Abstract
The increasing rate of biological invasions resulting from human transport or human-mediated changes to the environment have had devastating ecologic and public health consequences. The kissing bug, Triatoma infestans, has dispersed through the Peruvian city of Arequipa. The biological invasion of this insect has resulted in a public health crisis, putting thousands of residents of this city at risk of infection by Trypanosoma cruzi and subsequent development of Chagas disease. Here we show that populations of Tria. Infestans in geographically distinct districts within and around this urban center share a common recent evolutionary history although current gene flow is restricted even between proximal sites. The population structure among the Tria. Infestans in different districts is not correlated with the geographic distance between districts. These data suggest that migration among the districts is mediated by factors beyond the short-range migratory capabilities of Tria. Infestans and that human movement has played a significant role in the structuring of the Tria. Infestans population in the region. Rapid urbanization across southern South America will continue to create suitable environments for Tria. Infestans and knowledge of its urban dispersal patterns may play a fundamental role in mitigating human disease risk.
Keywords: Triatoma infestans, Chagas disease, vector, population genetics, genetic structure
Introduction
Invasive species can have devastating ecologic, economic, and public health consequences (Vitousek et al. 1996; Wilcove et al. 1998; Sala et al. 2000; Sax et al. 2005; Hooper et al. 2005; Pimentel et al. 2005). Anthropogenic activities have been responsible for species introductions through transportation to novel environments, habitat degradation and fragmentation (Tews et al. 2004; MacDougall & Turkington 2005), and the creation of novel landscapes such as urban environments (Grimm et al. 2008; Walsh et al. 2011). Rapid environmental alterations affect species differently; some species gain competitive advantages and thrive while others become locally extinct (Strayer et al. 2006; Barbosa et al. 2010; Morris 2010). The ramifications of anthropogenic action can be amplified beyond the initial impact by synergistic and cascading effects of new species interactions that exacerbate the impact on the community structure (Strayer et al. 2006; Scherber et al. 2010). For example, the disease-causing microbes vectored by an arthropod may expand into novel habitats due to the altered ecologies of the hosts, the microbes, or the vector itself (Mott et al. 1978; Lounibos 2002; Tompkins & Gleeson 2006; Suzan et al. 2009; Schaffner et al. 2009). Consequently, the population dynamics during the initial invasion and subsequent dispersal can be imperative to public health risk management (Crowl et al. 2008). In this study we examine the colonization pattern of Triatoma infestans, the primary vector of Chagas disease, in and around the city of Arequipa, Peru.
The Chagas disease system in Arequipa, Peru is ideal to study the dynamics of a recent invasion. The relatively recent establishment of Tria. infestans into the city of Arequipa (Bayer et al. 2009) has led to the transmission of Trypanosoma cruzi, the etiologic agent of Chagas disease, among domestic animals and humans (Levy et al. 2006; 2007; Bowman et al. 2008; Levy et al. 2011; Hunter et al. 2012). Currently, many thousands of residents of this city are at risk of contracting Chagas disease (Levy et al. 2006; Bowman et al. 2008). The emergence of Chagas disease in urban areas further emphasizes the public health importance of studies investigating the dynamic processes of invasion, colonization, and spread of disease-causing agents in urban systems.
Tria. infestans disperses actively by walking or flying as adults, although both occur over short spatial distances (< 2 km). Further, only adults in a low nutritional status or without access to blood meals are likely to use flight as a dispersal mechanism (Ceballos et al. 2005; Richer et al. 2007). The habitats created by the rapid development and abundant sources of blood from domestic animals and humans make the areas surrounding downtown Arequipa hospitable for an expanding population. Tria. infestans can also be dispersed passively through human travel and transportation of goods. Assisted by anthropogenic activities, Tria. infestans can travel much longer distances than it can by walking or flying.
The combination of active and passive dispersal can have dramatic effects on the population structuring of the insects. It is possible that Tria. infestans dispersed in Arequipa through a natural range expansion to progressively reach all of the areas in which it is currently found. Alternatively, Tria. infestans could have arrived through sporadic, longer-range migration events from the surrounding regions and subsequently spread through the city. Gene flow mediated only by natural walking or flying migrations of short distances should result in a pattern of isolation by distance (IBD). Passive, long-range migrations would lead to patterns of genetic diversity less strongly correlated with geographic distance, and perhaps more related to heavily traveled pathways.
In the present study, we used a population genetic modeling framework to assess the migratory history of recently established populations of Tria. infestans in Arequipa, Peru. We used microsatellite data to address broad but overlapping hypotheses. Specifically we sought to address if the Tria. infestans populations in the region are geographically structured or form one large interbreeding population, and if the extent of gene flow among populations is correlated with geographic distance. Each of the hypotheses leads to a distinct population genetic pattern under an idealized model, which when compared to the empirically observed pattern, can be used to infer the routes of dispersal. Spatial patterns of genetic diversity within populations are indicative of the origin, evolutionary history, and population dynamics, and allow the reconstruction of a population’s migratory history (Sakai et al. 2001; Gaudeul et al. 2011; Sloop et al. 2011; Bronnenhuber et al. 2011). Our results provide vital information about the dispersal patterns of a vector species that has put thousands of people at risk for infection with Tryp. cruzi.
Methods
Description of the study site
Arequipa, the second largest city in Peru, is located in the southern region in the foothills of the Andes Mountains (inset of Fig. 1). Arequipa has recently experienced an influx of migrants, many of whom live in rudimentary houses on hills surrounding the original urban center (Schuurman 1986; Paerregaard 1997). These houses, constructed primarily of stacked volcanic stone and bricks without mortar, are highly suitable habitat for Tria. infestans (Levy et al. 2006).
FIG. 1.
