1 may have a role in UV-B responses. Therefore, their function in UV-B responses was investigated. In plants, the first line of defense when exposed to UV-B is the synthesis of protective pigments like flavonoids and UV-B absorbing pigments. In our experiments, UV-B absorbing pigments levels increased in Col0, ddm1 and ros1 mutants after the UV-B treatment; however, when pigment levels were compared in baseline control conditions in the absence of UV-B, ddm1 mutants showed already elevated flavonoid levels similar to those in Col0 plants after the UV-B treatment. This demonstrates that arabidopsis plants deficient in chromatin remodeling are affected in the accumulation of UV-absorbing compounds, similarly as previously described in maize and arabidopsis chromatin remodeling deficient plants (Casati et al., 2006; Campi et al., 2012). In addition, ddm1 seedlings showed a significantly higher inhibition of root elongation by UV-B than Col0 plants; while ros1 roots were less affected by UV-B than those from Col0 plants. Together, these results suggest that ddm1 mutants are more sensitive to UV-B radiation than Col0 plants; whereas ros1 mutants are less responsive to this radiation.
In addition, we demonstrated that ddm1 mutants accumulated more damaged DNA after UV-B exposure compared to Col0 plants. Previous studies have shown that ddm1 plants have increased sensitivity to γ and UV-C radiation, they are susceptible to NaCl stress and are also deficient in DNA repair by methyl methane sulfonate (Shaked et al., 2006; Yao et al., 2012). Moreover, DDM1 participates in homologous recombination (Shaked et al., 2006). These data, in agreement with our results, demonstrate that DDM1 plays a role in response to DNA damage. The ddm1 mutants used in our experiments show high expression of the AtMu1 transposase, which is not expressed in the Col0 plants, demonstrating that these mutants have deficient methylation in some DNA regions (Singer et al., 2001). It is interesting that ddm1 plants constitutively express high levels of DNA repair enzymes, similarly as other mutants deficient in DNA repair (Yin et al., 2009; Liu et al., 2010a,b), suggesting that in all these mutants DNA repair-related genes were spontaneously induced. However, these increased expression levels do not correlate with increased DNA repair; therefore, DDM1 may participate directly in DNA repair, and not through the regulation of the expression of DNA repair genes. A comparison of mutants in DDM1 and MET1, a gene encoding a cytokine methyltransferase, suggested that DNA damage response is affected essentially by chromatin structure, while cytosine methylation was less critical (Shaked et al., 2006). Therefore, we suggest that DDM1 is important in chromatin remodeling during DNA repair of UV-B induced pyrimidine dimers.
In contrast, ddm1 plants show significantly lower levels of 8-oxodG than Col0 plants. DDM1 has been shown to increase meiotic recombination in both male and female lineages, but the effect is not equal throughout the genome (Melamed-Bessudo and Levy, 2012). In these mutants, euchromatic regions exhibit high rates of meiotic recombination, whereas no changes are found in heterochromatic centric and pericentric areas; demonstrating the involvement of DDM1 and chromatin remodeling in genome maintenance. DDM1 regulates histone H3 and DNA methylation; upon loss of DDM1 activity, a 70% reduction in DNA methylation is induced, promoting chromatin decondensation (Jeddeloh et al., 1999; Probst et al., 2003). Therefore, the DNA demethylation per se or altered chromatin remodeling could make the DNA more accessible to the BER repair system, as similarly suggested for homologous recombination enzymes (Melamed-Bessudo and Levy, 2012). Interestingly, the expression levels OGG1, an 8-oxoguanine DNA glycosylase of the BER, is similar in ddm1 and Col0 plants, so increased repair of 8-oxodG cannot be explained by changes in the activity of this enzyme. However, we cannot rule out that other glycosylases or repair enzymes may be up-regulated in the ddm1 mutants, for example by activation of silent genes from hypomethylated chromosomes.
On the other hand, in our experiments, ros1 showed less CPDs and 6-4PPs than Col0 plants after a UV-B treatment under light conditions; however, CPD accumulation after a 2 h recovery in the dark was higher in the mutants than in Col0. The results presented here show that transcripts for two photolyases, UVR2 (a CPD photolyase) and UVR3 (a 6-4PPs photolyase) are highly induced by UV-B in ros1, suggesting that the lower accumulation of photoproducts by UV-B may be due to increased photorepair in these mutants. This higher photorepair correlates with lower inhibition of primary root elongation by UV-B, suggesting that these mutants have higher UV-B tolerance than WT plants. On the contrary, ros1 plants accumulate elevated levels of 8-oxodG in the DNA; therefore, ROS1 may have a role in the repair of oxidative DNA damage. Interestingly, ROS1 is a DNA glycosylase that has been described to remove 5-meC and T mismatched to G (Morales-Ruiz et al., 2006), but its activity using oxidized bases as substrates has not been previously determined. Several ros1 suppressors have been identified, including replication protein A2 (RPA2A/ROR1) (Xia et al., 2006), DNA polymerase α (Liu et al., 2010b), DNA polymerase ε (Yin et al., 2009) and TOUSLED (Wang et al., 2007). These mutants release the TGS of 35S-NPTII and increase the expression of transcriptionally active information, but they do not change the DNA methylation state when mutated. All ros1 suppressors described above are sensitive to DNA damage, they respond to the damage with constitutive expression of DNA damage related genes, and most of them also have a high homologous recombination rate (Xia et al., 2006; Wang et al., 2007; Yin et al., 2009), suggesting that the silencing of chromatin is closely related with DNA replication, DNA repair and homologous recombination (Probst et al., 2009). However, with the exception that ros1 mutation increases the telomere length in arabidopsis (Liu et al., 2010b), ros1 mutants have not previously shown any differential response in DNA repair when compared to WT plants (Liu et al., 2010a). Our results suggest that in vivo, ROS1 may also have a role in the repair of 8-oxodG. Alternatively, a mutation in ROS1 may affect the expression of other glycosylases specific for 8-oxodG, similarly as determined for the UVR2 and UVR3 photolyases in this work. ros1 plants show altered levels of the other 5-meC glycosylases DML2, DML3 and DME1; thus, it is possible that this increase in the accumulation of 8-oxodG may be due to altered expression levels of different glycosylases in these mutants.
