Abstract
About half of patients with diffuse large B-cell lymphoma (DLBCL) do not respond to or relapse soon after the standard chemotherapy, indicating a critical need to better understand the specific pathways perturbed in DLBCL for developing effective therapeutic approaches. Mice deficient in the E3 ubiquitin ligase Smurf2 spontaneously develop B-cell lymphomas that resemble human DLBCL with molecular features of germinal center or post-germinal center B cells. Here we show that Smurf2 mediates ubiquitination and degradation of YY1, a key germinal center transcription factor. Smurf2 deficiency enhances YY1-mediated transactivation of c-Myc and B-cell proliferation. Furthermore, Smurf2 expression is significantly decreased in primary human DLBCL samples, and low levels of Smurf2 expression correlate with inferior survival in DLBCL patients. The Smurf2-YY1-c-Myc regulatory axis represents a novel pathway perturbed in DLBCL that suppresses B-cell proliferation and lymphomagenesis, suggesting pharmaceutical targeting of Smurf2 as a new therapeutic paradigm for DLBCL.
Introduction
In response to antigen stimulation, B cells undergo extensive proliferation to form germinal centers (GCs) in secondary lymphoid organs1. As a consequence of cell proliferation, mutagenic events may occur and target cancer-causing genes. In addition, B cells in GCs undergo distinct genetic processes to generate high-affinity antibodies, including somatic hypermutation (SHM) of the variable region of the immunoglobulin gene and class switch recombination (CSR) that changes immunoglobulin class. These processes can target non-immunoglobulin genes in the GC B cells, leading to genetic alterations that promote tumorigenesis2, 3, 4, 5, 6, 7, 8. To counteract these oncogenic effects, it has been postulated that tumor suppressors function to constrain the proliferation and survival of GC B cells at risk of malignant transformation. Identification of these specific tumor suppressors is critical to our understanding of malignancies originated in GCs.
Most non-Hodgkin’s lymphomas (NHLs) are derived from GC B cells or B cells that have passed through GCs9, 10. Diffuse large B-cell lymphoma (DLBCL) is the most common type of NHL, accounting for 30–40% of all new diagnoses11. Significant progress has been made in our understanding of the dysregulated pathways and genetic abnormalities that govern the development of DLBCL10, 12, 13. Current chemotherapy regimens using the combination of cyclophosphamide, doxorubicin, vincristine, and prednisone (CHOP), together with the anti-CD20 monoclonal antibody rituximab (R-CHOP), result in long-term remission in approximately 50% of DLBCL patients14. However, a significant fraction of DLBCL are still incurable, indicating that further understanding of the pathogenesis of this disease is needed in order to develop specific and effective therapeutic approaches.
Recently it has been shown that mice deficient in Smurf2 (Smad ubiquitination regulatory factor-2) spontaneously develop tumors including lymphomas of B-cell origin, indicating that Smurf2 functions as a tumor suppressor15, 16. It has been proposed that Smurf2 exerts its tumor suppressor function through its ability to maintain genomic integrity15 and regulate senescence16. In this report, we find that B-cell lymphomas developed in Smurf2-deficient mice resemble human DLBCL with molecular features of GC or post-GC B cells. We discover that Smurf2 ubiquitinates YY1, a master regulator of GC transcriptional program17, through which Smurf2 suppresses cell proliferation and c-Myc expression. This Smurf2-YY1-cMyc regulatory axis provides novel insight into lymphomagenesis in GC or post-GC B cells and is highly relevant in human DLBCL.
Results
B-cell lymphoma in Smurf2-deficient mice resembles DLBCL
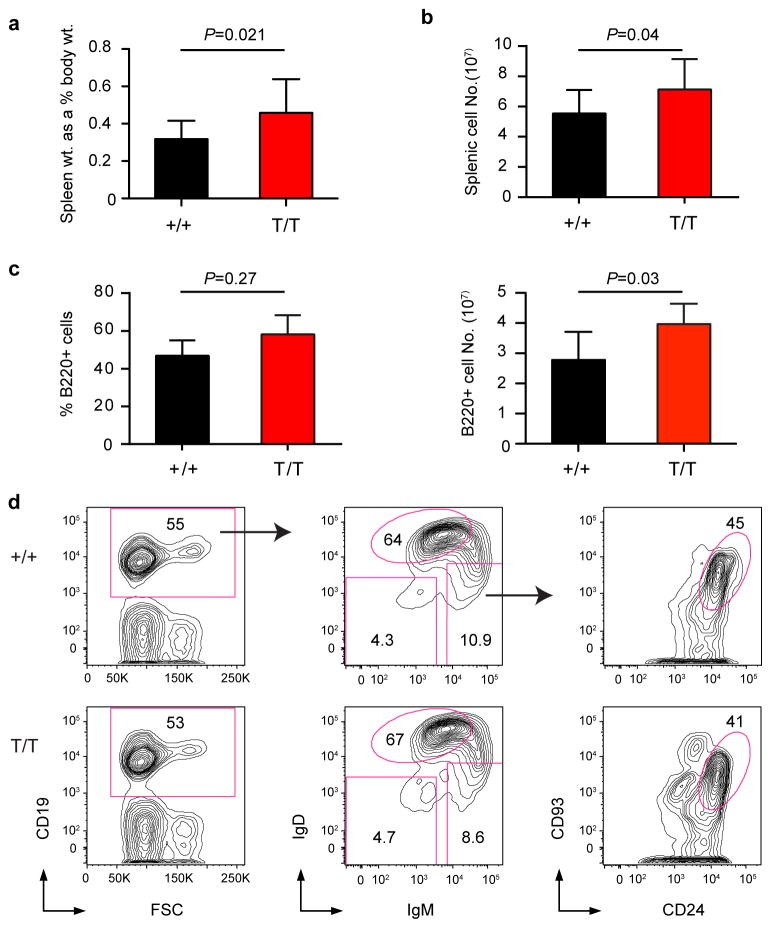
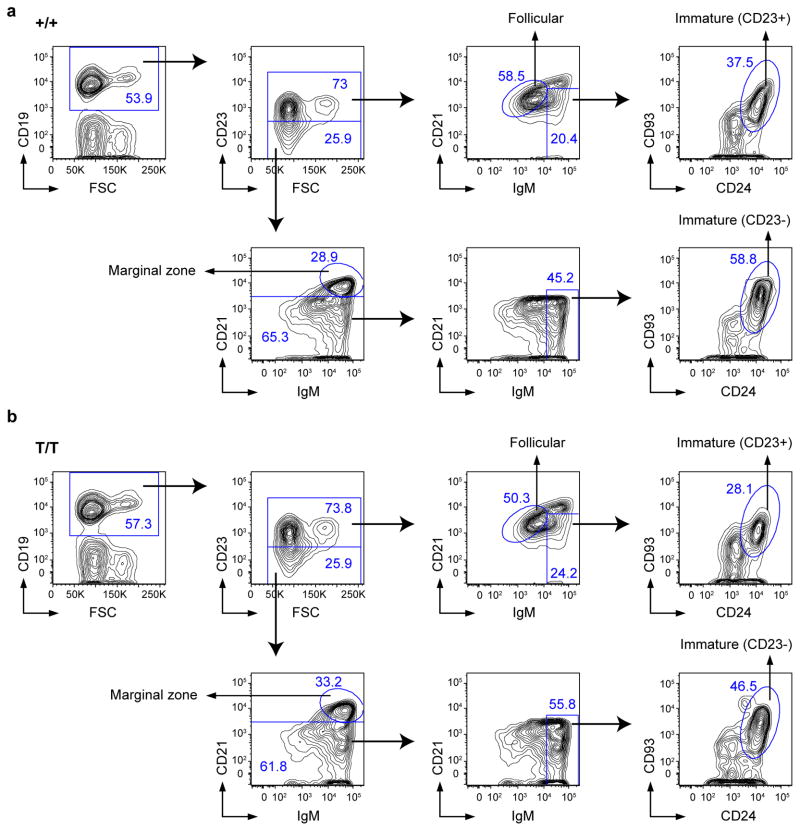
Previously we have shown that Smurf2-deficient (Smurf2T/T, T for the gene-trapped Smurf2 allele) or the heterozygous Smurf2+/T mice exhibit increased susceptibility to spontaneous tumorigenesis after 12 months of age, with the majority of tumors (72.7%) being lymphomas in spleen with a B-cell origin (i.e., B220+). All tumor-bearing Smurf2T/T or Smurf2+/T mice have enlarged spleens16, prompting us to characterize spleens in mice before malignancy. Compared with wild-type mice, an increase in spleen weight relative to body weight was found in 2-month old Smurf2T/T mice (Fig. 1a; 45.2% increase, P=0.021, Student t-test) or Smurf2+/T mice (Supplementary Fig. S1a; 22.5% increase, P=0.07, Student t-test). This increase in spleen weight was accompanied by an increase in total splenic cells (Fig. 1b) and B220+ B cells (Fig. 1c), meanwhile the frequency of splenic B220+ B cells remained unchanged in young Smurf2T/T compared to wild-type mice (Fig. 1c). Further, we analyzed B-cell development in bone marrow and spleen using flow cytometry. Between young Smurf2T/T and wild-type mice, we found no obvious difference in the frequencies of various B-cell sub-populations in bone marrow (Supplementary Fig. S2 and S3) and spleen (Fig. 1d, Fig. 2 and Supplementary Fig. S4), suggesting that B-cell development and differentiation are normal in Smurf2-deficient mice.
Figure 1. Characterization of splenic B cells.