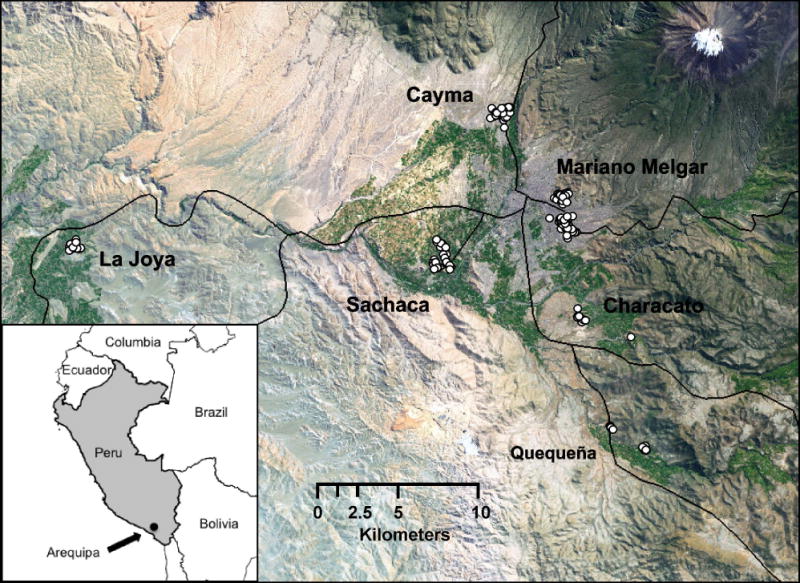
Map of the study area. The locations of sampled Tria. infestans individuals are represented as white dots. The roadways are represented as black grey lines. Inset shows the general location of the study area.
Sample Collection
Tria. infestans were collected between 2004 and 2009 from seven districts in the Arequipa region (Fig. 1, specific years and geographic coordinates in Table S1). Cayma, Mariano Melgar, and Paucarpata are urban districts in downtown Arequipa, Sachaca and Characato peri-urban districts at the edge of the city and La Joya and Quequeña are rural districts. The geographic distance between districts ranges from 1.8 km to 41 km (Table 2). The rural districts are separated by a rock-covered desert that have few or no vectors. The urban districts are connected by a continuous urban landscape with known vector populations. Peri-urban districts are separated from the urban districts by farmland.
Table 2.
FST values between all pairs of districts (below diagonal); Geographic distances (km) between districts (above diagonal).
| CA | CH | LJ | MM | PA | QU | SA | |
|---|---|---|---|---|---|---|---|
| CA | 14.634 | 30.309 | 7.065 | 8.595 | 24.652 | 10.345 | |
| CH | 0.08441* | 35.380 | 8.080 | 6.294 | 10.032 | 10.778 | |
| LJ | 0.03984* | 0.03432* | 33.772 | 33.806 | 41.056 | 25.006 | |
| MM | 0.05734* | 0.11086* | 0.05521* | 1.795 | 18.044 | 9.559 | |
| PA | 0.05325* | 0.15369* | 0.07292* | 0.04157* | 16.284 | 9.089 | |
| QU | 0.09822* | 0.13268* | 0.12172* | 0.16587* | 0.13298* | 18.951 | |
| SA | 0.02995 | 0.09387* | 0.03726* | 0.06722* | 0.03745* | 0.11636* |
P < 0.00238 [significance level of P (0.05) was adjusted for multiple comparisons]
Tria. infestans were collected during a recent vector control campaign in which vector control specialists applied deltamethrin to the interiors and exteriors of over 80,000 houses. Bugs that were flushed out by the insecticide were collected and later placed into individual tubes that were subsequently stored at -20 °C. Approximately 30 infested houses were chosen at random from each district and one insect was selected at random from among all the bugs collected from each selected house. Some districts had slightly smaller or larger sample sizes. Each sample was uniquely geocoded. The history of efforts to control Tria. Infestans in the region is complex. The current campaign is probably the first coordinated effort in the urban and peri-urban districts. La Joya was previously sprayed in 1996 {Delgado:2011ep}; there was possibly an undocumented control campaign in Quequeña in the 1980s. All districts in the study are currently under surveillance in the post-spray phase of the Ministry of Health’s control campaign.
Genetic data
DNA was extracted from two legs of each bug following the insect tissue protocol with extended lysis time from the Qiagen DNEasy Blood and Tissue Kit. A group of 13 microsatellites was chosen from a set of well-characterized markers (Table S2; (Garcia et al. 2004; Marcet et al. 2006). All microsatellite markers were amplified using PCR according to reaction conditions detailed in Table S2 using a fluorescently tagged forward primer (ABI dyes: 6-FAM, PET, VIC, or NED). Negative controls were run with each PCR reaction to guard against cross-contamination. Fragment sizing of the fluorescently tagged PCR product was done at the DNA Sequencing Center at the University of Pennsylvania (Applied Biosystems 3100 capillary sequencer and GeneMapper). All allele size electropherograms were visualized in PeakScanner (ABI), and any peaks that were ambiguous were rerun until clear peaks were obtained.
Population genetic analyses
The genetic diversity within each district was quantified by the mean number of alleles per locus, the average gene diversity over all loci, and the inbreeding coefficient, FIS. The mean observed heterozygosity and mean expected heterozygosity assuming Hardy-Weinberg equilibrium were calculated by averaging over all 13 loci. All calculations were performed in Arlequin v3.5 (Excoffier et al. 2005; Excoffier & Lischer 2010). A test of global linkage disequilibrium across all possible pairs of microsatellite markers was conducted using FSTAT v2.3.9.2 (Goudet 1995). The statistical significance level of all tests was adjusted for multiple comparisons (Bland & Altman 1995; Abdi 2007).