We have previously demonstrated that chromatin remodeling is essential during DNA repair by UV-B (Campi et al., 2012). In particular, because histone H3 and H4 acetylation is increased by UV-B (Casati et al., 2008), the effect of histone acetylation on DNA repair was previously analyzed, and our results demonstrated that when plants are pre-treated with curcumin, a histone acetylase inhibitor, DNA repair was impaired (Campi et al., 2012). Interestingly, in sdg26 mutants (SDG26 encodes a histone methyltransferase), a curcumin treatment previous to UV-B irradiation induced a significantly higher accumulation of CPDs than curcumin-treated WT plants. Therefore, a deficiency in the expression of a histone methyltransferase interferes directly or indirectly with the DNA damage repair mediated by histone acetylation, suggesting that both processes, histone acetylation and methylation, act synergistically during UV-B induced damage repair. In this manuscript, we show that enzymes that participate in DNA methylation are also important during DNA repair by UV-B, demonstrating that both genetic and epigenetic effects control DNA repair in plants.
Together, the results presented here demonstrate the participation of DDM1 and ROS1 in DNA repair after UV-B damage and oxidation. We propose that, in ddm1 mutants, DNA demethylation per se or altered chromatin remodeling could affect accessibility to DNA repair systems. On the contrary, we suggest that in ros1 mutants, lower accumulation of photoproducts is due to increased levels of photolyases by UV-B. Finally, ROS1, besides its demonstrated role as a 5-meC glycosylase, it could also act as an oxoproduct glycosidase.
Materials and methods
Plant material, growth conditions and irradiation protocols
The A. thaliana ecotype Columbia (Col0) was used for all the experiments. The T-DNA insertion mutants were obtained from the SALK T-DNA insertion mutant collection (Alonso et al., 2003). Mutants lines used are shown in Figures S1–S4 in Supplemental data. Arabidopsis plants were sown directly on soil and placed at 4°C in the dark. After 3 days, pots were transferred to a greenhouse and plants were grown at 22°C under a 16 h/8 h light/dark regime. Plants were exposed 4 h to UV-B radiation (315 nm) from UV-B bulbs (2 W m−2 UV-B and 0.65 W m−2 UV-A, Bio-Rad, Hercules, California) in a growth chamber, both in the presence or the absence of white light, and control plants were treated with the same plants covered with a polyester film (0.04 W m−2 UV-B, 0.4 W m−2 UV-A). Adult leaf samples from 4-weeks-old plants were collected immediately after irradiation, or 2 h after the end of the UV-B treatment, both under light and under dark conditions.
Identification of insertional T-DNA mutants
The genotype of plants with T-DNA constructs was determined using a PCR-based approach. Initial screening was performed using genomic DNA isolated from leaves by a modified cetyl-trimetyl-ammonium bromide (CTAB) method (Sambrook and Russel, 2001) and three combinations of primers. Two primers hybridize to specific genomic sequences (Table S1) and one primer is located inside the left border of the T-DNA. The presence or absence of the T-DNA insertion in the genes allowed the identification of homozygous, heterozygous and WT plants.
RT-PCR for expression analyses in the knockout T-DNA lines were carried out in a 25 μ l final volume containing 1X buffer Taq DNA polymerase, 3 mM MgCl2, 0.2 mM dNTP, 0.25 μ M of each primer, 0.625 U Taq DNA polymerase (Invitrogen, Carlsbad, California). Cycling were performed under the following conditions: 2 min denaturation at 95°C, 35 cycles of 10 s denaturation at 95°C, 15 s annealing at 57°C, 30 s amplification at 72°C and a final 7 min amplification at 72°C. RT-PCR products were separated on a 1% (w/v) agarose gel and stained with SYBR Safe (Invitrogen).
