Abstract
Dengue is an emerging disease in Nepal and was first observed as an outbreak in nine lowland districts in 2006. In 2010, however, a large epidemic of dengue occurred with 4,529 suspected and 917 serologically-confirmed cases and five deaths reported in government hospitals in Nepal. The collection of demographic information was performed along with an entomological survey and clinical evaluation of the patients. A total of 280 serum samples were collected from suspected dengue patients. These samples were subjected to routine laboratory investigations and IgM-capture ELISA for dengue serological identification, and 160 acute serum samples were used for virus isolation, RT-PCR, sequencing and phylogenetic analysis. The results showed that affected patients were predominately adults, and that 10% of the cases were classified as dengue haemorrhagic fever/ dengue shock syndrome. The genetic characterization of dengue viruses isolated from patients in four major outbreak areas of Nepal suggests that the DENV-1 strain was responsible for the 2010 epidemic. Entomological studies identified Aedes aegypti in all epidemic areas. All viruses belonged to a monophyletic single clade which is phylogenetically close to Indian viruses. The dengue epidemic started in the lowlands and expanded to the highland areas. To our knowledge, this is the first dengue isolation and genetic characterization reported from Nepal.
Keywords: Dengue fever, dengue 1 virus, epidemiology, Nepal
Introduction
Dengue fever (DF) is considered to be the most important of the arthorpod-borne viral disease in humans, and more than half of the world’s population in over 100 countries is at risk of infection [1]. Dengue hemorrhagic fever (DHF), the severe form of the disease, is endemic and frequently intensifies into epidemics in the countries of Southeast Asia (Laos, Cambodia, Vietnam, Singapore, Indonesia and Thailand), resulting in more cases and deaths [1, 2]. Dengue is also endemic in India and Pakistan and has recently emerged in Bangladesh (2000) and in Bhutan (2004) [3, 4].
In the Himalayan country of Nepal, the first case of dengue was reported in 2004 [5]. In 2006, an outbreak of dengue occurred there with 32 laboratory-confirmed cases. All four dengue virus (DENV) serotypes DENV-1, DENV-2, DENV-3 and DENV-4 were identified in nine districts of the lowland Terai region, but unfortunately without information on sequence analysis [6, 7]. From 2007, a few sporadic clinical cases were reported. Entomological studies carried out in 2009 identified larvae of Aedes aegypti (Ae. aegypti) in the Kathmandu and Lalitpur districts in Nepal [8]. Ae. aegypti is the primary vector of dengue, and it is likely that this vector is being introduced in this zone seasonally. In 2010, Nepal suffered its largest dengue epidemic. There had been no dengue virus isolation from a native Nepalese patient in the past. The main objective of the present study is to isolate the virus from Nepalese patients and to determine the origin of these viruses.
Methods
Patients
In 2010, a total of 280 suspected dengue patients from various district hospitals in Nepal were referred to the Sukraraj Tropical and Infectious Disease Hospital (STIDH) in Kathmandu, the capital city of Nepal. Clinical information was collected from patients who were admitted to the STIDH and their specific dengue disease, if present, was clinically identified as dengue fever (DF) or dengue haemorrhagic fever (DHF) based on the WHO classification (WHO, 1997). Initially, a patient was suspected to have DF when he or she had an acute onset of high fever lasting for 2–7 days and exhibited at least two of the following features: rashes, headache, arthralgia and leucopenia. A patient was suspected to have DHF when haemorrhagic manifestations such as ecchymosis, mucosal bleeding and plasma leakage were observed in addition to the features of DF. The suspected DF and DHF patients underwent further examinations for confirmation of the illness. A case was labeled as “probable” if the patient was found positive for the presence of IgM antibodies against dengue, and a case was labeled as “confirmed” when found positive by RT-PCR or virus isolation. Patients also underwent chest radiography and abdominal ultrasound tests in addition to hemoglobin count, hematocrit, and total blood and platelet monitoring as part of the clinical assessment.