Population genetic structure was assessed using four methods. The FST values between each pair of districts and the Analysis of Molecular VAriance (AMOVA) (Excoffier et al. 1992) were calculated in Arlequin. The proximity to downtown Arequipa was initially used to define the regional groups in AMOVA analyses such that urban districts (CA, MM, PA) were grouped separate from rural districts (CH, LJ, QU, SA). Post-hoc analyses using alternate regional groupings identified by genetic structure analyses were also generated. Statistical significance was determined by comparison to values generated from 10,000 permutations and adjustments for multiple comparisons when appropriate.
Cluster analyses were performed using the Bayesian clustering algorithm implemented in STRUCTURE v2.3.2 (Pritchard et al. 2000; Falush et al. 2007; Hubisz et al. 2009), assuming correlated allele frequencies, admixture, and no location data as a prior. Six iterations of the data were performed at each K = 2–15, the number of genetic clusters, with 100,000 Markov Chain Monte Carlo (MCMC) iterations and a 25% burn-in. STRUCTURE output was extracted using STRUCTURE HARVESTER (Earl & vonHoldt 2011), processed with CLUMPP (Jakobsson & Rosenberg 2007) to find the optimal alignment and remove “label-switching” differences of the six iterations, and plotted with Distruct (Rosenberg 2004). The Evanno method was used to calculate ΔK to infer the optimal number of clusters given the data (Evanno et al. 2005).
Isolation By Distance (IBD) was tested between districts and between pairs of individuals. District-level IBD was tested by comparing the FST matrix and the geographic distance matrix (both in Table 2) using a Mantel test with 1,000 permutations (Sokal & Rolf 1995; Peakall & Smouse 2006). IBD at the individual level was tested by comparing the matrix of pairwise geographic distances using GenAlEx (Peakall & Smouse 2006) between all pairs of individuals to either the matrix of genetic distance values calculated in GenePop (Raymond & Rousset 1995; Rousset 2008) (a statistic that approximates F/(1-F), (Rousset 2000)) or the matrix of pairwise codominant genetic distances (GenAlEx). Mantel tests of both genetic distance matrices against the geographic distance matrix as well as the natural log of geographic distance, which has been shown to improve the linearity (Rousset 1997), were performed in R (R Development Core Team 2012). The Mantel statistic was based on the Pearson’s product-moment correlation and significance of each Mantel test was determined by 10,000 permutations (Sokal & Rolf 1995).
Spatial autocorrelation was performed and visualized using Mantel correlograms in PASSaGE v.2 with 42 distance classes of equal distance (Rosenberg & Anderson 2011). Both genetic distance matrices (approximation of F/(1-F) and codominant genetic distance) were tested against geographic distances. The pairwise codominant genetic distance matrix was also used in a principle coordinates analysis in GenAlEx following the procedure described by Orloci (1978). The centroid of each district and its standard error were calculated and plotted in R to assess differentiation among districts.
Results
The 13 microsatellite loci were in linkage equilibrium and were sufficiently polymorphic to be informative in population genetic analyses. The number of alleles at each locus ranged from 4 to 11 and the mean number of alleles per locus was similar among districts (Table 1). Interestingly, the majority of alleles were found in most districts indicating a recent shared evolutionary or migratory history among districts. Significant departure from Hardy-Weinberg equilibrium was found in all seven districts as a consequence of considerable heterozygote deficiency. The districts showed statistically significant inbreeding (FIS > 0, p-val < 0.001) indicative of population substructure within each district. The global FIS for all of the districts combined was 0.39571, showing statistically significant substructure in the total population as well (Excoffier 2007).
Table 1.
Measures of genetic diversity in each district
| Location (Abbreviation) | Sample Size | Mean Num. Alleles/Locus | Observed Heterozygosity | Expected Heterozygosity | FIS |
|---|---|---|---|---|---|
| Cayma (CA) | 29 | 3.692 | 0.2441* | 0.5389 | 0.54894* |
| Characato (CH) | 28 | 3.385 | 0.3208* | 0.5002 | 0.34101* |
| LaJoya (LJ) | 29 | 3.462 | 0.3050* | 0.4811 | 0.37339* |
| Mariano Melgar (MM) | 29 | 3.615 | 0.2944* | 0.4668 | 0.36664* |
| Paucarpata (PA) | 28 | 3.923 | 0.3434* | 0.5179 | 0.35774* |
| Quequeña (QU) | 35 | 3.385 | 0.2768* | 0.4731 | 0.41583* |
| Sachaca (SA) | 34 | 3.846 | 0.3313* | 0.5144 | 0.35726* |
< 0.001
Gene flow among districts has been appreciably restricted resulting in significant population genetic structure among even very proximal districts (Table 2). Nearly all (20 out of 21) of the pairwise comparisons indicated significant Fst after correction for multiple comparisons. Only the districts of Sachaca and Cayma did not have significantly different frequencies of alleles (p=0.00554, a value greater than the value adjusted for multiple comparison p=0.00238). The pairwise FST values between these two districts and each of the other districts were also relatively low. Quequeña, a rural district about 20 kilometers from the city, appeared to be the most genetically distinct with relatively high FST values in all pairwise comparisons. Surprisingly, the most geographically distant district, La Joya, had low FST values when compared to the districts downtown. The year samples were collected did not have a measurable effect on population genetic structure among districts (Fig. S2).
The genetic differentiation among districts was also evident in principal component analyses (PCA, Fig. 2). Each district was centered in a distinct area of the PCA space and no two districts occupied the same area with one exception; the genetic diversity from the bugs sampled in Cayma and Sachaca, districts near downtown separated by 10.3 km, were similarly described in the first two axes of the PCA. These two districts also did not demonstrate statistically significant population genetic differentiation (Table 2). The contiguous districts, Mariano Melgar and Paucarpata, occupied distinct locations in the PCA space despite their geographic proximity. Interestingly, the genetic diversity in La Joya was similar to most other districts despite being geographically remote. The district of Quequeña was the only district that occupied two locations in the PCA space which correspond with the two distinct communities in the district: a northern, more recently established community near a thoroughfare and an isolated southern community founded centuries ago. The genetic diversity in the samples collected in the more connected community was similar to that in other districts, while the diversity in the samples collected in the older isolated community was distinct in terms of its placement in PCA space.