(a) Relative gross spleen weight to body weight of 2-month-old wild-type (+/+) and Smurf2T/T (T/T) mice (N=12). (b) Total live splenic cells (N=12) and (c) Percent and total live B220+ cells in spleens of 2-month-old wild-type and Smurf2T/T mice (N=6). Error bars in (a-c) are standard deviations. Student t-test is used for statistical analysis. (d) Representative FACS analysis of splenic B cells from 2-month-old wild-type and Smurf2T/T mice. Live cells (propidium iodide excluding) are displayed. The IgD++IgMint population, indicated by a circular gate in the middle panel, are follicular B cells. CD93+CD24+ cells, indicated by a circular gate in the right panel, are immature B cells. Frequency of each gated population as a percent of displayed cells is shown.
Figure 2. Characterization of B cells in the spleen of Smurf2-deficient mice.
Representative FACS analysis of splenic cells from 2-month old (a) wild-type (+/+) and (b) Smurf2T/T (T/T) mice. Live cells (propidium iodide excluding) are displayed. Sequential gating strategies are indicated by arrows. Frequency of each gated population as a percent of displayed cells is shown.
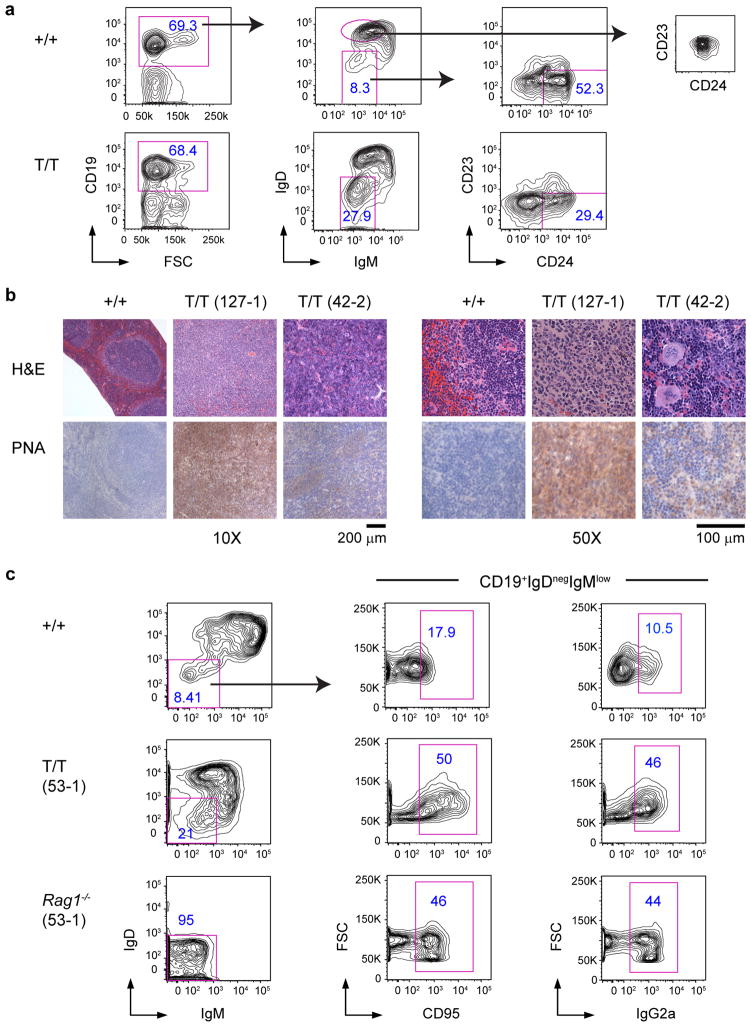
In lymphoma-bearing spleens of Smurf2-deficient mice, we observed a significant expansion (average 6.3-fold increase compared to wild-type mice) of IgDnegIgMlow B cells that were CD23 negative and heterogeneous for CD24 (Fig. 3a), suggesting a GC or post-GC phenotype. This expansion of IgDnegIgMlow B cells in tumor-bearing mice is in sharp contrast with young mice before malignancy, in which the frequency of the IgDnegIgMlow B-cell population was similar between young Smurf2-deficient and wild-type mice (Fig. 1d). Consistent with being GC-derived, these lymphomas showed positive staining for the GC markers peanut agglutinin (PNA) and CD95 (Fig. 3b,c), underwent class switch recombination to immunoglobulin γ2a (Fig. 3c and Supplementary Fig. S5) and exhibited increased somatic hypermutation in the immunoglobulin heavy chain (IgH) locus (Supplementary Table S1). Moreover, these lymphomas showed a gene expression pattern analogous to that in human DLBCL (Supplementary Fig. S6a). Collectively, these data indicate that lymphomas developed in Smurf2-deficient mice are GC or post-GC derived and have molecular features consistent with DLBCL18.
Figure 3. Characterization of B-cell lymphomas generated in Smurf2-deficient mice.
(a) Representative FACS analysis of splenic cells from lymphoma-bearing Smurf2T/T (T/T) and age-matched wild-type (+/+) mice. Frequency of each gated population as a percent of displayed cells is shown. CD23−CD24+ cells in the right panel are activated or previously activated B cells. The CD23 vs. CD24 plot for follicular B cells (IgD++IgMint) in wild-type mice is provided for comparison. (b) Representative H&E and PNA staining of lymphomas in spleen of Smurf2T/T mice and wild-type mouse spleen. Scale bars are 200 μm (10X) and 100 μm (50X). (c) Representative FACS analysis of CD19+ splenic B cells from a lymphoma-bearing Smurf2T/T mouse and a Rag1−/− recipient mouse injected with spleen cells from this lymphoma-bearing mouse are presented. An age-matched wild-type mouse is analyzed as a control, and CD19+IgDnegIgMlow population gated for CD95 or IgG2a is shown for comparison. Mice used in these studies were between 15 and 18 months of age.
We analyzed the genomic rearrangements of the IgH locus in representative lymphoma-bearing spleen samples, and detected a specific V(D)J rearrangement in some lymphoma samples (Supplementary Fig. S6b), consistent with a clonal origin of these lymphomas. The lack of detection of a rearrangement using the largest VH family (J588) in other lymphoma samples may reflect the presence of non-neoplastic cells in spleen samples, rearrangement with a different VH family, or oligoclonal origin. To further investigate tumorigenicity of Smurf2-deficient lymphomas, we injected splenic cells from lymphoma-bearing Smurf2-deficient mice into sub-lethally irradiated Rag1 knockout (Rag1−/−) mice. Rag1−/− recipient mice became moribund 6–8 weeks after injection. An extensive expansion of IgDnegIgMlow B cells was observed in the spleens of these Rag1−/− recipients (Fig. 3c). While injected splenic cells contained 1.2–1.3×105 IgDnegIgMlow B cells, 1.7–8.2×106 IgDnegIgMlow B cells were recovered in Rag1−/− mice, representing a 13 to 67-fold increase of this population. Furthermore, these IgDnegIgMlow B cells stained positively for CD95, consistent with a GC-derived phenotype (Fig. 3c). Positive staining for IgG2a confirmed that these tumors underwent class switch recombination (Fig. 3c). Given that the expanded population of IgDnegIgMlow B cells in Rag1−/− mice had the same GC B-cell phenotype as the injected tumor cells, and that the recipient Rag1−/− mice became moribund, these data are consistent with transfer of disease resembling DLBCL.
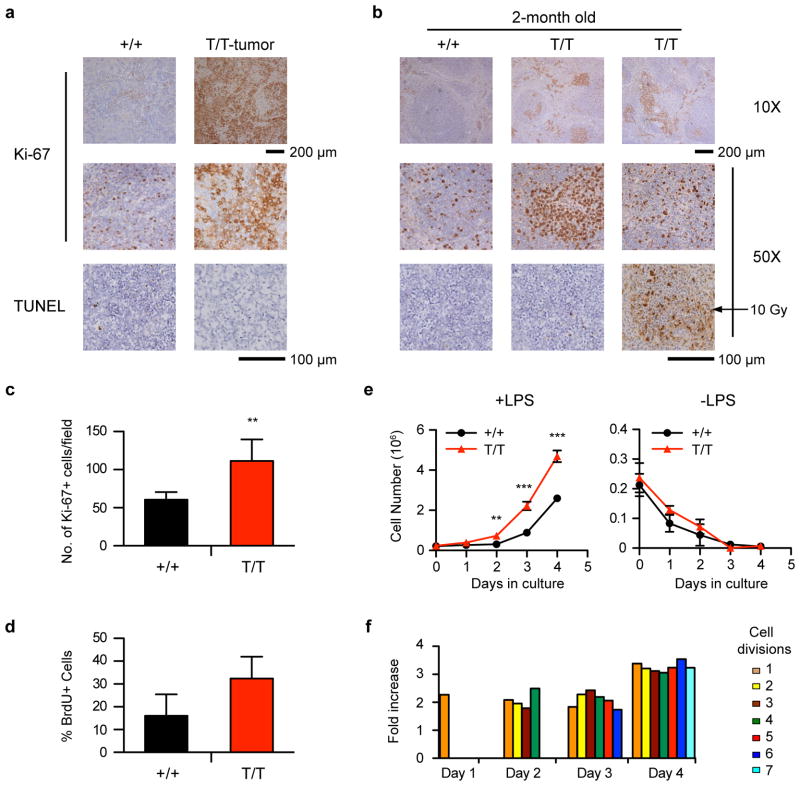
Enhanced proliferation in Smurf2-deficient splenic B cells
Lymphomas found in Smurf2-deficient mice showed an increase in the frequency of cells positive for the cell proliferation-associated antigen Ki-67 compared to spleen sections of age-matched wild-type mice (Fig. 4a). Interestingly, Ki-67 staining was increased in spleen, particularly white pulp, of 2-month old Smurf2T/T (Fig. 4b, c) or Smurf2+/T mice (Supplementary Fig. S1b) compared to wild-type littermates. Furthermore, increased bromodeoxyuridine (BrdU) incorporation was detected in B220+ splenic B cells (Fig. 4d) or total splenic cells (Supplementary Fig. S7a) of young Smurf2T/T mice compared to wild-type littermates. Taken together, these results suggest that increased cell proliferation observed in Smurf2-deficient splenic B cells and B-cell lymphomas is caused by reduced Smurf2 expression rather than as a consequence of tumorigenesis.