Blood samples
Blood specimens were collected from patients, kept at 4°C for less than 24 hours and stored at –70°C in Kathmandu. These samples were transported in dry ice from Nepal to the Institute of Tropical Medicine in Nagasaki, Japan for further processing. If a serum sample had been taken from a patient less than 7 days from the onset of fever, it was subjected to virus isolation. If the sample came from a patient who had fever of more than 7 days, it was tested for the presence of anti-dengue IgM antibodies by in-house IgM-capture ELISA. To rule out the presence of IgM antibodies against JE, the sample was also tested usingthe JE-DEN IgM combination ELISA kit (Panbio, Australia).
IgM-Capture ELISA
Serum samples obtained after the centrifugation of blood specimens were subjected to in-house IgM-capture ELISA for the detection of IgM antibodies against dengue following the procedures described previously [9, 10]. These samples were also examined to rule out the presence of IgM antibodies against Japanese encephalitis virus (JEV) using the Panbio JE-DEN combination ELISA kit (Panbio, Australia). Panbio units were computed for each sample according to the protocol of the manufacturer. If the value was greater than 11 JE or 11 DEN Panbio units, the sample was considered positive for IgM antibodies against JE or dengue, respectively and if less than 9 for either JE or dengue, the sample was considered negative for these antibodies.
Virus isolation
A volume of 10 µl from each acute serum sample was inoculated in a culture tube containing 70% confluent C6/36 cells. The cells were maintained in MEM with 2% FCS, incubated at 28°C for 7 days and observed daily for cytopathic effects. Infected culture fluid from each tube was collected and clarified by centrifugation, and the supernatant was stored at –80°C until use in the detection of dengue virus RNA by RT-PCR.
RNA extraction and RT-PCR
QIAamp viral RNA mini kit (QIAGEN, Hilden, Germany) was used to extract the genomic viral RNA from 140 µl of infected culture supernatants following the manufacturers’ protocol. RNA was eluted from the QIAspin columns in a final volume of 100 µl of elution buffer and stored at –70°C until use. Reverse transcription was performed using Superscript III reverse transcriptase (Invitrogen) and random hexamers or gene specific primer, DenV1NR. PCR was conducted in a reaction mixture containing TaKaRa LA Taq DNA polymerase (Takara Bio Inc., Otsu, Japan) and appropriate primers. The following primers were used: DENV consensus primers to detect the presence of dengue virus [11], serotype-specific primers [11] to identify the specific serotype if sample was found positive for dengue virus, and primers (Table 1) to amplify the part that contained the E protein-coding region and the NS5-3’-UTR (10125–10428, 304 bp). PCR was then carried out and the amplicons were analyzed by gel electrophoresis on a 2.0% agarose gel (Dotite) containing ethidium bromide (0.5 µg/ml). Millipore Ultrafree®-DA (MA, USA) was used to purify the amplicons to be used as a template for sequence analysis.
Table 1.
Primer used for gene amplification and sequencing.
| Name of Primers | Sequence | |
|---|---|---|
| E-region | ||
| For Amplification | DEN1-E1-F | ACATGCCATAGGAACATCCA |
| DEN1-E2-R | TCATTGGTGACAAAAATGCC | |
| For Sequencing | Den1Seq04F01429 | CGTCGGAAATACAGCTGACC |
| DEN1-E2-F | ATGGCTAGTCCACAAACAATGG | |
| Den1Seq05F01862 | GACCCAGCATGGAACTGTTT | |
| DEN1-E1-R | GTGATCCTAATACGACTACTTCCTG | |
| Den1Seq05R02439 | GTTCTCTGCCCTTCCAGTTG | |
| NS5-3'UTR region | ||
| For Amplification | Den1Seq24F09950 | CCATCACCAATGGATGACAA |
| Den1Seq25R10672 | TGCCTGGAATGATGCTGTAG | |
| For Sequencing | Den1Seq25F10372 | AAAATGAAGTCAGGCCGAAA |
| Den1Seq24R10457 | CAGCCTCCCAGGTTTTTACA | |
| For Reverse transcription | ||
| Name of Polynucleotide | Sequence | |
| DenV1NR | AGAACCTGTTGATTCAACAGCACCATTCCA | |
DNA sequencing and phylogenetic analysis
For phylogenetic analysis, the purified amplicons containing the E protein-coding and NS5-3’-UTR-spanning regions of the virus genome were sequenced using certain primers (Table 1) and the Big Dye Terminator Cycle Sequencing System. The products were analyzed using a 3100-Avant Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). MAFFT program version 6.864 [12] was used to align sequence data. PHYLIP package, version 3.69 [13] was used to construct the neighbor-joining phylogenetic tree [14] based on 2283 E region sequences. For NS5-3’-UTR region, Bayesian MCC tree was constructed using BEAST software version 1.6.2 [15]. Models were selected with the use of MrModeltest version 2.3 [16] and PAUP* version 4.0 beta 10 [17]. The trees were drawn using FigTree software, version 1.3.1 [18].