Quantitative RT-PCR
Total RNA was isolated from about 100 mg of tissue using the TRIzol reagent (Invitrogen) as described by the Manufacture's Protocol. The RNA was incubated with RNase-free DNase I (1 U/ml) following the protocol provided by the manufacturer to remove possible genomic DNA. Then, RNA was reverse-transcribed into first-strand cDNA using SuperScript II reverse transcriptase (Invitrogen) and oligo-dT as a primer. The resultant cDNA was used as a template for qPCR amplification in a MiniOPTICON2 apparatus (Bio-Rad), using the intercalation dye SYBRGreen I (Invitrogen) as a fluorescent reporter and Platinum Taq Polymerase (Invitrogen). Primers for each of the genes under study were designed using the PRIMER3 software (Rozen and Skaletsky, 2000) in order to amplify unique 150–250 bp products (Table S2 in Supplementary Material). Amplification conditions were carried out under the following conditions: 2 min denaturation at 94°C; 40 cycles at 94°C for 10 s, 57°C for 15 s, and 72°C for 30 s, followed by 10 min extension at 72°C. Three replicates were performed for each sample. Melting curves for each PCR were determined by measuring the decrease of fluorescence with increasing temperature (from 65 to 98°C). PCR products were run on a 2% (w/v) agarose gel to confirm the size of the amplification products and to verify the presence of a unique PCR product. Gene expressions were normalized to the A. thaliana calcium dependent protein kinase3 (CPK3, Table S2). The expression of this gene has been previously reported to remain unchanged by UV-B (Ulm et al., 2004).
DNA damage analysis
The induction of CPD, 6-4 photoproducts and 8-oxodG was determined using an assay described in detail previously (Stapleton et al., 1993), using monoclonal antibodies specific to CPDs (TDM-2), 6-4 photoproducts (64M-2) and 8-oxodG (N45.1 obtained from Cosmo Bio Co., Ltd., Japan). After the treatments, plant samples (0.1 g) were collected and immediately immersed in liquid nitrogen and stored at −80°C. The 1.5 μg (for CPD assays), 20 μ g (for 6-4 photoproduct assays) and 2 μg (for 8-oxodG assays) of the extracted DNA by a modified cetyl-trimetyl-ammonium bromide (CTAB) method was denatured in 0.3 M NaOH for 10 min and sextuplicate biological replicates were dot blotted onto a nylon membrane (Perkin Elmer Life Sciences, Waltham, Massachusetts). The membrane was incubated for 2 h at 80°C and then it was blocked in TBS (20 mM Tris-HCl, pH 7.6,137 mM NaCl) containing 5% dried milk for 1 h at room temperature or overnight at 4°C. The blot was then washed with TBS and incubated with the different antibodies (1:2000 in TBS) overnight at 4°C with agitation. Unbound antibody was washed away and secondary antibody (BioRad) conjugated to alkaline phosphatase (1:3000) was added. The blot was then washed several times followed by the addition of the detection reagents NBT and BCIP. Quantification was achieved by densitometry of the dot blot using ImageQuant software version 5.2. DNA concentration was fluormetrically determined using the Qubit dsDNA assay kit (Invitrogen), and checked in a 1% (w/v) agarose gels after quantification. DNA concentration was determined spectrophotometrically at 260 and 280 nm in the microplate reader (Biotek XS Power Wave) using the KC Junior computer program, and checked in a 1% (w/v) agarose gel after quantification.
Root length measurements
Petri dish-grown seedlings, surface-sterilized seeds were grown on MS growth medium and were held vertical in a growth chamber. Then, seedlings were UV-B treated for 2 h and kept in the absence of UV-B for 3 days. Plates were photographed before the treatment, and 24, 48, and 72 h after the end of the treatment, and the images were analyzed using the ImageJ program. Root lengths were determined by measuring the length of a line traced along the root.
Pigment measurements
UV-absorbing pigments (absorbance at 312 nm) were determined as described in Casati and Walbot (2004).
Statistical analysis
Statistical analysis was done using ANOVA models (Tukey test) using untransformed data with Sigma Stat 3.1.
Author contributions
Julia I. Qüesta, Julieta Fina and Paula Casati designed the experiments and analyzed the data. Julia I. Qüesta and Julieta Fina did the experiments. Paula Casati wrote the article. Julia I. Qüesta, Julieta Fina and Paula Casati edited the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Arabidopsis Biological Resource Center (ABRC, Columbus, OH) that provided seed stocks. This research was supported by FONCyT grants PICT 2007-00711 and 2010-00105 to Paula Casati. Paula Casati is a member of the Researcher Career of the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET).
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2013.00420/abstract
(A) Location of the T-DNA insertion in the ROS1 gene (SALK_135293 line). Exons are represented by blue boxes, introns by thin black lines and the UTR regions by light gray boxes. The T-DNA insertion is indicated as a triangle. (B) Analysis of the PCR products separated in 1% (w/v) agarose gels. The PCR reactions were done using genomic DNA from Col0 and SALK_135293 plants. Lanes 1 show the PCR products obtained for a WT plant using the SALK_135293 F and SALK_135293 R primers; while lanes 2 show the PCR products obtained for homozygous mutant plants using the Lb and SALK_13293 R primers. (C) Transcript levels were evaluated by RT-PCR followed by agarose gels on cDNAs obtained from RNA extracted from the mutant or WT lines. Amplifications were performed using ROS1 F and ROS1 R primers, which are specific for the ROS1 transcript. As a control, primers for the RPL10B transcript were used.
(A) Location of the T-DNA insertion in the ROS1 gene (SALK_045303 line). Exons are represented by blue boxes, introns by thin black lines and the UTR regions by light gray boxes. The T-DNA insertion is indicated as a triangle. (B) Analysis of the PCR products separated in 1% (w/v) agarose gels. The PCR reactions were done using genomic DNA from Col0 and SALK_045303 plants. Lanes 1 show the PCR products obtained for a WT plant using the SALK_045303 F and SALK_045303 R primers; while lanes 2 show the PCR products obtained for homozygous mutant plants using the Lb and SALK_045303 F primers. (C) Transcript levels were evaluated by RT-PCR followed by agarose gels on cDNAs obtained from RNA extracted from the mutant or WT lines. Amplifications were performed using ROS1 F and ROS1 R primers, which are specific for the ROS1 transcript. As a control, primers for the RPL10B transcript were used.
