Abstract
Arsenic and phosphorus are group 15 elements with similar chemical properties. Is it possible that arsenate could replace phosphate in some of the chemicals that are required for life? Phosphate esters are ubiquitous in biomolecules and are essential for life, from the sugar phosphates of intermediary metabolism to ATP to phospholipids to the phosphate backbone of DNA and RNA. Some enzymes that form phosphate esters catalyze the formation of arsenate esters. Arsenate esters hydrolyze very rapidly in aqueous solution, which makes it improbable that phosphorous could be completely replaced with arsenic to support life. Studies of bacterial growth at high arsenic:phosphorus ratios demonstrate that relatively high arsenic concentrations can be tolerated, and that arsenic can become involved in vital functions in the cell, though likely much less efficiently than phosphorus. Recently Wolfe-Simon et al. [1] reported the isolation of a microorganism that they maintain uses arsenic in place of phosphorus for growth. Here, we examine and evaluate their data and conclusions.
Keywords: arsenate, arsenic life, ester hydrolysis, phosphate
“Our children will starve without arsenic” wrote Joan Slonczewski, an award-winning science fiction writer and Yale-educated microbiologist. Her book Brain Plague [2] depicts the arsenic-contaminated world Prokaryon, where sentient microbes evolved not only to use but to require this normally toxic element. Recently Wolfe-Simon and coworkers [1] described the isolation of a microbe, GFAJ-1, from Mono Lake, CA, an environment that naturally contains high concentrations of arsenic. They assert that GFAJ-1 uses arsenic in place of phosphorus to sustain growth, a phenomenon reminiscent of the microbes described by Slonczewski. This study has generated significant commentary, often as anonymous electronic communications. In this essay, we will examine the data and the conclusions from that provocative publication. We emphasize that there are no additional outside data that substantiate or refute the findings in their study. Until their results are extended in their laboratory, or replicated in others, we can only analyze the available evidence from the perspective of the published literature.
Chemical considerations
First we will briefly review some background of the chemistry of arsenic and phosphorus as they relate to biology. Why does life on earth use primarily six elements: hydrogen, carbon, nitrogen, oxygen, phosphorus, and sulfur? Those six are among the most abundant and the lightest of elements, and they form strong chemical bonds with one another. Carbon in particular can form long chain polymers that are the building blocks of life. But these properties do not preclude the substitution of these elements with other less abundant elements in biological molecules, especially those lower in the same group of the periodic table. In general, elements in the same group have similar chemical properties, but their abundance varies inversely with their atomic number.
A rough analogy of the contrast between phosphorus and arsenic is the carbon-silicon dichotomy. Some algal diatoms use the group 14 element silicon instead of carbon for building exoskeletons [3]. So why is carbon, and not silicon, the backbone of most of the molecules of life? Could silicon substitute more widely for carbon? One reasonable explanation is the difference in bond lengths between carbon and silicon. Carbon-carbon single bonds are about 1.52 Å, while the siliconsilicon bond length is about 2.34 Å. Shorter bonds are stronger bonds, with bond length inversely related to bond strength and bond dissociation energy. This is why the silicon-silicon bond (230 kJ/mol) is weaker than the carboncarbon bond (356 kJ/mol), and why diamond, crystalline carbon with a Mohs hardness of 10, can scratch crystalline silicon, with a Mohs hardness of 7. To put it another way, longer bonds make for more brittle structures: picture the carbon-carbon polymer diamond, and the silicon dioxide polymer window glass. This does not mean that there could not be organisms where silicon replaces carbon, but those organisms would be more fragile.
A closer analogy is the sulfur-selenium similarity. Although the group 16 element selenium is not universally used in biology in place of sulfur, it readily substitutes for sulfur in amino acids such as selenocysteine and selenomethionine in spite of its longer bond length and inherent lower stability. In fact, selenium is an essential trace element because some enzymes use selenocysteine in their active site [4]. So, would it be reasonable to suppose that the group 15 element arsenic could replace phosphorus in some of the chemicals required for life? In biology, phosphorus is found primarily in the stable +5 oxidation state as phosphate and phosphate esters. The esters of pentavalent phosphorus are ubiquitous in biomolecules, from the sugar phosphates of intermediary metabolism to phospholipids to the phosphate backbone of DNA and RNA. Furthermore, the high energy phosphate ester bond of ATP is the foundation of cellular energy cycling. Arsenic, in contrast, has two biologically relevant oxidation states, +3 and +5, and it cycles between these states. Arsenite, its +3 oxidation state, is quite reactive and toxic, forming strong bonds of metallic character with thiols in proteins and small molecules. The more oxidized form, the much less toxic arsenate, has chemical similarities with phosphate.