Figure 4. Enhanced proliferation of splenic B cells in Smurf2-deficient mice.
(a) Representative Ki-67 and TUNEL staining of lymphomas in Smurf2T/T (T/T) mice. Staining of spleen sections of aged wild-type (+/+) mice is shown for comparison. (b) Representative Ki-67 and TUNEL staining of spleen sections of 2-month old wild-type and Smurf2T/T mice. TUNEL staining of spleen sections of an irradiated (10 Gy) mouse is shown for comparison. Scale bars are 200 μm (10X) and 100 μm (50X). (c) Quantitation of Ki-67 positive cells in 10 randomly selected fields in spleen sections of 2-month old wild-type and Smurf2T/T mice. (d) Analysis of BrdU incorporation in B220+ splenic cells of 2-month old wild-type and Smurf2T/T mice (N=3). (e) Splenic cells from 2-month old wild-type and Smurf2T/T mice are cultured with or without LPS. The number of B220+ viable cells (propidium iodide-negative) is determined by flow cytometry. Average of 3 independent experiments is shown. (f) The number of B220+ cells undergoing different numbers of cell division is determined by CFSE staining and flow cytometry. The ratio of the number of B220+ Smurf2T/T cells undergoing different cell divisions over that of wild-type cells in one representative experiment is presented. Error bars in (c–e) are standard deviations of at least 3 independent experiments. Student t-test is used to compare Smurf2T/T samples with wild-type samples. **: P<0.01 and ***: P<0.001.
To further analyze cell proliferation in splenic B cells, we cultured total spleen cells of 2-month old wild-type or Smurf2T/T mice with the B-cell mitogen lipopolysaccharide (LPS). In response to LPS, Smurf2T/T splenic B220+ B cells proliferated significantly better than wild-type cells (Fig. 4e), whereas cell viability was similar between them (Supplementary Fig. S7b). Using carboxyfluorescein succinimidyl ester (CFSE) to track cell divisions in cultured splenic B cells, we found that the number of Smurf2T/T splenic B cells undergoing successive cell divisions was significantly increased compared to wild-type cells (Fig. 4f and Supplementary Fig. S7c). Without LPS stimulation, cell death was similar between wild-type and Smurf2T/T B cells (Fig. 4e and Supplementary Fig. S7b). Furthermore, TUNEL staining for apoptosis was similar when spleen sections from 2-month old Smurf2T/T mice or lymphomas were compared to age-matched wild-type mice (Fig. 4a, b). Both observations suggest that apoptosis is unchanged in Smurf2-deficient cells. Collectively, these results indicate that cell proliferation is enhanced in Smurf2T/T splenic B cells, suggesting a possible mechanism underlying increased B-cell lymphomagenesis in Smurf2-deficient mice.
Elevated c-Myc expression in Smurf2-deficient mice
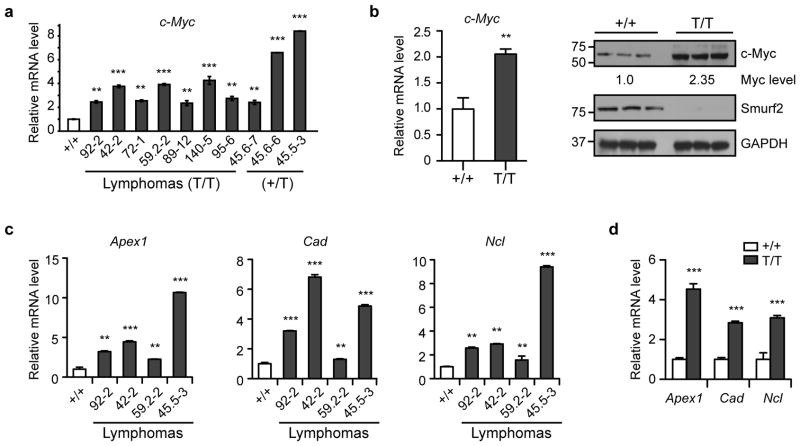
As up-regulation of c-Myc is frequently observed in B-cell lymphoma, and forced expression of c-Myc drives lymphomagenesis in mice19, 20, 21, it prompted us to examine c-Myc expression in lymphomas derived from Smurf2T/T or Smurf2+/T mice. The expression of c-Myc mRNA in these lymphomas was increased compared with spleens of wild-type mice of the similar age (Fig. 5a), while no IgH-Myc translocation was detected in lymphomas in Smurf2-deficient mice (Supplementary Fig. S8).
Figure 5. Elevated c-Myc expression in Smurf2-deficient mice.
(a) Quantitative RT-PCR analysis of c-Myc expression in lymphomas in Smurf2T/T (T/T) or Smurf2+/T (+/T) mice compared with spleen of aged wild-type (+/+) mice. (b) Quantitative RT-PCR and Western analyses of c-Myc expression in spleen of 2-month old wild-type and Smurf2T/T mice. Larger images of immunoblots are shown in Supplementary Figure S11a. Quantitative RT-PCR analysis of c-Myc target genes in (c) representative lymphomas from Smurf2T/T mice compared with spleen of aged wild-type mice, and (d) spleen of 2-month old wild-type and Smurf2T/T mice. Error bars are standard deviations of 3 independent experiments. Student t-test is used to compare Smurf2T/T samples with wild-type samples. **: P<0.01 and ***: P<0.001.
To understand whether c-Myc elevation underlies increased B-cell proliferation and lymphomagenesis in Smurf2-deficient mice, we examined c-Myc expression in young Smurf2T/T mice at the pre-neoplastic stage. We found an increase in c-Myc expression in spleen (Fig. 5b) and liver (Supplementary Fig. S9a, b) of 2-month old Smurf2T/T mice compared with age-matched wild-type mice, suggesting that Smurf2 deficiency leads to increased c-Myc expression. To further corroborate c-Myc mRNA elevation with Smurf2 deficiency, we examined the transcript levels of c-Myc transactivation targets Apex1, Cad and Ncl, which have been validated as c-Myc targets in multiple studies, but are not directly involved in cell proliferation (http://www.myccancergene.org/site/mycTargetDB.asp)22. We found increased transcript levels of these c-Myc targets in lymphomas (Fig. 5c) as well as spleen (Fig. 5d) and liver of young Smurf2T/T mice (Supplementary Fig. S9c).
Smurf2 ubiquitinates YY1 to regulate c-Myc expression
To investigate the underlying mechanism of Smurf2-mediated regulation of c-Myc expression, we searched for transcriptional regulators that have been shown to transactivate c-Myc and examined their potential as the ubiquitination targets of Smurf2. We reasoned that stabilization of such a transcriptional regulator in Smurf2-deficient mice could be responsible for elevated expression of c-Myc. YY1, which transactivates c-Myc23, 24 and is a central regulator of the GC B-cell-specific transcriptional program17, contains a PPDY motif that can potentially interact with the WW domains in Smurf2. We found that the protein levels of YY1 were increased in lymphomas derived from Smurf2T/T or Smurf2+/T mice (Fig. 6a) as well as in spleen (Fig. 6b) and liver (Supplementary Fig. S9b) of young Smurf2T/T mice compared with wild-type littermates. In contrast, the transcript level of YY1 was largely unchanged in Smurf2-deficient mice (Fig. 6b), suggesting a post-transcriptional regulation of YY1 by Smurf2. Supporting this notion, the protein half-life of YY1 was increased from 2.1-hr in wild-type splenic B cells to 4.4-hr in splenic B cells of Smurf2T/T mice (Fig. 6c), suggesting that Smurf2 deficiency leads to an increase in protein stability of YY1.
Figure 6. Smurf2 regulates the stability of YY1 protein.
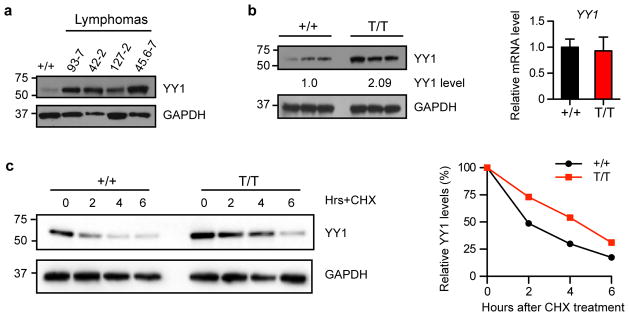
(a) Western analysis of YY1 expression in lymphomas compared with spleen of aged wild-type (+/+) mice. (b) Western and quantitative RT-PCR analyses of YY1 expression in spleens of 2-month old wild-type and Smurf2T/T (T/T) mice. Error bars are standard deviations of 3 independent experiments. (c) Stability of YY1 protein is determined in splenic B cells from 2-month old wild-type and Smurf2T/T mice after treatment with cycloheximide (CHX). The levels of YY1 protein at time 0 are set as 100%. Larger images of immunoblots are shown in Supplementary Figure S11b.
