A novel H7N9 influenza A virus first detected in March 2013 has since caused more than 130 human infections in China, resulting in 40 deaths1,2. Preliminary analyses suggest the virus is a reassortant of H7, N9 and H9N2 avian influenza viruses and carries some amino acids associated with mammalian receptor binding, raising concerns of a new pandemic1,3,4. However, neither the source populations of the H7N9 outbreak lineage nor the conditions for its genesis are fully known5. Through a combination of active surveillance, screening of virus archives, and evolutionary analyses, we find that H7 viruses likely transferred from domestic duck to chicken populations in China on at least two independent occasions. We show that they subsequently reassorted with enzootic H9N2 viruses to generate the H7N9 outbreak lineage, and a related previously unrecognised H7N7 lineage. The H7N9 outbreak lineage has spread over a large geographic region and is prevalent in chickens at live poultry markets (LPM), which are thought to be the immediate source of human infections. Whether the H7N9 outbreak lineage has, or will, become enzootic in China and neighbouring regions requires further investigation. The discovery here of a related H7N7 influenza virus in chickens that has the ability to experimentally infect mammals, suggests that H7 viruses may pose threats beyond the current outbreak. The continuing prevalence of H7 viruses in poultry could lead to the generation of highly pathogenic variants and further sporadic human infections, with a continued risk of the virus acquiring human-to-human transmissibility.
Following the initial reports of H7N9 influenza infection in humans, field surveillance was conducted during 4th–18th April in Wenzhou (Zhejiang Province, 500 km south of Shanghai) and Rizhao (Shandong Province, 600 km north of Shanghai), which both border the main outbreak region, and in Shenzhen (Guangdong Province, 1200 km south of Shanghai), an area that has not reported human cases (Supplementary Fig. 1). A total of 1,341 pairs of oropharyngeal and cloacal samples were collected from chickens, ducks, geese, pigeons, partridges and quail. A further 1006 faecal and water samples from LPMs, farms and wetlands were also collected (Supplementary Table 1). 388 haemagglutin-positive agents were isolated (10.5% of samples), of which 60 and 85 represented H7 and H9 influenza A viruses. The remaining positive isolates represented other subtypes of influenza A viruses or avian paramyxoviruses (Supplementary Table 1).
H7 influenza A viruses were only detected in Wenzhou and Rizhao and only in LPMs. All H7 isolates from Rizhao were H7N9 viruses, while those from Wenzhou were all H7N7 viruses, except for two duck isolates that were H7N2 and H7N3 viruses. All H9 isolates were H9N2 viruses (80 from LPMs, 5 from farms). At LPMs in Wenzhou the H7 virus was at its highest prevalence in chickens (10.1%; 46/457), followed by ducks (2.4%; 3/125) and pigeons (1.6%; 3/188). In Rizhao LPMs H7N9 viruses were only found in chickens (0.7%; 8/1113). Of the chicken isolates, 100% of H7N9, 65.3% of H7N7 and 94.8% of H9N2 viruses were from oropharyngeal swabs (Supplementary Table 1), suggesting that these H7N9 and H7N7 viruses might replicate in the upper respiratory tract of terrestrial poultry, similar to the enzootic H9N2 viruses6.
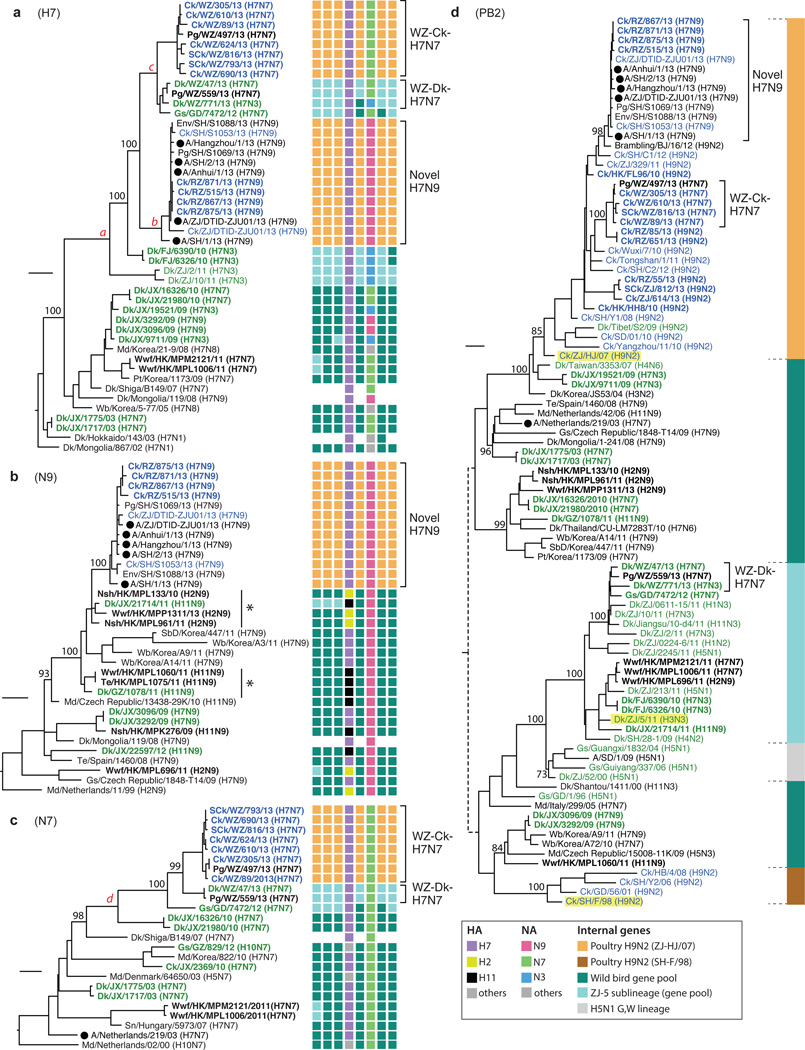