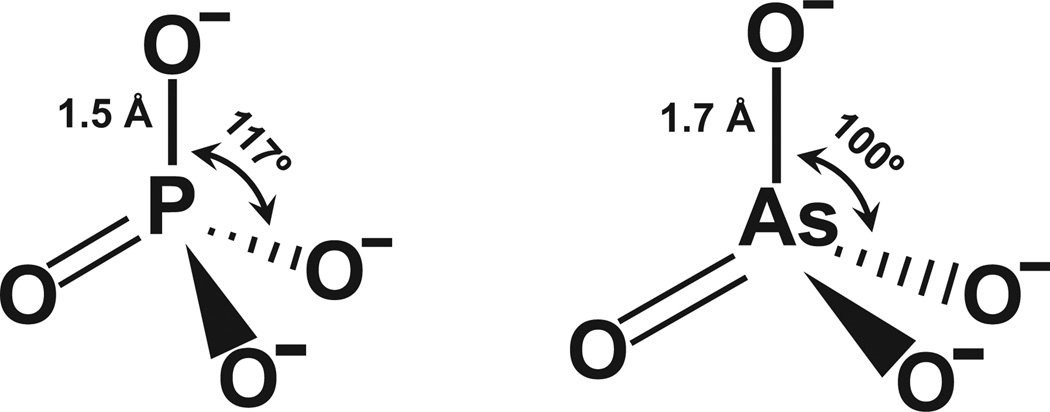
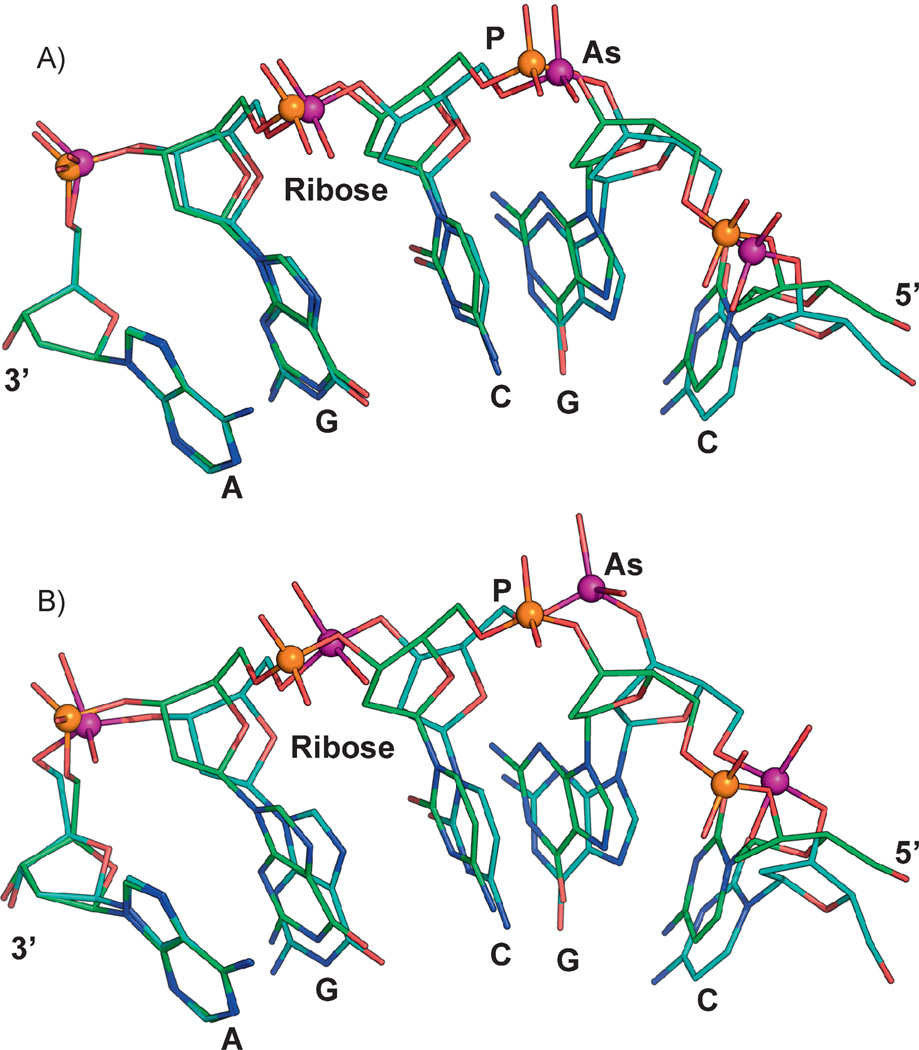
Arsenate is able to form esters that are similar to phosphate esters. However, these compounds are less stable – or to put it another way, more brittle – than the corresponding phosphate esters. This is at least in part a consequence of bond lengths: the P–O bond length in phosphates is about 1.5 Å [5], while the As–O bond of arsenates is about 1.6–2 Å (the bond lengths vary in different compounds, but arsenic generally makes longer bonds than phosphorus) (Fig. 1). Again, longer bonds are more easily broken, so arsenate esters hydrolyze more readily than phosphate esters. Another consideration is bond angles. The P–O bond angle is approximately 117°, while the As–O bond angle is about 100° (again these can vary in different compounds, but the arsenic angles are generally less obtuse than the phosphorus angles). Although no force field parameters have been determined for arsenic, we attempted to model “arsenic DNA”. In the Cambridge Structural Database, As–O distances and O–As–O angles vary from about 1.7 to 2.0 Å and 100° to 109°, respectively. In the Molecular Modeling Database (MMDB), which is part of the Entrez system, there are a number of arsenate structures. For example, two protein structures with bound arsenate ligands are PDB IDs (protein data bank identifiers) 3ENZ and 1TA4, which have As–O bond lengths of 1.7 and 1.93 Å, respectively, and which both have average bond angles of 109°. Because the bond angles and distances vary in different arsenate salts and esters, we chose two sets of initial parameters, with As–O bond distances and O–As–O angles of 1.7 Å and 109° (Fig. 2A) and 1.957 Å and 100.1° (Fig. 2B), and then energy minimized structures of a single-stranded 5-mer based on the crystal structure of B-DNA (PDB ID 1BNA). The resulting two models of “arsenic DNAs” provide a first approximation of what DNA would look like if all of the phosphate was substituted with arsenate. Comparison of the normal phosphate 5-mer with either arsenate structural models shows small but clear differences in the orientation and location of the bases. It is difficult to predict from this modeling how the changes would affect Watson-Crick DNA base pairing or base stacking, but even small differences could represent a challenge for the enzymes of replication or transcription. In the eons of time since the origin of life on earth, enzymes might have adapted to these constraints, but would they be able to switch back and forth between phosphate DNA and arsenate DNA? Even if the overall structure of DNA or RNA with arsenate substituting for phosphate were compatible with biological function, the instability of the arsenate esters that form the backbone of the nucleic acids would most likely present an insurmountable obstacle to formation of an arsenate backbone in DNA or RNA.
Figure 1.
Comparison of phosphate and arsenate anions. The bond angles and distances are approximated using averages from a number of different phosphate and arsenate structures.
Figure 2.
Molecular modeling of arsenate DNA. The structure of a B-DNA dodecamer (PDB id:1BNA) was used for constructing molecular models. The final model was subject to energy minimization using CHARMM [55]. Initial energy minimization was done with a steep descent algorithm (100 steps) and followed by an adopted basis Newton-Raphson method (1,000 steps). To construct models with arsenate replacing phosphate, the phosphorus atoms in 1BNA were modified to arsenic atoms. Similar changes were incorporated into the CHARMM toplogy and parameter file. In the CHARMM nucleic acid parameter file, the initial O–As distances and O–P–O angles were modified to either (A) 1.7 Å and 109.0° or (B) 1.957 Å and 100.1°. Energy minimization was carried out as described above. Other parameters such as torsion angles and force fields were not modified for these examples. A single strand with five repeating nucleotide units (5′-CGCGA-3’) is shown for both phosphate- (orange sphere) or arsenate- (purple sphere) containing molecules. The phosphate and arsenate models were superimposed to illustrate the difference between the two 5-mers, which are anchored by the adenine base at the 3’ end (left).