Vector collection
House-to-house surveys were conducted on a total of 256 houses in the pre-monsoon season, 825 in the monsoon season, and 545 for screening of larval breeding places, covering residential areas, hotels, market, industrial areas of Kathmandu and Lalitpur district from April 2009 to March 2010. For each house, the number and category of potential larval habitats with and without water were noted. Surveys were conducted by searching all accessible artificial containers containing water in and around the houses for the presence of Aedes aegypti and Ae. albopictus larvae and/or pupae. Altogether nine different types of habitat were investigated: discarded tires, plastic drums, metal drums, metal containers, plastic buckets, flower pots, mud pots, cement tanks, and plastic pots. The larvae and pupae of mosquitoes were collected by the dipper method. Aspiration collections of adults were attempted in all outdoor dwellings with the help of flash-lights in order to record speciesduring the morning hours (0700–0900 hrs) in those breeding habitats. A different collection bottle was used for each category of container in each locality, and the larvae were brought to the laboratory and reared [19] until the emergence of adults and identified on the basis of morphological characteristics using published taxonomic keys [20]. Altogether, 1,876 wet containers in 892 households were found in Kathmandu district, and 1,807 wet containers in 734 households were found in Lalitpur, Dhading and Tanahu districts as breeding habitats for Ae. albopictus larvae.
Statistical Analysis
Statistical analysis was conducted using the software SSPS 13.0 version.
Ethical Statement
The study did not require approval from an institutional review board because the blood samples came from patients undergoing medical treatment at Sukraraj Tropical and Infectious Diseases Hospital under the auspicious of the Ministry of Health and Population, Nepal. Through this hospital, the government not only helps in providing medical care for patients, but also gathers data from patients as part of health surveillance duties.
For the entomological study, permission from the owners/residents was obtained before mosquito specimens were collected from their premises or residences.
Results
Geographical distribution and seasonality
Dengue is an emerging disease in Nepal. A small outbreak was reported in this country in 2006, and sporadic cases were reported from 2007 to 2009. From August to December 2010, an unusual increase in the number of febrile patients was observed. The 2010 outbreak started at the beginning of August from Damauli city (altitude of 360 meters) in the Tanahu district and Dhangadi city in the Kailali district (altitude of 190 meters) (Fig. 1) [21]. It expanded immediately to Bharatpur city, Chitawan district. The number of cases increased rapidly during the first week of September, and in October the outbreak spread even further to neighboring districts of Nawalparashi and Butwal, and to the headquarters of Rupendehi district. Finally, by the end of October the disease appeared in the highland districts of Dhading and Kathmandu (altitude of 1090 and 1350 meters, respectively), previously known as “dengue-free” zones (Fig. 1). The outbreak peaked in November (after the rainy season had finished) and subsided during the dry winter season around the second week of December.
Fig. 1.
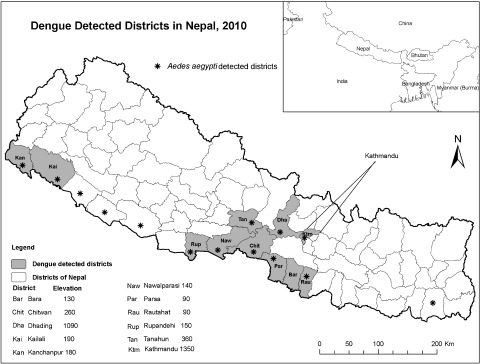
Dengue and Aedes aegypti confirmed districts in Nepal, 2010.