These samples were sequenced to investigate the evolutionary history of avian influenza viruses implicated in the current outbreak of H7N9 infections of humans and poultry. Full genome sequences were obtained for 34 H7N7, 4 H7N9 and 19 H9N2 isolates. The H7 and N7/9 genes of 16 mixed H7/H9 infections were sequenced (Supplementary Table 1) as were 3 H7N9 and 3 H7N7 samples that had multiple H9N2-like internal gene segments. The H7 haemagglutinin (HA) gene sequences of the H7N9 viruses isolated from chickens in Rizhao formed a tight monophyletic group (lineage ‘b’ in Fig. 1a) with previously reported human and avian viruses from the current H7N9 outbreak. This was most closely related to a group comprising mostly H7N7 viruses obtained from Wenzhou chickens, ducks and pigeons (lineage ‘c’ in Fig. 1a). All viruses isolated from chickens in these two groups had internal gene complexes that were closely related to those present in co-circulating H9N2 viruses.
Figure 1. Phylogenies of haemagglutinin, neuraminidase and PB2 genes.
Phylogenies of a, H7 haemagglutinin (n=46); b, N9 neuraminidase (n=34); c, N7 neuraminidase (n=25); d, PB2 (n=93) genes. Sequences reported in this study have their taxa names shown in bold. Genotypes of the influenza viruses are shown on the right (a, b, c) as eight coloured blocks representing each gene segment (from left to right: PB2, PB1, PA, HA, NP, NA, M, and NS; absent if the sequence is unavailable) with the colour indicating the subtype (for HA, NA) or lineage (internal genes; indicated by the solid vertical line in panel d) of that segment. Bootstrap support values (%) from 1,000 pseudo-replicates are shown for selected lineages. Support values for lineages ‘a’ – ‘d’ were all 100%. The scale bar to the left of each tree represents 0.01 substitutions/site. The ‘*’ in panel b denotes N9 sub-lineages linking the viruses of domestic ducks and wild birds. Host species are: Ck (chicken), SCk (silkie chicken), Dk (duck), SbD (spot-billed duck), Md (mallard), Gs (goose), Nsh (Northern shoveler), Wb (wild bird), Wwf (wild waterfowl), Te (common teal), Pg (pigeon), Pt (pintail). Geographic locations: SD (Shandong), ZJ (Zhejiang), JX (Jiangxi), GZ (Guizhou), GD (Guangdong), FJ (Fujian), HK (Hong Kong), WZ (Wenzhou), RZ (Rizhao) and SH (Shanghai). Viruses from different hosts are indicated by: humans, circles; chickens, blue names; domestic ducks or geese, green names.
To examine the genesis of these H7N9 and H7N7 viruses, we sequenced 197 archived isolates of H7, N9, N7 and H9N2 viruses, obtained during previous influenza surveillance between 2000–2013 in southern China (Supplementary Fig. 1). These sequences were analysed together with those obtained in our post-outbreak surveillance, plus all closely related sequences from public databases (see online Methods). H7 influenza viruses from East Asian migratory waterfowl were introduced into domestic ducks in China on multiple occasions in the last decade (Fig.1a, Supplementary Fig. 2). In 2009–10, H7 viruses with five NA subtypes were found in duck farms and LPMs in Jiangxi, suggesting an epidemiological bridge from migratory birds to sentinel farm ducks and then to market birds (Supplementary Fig. 2)7. These introductions to domestic birds persisted for less than two years, except for the introduction of H7N3 viruses (initially isolated in Fujian and Zhejiang in 2010–11) that led to the 2013 H7N9 outbreak lineage viruses and the H7N7 viruses from Wenzhou (lineage ‘a’ in Fig. 1a).