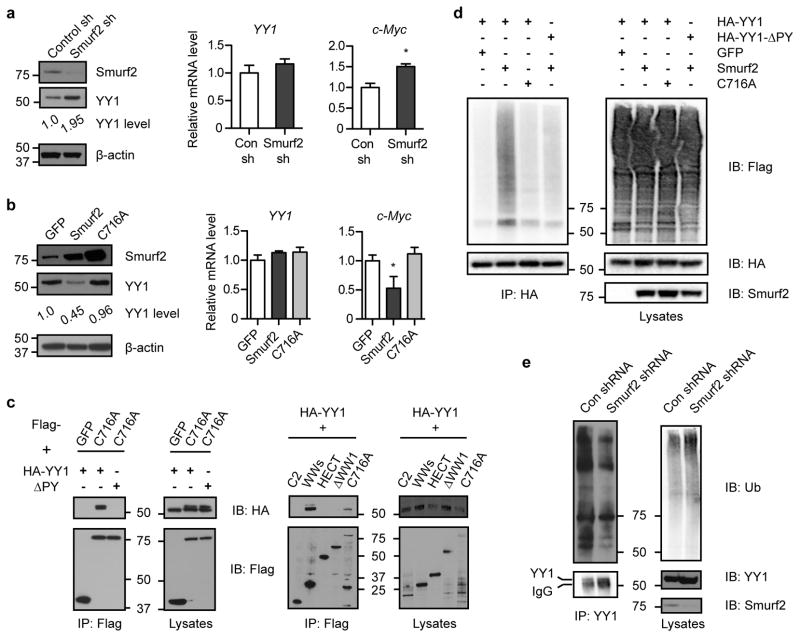
To further characterize Smurf2-mediated regulation of YY1, we stably expressed short-hairpin RNA (shRNA) targeting Smurf2 in a human DLBCL cell line (SUDHL-6), and found that down-regulation of Smurf2 led to an increase in YY1 protein (Fig. 7a). Conversely, ectopic expression of Smurf2 resulted in a reduction in the steady-state level of YY1 protein. In contrast, a ligase mutant C716A, in which the conserved cysteine at residue 716 is replaced by alanine to abolish its E3 ubiquitin ligase activity25, 26, 27, did not lead to changes in YY1 (Fig. 7b), indicating that regulation of YY1 bySmurf2 requires its ubiquitin ligase activity. Consistent with what we found in Smurf2-deficient mice (Fig. 5), c-Myc transcripts were regulated by Smurf2 (Fig. 7a, b). In contrast, YY1 transcripts were largely unchanged when Smurf2 expression was altered (Fig. 7a, b), consistent with the notion that Smurf2 regulates YY1 post-transcriptionally. These results led us to hypothesize that Smurf2 is the E3 ubiquitin ligase responsible for ubiquitination of YY1.
Figure 7. Smurf2 mediates ubiquitination of YY1.
(a) Smurf2 expression is knocked down by shRNA or (b) Smurf2 or ligase mutant C716A is ectopically expressed in human DLBCL cells SUDHL-6. The expression of YY1 or c-Myc is analyzed in Western and quantitative RT-PCR. Error bars are standard deviations of 3 independent experiments. Student t-test is used for statistical analysis. *: P<0.05. (c) Smurf2 interacts with YY1. 293T cells are transfected with indicated constructs, and immunoprecipitation (IP) with anti-Flag antibody is followed by immunoblotting (IB) with anti-HA antibody. Larger images of immunoblots are shown in Supplementary Figure S11c and S12. (d) Smurf2 ubiquitinates YY1. 293T cells are transfected with HA-YY1 (or ΔPY), 3xFlag-Ub and Smurf2 (or C716A). IP with anti-HA antibody is followed by IB with anti-Flag antibody to detect poly-ubiquitinated YY1. (e) Down-regulation of Smurf2 by shRNA results in decreased ubiquitination of endogenous YY1 in human DLBCL cell line SUDHL-6. IP with anti-YY1 antibody is followed by IB with anti-ubiquitin antibody to detect poly-ubiquitinated YY1. Larger images of immunoblots are shown in Supplementary Figure S13a.
To test this hypothesis, we first investigated whether Smurf2 interacts with YY1, as the C2-WW-HECT class of E3 ligases interacts with their protein substrates to catalyze ubiquitination. To limit potential degradation of YY1 by Smurf2, we used the ligase mutant C716A in co-immunoprecipitation with YY1. Smurf2 and YY1 were found to form a complex by co-immunoprecipitation, whereas deletion of the PPDY motif of YY1 (ΔPY) abolished the Smurf2-YY1 interaction (Fig. 7c). Furthermore, we found that three WW domains (i.e. WWs), but not the C2 or HECT domain of Smurf2 alone, were sufficient to interact with YY1. Deletion of the N-terminal WW domain (ΔWW1) abolished the interaction between Smurf2 and YY1 (Fig. 7c). Collectively, these results indicate that the Smurf2-YY1 interaction is mediated by the WW domains of Smurf2 and the PPDY motif of YY1.
We next investigated whether Smurf2 induces ubiquitination of YY1. Ubiquitination of YY1 was greatly induced in the presence of Smurf2 compared with a GFP control, whereas catalytically inactive C716A lost the ability to ubiquitinate YY1 (Fig. 7d and Supplementary Fig. S10). Furthermore, deletion of the PPDY motif in YY1 diminished Smurf2-mediated ubiquitination (Fig. 7d and Supplementary Fig. S10). Conversely, we used stably expressed shRNA to knockdown the expression of Smurf2 in SUDHL-6 cells, and found that down-regulation of Smurf2 resulted in decreased ubiquitination of endogenous YY1 (Fig. 7e). Collectively, these results indicate that Smurf2 is the E3 ubiquitin ligase responsible for ubiquitination of YY1.
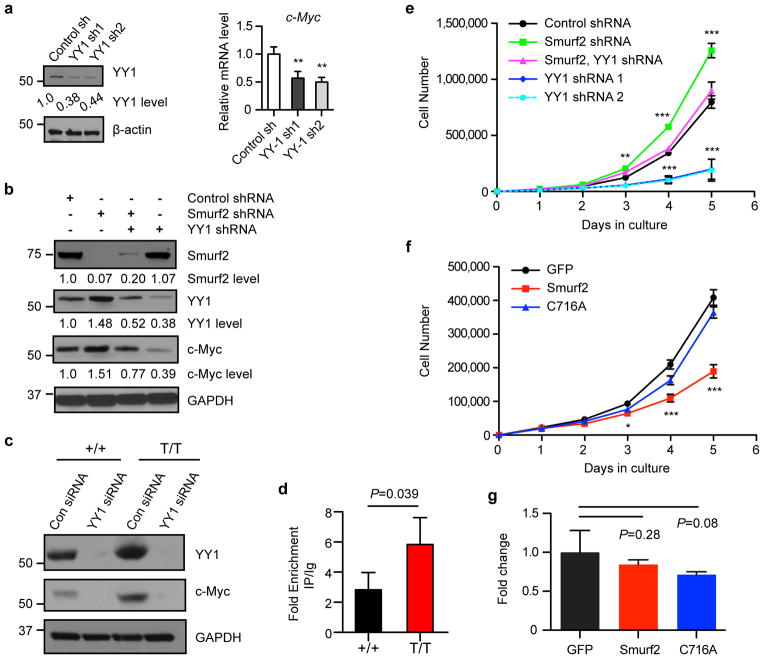
YY1 has been shown to bind to the human c-Myc promoter in a chromatin immunoprecipitation (ChIP) assay and to be sufficient to up-regulate c-Myc expression23, 24. We expressed shRNA to knockdown YY1 expression in SUDHL-6 cells, and found that c-Myc transcripts were decreased upon YY1 down-regulation (Fig. 8a), indicating that YY1 is required to regulate c-Myc expression. As shRNA knockdown of Smurf2 expression in SUDHL-6 cells led to elevation of c-Myc, further down-regulation of YY1 by shRNA resulted in decreased c-Myc expression (Fig. 8b), suggesting that YY1 is the critical mediator of Smurf2-regulated c-Myc expression. Further supporting this notion, knockdown of YY1 expression by siRNA led to down-regulation of c-Myc in primary B cells isolated from spleens of wild-type and Smurf2T/T mice (Fig. 8c). Moreover, ChIP analysis indicated that YY1 was bound to the c-Myc promoter in mouse spleen, and this binding was increased in spleen of Smurf2T/T mice compared with wild-type mice (Fig. 8d), consistent with the mechanism in which stabilization of YY1 in Smurf2-deficient cells up-regulates c-Myc expression.
Figure 8. Smurf2-YY1 regulates c-Myc expression and cell proliferation.
(a) Down-regulation of YY1 by shRNA (left image) leads to decreased c-Myc in human DLBCL cell line SUDHL-6 as determined by quantitative RT-PCR (right image). (b) Western analysis of c-Myc expression in SUDHL-6 cells after shRNA-mediated knockdown of Smurf2 or YY1 expression. (c) Knockdown of YY1 by siRNA leads to down-regulation of c-Myc in splenic B cells of wild-type (+/+) and Smurf2T/T (T/T) mice. (d) Chromatin immunoprecipitation (ChIP) assay of YY1 binding onto the c-Myc promoter in spleens of 2-month old wild-type and Smurf2T/T mice. Fold enrichment of ChIP with anti-YY1 antibody over IgG control is shown. (e) Growth curves of human DLBCL cell line SUDHL-6 after shRNA-mediated knockdown of Smurf2 or YY1 expression. (f) Growth curves and (g) apoptosis assay of SUDHL-6 cells after ectopic expression of Smurf2, ligase mutant C716A or GFP control. Error bars are standard deviations of 3 independent experiments. Cells with shRNA knockdown or Smurf2 expression were compared to control shRNA or GFP, respectively using Student t-test. *: P<0.05, **: P<0.01 and ***: P<0.001. Larger images of immunoblots are shown in Supplementary Figure S13b.