FIG. 2.
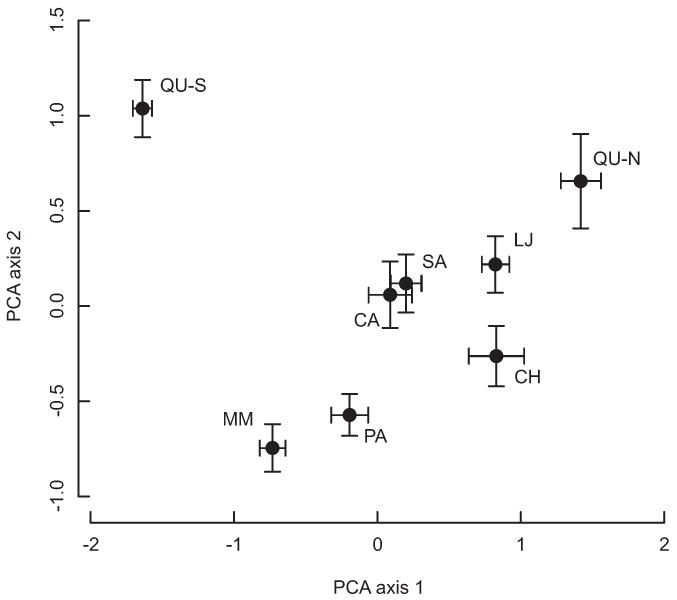
A principal components analysis (PCA) of the microsatellite data from each sampled district. The dots represent the centroid location of the individuals from each district along with the standard error on the estimate of the centroid. All districts occupy a unique location on the PCA plot except Sachaca and Cayma, which are also the pair of communities that have a nonsignificant FST (Table 2). Despite being geographically distant, the La Joya district occupies a similar location in PCA space to the other districts. The Quequeña district is split into two distinct groups based on sampling location with the northern location similar to the other districts while southern location is distinct. The first two principal component axes explain 42.2% of the total genetic variation.
Results from the AMOVA analyses confirmed that, despite the significant population genetic differentiation, the majority of the genetic diversity was found within each district (Table 3). However, significant genetic variation was found among the districts within groups regardless of the ‘regional’ groupings of districts in AMOVA analyses. The significant differences among districts were driven by the differences in the frequencies of alleles suggesting current limits to gene flow but a recent shared history among districts.
Table 3.
Population genetic structure of Tria. infestans within and between districts region (AMOVA)
| Groupings | Source of Variation | Variance Components | Fixation Indices | Percentage of Variation |
|---|---|---|---|---|
| [CA, MM, PA] [CH, LJ, QU, SA] | Among Groups | 0.02497 Va | FCT = 0.00844 | 0.84 |
| Among Districts within Groups | 0.27824 Vb* | FSC = 0.08039* | 7.97 | |
| Within Districts | 3.18299 Vc* | FST = 0.08815* | 91.18 | |
|
| ||||
| [CA, CH, LJ, MM, PA, SA] [QU] | Among Groups | 0.21287 Va | FCT = 0.05877 | 5.88 |
| Among Districts within Groups | 0.22629 Vb* | FSC = 0.06637* | 6.25 | |
| Within Districts | 3.18299 Vc* | FST = 0.12124* | 87.88 | |
The difference among districts in the frequency distribution of alleles was evident at all population clustering levels in STRUCTURE analyses (Fig. 3). Although the majority of the clusters were found in most districts, the most frequent population cluster differed across districts as did the frequencies of minority clusters. Those districts in the city center (Paucarpata and Mariano Melgar) were similar to each other at all values of K. The peri-urban districts (Sachaca, Cayma, and Characato) differed slightly in both composition and frequency of population clusters from the downtown areas, especially at low values of K. The rural districts are the most distinct from the city center (La Joya and Quequeña), although La Joya was much more similar to the urban districts.
FIG. 3.
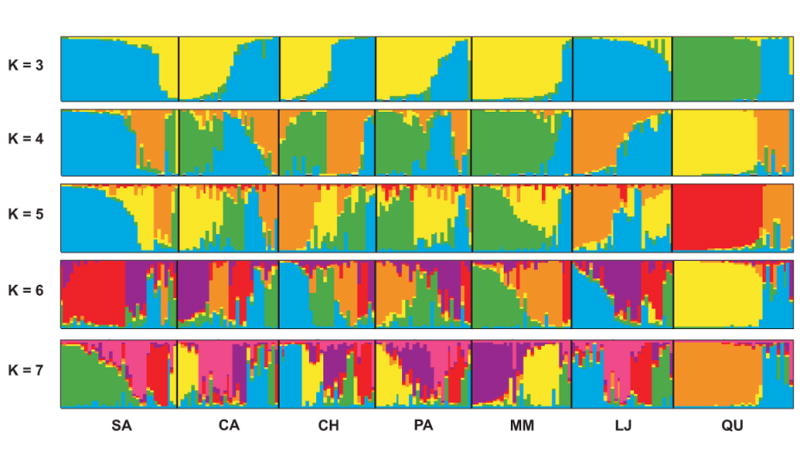
Proportional membership of each Tria. infestans sample in clusters for K = 3–7 as identified in STRUCTURE. Individuals from the Quequeña district are separated into two distinct groups at all values of K, one of which is comprised of individuals assigned to a cluster not found in other districts. The other districts are comprised of bugs from the same clusters but at different frequencies. The most likely estimate of K is 4, as determined by the ΔK method (Evanno et al. 2005).