Previous analyses have suggested that the N9 gene of the H7N9 outbreak lineage was derived from wild bird viruses in Europe and Korea3,4. However, our data shows that, for this gene, more closely related H11N9 and H2N9 viruses are found in migratory wild birds in Hong Kong in 2010/11 (‘*’ in Fig. 1b). Phylogenetic linkage between the viruses of these birds and those of domestic ducks in China can be observed at least twice (‘*’ in Fig. 1b) prior to the emergence of the N9 gene in the 2013 outbreak (Fig. 1b, Supplementary Fig. 3).
These data also allow us to reconstruct the genesis of the novel H7N7 lineage closely related to the H7N9 outbreak strain. The N7 of this lineage (‘d’ in Fig. 1c) arose from H7N7 viruses present in the domestic waterfowl of China since at least 2010 (Fig.1c, Supplementary Fig. 4). After entering domestic ducks, these early H7N7 viruses and H7N3 viruses (which had internal genes from a sub-lineage, ZJ-5, of the wild bird viral gene pool) co-circulated in these birds in eastern China during 2010–11 (Fig. 1) and gave rise to H7N7 viruses with ZJ-5 internal genes (WZ-Dk-H7N7; Fig. 1a). H7N7 viruses were also introduced to chickens and by reassortment obtained internal genes from co-circulating H9N2 viruses, generating the H7N7 viruses found in Wenzhou (WZ-Ck-H7N7; Figs 1–2).
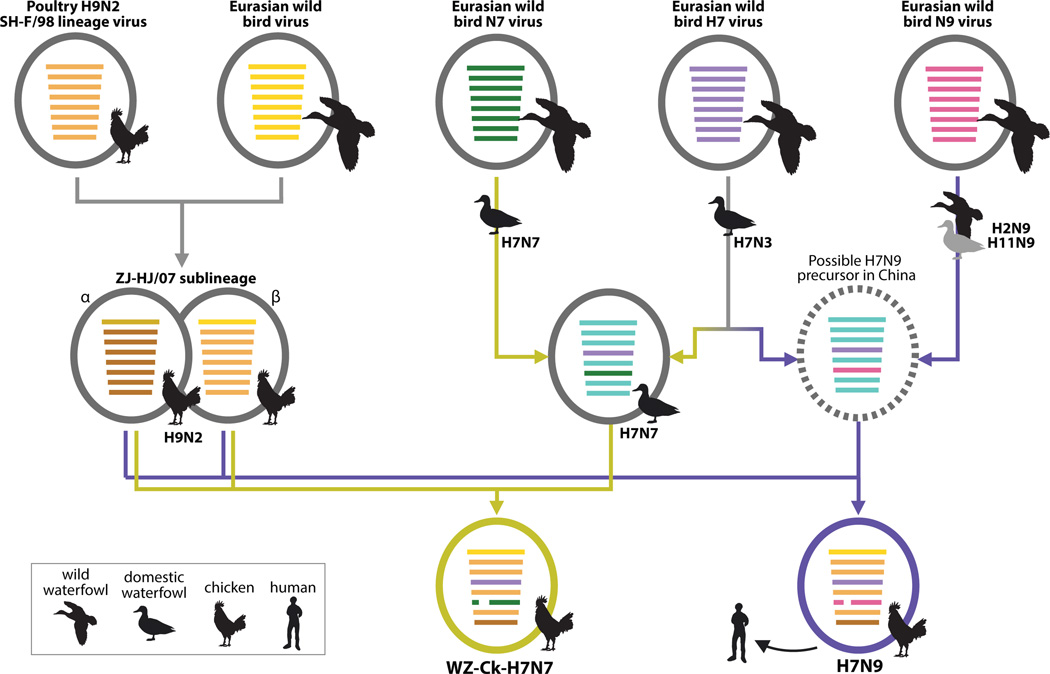
Figure 2. Evolutionary pathways of the H7N9 and H7N7 viruses.
Virus particles are represented by coloured ovals containing horizontal bars that represent the eight gene segments (from top to bottom: PB2, PB1, PA, HA, NP, NA, M, and NS). Segments in descendent viruses are coloured according to their corresponding source viruses (top row) to illustrate gene ancestry through reassortment events. Source viruses for a reassortment are adjacent to arrow tails; arrowheads point to the resulting reassortants. Bars coloured cyan indicate gene segments of the ZJ-5 sub-lineage of wild bird viruses. A broken bar in segment 6 (NA) indicates a stalk region deletion. The virus indicated by a broken oval represents a hypothetical reassortant.