(A, C) Location of the T-DNA insertion in the DDM1 gene (SALK_000590 (A) and SALK_093009 (C) lines). Exons are represented by blue boxes, introns by thin black lines and the UTR regions by light gray boxes. The T-DNA insertion is indicated as a triangle. (B) Analysis of the PCR products separated in 1% (w/v) agarose gels. The PCR reactions were done using genomic DNA from Col0 and SALK_000590 plants. Lanes 1 show the PCR products obtained for a WT plant using the SALK_000590 F and SALK_000590 R primers; while lanes 2 show the PCR products obtained for homozygous mutant plants using the Lb and SALK_000590 F primers. (D) Analysis of the PCR products separated in 1% (w/v) agarose gels. The PCR reactions were done using genomic DNA from Col0 and SALK_093009 plants. Lanes 1 show the PCR products obtained for a WT plant using the SALK_093009 F and SALK_093009 R primers; while lanes 2 show the PCR products obtained for homozygous mutant plants using the Lb and SALK_093009 R primers.
Expression of DDM1 transcripts in Col0 plants and ddm1 mutants. Transcript levels were evaluated by RT-PCR followed by agarose gels (A) and RT-qPCR (B) on cDNAs obtained from RNA extracted from the mutant or WT lines. Amplifications were performed using DDM1 F and DDM1 R primers, which are specific for the DDM1 transcript. As controls, primers for the RPL10B (A) and CPK3 (B) transcripts were used. (B) Expression values are relative to the values in Col0 plants. Data show mean values ± S.E.M. of at least three independent experiments.
Phenotypes of ddm1 mutants. (A) Images of 4-weeks-old Col0 and SALK_093009 plants. (B) Images of 6-weeks-old Col0 and SALK_093009 plants. (C) Analysis of AtMu1 transposase expression in Col0 and SALK_093009 plants. Transcript levels were evaluated by RT-PCR followed by agarose gels on cDNAs obtained from RNA extracted from the mutant or WT lines.
Relative expression of DNA repair transcripts by RT-qPCR in Col0, ddm1 (ddm1_000509 line) and ros1 (ros1_045303 line). Levels of UVR2, UVR3, UVR7, UVH1, UVH6, and OGG1 were assayed in arabidopsis plants that were irradiated with UV-B for 4 h (UV-B) or were kept under control conditions without UV-B (control). Expression values are relative to the values in Col0 plants under control conditions in the absence of UV-B. The CPK3 transcript was used as a control. Data show mean values ± S.E.M. of at least three independent experiments.
Relative expression of DNA glycosidase transcripts by RT-qPCR in ros1 plants. Levels of DML2, DML3 and DME1 were assayed in arabidopsis plants that were irradiated with UV-B for 4 h (UV-B) or were kept under control conditions without UV-B (control). Expression values are relative to the values in Col0 plants under control conditions in the absence of UV-B. The CPK3 transcript was used as a control. Data show mean values ± S.E.M. of at least three independent experiments.
Primers used for identification of homozygous mutant lines.
Primers used for RT-qPCR.
References
- Agius F., Kapoor A., Zhu J.-K. (2006). Role of the Arabidopsis DNA glycosylase_lyase ROS1 in active DNA demethylation. Proc. Natl. Acad. Sci. U.S.A. 103, 11796–11801 10.1073/pnas.0603563103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., Chen H., Shinn P., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657 10.1126/science.1086391 [DOI] [PubMed] [Google Scholar]
- Bender J. (2004). DNA methylation and epigenetics. Annu. Rev. Plant Biol. 55, 41–68 10.1146/annurev.arplant.55.031903.141641 [DOI] [PubMed] [Google Scholar]
- Bieza K., Lois R. (2001). An Arabidopsis mutant tolerant to lethal ultraviolet-B levels shows constitutively elevated accumulation of flavonoids and other phenolics. Plant Physiol. 126, 1105–1115 10.1104/pp.126.3.1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray C., West C. (2005). DNA repair mechanisms in plants: crucial sensors and effectors for the maintenance of genome integrity. New Phytol. 168, 511–528 10.1111/j.1469-8137.2005.01548.x [DOI] [PubMed] [Google Scholar]
- Britt A. B. (1996). DNA damage and repair in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 4, 75–100 10.1146/annurev.arplant.47.1.75 [DOI] [PubMed] [Google Scholar]
- Campi M., D'Andrea L., Emiliani J., Casati P. (2012). Participation of chromatin-remodeling proteins in the repair of ultraviolet-B-damaged DNA. Plant Physiol. 158, 981–995 10.1104/pp.111.191452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P., Campi M., Chu F., Suzuki N., Maltby D., Guan S., et al. (2008). Histone acetylation and chromatin remodeling are required for UV-B–dependent transcriptional activation of regulated genes in maize. Plant Cell 20, 827–842 10.1105/tpc.107.056457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P., Stapleton A. E., Blum J. E., Walbot V. (2006). Genome-wide analysis of high altitude maize and gene knockdown stocks implicates chromatin remodeling proteins in responses to UV-B. Plant J. 46, 613–627 10.1111/j.1365-313X.2006.02721.x [DOI] [PubMed] [Google Scholar]
- Casati P., Walbot V. (2004). Crosslinking of ribosomal proteins to RNA in maize ribosomes by UV-B and its effects on translation. Plant Physiol. 136, 3319–3332 10.1104/pp.104.047043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S. W. L., Henderson I. R., Jacobsen S. E. (2005). Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 6, 590–590 10.1038/nrg1664 [DOI] [PubMed] [Google Scholar]