Smurf2 and YY1 regulate proliferation of DLBCL cells
To understand the consequence of Smurf2-mediated ubiquitination of YY1 in lymphomagenesis, we investigated the Smurf2-YY1 axis in regulation of cell proliferation in human DLBCL cells. Consistent with our observation of increased cell proliferation in Smurf2-deficient mice, shRNA knockdown of Smurf2 expression inSUDHL-6 cells (Fig. 8b) led to enhanced cell proliferation (Fig. 8e). Down-regulation of YY1 by shRNA (Fig. 8b) resulted in decreased proliferation in SUDHL-6 cells, indicating that YY1 is a critical regulator of cell proliferation in DLBCL cells (Fig. 8e). Furthermore, down-regulation of YY1 in SUDHL-6 cells that already had shRNA knockdown of Smurf2 (Fig. 8b) alleviated the increase in cell proliferation mediated by Smurf2 deficiency (Fig. 8e), suggesting that Smurf2 regulates cell proliferation through YY1. Conversely, ectopic expression of wild-type Smurf2, but not the ligase mutant C716A, in SUDHL-6 cells (Fig. 7b) led to significantly decreased cell proliferation (Fig. 8f), while apoptosis was not significantly changed (Fig. 8g). Taken together, these results indicate the Smurf2-YY1 regulatory axis is critical in proliferation of lymphoma cells, providing a plausible mechanism underlying increased cell proliferation and lymphomagenesis in Smurf2-deficient mice.
Smurf2 expression correlates with DLBCL patient survival
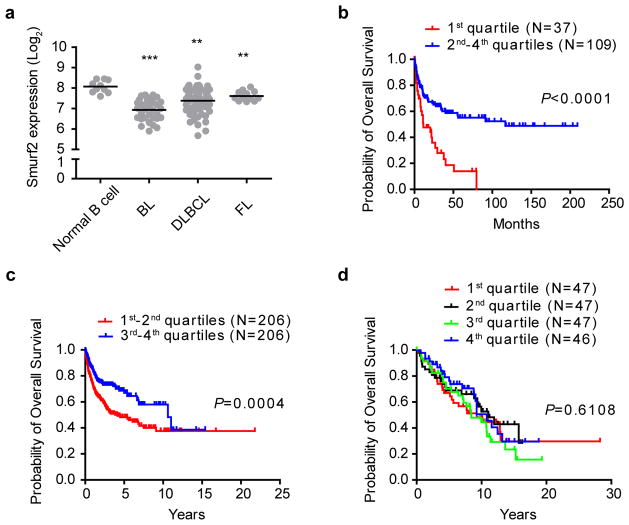
To investigate whether Smurf2 deficiency has clinical relevance in human lymphomagenesis, we analyzed Smurf2 expression in primary human lymphoma samples. In a published microarray dataset (GSE2350) that contains samples of human primary B-cell lymphoma and normal B cells28, we found that the expression of Smurf2 was significantly decreased in DLBCL, Burkitt’s lymphoma and follicular lymphoma compared to normal B cells (Fig. 9a).
Figure 9. The level of Smurf2 expression correlates with overall survival of human DLBCL patients.
(a) Dot plot of Smurf2 expression in normal B cells (N=10), Burkitt’s lymphoma (BL, N=33), DLBCL (N=60) and follicular lymphoma (FL, N=14). One-way ANOVA is used to compare tumors with normal B cells. **: P<0.01 and ***: P<0.001. Kaplan-Meier curves of overall survival of patients in human B-cell lymphoma datasets (b) GSE4475, (c) GSE10846 and (d) a follicular lymphoma dataset plotted according to the level of Smurf2 expression, with the 1st quartile having the lowest Smurf2 expression. The log-rank test is used for statistical analysis.
To further understand the relevance of decreased Smurf2 expression in human lymphomagenesis, we next investigated whether the level of Smurf2 expression correlates with clinical outcome. Smurf2 expression has been measured in three human primary B-cell lymphoma microarray datasets, each of which contains survival information of more than 100 patients29, 30, 31. Within each dataset, we divided lymphoma samples into four groups based on the level of Smurf2 expression, with the 1st quartile having the lowest Smurf2 expression. Survival analysis of dataset GSE4475 in Gene Expression Omnibus (GEO, http://www.ncbi.nil.nih.gov/geo), which contains DLBCL and some Burkitt’s lymphoma patients29, showed that overall survival of patients with the lowest Smurf2 expression (1st quartile) was significantly worse (P<0.0001, log-rank test) than that of patients with higher Smurf2 expression (2nd–4th quartile) (Fig. 9b). Similarly, a significantly poor survival prognosis (P=0.0004, log-rank test) was observed in patients with low level of Smurf2 expression (1st and 2nd quartiles) compared to patients with high level of Smurf2 expression (3rd and 4th quartiles) in an independent dataset GSE10846 in GEO (Fig. 9c), which contains only DLBCL patients30. Interestingly, we found no significant difference in overall survival of patients in a human follicular lymphoma dataset31 based on Smurf2 expression level (Fig. 9d), suggesting a specific role of Smurf2 deficiency in human DLBCL or possibly Burkitt’s lymphoma.
Certain clinical and molecular parameters have been shown to predict DLBCL patient survival32, 33, 34. Univariate analysis indicated a significant correlation between poor survival prognosis and age, Ann Arbor stage, molecular subtype, or International Prognostic Index (IPI) score. To determine whether Smurf2 expression can predict clinical outcomes independently of these known parameters, we carried out a multivariate Cox regression analysis, and found that low level of Smurf2 expression was an independent predictor of patient survival in both DLBCL cohorts (P=0.01 for GSE4475 and P=0.027 for GSE10846) (Table 1). These results indicate that Smurf2 expression is a valuable prognostic marker for DLBCL patient survival, and further suggest that Smurf2 is a potential therapeutic target for DLBCL.
Table 1.
Univariate and multivariate analyses of prognostic factors associated with DLBCL patient overall survival
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Variables | HR (95% CI)a | P | HR (95% CI)a | P |
| GSE4475 (N=102)b | ||||
| Agec | 2.35 (1.41–3.92) | 0.001 | 1.94 (0.97–3.90) | 0.062 |
| Ann Arbor stag d | 2.16 (1.28–3.65) | 0.004 | 2.60 (1.37–4.94) | 0.004 |
| Ki-67 scoree | 0.97 (0.55–1.72) | 0.912 | 1.26 (0.62–2.56) | 0.526 |
| Myc translocationf | 1.11 (0.68–1.81) | 0.68 | 1.38 (0.68–2.80) | 0.379 |
| Subtypeg | 2.26 (1.34–3.82) | 0.002 | 2.12 (1.10–4.07) | 0.025 |
| Smurf2 expressionh | 2.68 (1.64–4.39) | <0.001 | 2.48 (1.24–4.93) | 0.01 |
| GSE10846 (N=311)b | ||||
| Agec | 2.05 (1.47–2.86) | <0.001 | 1.41 (0.94–2.11) | 0.099 |
| Ann Arbor staged | 1.82 (1.31–2.52) | <0.001 | 0.95 (0.61–1.47) | 0.805 |
| Subtypeg | 2.81 (1.98–3.99) | <0.001 | 2.17 (1.49–3.16) | <0.001 |
| Revised IPI scorei | 3.20 (2.31–4.42) | <0.001 | 2.90 (1.84–4.58) | <0.001 |
| Smurf2 expressionj | 1.72 (1.28–2.42) | <0.001 | 1.51 (1.05–2.18) | 0.027 |
HR: hazard ration; CI: confidence interval;
Only patients with complete information in all variables were used;
Age: ≥60 vs. <60 years;
Ann Arbor stage: III and IV vs. I and II;
Ki-67 score: ≥95% vs. <95%;
Myc translocation: presence vs. absence;
Subtype: ABC vs. GCB;
Smurf2 expression: Q1 vs. Q2-Q4;
Revised IPI score: high (3, 4 and 5) vs. intermediate (1 and 2) and low (0);
Smurf2 expression: Q1+Q2 vs. Q3+Q4.
Discussion
In this study, we found that lymphomas developed in Smurf2-deficient mice have molecular features of GC or post-GC B cells and resemble human DLBCL. A significant decrease in Smurf2 expression is found in human DLBCL compared to normal B cells, indicating the clinical relevance of Smurf2 deficiency in human lymphomagenesis. Furthermore, our finding that low levels of Smurf2 expression correlate with poor survival prognosis in DLBCL patients suggests that Smurf2 is part of an important molecular pathway controlling lymphoma development. This pathway, identified in this study as the Smurf2-YY1-c-Myc regulatory axis, is novel and specific in suppression of B-cell proliferation and consequently lymphomagenesis. To our knowledge, Smurf2 is the first E3 ligase identified to ubiquitinate YY1. As a transcriptional regulator, YY1 plays a critical role in many biological processes including development, differentiation and proliferation, and has been implicated in oncogenesis35, 36. Specific to B cells, YY1 is a critical regulator of B-cell development37, and has recently been identified as a central regulator of the GC B-cell-specific transcriptional program17. The expression of YY1 is found to be increased in high-grade DLBCL or Burkitt’s lymphoma compared to low grade lymphoma or normal B cells38. Furthermore, elevated YY1 expression correlates with poor survival prognosis of DLBCL patients39, suggesting an oncogenic function for YY1 in human B-cell lymphomagenesis. The Smurf2-YY1-c-Myc regulatory axis is thus a useful prognostic predictor of clinical outcome in DLBCL patients and a potential therapeutic target for treatment of DLBCL. There is no evidence that the Smurf2 locus is deleted in human DLBCL40, 41 or Burkitt’s lymphoma42, 43, suggesting that epigenetic regulation probably plays an important role in Smurf2 deficiency in human lymphomagenesis. Moreover, we showed that restoration of Smurf2 expression in human DLBCL cells can suppress cell proliferation (Fig. 8f), providing an opportunity to design strategies to increase the expression of Smurf2 in lymphomas as a new therapeutic approach complementing current CHOP/R-CHOP regimens.