The lability of ATP, the currency of bioenergetics, with one, two, or three arsenates in place of phosphate is also a serious issue of concern. The short half-life of ADP-arsenate, which spontaneously hydrolyzes about 100,000 fold faster than ATP, is the primary source of the uncoupling action of arsenate in oxidative phosphorylation [6]. For AMP-diarsenate or adenosine triarsenate this issue is likely to be even more dire – to our knowledge these compounds have never been synthesized, probably because they are too labile to exist long enough to identify. Another point often overlooked is that arsenate is readily reduced to arsenite in the cytosol of cells, potentially adding to the instability of arsenylated compounds.
Biological considerations
Next we will briefly review some biological and biochemical aspects of arsenic and phosphorus. Since life can adapt to unusual chemistries, it is important to examine whether and how organisms have adapted to the presence of arsenic. Arsenic has never been shown to be required for any essential biological process. Unlike essential metals, arsenic does not serve a structural or catalytic role for any enzyme except for a few involved in arsenic detoxification (for reviews see [7, 8]). However, this section will illustrate several very important points: (i) extant enzymes do use arsenic, in some cases adventitiously in lieu of phosphate, and in other cases because pathways arose to cope with arsenic; (ii) arsenic can be used biologically as a source of energy; and (iii) arsenic can be incorporated into stable biomolecules.
Arsenic tolerance
As a result of the environmental ubiquity of arsenic, nearly every organism has evolved mechanisms to detoxify it [7]. Microbes in particular have a number of transporters and enzymes specific for arsenic (or the chemically related metalloid antimony) that facilitate cellular efflux, conferring arsenic tolerance [9], or transform arsenic into less toxic species [10, 11]. Thus, enzymes and transporters have evolved to recognize arsenic and catalyze reactions using it.
Arsenic as a source of energy
Not all enzyme systems that have arsenate as a substrate are for detoxification. Microbes have evolved enzymes to use arsenate as fuel (see reviews [12–14]). Arsenate is electrochemically positive, with an oxidation-reduction potential of +139 mV, which makes it useful as a terminal electron acceptor for dissimilatory respiratory chains in anaerobic organisms. By way of reference, the phosphate reduction potential is −690 mV, which is quite low and normally not considered to be a routine microbial electron acceptor, though phosphine gas evolution from sewage has been documented [15]. Ahmann et al. were the first to demonstrate that a microorganism can anaerobically respire arsenate and use it to produce ATP [16]. The ArrAB enzyme that catalyzes dissimilatory arsenate reduction is a heterodimer belonging to the dimethylsulfoxide (DMSO) reductase family of mononuclear molybdenum enzymes, but has evolved an active site that provides specificity for arsenate. Numerous microbes belonging to the low G + C Gram positives, and the (β-, γ-, and ε-proteobacteria phyla have been isolated and shown to utilize arsenate for anaerobic respiration, using acetate, lactate, pyruvate, glycerol, ethanol, or hydrogen as electron donors.
Microbes also have evolved the ability to carry out the reverse reaction, arsenite oxidation, perhaps as a defense since arsenate is less toxic than arsenite [12–14]. Probably more important physiologically is that arsenite can also serve as a source of electrons to generate energy. Santini et al. were the first to reproducibly demonstrate that a Rhizobium-like organism can grow with arsenite as sole energy source and CO2 as sole carbon source [17]. Other arsenite chemoautotrophs displaying similar growth capabilities have been identified as Sinorhizobium-Ensifer [18], Acidicauldus [19], and Alkalilimnicola ehrlichii [20], in addition two anaerobic arsenite oxidizers were identified as Azoarcus-and Sinorhizobium-like organisms [21]. Most recently, Kulp et al. isolated an Ectothiorhodospira sp., also from Mono Lake, CA, that could use arsenite as a reductant to fuel anoxygenic photosynthesis [22]. Phosphite oxidation by anaerobes has also been observed in sulfate reducing proteobacteria of the δ-subclass [23], but the PtxD enzyme of Desulfotignum phosphitoxidans that catalyzes this reaction [24] is unrelated to the arsenite oxidase enzyme [25].