- Choi Y., Gehring M., Johnson L., Hannon M., Harada J. J., Goldberg R. B., et al. (2002). DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in arabidopsis. Cell 110, 33–42 10.1016/S0092-8674(02)00807-3 [DOI] [PubMed] [Google Scholar]
- Eberharter A., Becker P. B. (2002). Histone acetylation: a switch between repressive and permissive chromatin. Second in review on chromatin dynamics. EMBO Rep. 3, 224–229 10.1093/embo-reports/kvf053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen J. A., Sweder K. S., Hanawalt P. C. (1995). Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 23, 2715–2723 10.1093/nar/23.14.2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz P., Soppe W., Schubert I. (2003). Heterochromatin in interphase nuclei of Arabidopsis thaliana. Chromosome Res. 11, 227–240 10.1023/A:1022835825899 [DOI] [PubMed] [Google Scholar]
- Friedberg E. C., Walker G. C., Siede W. (1995). DNA Repair and Mutagenesis. Washington, DC: American Society For Microbiology [Google Scholar]
- Gehring M., Huh J. H., Hsieh T. F., Penterman J., Choi Y., Harada J. J., et al. (2006). DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 124, 495–506 10.1016/j.cell.2005.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M., Reik W., Henikoff S. (2009). DNA demethylation by DNA repair. Trends Genet. 25, 82–90 10.1016/j.tig.2008.12.001 [DOI] [PubMed] [Google Scholar]
- Gendrel A.-V., Lippman Z., Yordan C., Colot V., Martienssen R. A. (2002). Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297, 1871–1873 10.1126/science.1074950 [DOI] [PubMed] [Google Scholar]
- Gerhardt K. E., Wilson M. I., Greenberg B. M. (1999). Tryptophan photolysis leads to a UVB-induced 66 kDa photoproduct of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) in vitro and in vivo. Photochem. Photobiol. 70, 49–56 [DOI] [Google Scholar]
- Gong Z., Morales-Ruiz T., Ariza R. R., Roldán-Arjona T., David L., Zhu J. K. (2002). ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111, 803–814 10.1016/S0092-8674(02)01133-9 [DOI] [PubMed] [Google Scholar]
- Hays J. B. (2002). Arabidopsis thaliana, a versatile model system for study of eukaryotic genome-maintenance functions. DNA Repair 1, 579–600 10.1016/S1568-7864(02)00093-9 [DOI] [PubMed] [Google Scholar]
- Hideg E., Jansen M. A. K., Strid A. (2013). UV-B exposure, ROS, and stress: inseparable companions or loosely linked associates? Trends Plant. Sci. 18, 107–115 10.1016/j.tplants.2012.09.003 [DOI] [PubMed] [Google Scholar]
- Hirochika H., Okamoto H., Kakutani T. (2000). Silencing of retrotransposons in arabidopsis and reactivation by the ddm1 mutation. Plant Cell 12, 357–369 10.1105/tpc.12.3.357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen M. A. K., Gaba V., Greenberg B. M. (1998). Higher plants and UV-B radiation: balancing damage, repair and acclimation. Trends Plant. Sci. 3, 131–135 10.1016/S1360-1385(98)01215-1 [DOI] [Google Scholar]
- Jeddeloh J. A., Bender J., Richards E. J. (1998). The DNA methylation locus DDM1 is required for maintenance of gene silencing in Arabidopsis. Genes Dev. 12, 1714–1725 10.1101/gad.12.11.1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeddeloh J. A., Stokes T. L., Richards E. J. (1999). Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 22, 94–97 10.1038/8803 [DOI] [PubMed] [Google Scholar]
- Jordan N. D., West J. P., Bottley A., Sheikh M., Furner I. (2007). Transcript pro?ling of the hypomethylated hog1 mutant of Arabidopsis. Plant Mol. Biol. 65, 571–586 10.1007/s11103-007-9221-4 [DOI] [PubMed] [Google Scholar]
- Kakutani T., Jeddeloh J. A., Flowers S. K., Munakata K., Richards E. J. (1996). Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc. Natl. Acad. Sci. U.S.A. 93, 12406–12411 10.1073/pnas.93.22.12406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S., Sakaguchi K. (2006). DNA repair in plants. Chem. Rev. 106, 753–766 10.1021/cr040482n [DOI] [PubMed] [Google Scholar]
- Klein H. L. (1997). RDH54, a RAD54 homologue in Saccharomyces cerevisiae, is required for mitotic diploid-specific recombination and repair and for meiosis. Genetics 147, 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y., Matsui A., Kawashima M., Kaminuma E., Ishida J., Morosawa T., et al. (2008). Identification of the candidate genes regulated by RNA-directed DNA methylation in Arabidopsis. Biochem. Biophys. Res. Commun. 376, 553–557 10.1016/j.bbrc.2008.09.046 [DOI] [PubMed] [Google Scholar]
- Landry L. G., Chapple C. C. S., Last R. L. (1995). Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiol. 109, 1159–1166 10.1104/pp.109.4.1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lario L. D., Ramirez-Parra E., Gutierrez C., Casati P., Spampinato C. P. (2011). Regulation of plant MSH2 and MSH6 genes in the UV-B induced DNA damage response. J. Exp. Bot. 62, 2925–2937 10.1093/jxb/err001 [DOI] [PubMed] [Google Scholar]