Increased susceptibility to B-cell lymphomagenesis in Smurf2-deficient mice is preceded by enhanced proliferation in splenic B cells. B cells undergo SHM and CSR in GCs to generate high-affinity antibodies. Both SHM and CSR are mutagenic and may target non-immunoglobulin genes. These “off-target” genetic alterations contribute to lymphomagenesis2, 3, 4, 5, 6, 7, 8. Enhanced proliferation would predispose Smurf2-deficient B cells to these mutagenic events, which ultimately contribute to lymphomagenesis. Consistent with this notion, Smurf2-deficient mice develop lymphomas at 12–15 months of age, most likely reflecting the infrequent generation of splenic GCs in un-immunized mice along with the accumulation of mutations with age. Recently it has been shown that Smurf2 regulates histone H2B monoubiquitination by targeting ring finger protein 20 (RNF20) for ubiquitination and degradation. Loss of Smurf2 leads to genomic instability through alterations in histone modification15, suggesting an important mechanism through which Smurf2 functions as a tumor suppressor. Our work now adds a novel dimension to the tumor suppressive functions of Smurf2.
As YY1 has been shown to transactivate c-Myc expression23, 24, our discovery of Smurf2-mediated ubiquitination and degradation of YY1 provides a novel mechanism by which Smurf2 regulates c-Myc expression and cell proliferation. Myc translocation is not common in DLBCL (5–10%), however, increased Myc expression is observed in ~30% of DLBCL and correlates with poor survival prognosis in DLBCL patients44. While no IgH-Myc translocation was found, lymphomas derived from Smurf2-deficient mice all showed increased expression of c-Myc. The Smurf2-YY1-c-Myc axis identified in the present study provides a plausible mechanism for increased Myc expression without chromosomal translocation in DLBCL.
As a master regulator of cell proliferation45, the oncogenic function of c-Myc in B-cell lymphomagenesis is well documented19, 20, 21, 46. In response to activated oncogenes, intrinsic tumor suppression mechanisms such as apoptosis and senescence are triggered to restrain oncogenic proliferation. With c-Myc, an additional layer of complexity is provided by the exquisite sensitivity of cells to different levels of c-Myc overexpression. High level of c-Myc overexpression increases cell proliferation and drives lymphomagenesis, but also induces apoptosis20, 21, 47. Suppression of apoptosis exacerbates c-Myc-induced lymphomagenesis, suggesting that apoptosis antagonizes c-Myc’s oncogenic activity21. Further, it has been recently found that low level of c-Myc elevation promotes cell proliferation without inducing apoptosis48. Consistent with this observation, we found that Smurf2-deficient mice showed low level (~2-fold) of c-Myc elevation (Fig. 5b) and enhanced proliferation in B cells without significant induction of apoptosis (Fig. 4 and Supplementary Fig. S7). In the absence of apoptosis, senescence likely becomes the critical tumor suppression mechanism to antagonize the oncogenic activity of Myc. A recent study shows that constitutive activation of c-Myc indeed activates senescence49. Smurf2 is an important regulator of senescence16, 50, 51, 52. In Smurf2-deficient cells, impaired senescence response leads to prolonged cell proliferation including in splenic cells16, suggesting that enhanced cell proliferation induced by low level of c-Myc activation coupled with an impaired senescence response drives lymphomagenesis in Smurf2-deficient mice. Smurf2 thus plays multiple roles in tumor suppression by maintaining chromatin landscape and genomic stability, suppressing cell proliferation and regulating the senescence response.
Methods
Smurf2-deficient mice
Smurf2-deficient mice as described previously16 have been backcrossed to C57BL/6 for >10 generations. Tissues were harvested for DNA, RNA and protein preparation, or fixed in 10% phosphate-buffered formalin. 5-μm paraffin sections were stained for Hematoxylin & Eosin, PNA (Vector Labs), TUNEL (Roche), B220 and Ki-67 (antibody sources and dilutions in Supplementary Table S2). For transplantation, 2×106 splenic cells from tumor-bearing mice were injected retro-orbitally into sub-lethally irradiated (3 Gy) Rag1−/− mice (Jackson Laboratories). All studies were carried out according to guidelines approved by the Institutional Animal Care and Use Committee of University of Massachusetts Medical School.
Flow cytometry
Spleens were collected from 2-month old mice of both sexes and ground between frosted glass slides. Bone marrow cells were harvested from long leg bones. After red blood cells were lysed, cells were filtered through 70-μm nylon mesh and incubated with anti-CD16/32 antibody (10 min on ice). Cells were incubated with primary antibodies for 20 min and washed three times with staining media (biotin-, flavin-, and phenol red-deficient RPMI-1640 medium with 10 mM pH7.2 HEPES, 0.02% sodium azide, 1 mM EDTA and 2% FBS). Antibodies (sources and dilutions in Supplementary Table S3) included B220-APC (RA3-6B2), CD19-PE-TR (ID3), CD21-FITC (7G6), CD23-biotin (B3B4), CD24-FITC (30-F1), CD43-PE (S7), CD93-PE-Cy7 (AA4.1), CD95-PE (15A7), CD117(c-kit)-PE-Cy5.5 (2B8), CD127-PE (A7R34), IgD-PE or -biotin (11–26), IgM-APC, -biotin or -PE-Cy7 (II/41), IgG2a-PE (m2a-15F8) and Ly6C-FITC (AL21). Biotin-stained cells were incubated with streptavidin-pacific blue (Invitrogen) for 15 min on ice, washed three times and resuspended in 1 μg/ml propidium iodide to exclude dead cells. Flow cytometry was performed on 5-laser, 18-detector LSR II (BD Biosciences), and data were analyzed using FlowJo (Treestar).
Splenic B-cell proliferation assays
Mouse spleen was collected and processed as described above. Single splenic cell suspension was treated with 10 μM carboxyfluorescein succinimidyl ester (Invitrogen) for 10 min at 37°C, and cultured in RPMI-1640 medium with 10% FBS, 100 μM MEM non-essential amino acids, 2 mM glutamine, and 50 μM 2-mercaptoethanol (Invitrogen)in a humidified chamber containing 5% CO2 at 37°C. Cells were treated with 5 μM lipopolysaccharide (Sigma) or left untreated. Cells were stained with anti-B220-APC antibody daily for 4 days. After being washed and re-suspended in 1 μg/ml propidium iodide, cells were analyzed by flow cytometry with BD FACSCaliber and FlowJo.
Mice were injected with 1 mg bromodeoxyuridine (BrdU, BD Biosciences) intraperitoneally. Spleen was collected 24 hours later and processed as described above. Splenic cells were fixed, and stained with anti-BrdU-FITC antibody using BrdU flow kit (BD Biosciences) and anti-B220-APC antibody. Data were collected by flow cytometry on BD FACSCaliber and analyzed using FlowJo.
Cycloheximde treatment
Mouse splenic cells were harvested and cultured as described above. Cells were treated with 15 μg/ml cycloheximide (Sigma), and cell lysates were collected every 2 hours post treatment.
Class switch recombination and somatic hypermutation assays
Transcripts of mouse IgH were amplified by quantitative RT-PCR as described53. Briefly, μ (IgM) transcripts were amplified using primers ImF and CmR. Post-switch transcripts were amplified using the following primers: ImF and Cg1R for γ1 (IgG1), ImF and Cg2aR for γ2a (IgG2a), ImF and Cg2bR for γ2b (IgG2b), ImF and Cg3R for γ3 (IgG3). Primers used are ImF: 5′-CTCTGGCCCTGCTTATTGTTG-3′; CmR: 5′-GAAGACATTTGGGAAGGACTGACT-3′; Cg1R: 5′-GGATCCAGAGTTCCAGGTCACT-3′; Cg2aR: 5′-GCCACATTGCAGGTGATGGA-3′; Cg2bR: 5′-CACTGAGCTGCTCATAGTGTAGAGTC-3′; and Cg3R: 5′-CTCAGGGAAGTAGCCTTTGACA-3′.
The frequency of somatic mutation in mouse IgH was determined as described53. Briefly, VH186.2 transcripts of mouse IgH μ isotype were amplified in RT-PCR using Phusion hot start high-fidelity Taq Polymerase (New England Biolabs) with primers VH186.2 (5′-TTCTTGGCAGCAACAGCTACA-3′) and CmR (5′-GAAGACATTTGGGAAGGACTGACT-3′). PCR products were cloned into pGEM-T-easy vector (Promega), and individual colonies were sequenced.
Clonal analysis of mouse B-cell lymphomas
Rearrangements of the variable region of mouse IgH were analyzed as described54. Briefly, genomic DNA was prepared from lymphoma-bearing spleen of Smurf2-deficient mice or spleen of wild-type mice using classic genomic DNA isolation kit (LamdaBiotech), and used in PCR with primers VH (J558FR3: 5′-CAGCCTGACATCTGAGGACTCTGC-3′) and 3′ of JH4 (JH4int: 5′-CTCCACCAGACCTCTCTAGACAGC-3′).
Analysis of IgH-Myc translocation
IgH-Myc translocation in mouse B-cell lymphomas was detected as described55. Briefly, genomic DNA isolated from lymphomas was used as template in nested PCR using expended long PCR system (Roche). Primers were: 5′-TGAGGACCAGAGAGGGATAAAAGAGAA-3′ and 5′-GGGGAGGGGGTGTCAAATAATAAGA-3′ (1st round); 5′-CACCCTGCTATTTCCTTGTTGCTAC-3′and 5′-GACACCTCCCTTCTACACTCTAAACCG-3′ (2nd round). Myc locus was amplified as controls with primers of 5′-GGGGAGGGGGTGTCAAATAATAAGA-3′ and 5′-GTGAAAACCGACTGTGGCCCTGGAA-3′. Two lymphoma samples (318 and 334) with IgH-Myc translocation56 were kindly provided by Dr. John Manis (Harvard Medical School) and used as positive controls.