The district of Quequeña was consistently different from the other districts at all values of K. The insect population was predominantly composed of individuals assigned to a unique cluster corresponding to the two sampling locations within the district. Two of the other peri urban districts, Sachaca and Characato, also have geographically distinct communities and similar internal geographic genetic structuring (Fig. S3). The internal substructure in these communities was not sufficient occupy two locations in the PCA space. Although the optimal value of K=4, as determined by the Evanno ΔK method, was not significantly more likely by the log-likelihood ratio test (Fig. S1), similar patterns were observed at most values of K.
The population genetic structure inferred from the genetic data does not follow a simple isolation by distance (IBD) model at all spatial scales. At course between–district scales, geographic distance was not correlated with genetic distance (r = -0.0001, R2 = 0.000128). Similarly, the geographic distance matrix describing the distance among individuals was not significantly correlated with either the F-statistic matrix (Mantel r = 0.1071, p < 0.0001) or the codominant genetic distance (Mantel r = 0.1497, p < 0.0001). However, there is significant spatial autocorrelation at distances less than 5 km with the strongest correlations at very short distances (Fig. 4). No spatial autocorrelation was detected at distances greater than 5 km (r ≈ 0).
FIG. 4.
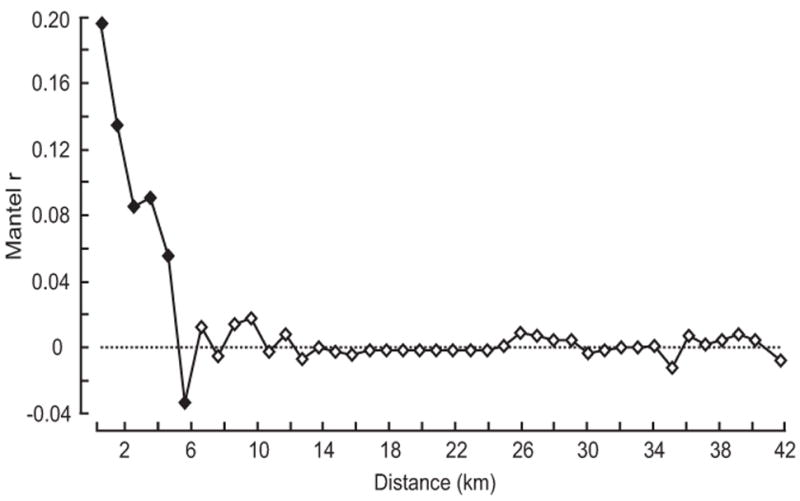
Spatial autocorrelation of genetic distance and geographic distance. There is significant spatial autocorrelation at distances less than 5 km. The spatial autocorrelation rapidly dissipates with distance and is not significant at distances greater than 5 km.
Discussion
Biological invasions have devastating ecologic, economic and public health consequences, and in many cases are due to the unprecedented movement of human populations and human-mediated changes in environmental landscapes (Vitousek et al. 1996). The kissing bug, Tria. infestans, has recently colonized the Peruvian city of Arequipa likely due to the rapid urbanization of the surrounding areas (Bayer et al. 2009). The biological invasion of this insect has resulted in a public health crisis, putting thousands of residents of this city at risk of infection by Tryp. cruzi and necessitating a large and expensive vector control campaign. Our analyses show that insect populations in the districts within and around this urban area are genetically structured. Further, geographic distance alone cannot explain this population genetic structuring, suggesting that a range expansion by means of Tria. infestans walking or flying cannot account for the shared evolutionary history among bugs in each district. While it is likely that several human, ecological, and historical factors with complex interactions have shaped the genetic structure in this system, the data presented here were consistent with the hypothesis that human movements have played a central role in the structuring of the Tria. infestans population in the region.
The microsatellite loci investigated in this study are variable even over the small spatial scales and the short temporal scales examined. However, without an empirically parameterized mutation model for each microsatellite, a reliable estimate of the time to the most recent common ancestor of Tria. infestans in the city, and thus the date of the initial invasion, cannot be obtained. Regardless, these microsatellite loci were sufficiently variable to investigate the current and historical patterns of gene flow in Arequipa. The majority of the alleles at all microsatellite loci were found in all districts suggesting a recent shared ancestry of all bugs or extensive gene flow.
Despite shared alleles among districts, bugs sampled from Arequipa are not a large panmictic population. Gene flow is limited such that the frequency of alleles differed significantly among districts and strong signatures of population genetic structure were observed in the differentiation of PCA centroids, STRUCTURE clustering analysis, and AMOVA analyses. Both the STRUCTURE clustering analysis and AMOVA analyses that demonstrated that although much of the genetic variation was contained within districts, a significant amount of variation was among-district (Table 3). Regardless of how districts were grouped into ‘regions’ in AMOVA analyses, among-district variation was always statistically significant while among-region variation was negligible, even when the most genetically distinct district, Quequeña, was in a grouping by itself.
Patterns of isolation by distance (IBD) were apparent at spatial scales relevant to the natural dispersal capabilities of Tria. infestans (≈ 1km). Spatial autocorrelation of genetic distances was apparent over short distances (< 4km) and pronounced at very short distances (< 1km). The spatial autocorrelation at short distances suggests that Tria. infestans can migrate among neighboring houses, which confirms previous observations in trapping studies (Levy et al. 2008).
The patterns of spatial autocorrelation and the population genetic structure of the Tria. infestans populations at among-district scales did not support the IBD expectation that genetic and geographic distances correlate as would be seen if bugs dispersed only through walking or flying. Some geographically distant districts were genetically similar to the populations in the city while others were distinct (Fig. 3). For example, the genetic distance between the two adjacent downtown districts (Mariano Melgar and Paucarpata) was similar in magnitude to the genetic distance between either district and La Joya, the most geographically distant district about 33.7 km away from both (Table 2). Similarly, the genetic distance between Characato and Paucarpata was relatively high (FST = 0.15369, separated by 6.3 km), while the genetic distance between Characato and La Joya was much lower (FST = 0.03432, separated by 35km). These data suggest that long- distance gene flow, well outside the migratory capacity of Tria. infestans, occurs frequently in the area. These data suggests a factor beyond physical separation significantly influenced gene flow across the region.