- Li J. Y., Oulee T. M., Raba R., Amundson R. G., Last R. L. (1993). Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. Plant Cell 5, 171–179 10.1105/tpc.5.2.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z., May B., Yordan C., Singer T., Martienssen R. (2003). Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 1:E67 10.1371/journal.pbio.0000067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Wang J., Miki D., Xia R., Yu W., He J., et al. (2010a). DNA replication factor C1 mediates genomic stability and transcriptional gene silencing in Arabidopsis. Plant Cell 22, 2336–2352 10.1105/tpc.110.076349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Ren X., Yin H., Wang Y., Xia R., Wang Y., et al. (2010b). Mutation in the catalytic subunit of DNA polymerase a influences transcriptional gene silencing and homologous recombination in Arabidopsis. Plant J. 61, 36–45 10.1111/j.1365-313X.2009.04026.x [DOI] [PubMed] [Google Scholar]
- Martínez-Macías M. I., Qian W., Miki D., Pontes O., Liu Y., Tang K., et al. (2012). A DNA 30 phosphatase functions in active DNA demethylation in Arabidopsis. Mol. Cell 45, 357–370 10.1016/j.molcel.2011.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazza C. A., Boccalandro H. E., Giordano C. V., Battista D., Scopel A. L., Ballaré C. L. (2000). Functional significance and induction by solar radiation of ultraviolet-absorbing sunscreens in field-grown soybean crops. Plant Physiol. 122, 117–125 10.1104/pp.122.1.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed-Bessudo C., Levy A. A. (2012). Deficiency in DNA methylation increases meiotic crossover rates in euchromatic but not in heterochromatic regions in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, E981–E988 10.1073/pnas.1120742109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirouze M., Reinders J., Bucher E., Nishimura T., Schneeberger K., Ossowski S., et al. (2009). Selective epigenetic control of retrotransposition in Arabidopsis. Nature 461, 427–430 10.1038/nature08328 [DOI] [PubMed] [Google Scholar]
- Mittelsten Scheid O., Probst A. V., Afsar K., Paszkowski J. (2002). Two regulatory levels of transcriptional gene silencing in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 99, 13659–13662 10.1073/pnas.202380499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura A., Yonebayashi S., Watanabe K., Toyama T., Shimada H., Kakutani T. (2001). Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411, 212–214 10.1038/35075612 [DOI] [PubMed] [Google Scholar]
- Morales-Ruiz T., Ortega-Galisteo A. P., Ponferrada-Marín M. I., Martínez-Macías M. I., Ariza R. R., Roldán-Arjona T. (2006). DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc. Natl. Acad. Sci. U.S.A. 103, 6853–6858 10.1073/pnas.0601109103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormrod D. P., Landry L. G., Conklin P. L. (1995). Short-term UV-B radiation and ozone exposure effects on aromatic secondary metabolite accumulation and shoot growth of flavonoid-deficient arabidopsis mutants. Physiol. Plant. 93, 602–610 10.1111/j.1399-3054.1995.tb05106.x [DOI] [Google Scholar]
- Ortega-Galisteo A. P., Morales-Ruiz T., Ariza R. R., Roldán-Arjona T. (2008). Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol. Biol. 67, 671–681 10.1007/s11103-008-9346-0 [DOI] [PubMed] [Google Scholar]
- Penterman J., Zilberman D., Huh J. H., Ballinger T., Henikoff S., Fischer R. L. (2007). DNA demethylation in the Arabidopsis genome. Proc. Natl. Acad. Sci. U.S.A. 104, 6752–6757 10.1073/pnas.0701861104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfluger J., Wagner D. (2007). Histone modifications and dynamic regulation of genome accessibility in plants. Curr. Opin. Plant Biol. 10, 645–652 10.1016/j.pbi.2007.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponferrada-Marín M. I., Roldán-Arjona T., Ariza R. R. (2009). ROS1 5-methylcytosine DNA glycosylase is a slow-turnover catalyst that initiates DNA demethylation in a distributive fashion. Nucleic Acids Res. 37, 4264–4274 10.1093/nar/gkp390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst A. V., Dunleavy E., Almouzni G. (2009). Epigenetic inheritance during the cell cycle. Nat. Rev. Mol. Cell Biol. 10, 192–206 10.1038/nrm2640 [DOI] [PubMed] [Google Scholar]
- Probst A. V., Fransz P. F., Paszkowski J., Mittelsten Scheid O. (2003). Two means of transcriptional reactivation within heterochromatin. Plant J. 33, 743–749 10.1046/j.1365-313X.2003.01667.x [DOI] [PubMed] [Google Scholar]
- Roldan-Arjona T., Ariza R. R. (2009). DNA demethylation, in DNA and RNA Modification Enzymes: Comparative Structure, Mechanism, Functions, Cellular Interactions and Evolution, ed Grosjean H. (Austin, TX: Landes Bioscience; ), 149–161 [Google Scholar]
- Rosa M., Von Harder M., Cigliano R. A., Schlögelhofer P., Mittelsten Scheida O. (2013). The Arabidopsis SWR1 chromatin-remodeling complex is important for DNA repair, somatic recombination, and meiosis. Plant Cell 25, 1990–2001 10.1105/tpc.112.104067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H. (2000). Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365–386 10.1385/1-59259-192-2:365 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russel D. W. (2001). Molecular Cloning – A Laboratory Manual. New York, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Shaked H., Avivi-Ragolsky N., Levy A. A. (2006). Involvement of the Arabidopsis SWI2/SNF2 chromatin remodeling gene family in DNA damage response and recombination. Genetics 173, 985–994 10.1534/genetics.105.051664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara M., Shita-Yamaguchi E., Buerstedde J. M., Shinagawa H., Ogawa H., Shinohara A. (1997). Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homologue of RAD54, RDH54/TID1, in mitosis and meiosis. Genetics 147, 1545–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer T., Yordan C., Martienssen R. A. (2001). Robertson's mutator transposons in A. thaliana are regulated by the chromatin-remodeling gene Decrease in DNA Methylation (DDM1). Genes Dev. 15, 591–602 10.1101/gad.193701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin R. K., Martienssen R. (2007). Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8, 272–285 10.1038/nrg2072 [DOI] [PubMed] [Google Scholar]