Cell culture and transfection
SUDHL-6 cells (kindly provided by Dr. Subbarao Bondada, University of Kentucky) werecultured in RPMI-1640 medium with 10% FBS in a humidified chamber containing 5%CO2 at 37°C. Lentiviral vectors pLenti-CMV-Smurf2-IRES-Puro, pLenti-CMV-C716A-IRES-Puro or pLenti-CMV-GFP-IRES-Puro were described previously51. Briefly, fragments of Smurf2 or GFP cDNA were inserted ito a lentiviral vector to express Smurf2 or GFP under the control of a CMV promoter. Lentiviral shRNA constructs targeting Smurf2 (V2LHS_10399), YY1 (V2LHS_219592, V2LHS_389741) and a non-silencing shRNA control (RHS4346) were purchased from Open Biosystems. Lentiviral packaging and infection were carried out as described51. Briefly, Lentiviral vectors were co-transfected with a plasmid (pMD2. VSV-G) encoding vesicular stomatitis virus glycoprotein (VSV-G) and a plasmid (pCMVdR8.74) encoding packaging proteins into 293T cells. VSV-G pseudotyped virus were collected 48 hr after transfection and used to infected target cells in the presence of 4 μg/ml polybrene (Sigma). Cells were FACS sorted for GFP+ cells (shRNA knockdown) or selected with puromycin (1 μg/ml, Sigma) for 1 week.
To analyze cell proliferation, sorted or selected cells were seeded at 5×103 per well in 6-well plates, harvested in triplicate and counted daily for 5 days using Z1Coulter Particle Counter (Beckman Coulter). Apoptosis was measured using Caspase-Glo 3/7 assay (Promega).
Splenic B cells were isolated using anti-B220-biotin antibody and anti-biotin beads with AutoMACS Pro (Miltenyi Biotec) and cultured with 5 μM LPS as described above. 3×106 cells were transfected with 10 pmol YY1 siRNA (sc-36864) or control siRNA (sc-37007, Santa Cruz Biotechnology) using mouse B cell Nucleofector kit on Nucelofector I device (Lonza) with program Z-01. Cell lysates were collected 48 hours after transfection.
Quantitative RT-PCR
Total RNA was isolated using RNeasy Mini kit (Qiagen) and reverse-transcribed using Superscript II (Invitrogen). Real-time PCR was performed using SYBR Green PCR kit (Bio-Rad). The following primers were used: Smurf2 (F: 5′-ATGAAGTCATTCCCCAGCAC-3′; R: 5′-AACCGTGCTCGTCTCTCTTC-3′), c-Myc (F: 5′-GGACAGTGTTCTCTGCC-3′; R: 5′-CGTCGCAGATGAAATAGG-3′), YY1 (F: 5′-TGAGAAAGCATCTGCACACC-3′; R: 5′-CGCAAATTGAAGTCCAGTGA-3′), Apex1 (F: 5′-GCTCCGTCAGACAAAGAAGG-3′; R: 5′-GCATTGGGAACATAGGCTGT-3′), Cad (F: 5′-TGGTCAGTTCATCCTCACTCC-3′; R: 5′-TACATGCCGTTCTCAGCTTG-3′), Ncl (F: 5′-TAAGGGTGAAGGTGGCTTTG-3′; R: 5′-CCTTGTGGCTTGAAGTCTCC-3′) and β-actin (F: 5′-GCTCTTTTCCAGCCTTCCTT-3′; R: 5′-GTGCTAGGAGCCAGAGCAGT-3′).
Western blot and co-immunoprecipitation analysis
Total cell lysates were collected using RIPA buffer (50 mM Tris–HCl pH 7.5, 150 mMNaCl, 1% Triton X-100, 0.1% SDS, 0.5% deoxycholic acid, 0.02% sodium azide) with freshly added complete protease inhibitors (Roche). Protein lysates (20 μg) were separated by SDS–PAGE Criterion X-gel (Bio-Rad) and transferred to nitrocellulose membranes (GE Osmonics). Immunoblots were analyzed by Western blotting and visualized using a Western lightening chemiluminescence detection kit (PerkinElmer). Primary antibodies were Smurf2, c-Myc, YY1, Flag, HA, ubiquitin, β-actin and GAPDH (antibody sources and dilutions in Supplementary Table S2). For co-immunoprecipitation, cells were lysed in NP40 lysis buffer (20 mM Tris–HCl, 150 mM NaCl, 2 nM EDTA, 1% Nonidet P-40) plus complete protease inhibitors (Roche). Lysates were incubated with anti-Flag M2 affinity gel (Sigma) overnight at 4°C. Immunoprecipitates were washed four times with NP40 lysis buffer, and analyzed in Western blot with anti-HA antibody.
Ubiquitination assay
293T cells were transfected with HA-YY1 (a gift of Dr. Yang Shi, Harvard Medical School), 3xFlag-ubiquitin (provided by Dr. Quan Lu, Harvard School of Public Health) and Smurf2. GFP and C716A were used as controls. Cells were treated with MG132 (20 μM, Sigma) for 2 hours and lysed in RIPA buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 0.5% deoxycholic acid, 0.02% sodium azide) plus 10 mM N-ethylmaleimide (Fisher Scientific). Cell lysates were incubated with anti-HA or anti-Flag affinity gel (Sigma) overnight at 4°C. Immunoprecipitates were washed with RIPA buffer three times, and analyzed in Western blot with anti-Flag or anti-HA antibody to detect ubiquitin conjugation. To detect ubiquitination of endogenous YY1 in SUDHL-6 cells, cell lysates were incubated with anti-YY1 antibody overnight at 4°C followed by incubation with protein A-agarose (Invitrogen). Poly-ubiquitinated YY1 was detected in Western blot using anti-ubiquitin antibody.
Chromatin immunoprecipitation assay
Mouse spleen was collected and processed as described above. Single cell suspension was cross-linked using 1% formaldehyde (Sigma) in RPMI-1640 medium (room temperature for 10 min). After neutralization with glycine (125 nM), cells were lysed in SDS lysis buffer. Chromatin was sonicated to fragments of ~500 bp, and Chromatin immunoprecipitation (ChIP) with anti-YY1 antibody or matched IgG was performed using ChromaFlash one step ChIP kit (Epigentek). After reverse of cross-linking (65°C for 3 hours), the c-Myc promoter region containing the YY1 binding site was amplified in quantitative PCR with primers: F: 5′-TCCCCAGCCTTAGAGAGACG-3′ and R: 5′-GGCTCCGGGGTGTAAACAGT-3′. Chromatin before ChIP was used as input.
Gene expression and statistical analyses
Microarray data (GSE2350, GSE4475 and GSE10846) were retrieved from GEO(http://www.ncbi.nil.nih.gov/geo). Expression of Smurf2 (log2 transformed) was analyzedwith dot plot, and one-way ANOVA was used for statistical analysis. Kaplan-Meier survival curves were plotted using GraphPad Prism 5.0, and log-rank test was used for statistical analysis. Univariate and multivariate Cox regression analyses were used to estimate the hazard ratios and 95% confidence intervals, and statistical significance was analyzed using SPSS Statistics 19 software (IBM).
Human DLBCL dataset GSE10846 contains samples from patients receiving CHOP or R-CHOP treatment. Revised International Prognostic Index (IPI) score was used to group these patients: low (0), intermediate (1 or 2) or high (3, 4 or 5)34. Some IPI variables were missing in some samples. If the missing IPI variables did not change the IPI grouping of a given sample, this sample was included in the analysis. Otherwise, the sample was excluded.
Data were presented as mean ± SD. Two-tailed and unpaired Student t-test was used for pairwise comparisons, with P<0.05 considered as statistically significant.
Supplementary Material
Acknowledgments
We thank Dr. John Manis for IgH and IgH-Myc PCR protocols, reagents and helpful discussion; Drs. Subbarao Bondada, Quan Lu, Madelyn Schmidt, Yang Shi, Janet Stavnezer and Robert Woodland for kindly providing reagents; Dr. Suyang Hao for pathological diagnosis of lymphoma samples; Dr. Madelyn Schmidt for advice on LPS stimulation and CFSE staining; Erin Cloherty and Laura Fineman for technical assistance. This work was supported by grants from the National Cancer Institute (R01CA131210) and The Ellison Medical Foundation (AG-NS-0347-06) to H.Z.
Footnotes
Conflict of Interest
The authors declare no competing financial interest.
Author Contributions
C.R, R.M.G and H.Z. developed the concept and designed the experiments; C.R., H.C., Y.K., R.M.G and H.Z. did the experiments and/or analyzed data; S.N.J. contributed reagents, intellectual input and editorial assistance; C.R. and H.Z. wrote the paper; H.Z. supervised the project and obtained funding.