(*) indicates Aedes aegypti positive districts and shaded areas indicate dengue positive districts. Numbers indicate the elevation (meter) of each district capital.
Clinical features
Of the 280 patients who were referred to and sought medical help at STIDH, 242 were admitted. Almost all of these patients manifested features of classical DF such as fever, headache and myalgia. Hemorrhagic manifestations characteristic of DHF were also noted in the form of ecchymosis, epistaxis, gum bleeding and gastrointestinal (GI) bleeding. Based on the clinical data, 20% (48/242) of the patients experienced GI bleeding, abdominal pain, vomiting and difficulty in movement in addition to the features of DF. Twelve percent (28/242) of the patients suffered coughs and difficulty in breathing. Unusual manifestations such as splenomegaly were also observed in 4% (9/242) of patients with DHF. Abdominal ultrasound showed that 15% (36/242) of the patients had hepatomegaly, while 8% (19/242) had mild to moderate ascitis. Some of the patients showed acalculus cholelithiasis and thickening of the gall bladder. Four percent (9/242) of the patients showed hepatosplenomegaly. One patient showed signs of acute pancreatitis. On chest X-ray, 4% of the patients with DHF showed pleural effusion that was mostly unilateral.
Incidence
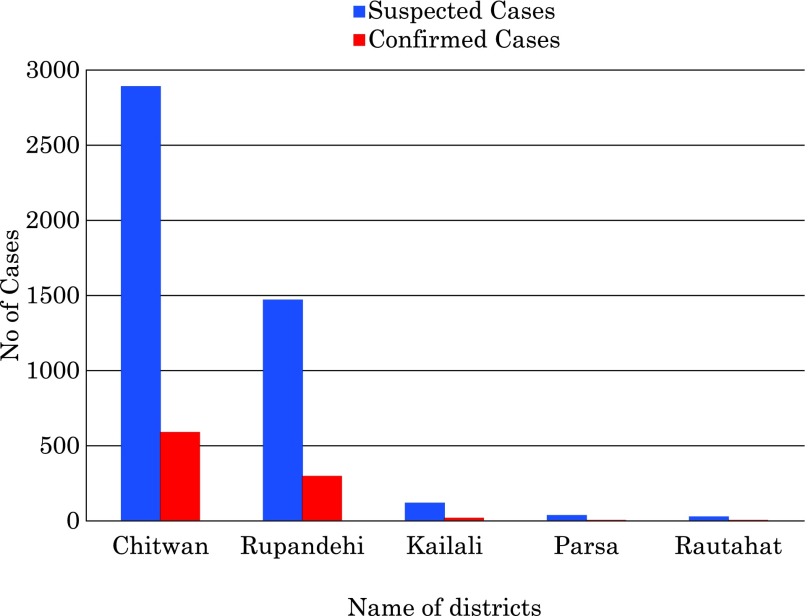
The Epidemiology Disease Control Division reported 4,529 suspected cases, 917 serologically-confirmed cases and five official deaths by the end of December. However, these figures were only from government hospitals across Nepal (Fig. 2). It is noted that the affected patients were predominately adults, the child adult ratio being 0.2:1. Most of the patients were diagnosed clinically as DF. But 10% of the patients in the present study admitted at STIDH were diagnosed clinically as DHF and were treated with blood and platelet transfusions.
Fig. 2.
Epidemiological curve showing the distribution of dengue cases in 2010.
Serological confirmation
A total of 120 serum samples collected after 7 days of fever were investigated using IgM-capture ELISA for dengue, and 35% were found to be positive for dengue. These samples were also investigated for JE, and all were found to be negative for anti-JE IgM.