- Soppe W. J. J., Jasencakova Z., Houben A., Kakutani T., Meister A., Huang M. S., et al. (2002). DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 21, 6549–6559 10.1093/emboj/cdf657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton A. E., Mori T., Walbot V. (1993). A simple and sensitive antibody-based method to measure UV-induced DNA damage in Zea mays. Plant Mol. Biol. Rep. 11, 230–236 10.1007/BF02669850 [DOI] [Google Scholar]
- Tong H., Leasure C. D., Hou X., Yuen G., Briggs W., He Z. H. (2008). Role of root UV-B sensing in Arabidopsis early seedling development. Proc. Natl. Acad. Sci. U.S.A. 105, 21039–21044 10.1073/pnas.0809942106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukahara S., Kobayashi A., Kawabe A., Mathieu O., Miura A., Kakutani T. (2009). Bursts of retrotransposition reproduced in Arabidopsis. Nature 461, 423–426 10.1038/nature08351 [DOI] [PubMed] [Google Scholar]
- Ulm R., Baumann A., Oravecz A., Mate Z., Adam E., Oakeley E. J., et al. (2004). Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 1397–1402 10.1073/pnas.0308044100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant I., Paszkowski J. (2007). Role of histone and DNA methylation in gene regulation. Curr. Opin. Plant Biol. 10, 528–533 10.1016/j.pbi.2007.06.008 [DOI] [PubMed] [Google Scholar]
- Vanyushin B. F., Ashapkin V. V. (2011). DNA methylation in higher plants: past, present and future. Biochim. Biophys. Acta 1809, 360–368 10.1016/j.bbagrm.2011.04.006 [DOI] [PubMed] [Google Scholar]
- Verbsky M. L., Richards E. J. (2001). Chromatin remodeling in plants. Curr. Opin. Plant Biol. 4, 494–500 10.1016/S1369-5266(00)00206-5 [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada J. P., Thomas J., Spillane C., Coluccio A., Hoeppner M. A., Grossniklaus U. (1999). Maintenance of genomic imprinting at the Arabidopsis medea locus requires zygotic DDM1 activity. Genes Dev. 13, 2971–2982 10.1101/gad.13.22.2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vongs A., Kakutani T., Martienssen R. A., Richards E. J. (1993). Arabidopsis thaliana DNA methylation mutants. Science 260, 1926–1928 10.1126/science.8316832 [DOI] [PubMed] [Google Scholar]
- Wang Y., Liu J., Xia R., Wang J., Shen J., Cao R., et al. (2007). The protein kinase TOUSLED is required for maintenance of transcriptional gene silencing in Arabidopsis. EMBO Rep. 8, 77–83 10.1038/sj.embor.7400852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R., Wang J., Liu C., Wang Y., Zhai J., Liu J., et al. (2006). ROR1/RPA2A, a putative replication protein A2, functions in epigenetic gene silencing and in regulation of meristem development in Arabidopsis. Plant Cell 18, 85–103 10.1105/tpc.105.037507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaish M. W., Colasanti J., Rothstein S. J. (2011). The role of epigenetic processes in controlling flowering time in plants exposed to stress. J. Exp. Bot. 62, 3727–3735 10.1093/jxb/err177 [DOI] [PubMed] [Google Scholar]
- Yao Y., Bilichek A., Golubov A., Kovalchuk I. (2012). ddm1 plants are sensitive to methyl methane sulfonate and NaCl stresses and are deficient in DNA repair. Plant Cell Rep. 31, 1549–1561 10.1007/s00299-012-1269-1 [DOI] [PubMed] [Google Scholar]
- Yin H., Zhang X., Liu J., Wang Y., He J., Yang T., et al. (2009). Epigenetic regulation, somatic homologous recombination, and abscisic acid signaling are influenced by DNA polymerase {epsilon} mutation in Arabidopsis. Plant Cell 21, 386–402 10.1105/tpc.108.061549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Kapoor A., Sridhar V. V., Agius F., Zhu J. K. (2007). The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Curr. Biol. 17, 54–59 10.1016/j.cub.2006.10.059 [DOI] [PubMed] [Google Scholar]
- Zhu J. K. (2009). Active DNA demethylation mediated by DNA glycosylases. Annu. Rev. Genet. 43, 143–166 10.1146/annurev-genet-102108-134205 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Location of the T-DNA insertion in the ROS1 gene (SALK_135293 line). Exons are represented by blue boxes, introns by thin black lines and the UTR regions by light gray boxes. The T-DNA insertion is indicated as a triangle. (B) Analysis of the PCR products separated in 1% (w/v) agarose gels. The PCR reactions were done using genomic DNA from Col0 and SALK_135293 plants. Lanes 1 show the PCR products obtained for a WT plant using the SALK_135293 F and SALK_135293 R primers; while lanes 2 show the PCR products obtained for homozygous mutant plants using the Lb and SALK_13293 R primers. (C) Transcript levels were evaluated by RT-PCR followed by agarose gels on cDNAs obtained from RNA extracted from the mutant or WT lines. Amplifications were performed using ROS1 F and ROS1 R primers, which are specific for the ROS1 transcript. As a control, primers for the RPL10B transcript were used.