The H9N2 viruses that contributed internal genes to the H7N9 and H7N7 lineages were formed by reassortment of the major H9N2 lineage in China (SH-F/98) with a Eurasian wild bird virus, from which the former acquired a PB2 segment, creating a separate sub-lineage (ZJ-HJ/07; Fig. 2; Supplementary Figs 5–10). The internal genes of the ZJ-HJ/07 sub-lineage form two distinct subgroups (marked ‘α’ and ‘β’ in Supplementary Figs 5–10), based on the PB2 segment phylogeny. Reassortment between α and β led to the internal gene cassettes of the H7N9 and H7N7 viruses having an NS gene from α and other internal genes from β. (Fig. 2).
Even though the H7N9 and H7N7 chicken isolates obtained their internal gene cassettes from similar H9N2 viruses, all of their six internal genes formed distinct sublineages by subtype (Fig. 1d, Supplementary Figs 5–10). This indicates that the H7N9 and H7N7 viruses resulted from two independent interspecies transmissions to chickens, most likely from domestic ducks, and subsequent reassortment events. Some H7N9 and H7N7 viruses had one or two internal genes that fell outside these sublineages and instead clustered with other co-circulating H9N2 chicken viruses (Supplementary Figs 5–10). Thus, further reassortments with other H9N2 viruses continued to occur in chickens, increasing the genetic diversity of the H7N9 and H7N7 viruses.
Both the H7N9 and H7N7 lineages have three major sources of their eight gene segments (Fig. 2). Although the H7N7 virus suggests a path of two consecutive reassortment events (yellow pathway arrows, Fig. 2), the evidence is not as clear for H7N9 viruses (purple pathway arrows, Fig. 2). For the two scenarios for the generation of the H7N9 outbreak lineage, molecular clock dating of divergence dates suggests: either a single reassortment event between mid-2011 and mid-2012, or a reassortment between H7 and N9 viruses in 2011 to generate an H7N9 ancestor, probably in ducks, followed by an interspecies transmission and a second reassortment with chicken H9N2 viruses from mid-2011 to late 2012 (Supplementary Fig. 11; substitution rates are shown in Supplementary Table 2). However alternative reassortant pathways and hosts cannot be definitively excluded due to the limited numbers of H7 and N9 viruses identified during outbreak surveillance, and it is possible for either the H7 or N9 viruses to have first reassorted with H9N2 viruses.
The H7N9 and H7N7 viruses have similar, but independent evolutionary origins. Their HA genes originated from H7 viruses that have been introduced to and established among the domestic ducks in China since 2010. At that time, these duck H7 viruses had internal genes from the ZJ-5 sub-lineage, as did H3N3, H1N2, and H11N3 duck viruses found in southern and eastern China during 2009–2013 (Fig. 1). An N9 virus that was a related precursor of the H7N9 viruses (Dk/JX/21714/11; H11N9), had polymerase genes from the ZJ-5 sub-lineage (Fig. 1b ‘*’, d), linking duck N9 viruses to the duck H7N3 precursors of the H7N9 lineage.
The HA of the H7N9 outbreak lineage (including viruses isolated from Rizhao, but excluding Shanghai/1/2013 & Ck/ZJ/DTID/ZJU01/2013) had the amino acid substitution Q235L/I (position 226 in H3 numbering) indicating that this substitution, which favours α2,6 sialic acid binding, appears to have originated in chickens. The equivalent substitution in chicken H9N2 viruses has been observed since the late 1990s and was linked to virus replication in the upper respiratory tract of birds6. The HAs of the H7N7 lineage from Wenzhou, and their closely related duck viruses (Fig. 1a) do not have this substitution. Of the other amino acid substitutions linked to α2,6 sialic acid binding seen in the H7N9 viruses (G195V, A146S; 186, 138 in H3 numbering)4, only G195V is seen in one H7N7 virus (Supplementary Fig. 12). However, two recent studies have shown that, despite these mutations, the H7 HA has limited binding to human receptors8,9. Both NAs of the H7N7 and H7N9 chicken viruses have overlapping deletions (20 amino acids in N7, positions 53–72; five amino acids in N9, positions 69–73 N9) in the stalk region, which are often observed in influenza virus lineages established in terrestrial poultry10. Duck H7 viruses (N7, N3 and N2) from Wenzhou did not have a stalk deletion. The E627K and D701N substitutions in PB2 normally seen in mammalian viruses11 are only found in human H7N9 isolates (but not in all) and not in the chicken H7N9 and H7N7 viruses obtained here. While it cannot be excluded, there is no evidence from these data to suggest that these viruses are mammalian adapted or that a mammalian intermediate host was involved in the human H7N9 infections.