The patterns of genetic similarities among districts are correlated with the degree to which districts are connected. The urban districts are highly connected to each other by the continuous urban environment that is hospitable to vectors and allows some degree of gene flow. These urban districts are also connected with the rural areas to varying degrees due to the road and highway system. The regular travel of people from rural areas to the city diversifies the urban area. The district of La Joya, though geographically remote, is situated along the main highway connecting Arequipa to the capital Lima, a road that is heavily traveled. Additionally, La Joya was sprayed with insecticide in 1996 (Delgado et al. 2011). It seems likely that the insect populations that re-emerged following control originated from the larger populations in the city, which were not treated with insecticide until a decade later and that the reemergence was facilitated by the district’s high level of connectedness.
The peri-urban and rural areas investigated (Characato, Sachaca, and Quequeña) are less connected to other areas by roads and the residents, may travel less frequently to the city. Further, there is internal socioeconomic stratification within these regions that is geographically demarcated leading to population genetic substructuring of Tria. infestans populations. For example, the district Quequeña is comprised of two main human communities: an older community in the southern region and a newer, more connected region, to the north. Tria. infestans samples from the isolated southern community were consistently assigned to a unique cluster in STRUCTURE at all values of K (Fig. 3) and formed a unique cluster or points in the PCA analysis (Fig. 2). The existence of a single, unique genetic cluster of Tria. infestans in the isolated human community suggests that there is little admixture in this population of vectors. The Tria. infestans samples from the northern community, however, were similar in allele presence and frequency with all other sampled communities and clustered with subpopulations from other districts in STRUCTURE and PCA analyses (Fig. 2 and Fig. 3). The genetic similarity of the Tria. infestans from the northern community to the bugs in the other districts suggested that human travel from this interconnected community to the city center or other districts resulted in the concomitant migration of bugs from other districts.
The differences in the degree of genetic relatedness among pairs of populations along with the lack of correlation between genetic and geographic distances suggested that human activity patterns are an important factor in the movement of Tria. infestans among districts. Further research into the volume of human movement among these communities is necessary to adequately address this hypothesis. Nevertheless, these findings emphasize the importance of urban ecological studies that investigate the interplay between ecological, historical, environmental, and human factors that shape the genetic structure of disease systems.
The processes that shape the patterns of gene flow in insects can be important in guiding vector control strategies. In particular, our results suggest that surveillance for the return of Tria. infestans following insecticide control should focus especially on highly-connected districts of cities. These hubs of human movement are both at higher risk of recolonization by Tria. infestans, and pose a greater risk to elimination campaigns as a whole, because they facilitate dispersal of vectors over a large geographical area. Previous studies that have aimed to optimize the ordering of vector control strategies, including timing and placement of insecticide application (Levy et al. 2010), need to be further expanded to consider dispersal of insects along road networks in addition to simple Euclidean distance. Additional studies on the finer scale of urban environments are necessary to improve surveillance and control of Tria. infestans and to eventually achieve the goal of disruption of transmission of Tryp. cruzi and prevent the accumulation of cases of Chagas disease in cities.
Supplementary Material
Acknowledgments
The Chagas Disease Working Group in Arequipa includes Fernando Málaga Chávez, Karina Oppe Álvarez, Andy Catacora Rospigliossi, Dr. Juan Cornejo del Carpio, Javier Quintanilla Calderón, Kate Levy, Malwina Niemierko, Corentin Barbu, and Renzo Salazar Sanchez.
The authors would like to express sincere gratitude to the Gerencia Regional de Salud en Arequipa (GERSA), Ministerio de Salud del Perú (MINSA), Dirección General de Salud de las Personas (DGSP), Estrategia Sanitaria Nacional de Prevención y Control de Enfermedades Metaxénicas y Otras Transmitidas por Vectores (ESNPCEMOTVS), Dirección General de Salud Ambiental (DIGESA), Gobierno Regional de Arequipa, Organización Panamericana de la Salud (OPS) and the Canadian International Development Agency (CIDA). The authors also thank Ellen Dotson, Paula Marcet, and Bob Wirtz. This work was supported by National Institutes of Health Grants P50 AI074285, K01 AI079162.
Footnotes
Data Accessibility
Microsatellite data generated in this study have been deposited to DRYAD and are accessible under doi:10.5061/dryad.25gn8
Author Contributions
The authors participated in the discussions that conceived and developed this study. J A-J, K B-M, V Q-M, and M L organized the field work; E F, J H and C K generated and analyzed the molecular data; E F, M L, and D B interpreted the results and led the manuscript writing; E F, C K, M L, and D B contributed with edits.