C-As compounds
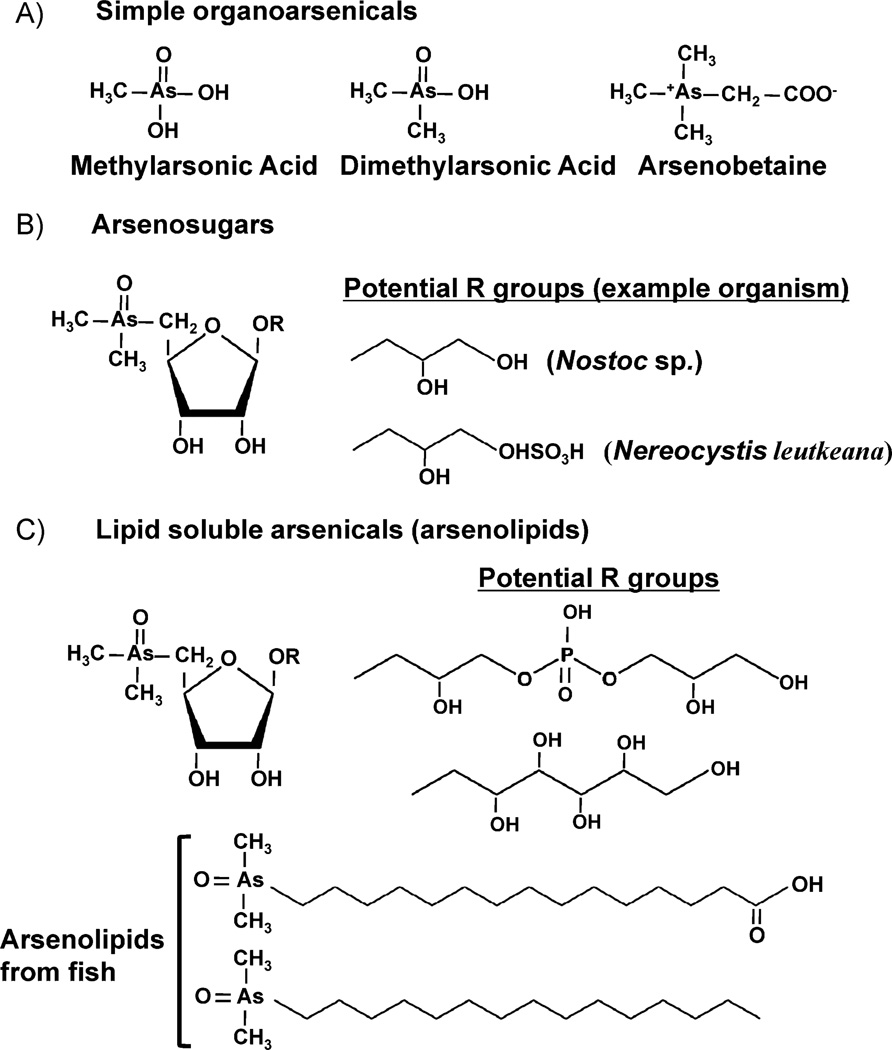
Even though C–As bonds (1.98 Å) are longer and have lower hydrolysis energy (264 kJ/mol) than C–P bonds (1.84 Å and 200 kJ/mol), both are quite robust. The enzymology of phosphonate hydrolysis is complex because of the difficulty of breaking the C–P bond [26]. Similarly, biomolecules with C–As bonds are quite stable [27] and easily isolated (Fig. 3) (see [28–30] for reviews of arsenic compounds found in nature). Arsenate predominates in oxygenated marine environments, and the majority of compounds with C–As bonds have been identified in marine organisms. Arsenic methylation is the most likely entry point into biosynthesis of more complicated compounds. Inside of cells arsenate is reduced to arsenite, which then undergoes an alternating series of oxidative methylations followed by reductions to form trimethylarsine gas, as proposed by Challenger [31]. These reactions are catalyzed by the enzyme As(III) S-adenosylmethionine methyltransferase (AS3MT or ArsM) [10, 11, 32]. The intermediate dimethylarsenate frequently builds up inside of cells and may be the substrate for incorporation into arsenobetaine and arsenolipids, which are common in marine animals. Biomethylation of arsenic has been suggested to play an important role in the global arsenic biogeocycle [10].
Figure 3.
Arsenic-containing biomolecules. Examples shown here progress from the simplest methylarsonic and dimethylarsinic acids (A), which are precursors to progressively more complex arsenlylated compounds such as arsenobetaine (A), arsenosugars (B), and various arsenolipids (C). Fish arsenolipid structures are courtesy of Kevin Francesconi.
Arsenosugars are a major class of arsenic compounds found in marine algae, which probably synthesize them [33]. Arsenolipids are analogs of neutral phospholipids and are found in a number of marine organisms. Dembitsky and Levitsky offer a robust review of arsenolipid and arsenosugar occurrence and biosynthesis, illustrating many of the more than 100 naturally occurring As-containing lipids that have been isolated from fungi, plants, lichens, invertebrates, and fish [34] (Fig. 3).
Arsenic as a substitute for phosphate
The clearest example of arsenate substituting for phosphate is the adventitious entry of arsenate into most cells catalyzed by phosphate transporters. Transporters for phosphate and arsenate were first identified and characterized in Escherichia coli [35]. Like most bacteria studied to date, E. coli has two phosphate transporters: a low affinity, high velocity phosphate transporter (phosphate inorganic transport, Pit), which is also thought to be a Zn2+ transporter [36] and a high affinity, lower velocity phosphate-specific transporter (phosphate-specific transport, Pst). Both will translocate phosphate across the cytoplasmic membrane, although, as their names suggest, the Pst transporter has a higher affinity for phosphate and lower affinity for arsenate [37, 38]. Nevertheless, arsenate is a competitive inhibitor of both transporters so that, when the ratio of arsenate to phosphate is ≥10, arsenate uptake is inevitable by either type of transporter [38, 39]. Thus, under the growth conditions described by Wolfe-Simon et al., where the arsenate to phosphate ratio in the medium was ≥10,000, arsenate would most likely be taken into cells of GFAJ-1 by phosphate transport systems [1].
After arsenate enters the cell, there is wide agreement that enzymes of metabolism that use phosphate can also use arsenate in alkylation, acylation or phosphorylation reactions [34, 40, 41]. Notably, ADP-arsenate can be used as a substrate for hexokinase, which normally synthesizes glucose 6-phosphate, the first intermediate in glycolysis [6]. The glucose 6-arsenate formed by hexokinase could then be converted to glucose 1-arsenate by phosphoglucomutase, demonstrating that intermediary metabolism can form arsenylated compounds [42]. However, in that study the rate constant for spontaneous hydrolysis of glucose 6-arsenate was determined to be 4 × 10−4 seconds−1, compared to 4 × 10−9 seconds−1 for glucose 6-phosphate. Thus, the arsenate ester spontaneously hydrolyzes 100,000-fold faster than the phosphate ester, and cells would effectively starve for lack of intermediates of metabolism. In oxidative phosphorylation, arsenate can substitute for phosphate in ATP synthesis in mitochondria [6, 43, 44] forming ADP-arsenate rather than ATP. ADP-arsenate is readily synthesized by mitochondria, and the Km and Vmax are roughly the same for incorporation of phosphate or arsenate into the trinucleotide. However, the first order rate constant for hydrolysis of ADP-arsenate by submitochondrial particles was determined to be about 70 minutes−1 compared with about 0.5 minutes−1 for ATP [6]. This suggests that organisms dependent on ADP-arsenate for energy would be very, very tired! Yet, these energetic considerations do not rule out the existence of arsenic life.
From the above discussion, it is clear that arsenicals can substitute for phosphorus compounds in many critical cellular biochemical and biosynthetic reactions, and should reasonably be expected to be incorporated into various cellular constituents. The use of arsenate or arsenite in these reactions is primarily adventitious, but both transporters and enzymes have evolved active sites that specifically recognize arsenate or arsenite for detoxification or incorporation into stable compounds. It is the lability of arsenate esters that limits the ability of earthly life to substitute arsenic for phosphorus – a critical dilemma of chemistry that would be problematical for living organisms to overcome.