Although the chicken H7N7 viruses carry only some of the molecular markers seen in the human H7N9 isolates (Supplementary Fig. 12), they may still have the potential to infect humans or mammals. To assess infectivity in mammals, two groups (n=6 each) of ferrets were inoculated with Chicken/Wenzhou/610/2013 (H7N7) at 104.5 or 106.5 plaque-forming units (PFU). Both groups shed virus from 2 days post-inoculation (dpi) (high dose) and 3 dpi (low dose) and virus could be detected in rectal swabs at 4 dpi in both groups (Supplementary Fig. 13). Infectious virus and positive nucleoprotein (NP) stained cells could be detected in the nasal turbinate, and also in the trachea, lungs and hilar lymph nodes, with marginally higher levels from the high dose group at 3 and 5 dpi (Supplementary Figs 13, 14). This shows that the H7N7 lineage viruses can cause significant infection in mammals under experimental conditions, although virus shedding is lower than those of the 2009 pandemic H1N1 and 2013 human H7N9 viruses12.
These findings provide a comprehensive picture of the creation and establishment of the H7N9 viruses that have infected humans. Domestic ducks appear to act as key intermediate hosts by acquiring and maintaining diverse influenza viruses from migratory birds, by facilitating the generation of different combinations of H7 and N9 or N7 subtype viruses, and by transmitting these viruses to chickens. After transmission, reassortment with enzootic H9N2 viruses formed the current H7N9 or H7N7 viruses seen in chickens. This likely led to outbreaks in chickens, resulting in the rapid spread of the novel reassortant H7N9 lineage through LPMs, which then became the source of human infections. The cessation of human infections after the closure of LPM13, following a precedent set during the Hong Kong H5N1 ‘bird flu’ incident in 199714, strongly supports this proposition.
The detection of H7N7 chicken viruses in Wenzhou that, like H7N9 viruses, have the potential to infect mammals suggests the current pandemic threat extends beyond the H7N9 virus. Even though human infections with the H7N9 virus appear to be under control, it is too early to know if this virus has been eradicated from chickens over a larger geographic region. It is possible that H7N9 or H7N7 viruses are still present and may become enzootic in poultry. To ultimately control H7N9 and related viruses, it is necessary to re-consider the management of LPMs in urban areas. Long-term influenza surveillance remains essential for early warning of novel reassortant viruses and interspecies transmission events.
Methods
Influenza virus surveillance in Zhejiang, Shandong and Guangdong provinces
Live poultry markets (LPM), poultry farms and a wetland were surveyed for influenza viruses. If accessible, paired oropharyngeal and cloacal samples were taken from birds, otherwise, samples from isolated individual faecal droppings were collected. Some drinking water samples were also collected. Two cities that flanked the initial outbreak around Shanghai and a more distant city were surveyed (Supplementary Fig. 1) as these locations might have viruses related to the outbreak and/or viruses similar to those that led to the genesis of the outbreak virus. Sampling was conducted immediately prior to the closure of markets and/or culling of the birds.
Sampling was conducted at LPM, farms and a wetland in Wenzhou, Zhejiang, a city approximately 500 km south of Shanghai, from 7th – 10th April 2013. Poultry from LPM and farms were sampled at Rizhao, Shandong, a city approximately 600kms north of Shanghai, on the 17th and 18th April, 2013. Sampling was also carried out at LPM in Shenzhen, a major urban centre adjoining Hong Kong and approximately 1200 km south of Shanghai, on the 4th, 6th and 7th April, 2013. Details of the numbers and types of poultry sampled are given in Supplementary Table 1. After collection, each swab was placed in transport medium (M199) with antibiotics and kept in a cool box before and during shipping to the analysis laboratory. Swab materials were inoculated into 9 to 10-day-old embryonated chicken eggs and incubated for 48 hours at 37°C. Haemagglutinin positive isolates were harvested and further subtyped by haemmaglutination and neuraminidase inhibition (HAI and NAI) assays using a panel of reference antisera as described previously22. Standard precautions were taken to avoid cross contamination of samples. Original samples positive for H7 were confirmed with rapid diagnostic real-time RT-PCR for the presence of H7 and N9 gene segments (see below). All H7 and H9 influenza isolates from the Wenzhou markets (n=78, Supplementary Table 1), and all H7 isolates (n=8) and 18 of the 51 H9 isolates from the Rizhao markets and farms were selected for full genome sequencing.
Selection of archived samples collected from 2000–2013
Archived H7 (n=293, from 45 sampling occasions) and N9 (n=202, from 27 sampling occasions) viruses isolated during 2000 to 2013 from our avian influenza surveillance conducted in Fujian, Guangdong, Guangxi, Guizhou, Jiangxi, Yunnan and Hong Kong (Supplementary Fig. 1) were isolated using embryonated eggs and identified by standard HAI and NAI assays as described above. A total of 42 N9 isolates (1 H1, 5 H2, 1 H3, 1 H4, 11 H7, 4 H10, 19 H11) and a further 57 H7 (1 N1, 26 N3, 4 N6, 23 N7, 3 N8) isolates were selected for full genome sequencing. For 63 sampling occasions, all H7 and N9 isolates were sequenced. For two occasions from Fujian, 2/5 and 3/6 isolates were sequenced. For six occasions from Jiangxi, 3/5, 3/7, 4/10, 6/11, 3/33 and 10/171 were sequenced. This was based on at least one virus from each HAI and NAI titre group (i.e. those that had similar values from these assays). A further 98 H9N2 viruses isolated from Hong Kong retail poultry markets were sequenced.