References
- Abdi H. The Bonferonni and Šidák Corrections for Multiple Comparisons. In: Salkind NJ, editor. Encyclopedia of Measurement and Statistics. Sage Publications, Inc; 2007. p. 1136. [Google Scholar]
- Barbosa NPU, Fernandes GW, Carneiro MAA, Junior LAC. Distribution of non-native invasive species and soil properties in proximity to paved roads and unpaved roads in a quartzitic mountainous grassland of southeastern Brazil (rupestrian fields) Biological Invasions. 2010;12:3745–3755. [Google Scholar]
- Bayer AM, Hunter GC, Gilman RH, et al. Chagas Disease, Migration and Community Settlement Patterns in Arequipa, Peru. Plos Neglected Tropical Diseases. 2009;3 doi: 10.1371/journal.pntd.0000567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bland JM, Altman DG. Statistics notes: Multiple significance tests: the Bonferroni method. British Medical Journal. 1995;310:170–170. doi: 10.1136/bmj.310.6973.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman NM, Kawai V, Levy MZ, et al. Chagas Disease Transmission in Periurban Communities of Arequipa, Peru. Clinical Infectious Diseases. 2008;46:1822–1828. doi: 10.1086/588299. [DOI] [PubMed] [Google Scholar]
- Brenière SF, Lopez J, Vargas F, Barnabé C. Genetic variability and microdistribution of Triatoma infestans genotypes and Trypanosoma cruzi clones in Arequipa region (Peru) Memorias Do Instituto Oswaldo Cruz. 1997;92:401–408. doi: 10.1590/s0074-02761997000300018. [DOI] [PubMed] [Google Scholar]
- Bronnenhuber JE, Dufour BA, Higgs DM, Heath DD. Dispersal strategies, secondary range expansion and invasion genetics of the nonindigenous round goby, Neogobius melanostomus, in Great Lakes tributaries. Molecular Ecology. 2011;20:1845–1859. doi: 10.1111/j.1365-294X.2011.05030.x. [DOI] [PubMed] [Google Scholar]
- Ceballos LA, Vazquez-Prokopec GM, Cecere MC, Marcet PL, Gurtler RE. Feeding rates, nutritional status and flight dispersal potential of peridomestic populations of Triatoma infestans in rural northwestern Argentina. Acta Tropica. 2005;95:149–159. doi: 10.1016/j.actatropica.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Crowl TA, Crist TO, Parmenter RR, Belovsky G, Lugo AE. The spread of invasive species and infectious disease as drivers of ecosystem change. Frontiers in Ecology and the Environment. 2008;6:238–246. [Google Scholar]
- Delgado S, Neyra RC, Machaca VRQ, et al. A History of Chagas Disease Transmission, Control, and Re-Emergence in Peri-Rural La Joya, Peru. PLoS Neglected Tropical Diseases. 2011;5:e970. doi: 10.1371/journal.pntd.0000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl DA, vonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources. 2011;4:359–361. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L. Analysis of Population Subdivision. In: Balding DJ, Bishop M, Cannings C, editors. Handbook of Statistical Genetics. John Wiley & Sons-IEEE Press; New York, NY: 2007. pp. 980–1013. [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Scheider S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47. [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of Molecular Variance Inferred From Metric Distances Among Dna Haplotypes - Application to Human Mitochondrial-Dna Restriction Data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia BA, Zheng L, Rosas ARPDE, Segura EL. Isolation and characterization of polymorphic microsatellite loci in the Chagas’ disease vector Triatoma infestans Hemiptera: Reduviidae. Molecular Ecology Notes. 2004;4:568–571. [Google Scholar]
- Gaudeul M, Giraud T, Kiss L, Shykoff JA. Nuclear and chloroplast microsatellites show multiple introductions in the worldwide invasion history of common ragweed, Ambrosia artemisiifolia. PLoS ONE. 2011;6:e17658. doi: 10.1371/journal.pone.0017658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J. FSTAT (Version 1.2): A Computer Program to Calculate F-Statistics. Journal of Heredity. 1995;86:485–486. [Google Scholar]
- Grimm NB, Faeth SH, Golubiewski NE, et al. Global Change and the Ecology of Cities. Science. 2008;319:756–760. doi: 10.1126/science.1150195. [DOI] [PubMed] [Google Scholar]
- Hooper DU, Chapin FS, Ewel JJ, et al. Effects of biodiversity on ecosystem functioning: A consensus of current knowledge. Ecological Monographs. 2005;75:3–35. [Google Scholar]
- Hubisz MJ, Falush D, Stephens M, Pritchard JK. Inferring weak population structure with the assistance of sample group information. Molecular Ecology Resources. 2009;9:1322–1332. doi: 10.1111/j.1755-0998.2009.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter GC, Borrini-Mayorí K, Juárez JA, et al. A Field Trial of Alternative Targeted Screening Strategies for Chagas Disease in Arequipa, Peru. Plos Neglected Tropical Diseases. 2012;6:e1468. doi: 10.1371/journal.pntd.0001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Levy MZ, Bowman NM, Kawai V, et al. Periurban Trypanosoma cruzi-infected Triatoma infestans, Arequipa, Peru. Emerging Infectious Diseases. 2006;12:1345–1352. doi: 10.3201/eid1209.051662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy MZ, Chavez FSM, del Carpio JGC, et al. Rational spatio-temporal strategies for controlling a Chagas disease vector in urban environments. Journal of the Royal Society Interface. 2010:1061–1070. doi: 10.1098/rsif.2009.0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy MZ, Kawai V, Bowman NM, et al. Targeted screening strategies to detect Trypanosoma cruzi infection in children. Plos Neglected Tropical Diseases. 2007;1:e103. doi: 10.1371/journal.pntd.0000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy MZ, Quíspe-Machaca VR, Ylla-Velasquez JL, et al. Impregnated netting slows infestation by Triatoma infestans. The American Journal of Tropical Medicine and Hygiene. 2008:e1002801. [PMC free article] [PubMed] [Google Scholar]
- Levy MZ, Small DS, Vilhena DA, et al. Retracing micro-epidemics of Chagas disease using epicenter regression. PLoS Computational Biology. 2011;7:e1002146. doi: 10.1371/journal.pcbi.1002146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoGiudice K, Ostfeld RS, Schmidt KA, Keesing F. The ecology of infectious disease: Effects of host diversity and community composition on Lyme disease risk. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:567–571. doi: 10.1073/pnas.0233733100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lounibos LP. Invasions by insect vectors of human disease. Annual review of entomology. 2002;47:233–266. doi: 10.1146/annurev.ento.47.091201.145206. [DOI] [PubMed] [Google Scholar]