References
- 1.Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 2.Pasqualucci L, et al. BCL-6 mutations in normal germinal center B cells: evidence of somatic hypermutation acting outside Ig loci. Proc Natl Acad Sci U S A. 1998;95:11816–11821. doi: 10.1073/pnas.95.20.11816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pasqualucci L, et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- 4.Staszewski O, Baker RE, Ucher AJ, Martier R, Stavnezer J, Guikema JE. Activation-induced cytidine deaminase induces reproducible DNA breaks at many non-Ig Loci in activated B cells. Mol Cell. 2011;41:232–242. doi: 10.1016/j.molcel.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robbiani DF, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robbiani DF, et al. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol Cell. 2009;36:631–641. doi: 10.1016/j.molcel.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen HM, Peters A, Baron B, Zhu X, Storb U. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science. 1998;280:1750–1752. doi: 10.1126/science.280.5370.1750. [DOI] [PubMed] [Google Scholar]
- 8.Liu M, et al. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- 9.Allen CD, Okada T, Cyster JG. Germinal-center organization and cellular dynamics. Immunity. 2007;27:190–202. doi: 10.1016/j.immuni.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lenz G, Staudt LM. Aggressive lymphomas. N Engl J Med. 2010;362:1417–1429. doi: 10.1056/NEJMra0807082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.A clinical evaluation of the International Lymphoma Study Group classification of non-Hodgkin’s lymphoma. The Non-Hodgkin’s Lymphoma Classification Project. Blood. 1997;89:3909–3918. [PubMed] [Google Scholar]
- 12.Schneider C, Pasqualucci L, Dalla-Favera R. Molecular pathogenesis of diffuse large B-cell lymphoma. Semin Diagn Pathol. 2011;28:167–177. doi: 10.1053/j.semdp.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nogai H, Dorken B, Lenz G. Pathogenesis of non-Hodgkin’s lymphoma. J Clin Oncol. 2011;29:1803–1811. doi: 10.1200/JCO.2010.33.3252. [DOI] [PubMed] [Google Scholar]
- 14.Coiffier B. Diffuse large cell lymphoma. Curr Opin Oncol. 2001;13:325–334. doi: 10.1097/00001622-200109000-00003. [DOI] [PubMed] [Google Scholar]
- 15.Blank M, Tang Y, Yamashita M, Burkett SS, Cheng SY, Zhang YE. A tumor suppressor function of Smurf2 associated with controlling chromatin landscape and genome stability through RNF20. Nat Med. 2012;18:227–234. doi: 10.1038/nm.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramkumar C, et al. Smurf2 regulates the senescence response and suppresses tumorigenesis in mice. Cancer Res. 2012;72:2714–2719. doi: 10.1158/0008-5472.CAN-11-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Green MR, et al. Signatures of murine B-cell development implicate Yy1 as a regulator of the germinal center-specific program. Proc Natl Acad Sci U S A. 2011;108:2873–2878. doi: 10.1073/pnas.1019537108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morse HC, 3rd, et al. Bethesda proposals for classification of lymphoid neoplasms in mice. Blood. 2002;100:246–258. doi: 10.1182/blood.v100.1.246. [DOI] [PubMed] [Google Scholar]
- 19.Dalla-Favera R, Martinotti S, Gallo RC, Erikson J, Croce CM. Translocation and rearrangements of the c-myc oncogene locus in human undifferentiated B-cell lymphomas. Science. 1983;219:963–967. doi: 10.1126/science.6401867. [DOI] [PubMed] [Google Scholar]
- 20.Langdon WY, Harris AW, Cory S, Adams JM. The c-myc oncogene perturbs B lymphocyte development in E-mu-myc transgenic mice. Cell. 1986;47:11–18. doi: 10.1016/0092-8674(86)90361-2. [DOI] [PubMed] [Google Scholar]
- 21.Soucek L, Evan GI. The ups and downs of Myc biology. Curr Opin Genet Dev. 2010;20:91–95. doi: 10.1016/j.gde.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeller KI, Jegga AG, Aronow BJ, O’Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 2003;4:R69. doi: 10.1186/gb-2003-4-10-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riggs KJ, et al. Yin-yang 1 activates the c-myc promoter. Mol Cell Biol. 1993;13:7487–7495. doi: 10.1128/mcb.13.12.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu KW, et al. The activated Notch1 receptor cooperates with alpha-enolase and MBP-1 in modulating c-myc activity. Mol Cell Biol. 2008;28:4829–4842. doi: 10.1128/MCB.00175-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kavsak P, et al. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell. 2000;6:1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 26.Lin X, Liang M, Feng XH. Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-beta signaling. J Biol Chem. 2000;275:36818–36822. doi: 10.1074/jbc.C000580200. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Chang C, Gehling DJ, Hemmati-Brivanlou A, Derynck R. Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc Natl Acad Sci U S A. 2001;98:974–979. doi: 10.1073/pnas.98.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 29.Hummel M, et al. A biologic definition of Burkitt’s lymphoma from transcriptional and genomic profiling. N Engl J Med. 2006;354:2419–2430. doi: 10.1056/NEJMoa055351. [DOI] [PubMed] [Google Scholar]
- 30.Lenz G, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008;359:2313–2323. doi: 10.1056/NEJMoa0802885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dave SS, et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med. 2004;351:2159–2169. doi: 10.1056/NEJMoa041869. [DOI] [PubMed] [Google Scholar]
- 32.Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 33.Wright G, Tan B, Rosenwald A, Hurt EH, Wiestner A, Staudt LM. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci U S A. 2003;100:9991–9996. doi: 10.1073/pnas.1732008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sehn LH, et al. The revised International Prognostic Index (R-IPI) is a better predictor of outcome than the standard IPI for patients with diffuse large B-cell lymphoma treated with R-CHOP. Blood. 2007;109:1857–1861. doi: 10.1182/blood-2006-08-038257. [DOI] [PubMed] [Google Scholar]
- 35.Castellano G, et al. The involvement of the transcription factor Yin Yang 1 in cancer development and progression. Cell Cycle. 2009;8:1367–1372. doi: 10.4161/cc.8.9.8314. [DOI] [PubMed] [Google Scholar]
- 36.Shi Y, Lee JS, Galvin KM. Everything you have ever wanted to know about Yin Yang 1. Biochim Biophys Acta. 1997;1332:F49–66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- 37.Liu H, et al. Yin Yang 1 is a critical regulator of B-cell development. Genes Dev. 2007;21:1179–1189. doi: 10.1101/gad.1529307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castellano G, et al. Yin Yang 1 overexpression in diffuse large B-cell lymphoma is associated with B-cell transformation and tumor progression. Cell Cycle. 2010;9:557–563. doi: 10.4161/cc.9.3.10554. [DOI] [PubMed] [Google Scholar]
- 39.Sakhinia E, et al. Clinical quantitation of diagnostic and predictive gene expression levels in follicular and diffuse large B-cell lymphoma by RT-PCR gene expression profiling. Blood. 2007;109:3922–3928. doi: 10.1182/blood-2006-09-046391. [DOI] [PubMed] [Google Scholar]
- 40.Lenz G, et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc Natl Acad Sci U S A. 2008;105:13520–13525. doi: 10.1073/pnas.0804295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bea S, et al. Diffuse large B-cell lymphoma subgroups have distinct genetic profiles that influence tumor biology and improve gene-expression-based survival prediction. Blood. 2005;106:3183–3190. doi: 10.1182/blood-2005-04-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salaverria I, et al. Chromosomal alterations detected by comparative genomic hybridization in subgroups of gene expression-defined Burkitt’s lymphoma. Haematologica. 2008;93:1327–1334. doi: 10.3324/haematol.13071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scholtysik R, et al. Detection of genomic aberrations in molecularly defined Burkitt’s lymphoma by array-based, high resolution, single nucleotide polymorphism analysis. Haematologica. 2010;95:2047–2055. doi: 10.3324/haematol.2010.026831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horn H, et al. MYC status in concert with BCL2 and BCL6 expression predicts outcome in diffuse large B-cell lymphoma. Blood. 2013;121:2253–2263. doi: 10.1182/blood-2012-06-435842. [DOI] [PubMed] [Google Scholar]
- 45.Dang CV. MYC, microRNAs and glutamine addiction in cancers. Cell Cycle. 2009;8:3243–3245. doi: 10.4161/cc.8.20.9522. [DOI] [PubMed] [Google Scholar]
- 46.Harris AW, Pinkert CA, Crawford M, Langdon WY, Brinster RL, Adams JM. The E mu-myc transgenic mouse. A model for high-incidence spontaneous lymphoma and leukemia of early B cells. J Exp Med. 1988;167:353–371. doi: 10.1084/jem.167.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evan G, Littlewood T. A matter of life and cell death. Science. 1998;281:1317–1322. doi: 10.1126/science.281.5381.1317. [DOI] [PubMed] [Google Scholar]
- 48.Murphy DJ, et al. Distinct thresholds govern Myc’s biological output in vivo. Cancer Cell. 2008;14:447–457. doi: 10.1016/j.ccr.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Campaner S, et al. Cdk2 suppresses cellular senescence induced by the c-myc oncogene. Nat Cell Biol. 2010;12:54–59. 51–14. doi: 10.1038/ncb2004. [DOI] [PubMed] [Google Scholar]
- 50.Zhang H, Cohen SN. Smurf2 up-regulation activates telomere-dependent senescence. Genes Dev. 2004;18:3028–3040. doi: 10.1101/gad.1253004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang H, Teng Y, Kong Y, Kowalski PE, Cohen SN. Suppression of human tumor cell proliferation by Smurf2-induced senescence. J Cell Physiol. 2008;215:613–620. doi: 10.1002/jcp.21337. [DOI] [PubMed] [Google Scholar]
- 52.Kong Y, Cui H, Zhang H. Smurf2-mediated ubiquitination and degradation of Id1 regulates p16 expression during senescence. Aging Cell. 2011;10:1038–1046. doi: 10.1111/j.1474-9726.2011.00746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 54.Jolly CJ, Klix N, Neuberger MS. Rapid methods for the analysis of immunoglobulin gene hypermutation: application to transgenic and gene targeted mice. Nucleic Acids Res. 1997;25:1913–1919. doi: 10.1093/nar/25.10.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kovalchuk AL, Muller JR, Janz S. Deletional remodeling of c-myc-deregulating chromosomal translocations. Oncogene. 1997;15:2369–2377. doi: 10.1038/sj.onc.1201409. [DOI] [PubMed] [Google Scholar]
- 56.Wang JH, et al. Oncogenic transformation in the absence of Xrcc4 targets peripheral B cells that have undergone editing and switching. J Exp Med. 2008;205:3079–3090. doi: 10.1084/jem.20082271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.