Genomic sequencing
All selected isolates had all eight segments sequenced either using Sanger sequencing on an ABI 3730 genetic analyzer (Applied Biosystems) or high-throughput next-generation sequencing on a Genome Sequencer Junior (Roche). Where a sample contained more than one copy of any segment (indicating a mixed infection of two viruses), only sequences of the HA and NA sequences that could be explicitly defined or have been confirmed by single clonal Sanger sequencing were used in subsequent phylogenetic analyses.
Rapid diagnostic real-time RT-PCR
For rapid determination of the presence of any H7 and N9 viruses, primers specific for these subtypes were designed, based on the alignment of all H7 and N9 gene sequences available from GenBank on 1st April 2013. Viral RNA was extracted using QIAamp viral RNA minikit (QIAGEN) and cDNAs were synthesized with primer Uni12 using Transcriptor High Fidelity cDNA synthesis Kit (Roche). Copy numbers of the H7 HA or the N9 NA segment were determined on a LightCycler® 480 Real-Time PCR System (Roche) using LightCycler® FastStart DNA MasterPLUS SYBR Green I Kit (Roche). Primers were H7-765 (GTT TCA ATG GGG CHT TCA TAG C), H7-995R (ACA TTC TTC ATC CCT GTW GC), N9-160 (CAA GCC AAA CAA TAA TAA ACA A) and N9-293R (CAT GAA TTT ATA GTA CAG AGY CCT).
Sequence collection and alignment
All previously published influenza A virus sequences were collated from GenBank on 5th April, 2013. Seven full genome sequences of H7N9 influenza viruses from the outbreak in China were downloaded from GISAID (http://gisaid.org) and two from GenBank (http://www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html). Acknowledgement of the sources of the GISAID sequences is given in Supplementary Table 4. These database sequences were combined with the sequences generated in this study (Supplementary Table 3) for further analysis. Sequences of each gene segment were aligned using MUSCLE v3.523 with manual adjustment.
Phylogenetic analysis
Preliminary global phylogenetic analysis of all sequences was done using RAxML v7.6.817. This identified the major Eurasian lineage that contains the H7N9 human isolates and the H7N7 and H7N9 isolates sequenced here. The major Eurasian lineage was reduced to approximately 1,000 sequences. Care was taken to ensure all sequences closely related to the H7N9 outbreak isolates and the novel H7N7 viruses were retained (Supplementary Table 5). Maximum likelihood phylogenies were reconstructed using the GTR+Γ nucleotide substitution model in PhyML v3.016. Topological robustness was assessed by a Shimodaira–Hasegawa approximate likelihood ratio test (SH-like) branch test15. The phylogenies for H7, N9, N7, PB2, PB1, PA, NP, M and NS segments are shown in Supplementary Figs 2–10. Smaller trees, with only essential taxa are presented in Fig. 1 with bootstrap support values from 1,000 pseudo-replicates to indicate confidence of the topology.
Molecular dating
The evolutionary timescales of the H7N7 and H7N9 phylogenies were inferred by a molecular clock model that was calibrated using the virus sampling dates. Gene sequences of the H7N9 and H7N7 viruses were analysed with a selected set of closely related viruses. A relaxed clock model with uncorrelated lognormal distribution (UCLD)24, tree topologies and other evolutionary parameters were jointly estimated using the Bayesian Markov Chain Monte Carlo (BMCMC) method implemented in BEAST v1.7.518. The SRD06 nucleotide substitution model was employed25. A Bayesian skyride model with time-aware smoothing was used26 and a uniform prior and random walk operator was assigned to virus taxa whose sampling dates were known only to the nearest year. Multiple MCMC trajectories were computed and combined, giving from 2 to 10 × 108 total steps for each data set, with sampling every 1,000 steps. Convergence, at effective sample sizes > 200, of relevant parameters were assessed in Tracer v1.527. Maximum clade credibility (MCC) trees with branches scaled by time were summarised, and the time of divergence (tDiv) and time of most recent common ancestor (tMRCA) of the H7N9 and H7N7 viruses were obtained as illustrated in Supplementary Fig. 11. All data sets were screened to exclude mosaic sequences, as previously described28, before molecular clock and further evolutionary analyses were undertaken.