Virus isolation and serotyping
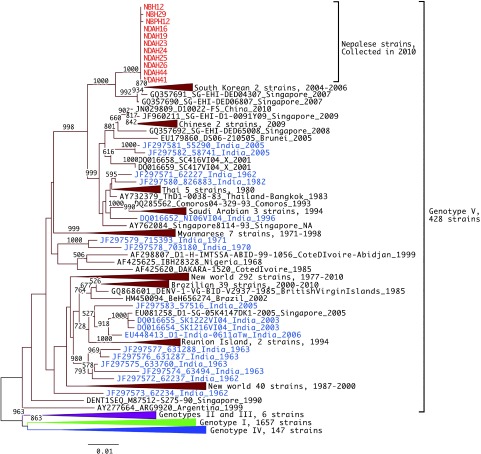
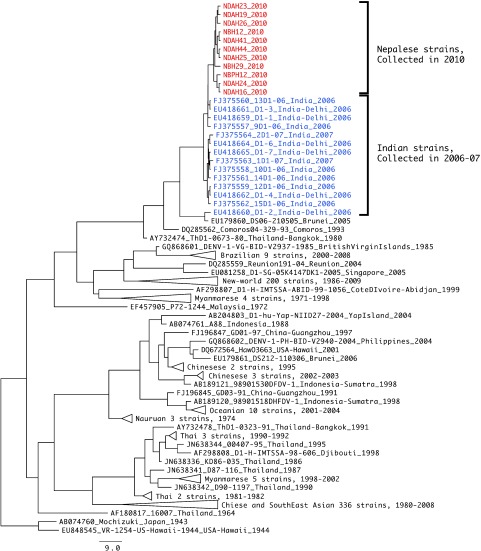
Only 160 acute serum samples were available for virus isolation in C6/36 cells. Twenty one (13%) of these samples were found to be positive for dengue after the culture fluids of the cells inoculated with these samples were subjected to RT-PCR in the presence of dengue consensus primers. Our genetic characterization of dengue viruses (all DENV-1, Table 2) isolated from patients in four major outbreak areas of Nepal suggests that the DENV-1 strains responsible for the 2010 epidemic originated from India. We read sequences of eleven strains. Fig. 3 and Fig. 4 show the phylogenetic tree drawn from Nepalese strains representing major epidemic districts.
Table 2.
List of isolates with clinical information of patients and GeneBank accession numbers of registered nucleotide sequences of E and NS5-3’UTR regions.
| ID | Age/Sex | Onset | Serotype | City/District | E region | NS5-3'UTR | |
|---|---|---|---|---|---|---|---|
| 1 | NDaH14-10 | 12/F | 11/08/2010 | D1 | Damauli/Tanahu | ||
| 2 | NDaH15-10 | 11/M | 11/08/2010 | D1 | Damauli/Tanahu | ||
| 3 | NDaH16-10 | 8/M | 11/08/2010 | D1 | Damauli/Tanahu | JF754983 | JF754994 |
| 4 | NDaH17-10 | 15/M | 11/08/2010 | D1 | Damauli/Tanahu | ||
| 5 | NDaH19-10 | 21/F | 11/08/2010 | D1 | Damauli/Tanahu | JF754984 | JF754995 |
| 6 | NDaH23-10 | 5/F | 11/08/2010 | D1 | Damauli/Tanahu | JF754985 | JF754996 |
| 7 | NDaH24-10 | 36/F | 11/08/2010 | D1 | Damauli/Tanahu | JF754986 | JF754997 |
| 8 | NDaH25-10 | 24/F | 11/08/2010 | D1 | Damauli/Tanahu | JF754987 | JF754990 |
| 9 | NDaH26-10 | 56/F | 11/08/2010 | D1 | Damauli/Tanahu | JF754988 | JF754998 |
| 10 | NDaH41-10 | 67/F | 11/08/2010 | D1 | Damauli/Tanahu | JF754999 | |
| 11 | NDaH44-10 | 8/F | 12/08/2010 | D1 | Damauli/Tanahu | JF754989 | JF755000 |
| 12 | NBpH12-10 | 23/F | 22/09/2010 | D1 | Nawalparashi/Chitwan | JF754982 | JF754993 |
| 13 | NBpH13-10 | 26/F | 22/09/2010 | D1 | Nawalparashi/Chitwan | ||
| 14 | NBH12-10 | 17/F | 13/10/2010 | D1 | Bharatpur/Chitwan | JF754980 | JF754991 |
| 15 | NBH29-10 | 30/F | 13/10/2010 | D1 | Bharatpur/Chitwan | JF754981 | JF754992 |
| 16 | NBH33-10 | 19/M | 13/10/2010 | D1 | Bharatpur/Chitwan | ||
| 17 | NBH23-10 | 26/M | 13/10/2010 | D1 | Bharatpur/Chitwan | ||
| 18 | NBH36-10 | 38/M | 13/10/2010 | D1 | Bharatpur/Chitwan | ||
| 19 | NBH15-10 | 20/M | 14/10/2010 | D1 | Bharatpur/Chitwan | ||
| 20 | NBH44-10 | 29/M | 14/10/2010 | D1 | Bharatpur/Chitwan | ||
| 21 | NDbH20-10 | 68/M | 22/10/2010 | D1 | Dhadingbeshi/Dhading | JF800928 | JF800929 |
Fig. 3.