(A) Location of the T-DNA insertion in the ROS1 gene (SALK_045303 line). Exons are represented by blue boxes, introns by thin black lines and the UTR regions by light gray boxes. The T-DNA insertion is indicated as a triangle. (B) Analysis of the PCR products separated in 1% (w/v) agarose gels. The PCR reactions were done using genomic DNA from Col0 and SALK_045303 plants. Lanes 1 show the PCR products obtained for a WT plant using the SALK_045303 F and SALK_045303 R primers; while lanes 2 show the PCR products obtained for homozygous mutant plants using the Lb and SALK_045303 F primers. (C) Transcript levels were evaluated by RT-PCR followed by agarose gels on cDNAs obtained from RNA extracted from the mutant or WT lines. Amplifications were performed using ROS1 F and ROS1 R primers, which are specific for the ROS1 transcript. As a control, primers for the RPL10B transcript were used.
(A, C) Location of the T-DNA insertion in the DDM1 gene (SALK_000590 (A) and SALK_093009 (C) lines). Exons are represented by blue boxes, introns by thin black lines and the UTR regions by light gray boxes. The T-DNA insertion is indicated as a triangle. (B) Analysis of the PCR products separated in 1% (w/v) agarose gels. The PCR reactions were done using genomic DNA from Col0 and SALK_000590 plants. Lanes 1 show the PCR products obtained for a WT plant using the SALK_000590 F and SALK_000590 R primers; while lanes 2 show the PCR products obtained for homozygous mutant plants using the Lb and SALK_000590 F primers. (D) Analysis of the PCR products separated in 1% (w/v) agarose gels. The PCR reactions were done using genomic DNA from Col0 and SALK_093009 plants. Lanes 1 show the PCR products obtained for a WT plant using the SALK_093009 F and SALK_093009 R primers; while lanes 2 show the PCR products obtained for homozygous mutant plants using the Lb and SALK_093009 R primers.
Expression of DDM1 transcripts in Col0 plants and ddm1 mutants. Transcript levels were evaluated by RT-PCR followed by agarose gels (A) and RT-qPCR (B) on cDNAs obtained from RNA extracted from the mutant or WT lines. Amplifications were performed using DDM1 F and DDM1 R primers, which are specific for the DDM1 transcript. As controls, primers for the RPL10B (A) and CPK3 (B) transcripts were used. (B) Expression values are relative to the values in Col0 plants. Data show mean values ± S.E.M. of at least three independent experiments.
Phenotypes of ddm1 mutants. (A) Images of 4-weeks-old Col0 and SALK_093009 plants. (B) Images of 6-weeks-old Col0 and SALK_093009 plants. (C) Analysis of AtMu1 transposase expression in Col0 and SALK_093009 plants. Transcript levels were evaluated by RT-PCR followed by agarose gels on cDNAs obtained from RNA extracted from the mutant or WT lines.
Relative expression of DNA repair transcripts by RT-qPCR in Col0, ddm1 (ddm1_000509 line) and ros1 (ros1_045303 line). Levels of UVR2, UVR3, UVR7, UVH1, UVH6, and OGG1 were assayed in arabidopsis plants that were irradiated with UV-B for 4 h (UV-B) or were kept under control conditions without UV-B (control). Expression values are relative to the values in Col0 plants under control conditions in the absence of UV-B. The CPK3 transcript was used as a control. Data show mean values ± S.E.M. of at least three independent experiments.
Relative expression of DNA glycosidase transcripts by RT-qPCR in ros1 plants. Levels of DML2, DML3 and DME1 were assayed in arabidopsis plants that were irradiated with UV-B for 4 h (UV-B) or were kept under control conditions without UV-B (control). Expression values are relative to the values in Col0 plants under control conditions in the absence of UV-B. The CPK3 transcript was used as a control. Data show mean values ± S.E.M. of at least three independent experiments.
Primers used for identification of homozygous mutant lines.
Primers used for RT-qPCR.