Analysis of Wolfe-Simon et al
The Wolfe-Simon et al. study [1] attempts to examine how a bacterial cell might tolerate high levels of arsenic, a frequent observation in the literature that has never been completely explained. In discussing their observations, we will often refer to figures and tables in their study (e.g. W-S Fig. 1B) [1].
The most compelling data presented are those shown in W-S Fig. 1B, where microscopic cell counts showed increasing cell numbers when either phosphate (−As/+P cells) or arsenate (+As/−P cells) was provided. Comparatively little growth was observed in the absence of either oxyanion (−As/−P cells), indicating insufficient contaminating phosphate to support substantial growth. W-S Table S1 of Supporting Information showed that this background phosphate did not substantially exceed ~3 µM. The authors describe a 20-fold increase in cell number in the presence of arsenate, but W-S Fig. 1B does not support that description. Approximating from the graph, the initial cell count was ~106 cells/mL, which increased to 2 × 106 cells/mL in the −As/−P culture, or one round of cell division. The +As/−P culture increased to approximately 2 × 107 cells/mL after 7–8 days. The −As/ + P culture increased to approximately 2 × 108 cells/mL after 4–5 days. These values represent a maximum of about four cell doublings with arsenate, or a net of three doublings. At face value, these data support the authors’ conclusions that the cells of GFAJ-1 can use arsenate to replace phosphate for growth. If not, how else can a net of three cell divisions with 40 mM arsenate be explained? Does this in fact represent the replacement of phosphate with arsenate in the reactions of intermediary metabolism, phospholipids biosynthesis, DNA replication and transcription? Are there alternative explanations?
One trivial explanation would be that the arsenate added to the growth medium contained sufficient phosphate for a few rounds of cell division. The authors do not identify the commercial source of the arsenate used in this study, nor do they show an analysis of the arsenate, but a 0.1% contamination of 40 mM arsenate with phosphate might be sufficient to allow for growth. If the cells were scavenging phosphate from the arsenate, it might explain why the cells grew better in 40 than 5 mM arsenate. Yet, the ICP-MS analysis given in W-S Table S1 of Supporting Information indicates that there is no more phosphate in the +As/−P culture medium than in the −As/−P culture medium, so perhaps we need to consider other alternatives.
The GFAJ-1 cells may have adopted phosphate-sparing reactions in response to phosphorus starvation. Synthesis of sulfolipids to spare phosphate in phospholipids is well documented. In some environmentally prominent bacteria such as Prochlorococcus that live under sustained phosphate-limiting stress conditions, sulfolipids appear to be the dominant membrane constituent regardless of how much phosphate is provided experimentally [45]. Bacillus subtilis degrades cell wall teichoic acid to recycle phosphate [46] and, in response to limiting phosphate, can replace phospholipids with sulfolipids [47], as does Rhodobacter sphaeroides [48]. Thus, there is significant precedent to suggest that similar phosphate-sparing reactions might occur with the cell wall and membrane of the Gram positive Halomonas GFAJ-1. We also note, however, that the −As/−P culture medium used in the Wolfe-Simon et al. experiments contained enough sulfate that this type of phosphate sparing activity might have been expected to occur in the absence of added phosphate or arsenate and thus cannot explain the arsenate effect per se.
Another alternative is that the cells might be able to spare phosphate by substituting arsenic for phosphorus in biosynthetic and metabolic reactions. This need not be efficient but only sufficient for a few rounds of cell division. Phosphorus-rich macromolecules in the cell include DNA, RNA, and phospholipids. Of these, phospholipids are probably the easiest to replace. In W-S Table 2, the authors show by labeling that 1–5% of the arsenic appears in the lipid fraction. Arsenolipids are well documented (vida supra) and potentially could partially replace phospholipids. This in turn could lead to reduced membrane integrity, which may have contributed to the cell swelling observed in W-S Fig. 1C. The authors show (W-S Fig. 1A) that light scattering also increases, but the differential between cells grown in +As/−P medium versus −As/+P medium is only about 0.17– 0.25 OD units compared with the 10-fold difference in cell number observed in W-S Fig. 1B. The time course of light scattering increase with −As/+P medium corresponds to the increase in cell number, but the time course in +As/−P medium is much slower, such that the increase in culture optical density (i.e. swelling) continues for many days after cell division ceases. The scanning electron micrograph image (W-S Fig. 1E) and the increasing optical density (W-S Fig. 1A) are consistent with each other and indicate that the cells are experiencing some sort of abnormal growth or stress. This may derive from arsenicassociated production of reactive oxygen species and heat shock induction [49]. Heat shock related responses have also been associated with phosphate starvation [50]. The cells in the transmission electron micrograph have large inclusions. Bacteria generally do not have vacuoles, but they do store carbon in the form of poly-β-hydroxybutyrate (PHB) granules, which accumulate to high levels when carbon is abundant but some other macronutrient is lacking [51, 52], in this instance phosphorus. Consistent with the authors’ suggestion, cell swelling has also been correlated with PHB accumulation in Alcaligenes eutrophus [51, 52].