Phylogenetic tree of DENV-1 E region. Representative analysis of E region, 1,485 bps, 2,238 strains. Red and blue indicate Nepalese strains and Indian strains, respectively. The tree was constructed after 1,000 replicates of bootstrap analysis using neighbor joining method. Bootstrap values (%) greater than 50% are shown above branches. Labels of strains conform to the following format: (GenBank accession nos)_(Strain name)_(Country-region)_(Year of isolation). “NA” means that the information is not available.
Fig. 4.
Phylogenetic tree of DENV-1 E NS5-3’UTR spanning region. Maximum clade credibility (MCC) tree of NS5-3’UTR spanning region, 304 bps, of 579 strains with uncorrelated relaxed clock, GTR+G+I model.
Nucleotide sequence and phylogenetic analysis
DNA sequencing of the E-coding region and the NS5-3’UTR was done on 11 DENV-1 isolates representing the major epidemic districts in Nepal (Table 2). The nucleotide sequences of the E region and the NS5-3’UTR were compared with other sequences published in GenBank. Phylogenetic trees generated for the E region and the NS5-3’UTR are shown in Figure 4, respectively. The phylogenetic tree on E region shows that all the eleven Nepalese strains formed a monophyletic clade and that they belonged to genotype V, a cosmopolitan genotype containing American, West African and Asian strains. In the phylogenetic tree for the NS5-3’-UTR region, Nepalese strains are shown to form a subcluster with strains isolated from India between 2006 and 2007.
Vector
Aedes aegypti has been found in most of the epidemic sites (Fig. 1), suggesting that this mosquito species is the principal vector of the dengue virus. A total of 16 Ae. albopictus larvae were recorded in pre-monsoon (2009), 1,221 in monsoon (2009), and 179 in post-monsoon seasons (2010) from 892 houses in Kathmandu and 734 in Lalitpur district. Positivity of Ae. albopictus larvae was recorded in discarded tires lying outdoors at all the habitats searched for mosquitoes. However, immature stage Ae. albopictus were also observed in metal drums and in metal containers in Lalitpur district. We confirmed the presence of the mosquito at Dhading and Tanahu for the first time during the epidemic in 2010.
Discussion
In this paper, we reported the first isolation of dengue virus strains from patients in Nepal during the 2010 epidemic. All of the strains belong to DENV-1, and the analysis of the E-coding region of the genome provided insights in understanding the status of dengue in this country. It was during this epidemic that the affected areas expanded for the first time and included the highlands of Nepal. This phenomenon may be related to climate changes attributable to global warming. In fact, the information in this report raises concern that new areas may become susceptible to dengue outbreaks in the near future.
The eleven DENV-1 strains isolated in this study formed a monophyletic clade. This suggests that a DENV-1 population, possibly circulating in the Indian subcontinent, was introduced into the Nepali lowlands and subsequently spread to the highlands during the outbreak. Nepal shares a long open border with India, and Nepalese people visit India frequently for jobs and business opportunities. Movement of people between the two countries is facilitated by the fact that no visa is required. In the 2010 outbreak, it is likely that migrant workers infected with dengue virus in India returned to Nepal and subsequently transmitted the disease to the local community through the mosquito vector. Moreover, the DENV-1 outbreak in Nepal coincided with that in neighboring India [22] and Pakistan [23], indicating that the DENV-1 outbreak in Nepal was initially caused by virus strains from India. However, information from India is still limited.