Genome substitutions
The ancestral nucleotide sequence at each internal node of the ML tree was reconstructed by the ML joint method21 implemented in HYPHY v2.020. The sequences at both ends of a tree branch were compared, and the differences represent the substitutions that occurred along that branch. Substitutions in the positively selected codons identified by the mixed effects model of evolution (MEME)19 in Datamonkey29 are highlighted in Supplementary Fig. 12.
Animal infection experiments
Four- to five-month-old female Angora ferrets (Mustela putorius furo) were obtained through a laboratory ferret breeding program at the Wuxi Sangosho Co. Ltd. and confirmed to be influenza virus free by virus isolation in Madin-Darby Canine Kidney (MDCK) cells (ATCC) from nasal washes and rectal swabs and sero-negative by HAI assay against contemporary swine and human influenza viruses (swine: H1 and H3; human: seasonal H1N1, H3N2 and influenza B), and avian H5N1 and H9N2 that are enzootic in China, and the chicken H7N7 virus to be tested in this study. A minimum number of animals necessary to obtain reproducible results were used according to the ethical guidelines. Ferrets were randomly allocated across the treatment groups, to which investigators were not blinded.
A group of six ferrets were intranasally inoculated with a dose of 106.5 plaque-forming units (PFU; or 106.8 50% tissue culture infectious dose, TCID50) of the Chicken/Wenzhou/610/2013 (H7N7) virus (high dose group) and a second group were inoculated with 104.5 PFU (or 104.8 TCID50) of viruses (low dose group). Nasal and rectal washes were taken daily from each individual and titrated using standard TCID50 assays. At 3 and 5 days post-inoculation (dpi), three ferrets from each group were euthanatized and tissues from each major organ (pulmonary lobes, nasal turbinate, upper and lower trachea, and hilar lymph nodes) were collected for virus titration (TCID50 assays), RNA extraction and quantitative RT-PCR, and studies of tissue pathology (hematoxylin and eosin and viral nucleoprotein staining), following protocols described previously12. All animal experiment protocols were reviewed and approved by the Institutional Ethical Review Board (IERB) of Shantou University Medical College (Ref No. SUMC2013-111) and The University of Hong Kong Committee on the Use of Live Animals in Teaching & Research. All experiments with H7N7 and H7N9 viruses were conducted in bio-safety level 3 (BSL3) facilities, using enhanced BSL3 practices for the animal work and following practices in the approved institutional guidelines.
Statistical analysis
Statistical significance for the virus isolation rate in the paired oropharyngeal and cloacal swabs, and the different species of domestic birds were determined by Pearson’s χ2 analysis. Comparisons of viral load for the ferret studies were determined by Mann-Whitney and Student’s t test.
Supplementary Material
Acknowledgements
We thank our colleagues from the Joint Influenza Research Centre (SUMC/HKU) and the State Key Laboratory of Emerging Infectious Diseases for their technical assistance. This study was supported by the National Institutes of Health (National Institute of Allergy and Infectious Diseases contract HSN266200700005C), Li Ka Shing Foundation, the Area of Excellence Scheme of the University Grants Committee of the Hong Kong SAR (grant AoE/M-12/06), Shenzhen Peacock Plan High-End Talents Program (KQTD201203), the University Development Fund (HKU) and the Innovation and Technology Commission of the Hong Kong Government. T.T.-Y.L. was supported in part by a Newton International Fellowship of the Royal Society. Metabiota's involvement was supported by the U.S. Agency for International Development (USAID) Emerging Pandemic Threats Program, PREDICT project, under the terms of Cooperative Agreement Number GHN-A-OO-09-00010-00. The research leading to these results has received funding from the European Union Seventh Framework Programme [FP7/2007–2013] under Grant Agreement no. 278433-PREDEMICS, ERC Grant agreement no. 260864 and the Wellcome Trust (grant 092807) to A.R. and S.J.L.
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contributions Y.G., H.Z., T.T-Y.L. conceived the study; J.W., Y.S., B.Z., L.D., C.Y-H.L., W.H., Z.O., X.C. conducted surveillance; H.Z., J.W., C-L.C., C.M., L.L., Y.C., L.Z., H.L., Y.L., A.F., D.J.K. performed virus isolation, sequencing and animal experiments; T.T-Y.L., A.R., O.G.P., H.Z., D.K.S., S.J.L., L.L.M.P., J.S.M.P., G.M.L., Y. Shu, R.G.W, R.J.W., Y.G. contributed to the analysis; D.K.S., T.T-Y.L. wrote the manuscript; Y.G., H.Z., O.G.P., A.R. edited the manuscript.
Author Information All sequences generated by this study have been deposited in GenBank under accession numbers KF258943 - KF260956 and KF297287 - KF297322 (Supplementary Table 3).