- MacDougall AS, Turkington R. Are invasive species the drivers or passengers of change in degraded ecosystems? Ecology. 2005;86:42–55. [Google Scholar]
- Marcet PL, Lehmann T, Groner G, et al. Identification and characterization of microsatellite markers in the Chagas disease vector Triatoma infestans (Heteroptera: Reduviidae) Infection, Genetics and Evolution. 2006;6:32–37. doi: 10.1016/j.meegid.2005.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RJ. Anthropogenic impacts on tropical forest biodiversity: a network structure and ecosystem functioning perspective. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2010;365:3709–3718. doi: 10.1098/rstb.2010.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott KE, Muniz TM, Lehman JS, et al. House construction, triatomine distribution, and household distribution of seroreactivity to Trypanosoma cruzi in a rural community in northeast Brazil. The American Journal of Tropical Medicine and Hygiene. 1978;27:1116–1122. doi: 10.4269/ajtmh.1978.27.1116. [DOI] [PubMed] [Google Scholar]
- Orlóci L. Multivariate Analysis in Vegetation Research. Springer: 1978. [Google Scholar]
- Paerregaard K. Linking separate worlds: Urban migrants and rural lives in Peru 1997 [Google Scholar]
- Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel D, Zuniga R, Morrison D. Update on the environmental and economic costs associated with alien-invasive species in the United States. Ecological Economics. 2005;52:273–288. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of Population Structure Using Multilocus Genotype Data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing 2012 [Google Scholar]
- Raymond M, Rousset F. GENEPOP (version 1.2): Population genetics software for exact tests and ecumenicism. Journal of Heredity. 1995;86:248–249. [Google Scholar]
- Richer W, Kengne P, Cortez MR, et al. Active dispersal by wild Triatoma infestans in the Bolivian Andes. Tropical Medicine & International Health. 2007;12:759–764. doi: 10.1111/j.1365-3156.2007.01846.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg MS, Anderson CD. PASSaGE: pattern analysis, spatial statistics and geographic exegesis. Version 2. Methods in Ecology and Evolution. 2011;2:229–232. [Google Scholar]
- Rosenberg NA. distruct: a program for the graphical display of population structure. Molecular Ecology Notes. 2004;4:137–138. [Google Scholar]
- Rousset F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics. 1997;145:1219–1228. doi: 10.1093/genetics/145.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F. Genetic differentiation between individuals. Journal of Evolutionary Biology. 2000;13:58–62. [Google Scholar]
- Rousset F. Genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Molecular Ecology Resources. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- Sakai AK, Allendorf FW, Holt JS, et al. The population biology of invasive species. Annual Review of Ecology and Systematics. 2001;32:305–332. [Google Scholar]
- Sala OE, Chapin FS, Armesto JJ, et al. Biodiversity - Global biodiversity scenarios for the year 2100. Science. 2000;287:1770–1774. doi: 10.1126/science.287.5459.1770. [DOI] [PubMed] [Google Scholar]
- Sax DF, Stachowicz JJ, Gaines SD. Species invasions. Sinauer Associates Inc; 2005. [Google Scholar]
- Schaffner F, Kaufmann C, Hegglin D, Mathis A. The invasive mosquito Aedes japonicus in Central Europe. Medical and Veterinary Entomology. 2009;23:448–451. doi: 10.1111/j.1365-2915.2009.00825.x. [DOI] [PubMed] [Google Scholar]
- Scherber C, Eisenhauer N, Weisser WW, et al. Bottom-up effects of plant diversity on multitrophic interactions in a biodiversity experiment. Nature. 2010;468:553–556. doi: 10.1038/nature09492. [DOI] [PubMed] [Google Scholar]
- Schuurman F. John Turner revisited: an intra-urban migration model for colonial-type cities in Latin America. Journal of Economic and Social Geography. 1986;77:221–230. doi: 10.1111/j.1467-9663.1986.tb00824.x. [DOI] [PubMed] [Google Scholar]
- Sloop CM, Ayres DR, Strong DR. Spatial and temporal genetic structure in a hybrid cordgrass invasion. Heredity. 2011;106:547–556. doi: 10.1038/hdy.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal R, Rolf J. Biometry: the principles and practice of statistics in biological research. W. H. Freeman and Company; New York, NY: 1995. [Google Scholar]
- Strayer DL, Eviner VT, Jeschke JM, Pace ML. Understanding the long-term effects of species invasions. Trends in Ecology & Evolution. 2006;21:645–651. doi: 10.1016/j.tree.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Suzan G, Marce E, Giermakowski JT, et al. Experimental Evidence for Reduced Rodent Diversity Causing Increased Hantavirus Prevalence. PLoS ONE. 2009;4 doi: 10.1371/journal.pone.0005461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tews J, Brose U, Grimm V, et al. Animal species diversity driven by habitat heterogeneity/diversity: the importance of keystone structures. Journal of Biogeography. 2004;31:79–92. [Google Scholar]
- Tompkins DM, Gleeson DM. Relationship between avian malaria distribution and an exotic invasive mosquito in New Zealand. Journal of the Royal Society of New Zealand. 2006;36:51–62. [Google Scholar]
- Vitousek P, DAntonio C, Loope L, Westbrooks R. Biological invasions as global environmental change. American Scientist. 1996;84:468–478. [Google Scholar]
- Walsh JF, Molyneux DH, Birley MH. Deforestation: effects on vector-borne disease. Parasitology. 2011;106:S55–S75. doi: 10.1017/s0031182000086121. [DOI] [PubMed] [Google Scholar]
- Wilcove D, Rothstein D, Dubow J, Phillips A. Quantifying Threats to Imperiled Species in the United States. Bioscience. 1998;48:607–615. [Google Scholar]
- With KA. The landscape ecology of invasive spread. Conservation Biology. 2002;16:1192–1203. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