Goncalvez et al. [24] previously divided DENV-1 into five genotypes. From the clustering pattern on the phylogenetic tree of the E region, all the eleven Nepalese strains belong to genotype V, a cosmopolitan genotype containing American, West African and Asian strains. In particular, the Nepalese strains are close to the Asian subcluster. Patil et al. [25] and Domingo et al. [26] reported that Indian isolates form phylogenetic clusters. However, the Nepalese strains were phylogenetically separate from these Indian clusters. By using the NS5-3’-UTR region, Nepalese strains form a subcluster with strains isolated from India in 2006–2007 (Fig. 4). As more data from India are obtained in the future, it will become easier to elucidate the ecology of DENV in the Trans-Himalayan countries. The increasing movement of people between Nepal and India may allow a more frequent introduction of the dengue viruses in Nepal. In addition, the population of cities in Nepal is increasing, particularly Kathmandu, a rapidly growing urban center with 4 million inhabitants at present. This city is also suffering from limited infrastructure, scarcity of water and poorly planned housing. Moreover, it reported that the average temperature of Kathmandu is continuously rising [27], thus potentially facilitating further successful vector mosquito infestations in the highland areas of Nepal.
Nepal may be at a higher risk of DF/DHF outbreaks in future post-monsoon seasons. For this reason, the government of Nepal has recently identified dengue as an important emerging disease, and sanitation, mosquito control and health education programs are being conducted accordingly.
Acknowledgments
We thank Dr. Corazon C Buerano for critically reading this manuscript. We also thank the staff and of the Sukraraj Tropical and Infectious Disease Hospital, the district Hospitals of Dhading, Damauli and Bharatpur, the sub-regional hospitals, and the Ministry of Health, EDCD, Nepal for their support in this study. This work was supported by a Grant-in-Aid for Scientific Research (KAKENHI) No. 21256004, and Global COE program, MEXT, Japan; and the Health Labor Sciences Research Grant and the Program of Founding Research Centers for Emerging and Reemerging Infectious Diseases, JST, Japan.
References
- 1.World Health Organization Dengue guidelines for diagnosis, treatment, prevention and control. 2009. Available at: http://www.who.int/rpc/guidelines/9789241547871/en/ [PubMed]
- 2.Guzman MG, Halstead SB, Artsob H, Buchy P, Farrar J, Gubler DJ, Hunsperger E, Kroeger A, Margolis HS, Martínez E, Nathan MB, Pelegrino JL, Simmons C, Yoksan S, Peeling RW. Dengue: a continuing global threat. Nat Rev Microbiol 2010; 8: S7–S16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rahman M, Rahman K, Siddque AK, Shoma S, Kamal AH, Ali KS, Nisaluk A, Breiman RF. First outbreak of dengue hemorrhagic fever, Bangladesh. Emerg Infect Dis 2002; 8: 738–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dorji T, Yoon IK, Holmes EC, Wangchuk S, Tobgay T, Nisalak A, Chinnawirotpisan P, Sangkachantaranon K, Gibbons RV, Jarman RG. Diversity and origin of dengue virus serotypes 1, 2, and 3, Bhutan. Emerg Infect Dis 2009; 15: 1630–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pandey BD, Rai SK, Morita K, Kurane I. First case of dengue in Nepal. Nepal Med Coll J 2004; 6: 157–159 [PubMed] [Google Scholar]
- 6.Pandey BD, Morita K, Khanal SR, Takasaki T, Miyazaki I, Ogawa T, Inoue S, Kurane I. Dengue virus, Nepal. Emerg Infect Dis 2008; 14: 514–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malla S, Thakur GD, Shrestha SK, Banjeree MK, Thapa LB, Gongal G, Ghimire P, Upadhyay BP, Gautam P, Khanal S, Nisaluk A, Jarman RG, Gibbons RV. Identification of all dengue serotypes in Nepal. Emerg Infect Dis 2008; 14: 1669–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gautam I, Dhimal MN, Shrestha SR, Tamrakar AS. First Record of Aedes aegypti (L.) Vector of Dengue Virus from Kathmandu, Nepal. J Natural History Museum 2009; 24: 154–164 [Google Scholar]