RNA undergoes rapid and continual turnover and thus is another potential rich intracellular source of phosphate. GFAJ-1 growing in −As/+P medium has a large pool of ribosomal RNA, as expected and as evidenced by the heavily staining lower bands in W-S Fig. 2A, lane 3. The lack of visible rRNA in the +As/−P cells (W-S Fig. 2A, lane 2) is particularly revealing. If the cells were growing normally and not in death phase, the ribosomal 23S and 16S RNAs should have been robustly evident regardless of growth stage [53]; at a minimum, cellular rRNA content should at least equal DNA content, and thus if DNA is visible in the agarose gel then rRNA should be as well. Horiuchi et al. showed that phosphate-starved E. coli degraded RNA, presumably as a phosphate-sparing attempt [54]. Thus the conspicuous absence of rRNA in +As/−P GFAJ-1 would not be inconsistent with the idea that the cells were recycling RNA phosphorus for DNA synthesis. In combination with the phosphate-sparing effect of incorporating arsenate into membrane lipids, cannibalizing RNA phosphorus in GFAJ-1 could provide the cells with enough phosphate to carry out three cell divisions if the phosphorus was primarily directed to DNA replication. This would be a short term survival strategy, and would soon become fatal as the cells eventually lose the ability to synthesize protein for lack of mRNA templates and ribosomes. How might this explain arsenate-associated growth in the Wolfe-Simon et al. study? In control cells, resynthesis of phosphate-containing molecules would re-use phosphate to rebuild molecules degraded as part of normal turnover or in biosynthetic reactions geared toward cell division. When arsenate is available as an abundant substrate replacement, albeit temporarily, phosphate could potentially be diverted. This pool of diverted phosphate could eventually (7–8 days) be used in biosynthetic reactions that are essential for cell division and for which there can be no replacement, either short term or long term (e.g. DNA synthesis). This scenario would support the concept that for some reactions and cell components arsenic can, at least temporarily, replace phosphorus (perhaps in futile cycles), and is consistent with the literature reviewed above.
The arsenic content in the DNA of +As/−P GFAJ-1 is a critical measure for assessing the replacement of phosphorus by arsenic. Wolfe-Simon et al. analyzed this using ICP-MS, NanoSIMS and X-ray absorption spectroscopy (XAS). The ICP-MS assessment is complicated since the culture treatments cannot be normalized to facilitate accurate comparison. It is also worth noting that the nucleic acid preparation was not washed with 70% ethanol, which is used to remove salts that co-precipitate with nucleic acids. Therefore, it is possible that the arsenic found with the purified DNA was an artifact of preparation. To support their theory, the authors used XAS to assess the neighboring atoms of the arsenic. They demonstrated that the first neighbor shell around arsenic in the +As/−P cells consisted of four oxygen ligands, i.e. arsenate or an arsenate ester. The second shell atoms were not unambiguously identified, but the authors suggest that the results are consistent with the As–O–C bonds found in the DNA backbone. However, extreme caution should be taken when assigning long range structure from XAS data since free arsenate adventitiously carried but bound in an ordered manner to DNA could show similar long range structure. This could also account for the results presented by Wolfe-Simon et al. (Timothy Stemmler, Wayne State University School of Medicine, personal communication).
In the NanoSIMS analyses of agarose gel slices, there was “significant uncertainty” in the determination of phosphorus and arsenic in the gel slices. Presumably carbon from the agarose could also confuse normalization, and so it is difficult to make relative comparisons between the samples. Regardless of the technical issues in these analyses, as well as those elsewhere in the study, we suggest that the arsenic to phosphorus ratios in both types of analyses are too low to support the premise that arsenic was stably incorporated into DNA. After three cell divisions, arsenic should have significantly dominated phosphorus in the DNA. This was not the case and in all assays the phosphorus content of the DNA decreased 55- to > 100-fold in the +As/−P cells compared to −As/+P cells (e.g. aqueous DNA extracts, phenol fraction). In contrast, the arsenic content actually decreased (aqueous DNA extracts) or increased only marginally (phenol fraction). It is conceivable that GFAJ-1 cells attempted to incorporate arsenate into nucleotides prior to DNA replication or repair. However, the instability of the arsenate ester backbone would result in spontaneous cleavage of the DNA, as perhaps suggested by the faster electrophoretic mobility of DNA from the +As/−P cells. The DNA standards were not identified, but the DNA from +As/−P cells (W-S Fig. 2A) appeared to be of lower molecular weight compared to −As/+P cells, consistent with the possibility that spontaneous hydrolysis of incorporate arsenylated nucleotides resulted in the equivalent of DNA nicking, yielding smaller DNA fragments.
In summary, we have highlighted aspects of the chemistry and biology of arsenic that are compatible with arsenic substitution for phosphate in living organisms. The literature supports this, with much of the biochemistry being similar for phosphates or arsenates. It is primarily the extremely rapid rate of spontaneous hydrolysis of arsenate esters that would make small molecules such as sugar arsenates unstable and arsenate DNA or RNA rapidly fall apart. Consequently, the latter consideration indicates that arsenate life seems unlikely. However, we draw attention to the fact that there are really no positive data that directly challenge the general conclusion drawn by Wolfe-Simon et al. Our efforts here were aimed primarily at providing an objective assessment of the data and the concept of arsenic replacing phosphorus, in some cases offering alternative explanations for, and interpretations of, their data.
Acknowledgments
We thank Timothy Stemmler, Wayne State University and Chris Rensing, University of Arizona for helpful discussions and expert opinions, and Kevin Francesconi, University of Graz for supplying the fish arsenolipid structures. This study was supported by US National Institutes of Health Grant GM55425 to BPR and by National Science Foundation MCB-0817170 to TRM.