The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
References
- 1.Gao R, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl. J Med. 368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. Number of confirmed human cases of avian influenza A(H7N9) reported to World Health Organization. 2013 http://www.who.int/influenza/human_animal_interface/influenza_h7n9/08_ReportWebH7N9Numberpdf.
- 3.Liu D, et al. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet. 2013;381:1926–1932. doi: 10.1016/S0140-6736(13)60938-1. [DOI] [PubMed] [Google Scholar]
- 4.Kageyama T, et al. Genetic analysis of novel avian A(H7N9) influenza viruses isolated from patients in China, February to April 2013. Euro surveill. 2013;18:20453. [PMC free article] [PubMed] [Google Scholar]
- 5.Hvistendahl M, Normile D, Cohen J. Influenza. Despite large research effort, H7N9 continues to baffle. Science. 2013;340:414–415. doi: 10.1126/science.340.6131.414. [DOI] [PubMed] [Google Scholar]
- 6.Guo YJ, et al. Characterization of the pathogenicity of members of the newly established H9N2 influenza virus lineages in Asia. Virology. 2000;267:279–288. doi: 10.1006/viro.1999.0115. [DOI] [PubMed] [Google Scholar]
- 7.Duan L, et al. Influenza virus surveillance in migratory ducks and sentinel ducks at Poyang Lake, China. Influenza Other Respi. Viruses. 2011;5(Suppl 1):65–68. [PubMed] [Google Scholar]
- 8.Tharakaraman K, et al. Glycan Receptor Binding of the Influenza A Virus H7N9 Hemagglutinin. Cell. 2013;153:1486–1493. doi: 10.1016/j.cell.2013.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiong X, et al. Receptor binding by an H7N9 influenza virus from humans. Nature. 2013;499:496–499. doi: 10.1038/nature12372. [DOI] [PubMed] [Google Scholar]
- 10.Cheung CL, et al. Establishment of influenza A virus (H6N1) in minor poultry species in southern China. J. Virol. 2007;81:10402–10412. doi: 10.1128/JVI.01157-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamada S, et al. Biological and structural characterization of a host-adapting amino acid in influenza virus. PLoS Pathog. 2010;6:e1001034. doi: 10.1371/journal.ppat.1001034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu H, et al. Infectivity, Transmission, and Pathology of Human-Isolated H7N9 Influenza Virus in Ferrets and Pigs. Science. 2013;341:183–186. doi: 10.1126/science.1239844. [DOI] [PubMed] [Google Scholar]
- 13.Xu J, Lu S, Wang H, Chen C. Reducing exposure to avian influenza H7N9. Lancet. 2013;381:1815–1816. doi: 10.1016/S0140-6736(13)60950-2. [DOI] [PubMed] [Google Scholar]
- 14.Shortridge KF, et al. Interspecies transmission of influenza viruses: H5N1 virus and a Hong Kong SAR perspective. Vet. Microbiol. 2000;74:141–147. doi: 10.1016/s0378-1135(00)00174-7. [DOI] [PubMed] [Google Scholar]
- 15.Anisimova M, Gil M, Dufayard JF, Dessimoz C, Gascuel O. Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst. Biol. 2011;60:685–699. doi: 10.1093/sysbio/syr041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 17.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 18.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murrell B, et al. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012;8:e1002764. doi: 10.1371/journal.pgen.1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 21.Pupko T, Pe'er I, Shamir R, Graur D. A fast algorithm for joint reconstruction of ancestral amino acid sequences. Mol. Biol. Evol. 2000;17:890–896. doi: 10.1093/oxfordjournals.molbev.a026369. [DOI] [PubMed] [Google Scholar]
- 22.Huang K, et al. Establishment and lineage replacement of H6 influenza viruses in domestic ducks in southern China. J. Virol. 2012;86:6075–6083. doi: 10.1128/JVI.06389-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drummond AJ, Ho SY, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shapiro B, Rambaut A, Drummond AJ. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol. Biol. Evol. 2006;23:7–9. doi: 10.1093/molbev/msj021. [DOI] [PubMed] [Google Scholar]
- 26.Minin VN, Bloomquist EW, Suchard MA. Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Mol. Biol. Evol. 2008;25:1459–1471. doi: 10.1093/molbev/msn090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rambaut A, Drummond AJ. Tracer v1.5. 2007 http://tree.bio.ed.ac.uk/software/tracer/ [Google Scholar]
- 28.Lam TT, et al. Systematic phylogenetic analysis of influenza A virus reveals many novel mosaic genome segments. Infect. Genet. Evol. 2013;18:367–378. doi: 10.1016/j.meegid.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 29.Delport W, Poon AF, Frost SD, Kosakovsky Pond SL. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics. 2010;26:2455–2457. doi: 10.1093/bioinformatics/btq429. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.