- 9.Inoue S, Alonzo MT, Kurosawa Y, Mapua CA, Reyes JD, Dimaano EM, Alera MT, Saito M, Oishi K, Hasebe F, Matias RR, Natividad FF, Morita K. Evaluation of a dengue IgG indirect enzyme-linked immunosorbent assay and a Japanese encephalitis IgG indirect enzyme-linked immunosorbent assay for diagnosis of secondary dengue virus infection. Vector Borne Zoonotic Dis 2010; 10: 143–150 [DOI] [PubMed] [Google Scholar]
- 10.Natividad FF, Daroy ML, Alonzo MT, Matias RR, Suarez LA, Inoue S. Use of IgM-capture ELISA for confirmation of Japanese encephalitis infections in the Philippines. Southeast Asian J Trop Med Public Health 2006; 37: 136–139 [PubMed] [Google Scholar]
- 11.Morita K, Tanaka M, Igarashi A. Rapid identification of dengue virus serotypes by using polymerase chain reaction. J Clin Microbiol 1991; 29: 2107–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 2002; 30: 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felsenstein J. PHYLIP—Phylogeny Inference Package (Version 3.2). Cladistics 1989; 5: 164–166 [Google Scholar]
- 14.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987; 4: 406–425 [DOI] [PubMed] [Google Scholar]
- 15.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 2007; 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nylander JAA.MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University. 2004. MrModeltest version 2.3 http://www.abc.se/~nylander/mrmodeltest2/mrmodeltest2.html.
- 17.Swofford DL. “PAUP* beta version. Phylogenetic analysis using parsimony (*and other methods).” 2002. Sinauer Associated, Sunderland, MA. PAUP* version 4.0 beta 10.
- 18.FigTree software, version 1.3.1. Available at: http://tree.bio.ed.ac.uk/software/figtree/
- 19.Collins DL. Manual for mosquito rearing and experimental technique. Am Mosq Control Assoc Bull 1970; 5: 190 [Google Scholar]
- 20.Darsie RF, Jr, Pradhan SP. The mosquitoes of Nepal: Their identification, distribution and biology. Mosquito Systematic 1990; 22 (2): 69–130 [Google Scholar]
- 21.Sharma SP. Dengue outbreak affects more than 7000 people in Nepal. BMJ 2010; 341: c5496. [DOI] [PubMed] [Google Scholar]
- 22.Anoop M, Mathew AJ, Jayakumar B, Issac A, Nair S, Abraham R, Anupriya MG, Sreekumar E. Complete genome sequencing and evolutionary analysis of dengue virus serotype 1 isolates from an outbreak in Kerala, South India. Virus Genes 2012; 45(1): 1–13 [DOI] [PubMed] [Google Scholar]
- 23.Mahmood N, Rana MY, Qureshi Z, Mujtaba G, Shaukat U. Prevalence and molecular characterization of dengue viruses serotypes in 2010 epidemic. Am J Med Sci 2012; 343(1): 61–64 [DOI] [PubMed] [Google Scholar]
- 24.Goncalvez AP, Escalante AA, Pujol FH, Ludert JE, Tovar D, Salas RA, Liprandi F. Diversity and evolution of the envelope gene of dengue virus type 1. Virology 2002; 303: 110–119 [DOI] [PubMed] [Google Scholar]
- 25.Patil JA, Cherian S, Walimbe AM, Patil BR, Sathe PS, Shah PS, Cecilia D. Evolutionary dynamics of the American African genotype of dengue type 1 virus in India (1962–2005). Infect Genet Evol 2011; 11: 1443–1448 [DOI] [PubMed] [Google Scholar]
- 26.Domingo C, Palacios G, Jabado O, Reyes N, Niedrig M, Gascón J, Cabrerizo M, Lipkin WI, Tenorio A. Use of a short fragment of the C-terminal E gene for detection and characterization of two new lineages of dengue virus 1 in India. J Clin Microbial 2006; 44: 1519–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lohani SN. Climate change in Nepal—shall we wait until bitter consequences? J Agr Environ 2007; 8: 38–45 [Google Scholar]