References
- 1.Wolfe-Simon F, Blum JS, Kulp TR, Gordon GW, et al. A bacterium that can grow by using arsenic instead of phosphorus. Science. 2011 doi: 10.1126/science.1197258. in press. [DOI] [PubMed] [Google Scholar]
- 2.Slonczewski J. Brain Plague. New York: TOR Science Fiction; 2000. [Google Scholar]
- 3.Brzezinski MA. Mining the diatom genome for the mechanism of biosilicification. Proc Natl Acad Sci USA. 2008;105:1391–1392. doi: 10.1073/pnas.0711994105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stadtman TC. Biosynthesis and function of selenocysteine-containing enzymes. J Biol Chem. 1991;266:16257–16260. [PubMed] [Google Scholar]
- 5.Harkins PC, Petersson G, Haake P. Distortion of O-P-O bond angles in phosphorus monoanions: ab initio studies. J Inorg Biochem. 1996;61:25–41. [Google Scholar]
- 6.Moore SA, Moennich DM, Gresser MJ. Synthesis and hydrolysis of ADP-arsenate by beef heart submitochondrial particles. J Biol Chem. 1983;258:6266–6271. [PubMed] [Google Scholar]
- 7.Rensing C, Rosen BP. Heavy metals cycles (arsenic, mercury, selenium, others) In: Schaechter M, editor. Encyclopedia of Microbiology. Oxford, UK: Elsevier; 2009. pp. 205–219. [Google Scholar]
- 8.Bhattacharjee H, Rosen BP. Arsenic metabolism in prokaryotic and eukaryotic microbes. In: Nies DH, Silver S, editors. Molecular Microbiology of Heavy Metals. Heidelberg, New York: Springer-Verlag; 2007. pp. 371–406. [Google Scholar]
- 9.Fu H, Jiang X, Rosen BP. Metalloid transport systems. In: Sun H, editor. Biological Chemistry of Arsenic, Antimony and Bismuth. Hoboken, NJ: John Wiley & Sons, Ltd; 2011. pp. 181–207. [Google Scholar]
- 10.Qin J, Lehr CR, Yuan C, Le XC, et al. Biotransformation of arsenic by a Yellowstone thermoacidophilic eukaryotic alga. Proc Natl Acad Sci USA. 2009;106:5213–5217. doi: 10.1073/pnas.0900238106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin J, Rosen BP, Zhang Y, Wang G, et al. Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc Natl Acad Sci USA. 2006;103:2075–2080. doi: 10.1073/pnas.0506836103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stolz JF, Basu P, Santini JM, Oremland RS. Arsenic and selenium in microbial metabolism. Annu Rev Microbiol. 2006;60:107–130. doi: 10.1146/annurev.micro.60.080805.142053. [DOI] [PubMed] [Google Scholar]
- 13.Oremland RS, Stolz JF. The ecology of arsenic. Science. 2003;300:939–944. doi: 10.1126/science.1081903. [DOI] [PubMed] [Google Scholar]
- 14.Mukhopadhyay R, Rosen BP, Phung LT, Silver S. Microbial arsenic: from geocycles to genes and enzymes. FEMS Microbiol Rev. 2002;26:311–325. doi: 10.1111/j.1574-6976.2002.tb00617.x. [DOI] [PubMed] [Google Scholar]
- 15.Dévai I, Felfoldy L, Wittner I, Plosz S. Detection of phosphine: new aspects of the phosphorus cycle in the hydrosphere. Nature. 1988;333:343–345. [Google Scholar]
- 16.Ahmann D, Roberts AL, Krumholz LR, Morel FM. Microbe grows by reducing arsenic. Nature. 1994;371:750. doi: 10.1038/371750a0. [DOI] [PubMed] [Google Scholar]
- 17.Santini JM, Sly LI, Schnagl RD, Macy JM. A new chemolithoautotrophic arseniteoxidizing bacterium isolated from a gold mine: phylogenetic, physiological, and preliminary biochemical studies. Appl Environ Microbiol. 2000;66:92–97. doi: 10.1128/aem.66.1.92-97.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lugtu RT, Choi SC, Oh YS. Arsenite oxidation by a facultative chemolithotrophic bacterium SDB1 isolated from mine tailing. J Microbiol. 2009;47:686–692. doi: 10.1007/s12275-009-0279-3. [DOI] [PubMed] [Google Scholar]
- 19.D’Imperio S, Lehr CR, Breary M, McDermott TR. Autecology of an arsenite chemolithotroph: sulfide constraints on function and distribution in a geothermal spring. Appl Environ Microbiol. 2007;73:7067–7074. doi: 10.1128/AEM.01161-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoeft SE, Blum JS, Stolz JF, Tabita FR, et al. Alkalilimnicola ehrlichii sp. nov., a novel, arsenite-oxidizing haloalkaliphilic gammapro-teobacterium capable of chemoautotrophic or heterotrophic growth with nitrate or oxygen as the electron acceptor. Int J Syst Evol Microbiol. 2007;57:504–512. doi: 10.1099/ijs.0.64576-0. [DOI] [PubMed] [Google Scholar]
- 21.Rhine ED, Phelps CD, Young LY. Anaerobic arsenite oxidation by novel denitrifying isolates. Environ Microbiol. 2006;8:899–908. doi: 10.1111/j.1462-2920.2005.00977.x. [DOI] [PubMed] [Google Scholar]
- 22.Kulp TR, Hoeft SE, Asao M, Madigan MT, et al. Arsenic(III) fuels anoxygenic photosynthesis in hot spring biofilms from Mono Lake, California. Science. 2008;321:967–970. doi: 10.1126/science.1160799. [DOI] [PubMed] [Google Scholar]
- 23.Schink B, Friedrich M. Phosphite oxidation by sulphate reduction. Nature. 2000;406:37. doi: 10.1038/35017644. [DOI] [PubMed] [Google Scholar]
- 24.Simeonova DD, Wilson MM, Metcalf WW, Schink B. Identification and heterologous expression of genes involved in anaerobic dissimilatory phosphite oxidation by Desulfotignum phosphitoxidans. J Bacteriol. 192:5237–5244. doi: 10.1128/JB.00541-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellis PJ, Conrads T, Hille R, Kuhn P. Crystal structure of the 100 kDa arsenite oxidase from Alcaligenes faecalis in two crystal forms at 1.64 A and 2.03 A. Structure. 2001;9:125–132. doi: 10.1016/s0969-2126(01)00566-4. [DOI] [PubMed] [Google Scholar]
- 26.Chen CM, Ye QZ, Zhu ZM, Wanner BL, et al. Molecular biology of carbon-phosphorus bond cleavage. Cloning and sequencing of the phn (psiD) genes involved in alkylphosphonate uptake and C-P lyase activity in Escherichia coli B. J Biol Chem. 1990;265:4461–4471. [PubMed] [Google Scholar]
- 27.Committee on Medical Biological Effects of Environmental Pollutants NRC. Arsenic: Medical and Biological Effects of Environmental Pollutants. Washington, DC: National Academies Press; 1977. [PubMed] [Google Scholar]
- 28.Andreae MO. Determination of arsenic species in natural waters. Anal Chem. 1977;49:820–823. doi: 10.1021/ac50014a037. [DOI] [PubMed] [Google Scholar]
- 29.Edmonds JS, Francesconi KA. Transformations of arsenic in the marine environment. Experientia. 1987;43:553–557. doi: 10.1007/BF02143584. [DOI] [PubMed] [Google Scholar]
- 30.Cullen WR, Reimer KJ. Environmental arsenic chemistry. Chem Rev. 1989;89:713–764. [Google Scholar]
- 31.Challenger F. Biological methylation. Adv Enzymol Relat Subj Biochem. 1951;12:429–491. doi: 10.1002/9780470122570.ch8. [DOI] [PubMed] [Google Scholar]
- 32.Lin S, Shi Q, Nix FB, Styblo M, et al. A novel S-adenosyl-L-methionine:arsenic(III) methyltransferase from rat liver cytosol. J Biol Chem. 2002;277:10795–10803. doi: 10.1074/jbc.M110246200. [DOI] [PubMed] [Google Scholar]
- 33.Francesconi KA, Kuehnelt D. Arsenic compounds in the environment. In: Frankenberger WT Jr, editor. Environmental Chemistry of Arsenic. New York: Dekker; 2002. pp. 51–94. [Google Scholar]
- 34.Dembitsky VM, Levitsky DO. Arsenolipids. Prog Lipid Res. 2004;43:403–448. doi: 10.1016/j.plipres.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 35.Rosenberg H, Gerdes RG, Chegwidden K. Two systems for the uptake of phosphate in Escherichia coli . J Bacteriol. 1977;131:505–511. doi: 10.1128/jb.131.2.505-511.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsieh YJ, Wanner BL. Global regulation by the seven-component Pi signaling system. Curr Opin Microbiol. 13:198–203. doi: 10.1016/j.mib.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Willsky GR, Bennett RL, Malamy MH. Inorganic phosphate transport in Escherichia coli: involvement of two genes which play a role in alkaline phosphatase regulation. J Bacteriol. 1973;113:529–539. doi: 10.1128/jb.113.2.529-539.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willsky GR, Malamy MH. Characterization of two genetically separable inorganic phosphate transport systems in Escherichia coli. J Bacteriol. 1980;144:356–365. doi: 10.1128/jb.144.1.356-365.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willsky GR, Malamy MH. Effect of arsenate on inorganic phosphate transport in Escherichia coli. J Bacteriol. 1980;144:366–374. doi: 10.1128/jb.144.1.366-374.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dixon HBF. The biochemical action of arsonic acids especially as phosphate analogues. Adv Inorg Chem. 1997;44:191–227. [Google Scholar]
- 41.Itada N, Cohn M. The transfer of oxygen from arsenate-180 to phosphate in arsenate-stimulated adenosine triphosphatase reactions. J Biol Chem. 1963;238:4026–4031. [PubMed] [Google Scholar]
- 42.Long JW, Ray WJ., Jr Kinetics and thermodynamics of the formation of glucose arsenate. Reaction of glucose arsenate with phosphoglucomutase. Biochemistry. 1973;12:3932–3937. doi: 10.1021/bi00744a023. [DOI] [PubMed] [Google Scholar]
- 43.Gresser MJ. ADP-arsenate. Formation by submitochondrial particles under phosphorylating conditions. J Biol Chem. 1981;256:5981–5983. [PubMed] [Google Scholar]
- 44.Chan TL, Thomas BR, Wadkins CL. The formation and isolation of an arsenylated component of rat liver mitochondria. J Biol Chem. 1969;244:2883–2890. [PubMed] [Google Scholar]
- 45.Van Mooy BA, Rocap G, Fredricks HF, Evans CT, et al. Sulfolipids dramatically decrease phosphorus demand by picocyanobacteria in oligotrophic marine environments. Proc Natl Acad Sci USA. 2006;103:8607–8612. doi: 10.1073/pnas.0600540103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Merad T, Archibald AR, Hancock IC, Harwood CR, et al. Cell wall assembly in Bacillus subtilis: visualization of old and new wall material by electron microscopic examination of samples stained selectively for teichoic acid and teichuronic acid. J Gen Microbiol. 1989;135:645–655. doi: 10.1099/00221287-135-3-645. [DOI] [PubMed] [Google Scholar]
- 47.Minnikin DE, Abdolrahimzadeh H, Baddiley J. Variation of polar lipid composition of Bacillus subtilis (Marburg) with different growth conditions. FEBS Lett. 1972;27:16–18. doi: 10.1016/0014-5793(72)80398-3. [DOI] [PubMed] [Google Scholar]
- 48.Benning C, Huang ZH, Gage DA. Accumulation of a novel glycolipid and a betaine lipid in cells of Rhodobacter sphaeroides grown under phosphate limitation. Arch Biochem Biophys. 1995;317:103–111. doi: 10.1006/abbi.1995.1141. [DOI] [PubMed] [Google Scholar]
- 49.Del Razo LM, Quintanilla-Vega B, Brambila-Colombres E, Calderon-Aranda ES, et al. Stress proteins induced by arsenic. Toxicol Appl Pharmacol. 2001;177:132–148. doi: 10.1006/taap.2001.9291. [DOI] [PubMed] [Google Scholar]
- 50.Summers ML, Elkins JG, Elliott BA, McDermott TR. Expression and regulation of phosphate stress inducible genes in Sinorhizobium meliloti. Mol Plant-Microbe Interact. 1998;11:1094–1101. doi: 10.1094/MPMI.1998.11.11.1094. [DOI] [PubMed] [Google Scholar]
- 51.Mothes G, Skinfill Rivera I, Babel W. Competition between β-ketothiolase and citrate synthase during poly(β-hydroxybu tyrate) synthesis in Methylobacterium rhodesianum. Arch Microbiol. 1997;166:405–410. doi: 10.1007/BF01682987. [DOI] [PubMed] [Google Scholar]
- 52.Mansfield DA, Anderson AJ, Naylor LA. Regulation of PHB metabolism in Alcaligenes eutrophus. Can J Microbiol. 1995;41:44–49. [Google Scholar]
- 53.Ramagopal S. Metabolic changes in ribosomes of Escherichia coli during prolonged culture in different media. Eur J Biochem. 1984;140:353–361. doi: 10.1111/j.1432-1033.1984.tb08108.x. [DOI] [PubMed] [Google Scholar]
- 54.Horiuchi T, Horiuchi S, Mizuno D. Degradation of ribonucleic acid in Escherichia coli in phosphorus-deficient culture. Biochim Biophys Acta. 1959;31:570–572. doi: 10.1016/0006-3002(59)90044-7. [DOI] [PubMed] [Google Scholar]
- 55.Brooks BR, Brooks CL, III, Mackerell AD, Jr, Nilsson L, et al. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]